Abstract
The rise of bacterial variants in the presence of lytic phages has been one of the basic grounds for evolution studies. However, there are incongruent results among different studies investigating the effect of phage resistance acquisition on bacterial fitness and virulence. We used experimental evolution to generate three classes of Pseudomonas aeruginosa variants under selective pressure from two different homogeneous phage environments and one heterogeneous phage environment. The fitness and virulence determinants of the variants, such as growth, motility, biofilm formation, resistance to oxidative stress, and the production of siderophores and chromophores, changed significantly compared to the control. Variants with similar colony morphology that were developed through different phage treatments have different phenotypic traits. Also, mRNA transcription for genes associated with certain phenotypic traits changed significantly; however, sequencing did not reveal any point mutations in selected gene loci. Furthermore, the appearance of small colony variants and melanogenic variants and the increase in pyocyanin and pyoverdin production for some variants are believed to affect the virulence of the population. The knowledge gained from this study will fundamentally contribute to our understanding of the evolutionary dynamics of bacteria under phage selective pressure which is crucial to the efficient utilization of bacteriophages in medical contexts.
INTRODUCTION
Pseudomonas aeruginosa is a metabolically versatile gammaproteobacterium which inhabits terrestrial, aquatic, animal, human, and plant host-associated environments (1). This pathogen is endowed with a fairly large genome (2) containing genes for many different virulence factors and regulatory mechanisms, allowing it to adapt to hostile environments (3). Moreover, changing environmental conditions result in rapid diversification of P. aeruginosa genotype to produce a range of morphologically distinct phenotypic variants (4, 5) through processes such as phase variation (6) and adaptive mutations (7). This adaptability is believed to give the variants a selective advantage under unfavorable conditions. Clinical isolates of Pseudomonas have been reported to display a high degree of phenotypic diversity (8–10), especially those isolated from antibiotic treated patients. As a result of its high adaptability (11) and intrinsic antibiotic resistance (12), infections caused by P. aeruginosa are, in many instances, difficult to eradicate and can become persistent or even chronic (10), earning this bacterium a reputation as a paradigm of bacterial resistance (13).
By selecting for phage resistance, bacteriophages have been identified as agents that can drive the emergence of P. aeruginosa variants due to the strong selective pressure that they exert on the host community (14, 15). At an estimated total of 1031, bacteriophages are the most abundant biological agent on earth (16) and, based on their prevalence and effects on prokaryotic communities, they can play a significant role in bacterial survival, activity, and evolution (17). The rise of variants in the presence of lytic phage has been one of the foundations for evolution studies (18–25); however, this is also one of the reasons why phage therapy has always been considered with skepticism by the medical community (26). The current rise of antibiotic-resistant bacteria is forcing researchers to look for alternative treatments, and phage therapy has proven to be promising in several animal case studies (27–31), as well as in human clinical trials (32, 33). The emergence of phage-resistant variants is expected in many applications of phage therapy; therefore, the understanding of the evolutionary biology of bacterium-phage interactions, specifically, the characteristics of the resistant population, is of utmost importance for the successful use of phages.
The virulence of Pseudomonas is caused by the production of several extracellular (type II and type III secreted enzymes, pyocyanin, and siderophores such as pyoverdin) and cell-associated (flagella and type IV pili) virulence factors (34, 35). If phage predation selects for variants with alterations in any of the genes that are involved in biogenesis or regulation of these virulence determinants, the resulting phage-resistant variants could potentially exhibit altered levels of virulence. Literature reports of the high in vivo persistence of antibiotic-resistant variants of P. aeruginosa (8, 36–39) further highlight this concern.
Current reports investigating the cost of acquiring phage resistance on host systems are incongruent. Although some studies report a fitness cost associated with resistance (15, 20, 21), others report no association between phage resistance and host fitness (i.e., bacterial growth and motility) (24, 40–42). Because there are multiple paths to resistance (43–45), even if the host fitness is not affected, other phenotypic and virulence determinants may have been altered as a result of phage resistance. However, well-controlled studies on how other phenotypic and virulence traits are affected are scarce (46, 47). Moreover, the majority of published studies focus on single phage-host systems, whereas multiple-phage systems are more common in natural settings and have more applications relevant to the therapeutic use of phage (46, 47).
The purpose of this study was to investigate the effect of development of phage resistance on selected phenotypic traits and virulence determinants of P. aeruginosa. Three classes of P. aeruginosa variants were developed via experimental evolution under selective pressure from two homogeneous (class I and II) phage treatments and a heterogeneous (class III) phage treatment. We investigated various phenotypic traits of the P. aeruginosa variants, along with mRNA transcription for genes associated with these traits, and searched for point mutations in selected loci via sequencing. The phages (E79 and PP7) were chosen to target different host receptors in order to decrease the likelihood of cross-resistance. Variants with the same colony morphotype that developed under different phage treatments exhibited different phenotypic traits, suggesting the cost of phage resistance to be context dependent.
MATERIALS AND METHODS
Isolation of phage-resistant variants.
The bacteriophages used in the present study are listed in Table 1. P. aeruginosa PAO1, a wound isolate (48), was inoculated into 5 ml of Trypticase soy broth (TSB) supplemented with 0.5% glucose and grown (37°C, 150 rpm) to an optical density at 600 nm (OD600) of 0.05. A 10-μl inoculum of the bacterial culture was mixed with either 10 μl of a single phage (PP7 or E79, 109 PFU/ml), 10 μl of a 1:1 mixture of PP7 and E79 phages, or 10 μl of SMG (saline magnesium gelatin) phage buffer (NaCl at 5.8 g/liter, MgSO4 at 0.96 g/liter, 1 M Tris [50 ml], and gelatin at 0.1 g/liter [pH 8.0]) as a control and spread on a fresh lysogeny broth (LB) agar plate. The plates were incubated at 37°C for 4 days in a humidified incubator. A total of 20 samples, 5 samples from each plate, were collected nondestructively with an inoculating loop every 24 h (or as colonies emerged) and streaked on fresh LB agar plates. From these plates, single isolated colonies were selected, grown in 200 μl of TSB plus 0.5% glucose (OD600 = 0.2 to 0.3), and used to prepare frozen stocks. All characterization experiments were initially performed with these freshly grown cultures and repeated with cultures grown from the frozen glycerol stocks. The isolates were verified for contamination from other bacteria using PCR with primers directed against 16S ribosomal DNA (rDNA) (PA-SS-F [GGGGGATCTTCGGACCTCA] and PA-SS-R [TCCTTAGAGTGCCCACCCG]) (49). The isolation and characterization experiments were repeated independently three times to confirm the repeatability of the observed trends. The isolates were passed through 100 generations, and their phage susceptibility was tested after every second generation. The details of this method are presented in the supplemental material.
Table 1.
Bacteriophages used in this studya
| Phage (catalog no.) | Family | Genome (size) | Host receptor(s) | Morphotype |
|---|---|---|---|---|
| PP7 (HER 369) | Leviviridae | ssRNA (3588 bp) | Pili | Icosahedral, tail-less |
| E79 (HER 359) | Myoviridae (PB-1-like viruses) | dsDNA (65 kbp) | LPS | Long contractile tail (70-nm head, 138-nm tail) |
The source for these bacteriophages was the Félix d'Hérelle Reference Center for Bacterial Viruses, Universite Laval, Quebec, Canada.
Quantifying the frequency of emergence of phage resistance.
The frequency of formation of resistant bacteria for pure phage and phage mixture treatments was quantified according to the method described by Carlson (50). Briefly, phage was mixed with its host (phage/bacterium ratio of 103 to 104) and spread onto an LB agar plate. The number of bacterial colonies that grew after 24 h of incubation at 37°C was divided by the number of colonies of the control (bacteria not mixed with phage) to calculate the frequency of resistance.
Phenotypic characterization.
A planktonic culture of the ancestral P. aeruginosa PAO1 prepared from a single colony from a fresh LB agar plate was used in all characterization experiments as a reference.
Colony morphology.
To investigate medium-dependent colony morphology, Pseudomonas variants were grown in TSB plus 0.5% glucose (OD600 = 0.2 to 0.3) and spotted onto the following culture media using a replica plater: LB agar, LB agar plus 5% sheep blood, LB agar plus 4 mM FeSO4, and LB agar plus 40 µg/ml of Congo red and 20 µg/ml of Coomassie brilliant blue (Sigma-Aldrich). The plates were incubated (24 h, 37°C), and the colonies were observed with light microscopy and classified according to appearance.
Phage susceptibility assay.
Aliquots (1 μl) from the liquid culture of each variant (OD600 = 0.2 to 0.3) were mixed with 2 ml of 0.5% LB agar and spread onto 1.5% LB agar plates. The ancestral forms of PP7 and E79 (109 PFU/ml) were spotted (1 μl) onto the soft agar layer, incubated at 37°C overnight, and inspected for plaque formation.
Motility assays (swimming, swarming, and twitching).
Motility agar was prepared using nutrient broth plus 0.3% agar (for swimming), 1.5% agar (for twitching), and 0.5% agar (for swarming) and then inoculated with 1 μl of liquid culture below the agar surface (for swimming), at the interface between the agar and the petri dish (for twitching), and on the agar surface (for swarming). Plates were incubated at 37°C, and the diameter of the motility zone around the point of inoculation was measured on the swimming and swarming plates after both 20 and 40 h of incubation. The twitching plates were incubated for 24 h at 37°C, after which the twitching zone at the agar-plate interface was visualized by crystal violet staining, and the twitching zone diameter was recorded.
Growth rate and biofilm formation.
Mutants were inoculated into 200 μl of TSB in a 96-well microtiter plate (polystyrene; Falcon) and incubated for 24 h (37°C, 150 rpm). The OD600 was monitored with a microplate reader (Infinite M200 Pro; Tecan, Switzerland) and recorded every 5 min. From these recordings, the population growth rate (maximal rate of change of OD600 during log phase) and final yield (value of OD600 after 24 h) were recorded as growth determinants. The wells were then washed three times with phosphate-buffered saline (PBS), and the attached biomass was quantified using a crystal violet assay (51).
Antibiotic susceptibility and resistance to oxidative stress.
Antibiotic susceptibility of the variants was tested using the agar dilution method (52). Both tobramycin and streptomycin were used at a maximum concentration of 100 μg/ml and, for each antibiotic, six serial 1:1 dilutions were tested. To study the resistance of the variants to oxidative stress, the agar dilution method was used with hydrogen peroxide at a maximum concentration of 200 mM and four serial 1:1 dilutions were tested.
Production of exoproducts.
(i) For pyocyanin, pyoverdin, and pyorubin, pigments were extracted and assayed as previously described by Palumbo (53). Briefly, the technique involved chloroform extraction of cell-free supernatants of 3-day-old cultures to separate pyocyanin, followed by aqueous extraction of the medium to remove pyoverdin and pyorubrin together. Pyocyanin was extracted from chloroform into 0.2 M aqueous HCl and quantified by measuring the absorbance at 525 nm. Pyorubin, present in the aqueous extracts, was also quantified by measuring the absorbance at 525 nm. Fluorescence measurements (excitation/emission wavelengths, 405/465 nm) of the aqueous extracts were performed to quantify the pyoverdin concentration. To account for the difference in growth rates, the OD600 values for all of the bacterial cultures were measured before extracting the siderophores and pigments from their supernatants. The values obtained from spectrophotometric and fluorescence measurements were then divided by the corresponding OD600 for each variant. (ii) For pyomelanin, variants were spotted onto furunculosis agar (tryptone, 10 g; yeast extract, 5 g; l-tyrosine, 1 g; NaCl, 2 to 5 g; and agar, 15 g [per liter]) (54) to enhance the production of pyomelanin. (iii) For rhamnolipids, a 50-μl aliquot of the supernatants from 3-day-old cultures of variants was added to holes punched into LB agar plates supplemented with cetyltrimethylammonium bromide (CTAB; 0.2 g/liter) and methylene blue (MB; 0.005 g/liter) (55). The plates were incubated for 24 h at 37°C, followed by incubation at room temperature for 5 days, after which the diameter of the halo formed around the point of inoculation was measured as an indication of the amount of the rhamnolipid present in the supernatant. To account for the difference in growth rates, the OD600 values for all of the bacterial cultures were measured. The values obtained from CTAB plates were then divided by the corresponding OD600 for each variant.
RNA extraction, cDNA synthesis, and comparative qPCR.
Selected variants were grown in TSB (OD600 = 0.2 to 0.3). The total RNA was extracted using a Direct-Zol RNA miniprep kit (Zymo Research). The expression of target genes was quantified using two-step quantitative reverse transcription-PCR (RT-qPCR) analysis using a high-capacity cDNA reverse transcription kit (Applied Biosystems). RT-qPCR was performed with an ABI Prism 7900 HT thermal cycler (Applied Biosystems) using a Power SYBR green PCR master mix (Applied Biosystems). The results were analyzed using the threshold cycle method (56) with rpoD (sigma factor RpoD) as an endogenous reference. All kits were used according to the manufacturer's instructions. The oligonucleotide primers used for RT-qPCR are listed in Table S1 in the supplemental material. Three independent isolated RNA samples were analyzed for each data point.
Preparation of genomic DNA, PCR, and sequencing.
GenElute bacterial genomic DNA purification kit (Sigma-Aldrich) was used to isolate bacterial genomic DNA. Oligonucleotide primers (see Table S2 in the supplemental material) were designed using Primer3 Plus (57) based on the published PAO1 genome sequence. Sequencing of PCR products was carried out by using Applied Biosystems 3730xl DNA analyzer technology at the McGill University and the Génome Québec Innovation Centre. The sequences were aligned against the registered P. aeruginosa PAO1 genome sequences using CLC sequence viewer (v.6; CLC Bio A/S).
Statistical analysis.
Results are reported in terms of population mean ± the 95% confidence intervals. The significance of the difference between phenotypic features of variants and control was analyzed by using the Student t test with no additional correction. Significance of difference between the variant groups was analyzed using single-factor analysis of variance (ANOVA). The statistical analysis was performed using Statistica 8.0 (Stat Soft, Inc., San Jose, CA), and P values of <0.05 were considered significant.
RESULTS
Isolation of P. aeruginosa PAO1 variants.
P. aeruginosa variants were categorized based on the type of phage challenge: PP7 phage (class I), E79 phage (class II), 1:1 mixture of both phages (class III), and no-phage challenge (control) with 20 variants in each class. The frequency of resistance was independently determined to be (1.3 ± 0.92) × 10−7 CFU for the phage mixture and (8.2 ± 2.5) × 10−6 CFU for each of the single phages; these figures are the average of three experiments and are presented as orders of magnitude. The strain was confirmed to be P. aeruginosa using 16S rDNA PCR for all isolates.
Phage susceptibility.
The variants were spot tested with the ancestral form of each phage. Both PP7 and E79 formed clear plaques, indicating full lysis, on the bacterial lawns of the ancestral form of P. aeruginosa PAO1 and on all of the isolates of the control group. Table S3 in the supplemental material presents a summary of the results for the susceptibility test; no cross-resistance was observed. Most of the PP7-resistant cells were completely resistant, whereas most of the E79-resistant cells showed partial resistance, indicated by a decrease of bacterial density where the phage was inoculated. To assess reversion to phage sensitivity, variants were serially passaged in TSB. Isolates from classes I and III were observed to be more resistant to reversion to phage sensitivity. It took 64 (class I) and 78 (class III) generations for half of the variants to revert to phage susceptibility. This value dropped to 34 generations for class II isolates. The numbers reported are the average of duplicate experiments rounded to the nearest digit.
Colony morphology.
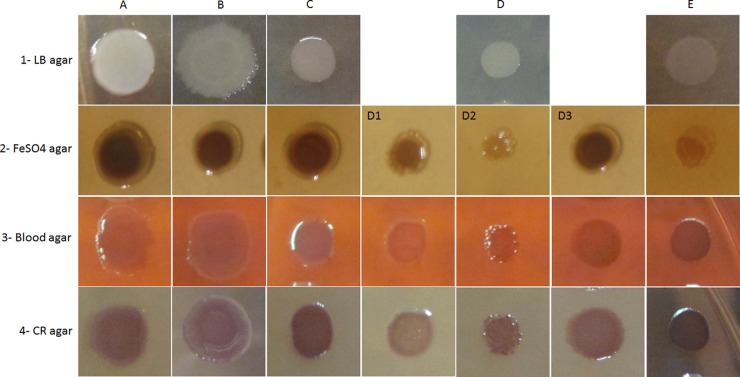
The variants were categorized into five groups based on their colony morphology on different solid media after 24 h of incubation (Fig. 1). In general, the colony morphologies on LB agar could be classified as: glossy with diffuse edges (group A), glossy with round edges (group B), small glossy colonies (group C), small nonglossy colonies (group D, subdivided into three groups based on their morphology on other nutrient media), and small colonies producing brown pigment in the surrounding agar (group E). The variants classified as small colony variants (SCVs) had a 24-h colony diameter of <4.5 mm. In contrast, the colony diameters for classes A and B were ∼8 mm. Colonies of the control group all displayed a morphotype similar to group A (large, spreading, grayish colonies with dark centers and translucent irregular edges), typical of the colonial morphology of the ancestral form of P. aeruginosa PAO1. Group E colonies had small, circular, and translucent colonies, and the surrounding agar became a characteristic coffee-brown color, indicative of pyomelanin production. Closer inspection revealed the colonies themselves to be grayish on LB medium and light brown on FeSO4 medium. Some variants in group D exhibited light brown or colorless colonies on FeSO4 medium indicating that iron uptake was likely compromised, whereas wild-type and control colonies typically showed a brown coloration in high-iron medium. Thus, group D was subdivided into group D1 (faint brown color in FeSO4 medium and faint pink in blood agar), group D2 (colorless colonies on FeSO4-supplemented plates), and group D3 (dark brown color on FeSO4-supplemented plates). Moreover, colonies in group D1 had a light pink color in Congo red (CR)-supplemented media, indicating biofilm formation could be affected since the red color accumulated over time by a colony due to EPS-dependent CR binding serves as a surrogate for assessment of EPS production. Table 2 provides a quantitative presentation of the effect of each phage treatment on the diversification of the colony morphology in each class of isolates. It is noteworthy that some colony types uniquely emerged from specific phage treatments, e.g., morphotype C from phage PP7 or morphotypes D3 and E from phage E79.
Fig 1.
Types of colony morphology of P. aeruginosa PAO1 phage-resistant variants on LB agar (row 1), LB agar plus 4 mM FeSO4 (row 2), LB agar plus 5% sheep blood (row 3), or LB agar with Congo red (CR) (row 4). Columns: A, glossy colony with diffuse edges; B, glossy colony with smooth edges, on CR agar the color is concentrated in the center of colony; C, small glossy colony, dark color on CR agar; D, small nonglossy colony; D1, light brown on FeSO4 agar and light pink on blood agar, light rose on CR agar; D2, colorless on FeSO4 agar; D3, dark rose on CR agar; E, small nonglossy colony producing brown pigments, faint color on FeSO4 agar.
Table 2.
Percentage of each colony group in the three classes of isolates based on the type of phage challenge
| Class or type (description) | % of each colony morphotype for each class of isolatesa |
||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D1 | D2 | D3 | E | |
| I (PP7 phage) | 50 | 35 | 10 | 5 | |||
| II (E79 phage) | 15 | 35 | 35 | 15 | |||
| III (phage mixture) | 5 | 10 | 15 | 15 | 10 | 40 | 5 |
| Control (no phage) | 100 | ||||||
The number of colonies in each class was 20. Colony morphologies of P. aeruginosa PAO1 phage-resistant variants: A, glossy colony with diffuse edges; B, glossy colony with smooth edges; C, small glossy colony, dark color on CR agar; D, small nonglossy colony; D1, light brown on FeSO4 agar and light pink on blood agar, light rose on CR agar; D2, colorless on FeSO4 agar; D3, dark rose on CR agar; E, small nonglossy colony producing brown pigments, faint color on FeSO4 agar.
Growth rate and biofilm formation.
The measured OD600 was used to calculate the growth rate (ΔOD600/Δt) at different time points during the logarithmic growth phase. These data were used to calculate the maximum growth rate during log phase. Furthermore, biofilm formation was quantified by measuring the optical density of solubilized crystal violet in absolute ethanol at 570 nm. This value was divided by the OD600 value measured for the planktonic bacteria in each well to account for the difference in growth rates of the variants. These data are presented in Table 3 and Fig. S1 in the supplemental material. Variants in classes II (E79 resistant) and III (PP7 and E79 resistant) exhibited lower growth rates and yields (Table 3; see also Fig. S1a to d in the supplemental material) compared to the control. The growth rate and yield for various colony morphotypes in each class were analyzed in Fig. S1b and d in the supplemental material. Interestingly, isolates that have the same colony morphotypes but have evolved through different evolutionary contexts (selective pressure from different phage environments) appear to have different growth determinants. Figure S1b shows that all of the colony groups in class III have lower growth rates compared to the same colony groups in class I. When biofilm formation was quantified, only class I isolates (PP7-resistant) showed significantly less biofilm formation (Table 3; see also Fig. S1e and f in the supplemental material). It is noteworthy that all of the colony morphotypes in this class formed less biofilm compared to the same colony morphotypes in classes I and III (see Fig. S1f in the supplemental material). Biofilms were observed to predominantly form at the air-liquid interface, although a number of the class II variants also form biofilms at the bottom of the wells. Another interesting observation from Fig. S1f is that, although evolved through the same evolutionary process (i.e., same phage treatment), colony group E displays significantly less biofilm formation compared to the other colony groups in both classes II and III. ANOVA revealed no statistically significant variation within the classes between different colony morphotypes for growth and biofilm formation. As expected based on the colony morphologies on CR plates, variants with colony morphotype D1 were less capable of forming biofilms (data not shown).
Table 3.
Values of fitness and virulence determinants for the P. aeruginosa phage-resistant variants categorized based on phage treatmenta
| Parameterb | Avg (±95% CI) |
|||
|---|---|---|---|---|
| Control | Phage challengec |
|||
| Class I | Class II | Class III | ||
| Max growth rate (ΔOD600/Δt) | 8.6 × 10−3 (3 × 10−3) | 7.5 × 10−3 (2 × 10−3) | 3.9 × 10−3 (1 × 10−3) | 4 × 10−3 (1 × 10−3) |
| Final yield (OD600) | 1.58 (4 × 10−2) | 1.51 (6 × 10−2) | 1.21 (0.2) | 1.22 (0.2) |
| Biofilm formation (OD570)* | 1.22 (0.3) | 0.46 (0.3) | 1.42 (0.4) | 1.15 (0.5) |
| Twitching motility (mm)* | 23.65 (1.4) | 9.98 (1.9) | 18.78 (1.9) | 10.58 (2.5) |
| Swimming motility (mm) (20–40 h)* | 45.9 (5.6) to 83.7 (7.3) | 44.7 (3.9) to 88.6 (7.9) | 33.6 (3.9) to 62.9 (6.4) | 33.7 (3.6) to 62.3 (4.6) |
| Swarming motility (mm) (20–40 h)* | 103.5 (25.7) to 128.6 (14.2) | 80.2 (19.8) to 83.2 (17.7) | 42.2 (12.2) to 107.5 (17.4) | 53.8 (18.2) to 65.4 (20.3) |
| Pyocyanin (OD525)* | 0.015 (3 × 10−3) | 0.017 (4 × 10−3) | 0.027 (0.011) | 0.022 (6 × 10 −3) |
| Pyoverdin (ex/em 405/465 nm)* | 1.8 × 105 (4.4 × 104) | 2.3 × 105 (4.3 × 104) | 2.1 × 105 (4.3 × 104) | 2.5 × 105 (4.5 × 104) |
| Pyorubin (OD525)* | 0.10 (0.058) | 0.0975 (0.039) | 0.139 (0.082) | 0.1909 (0.07) |
| Pyomelanin (mm)* | – | – | + | + |
| Rhamnolipids* | 1.89 (0.5) | 3.07 (0.3) | 1.34 (0.5) | 1.57 (0.5) |
| H2O2 MIC (nM) | 14.47 (2.2) | 27.34 (4.4) | 33.33 (6.7) | 19.44 (4.6) |
| Streptomycin MIC (mg/ml) | 10.3 (2.7) | 12.8 (2.5) | 7.3 (3.8) | 11.8 (3.4) |
| Tobramycin MIC (mg/ml) | 12.8 (2.5) | 6.8 (1.6) | 12.1 (3.3) | 10.3 (2.7) |
Each value represents the class average (±95% confidence interval [CI]). Boldface numbers represent statistically significant differences from the control (threshold, P < 0.05). Values of fitness and virulence determinants for the P. aeruginosa phage-resistant variants are categorized based on phage treatment.
*, the indicated parameter values have been divided by the growth level (OD600) to correct for different growth rates. ex/em, excitation wavelength/emission wavelength.
Class I, variants resistant to phage PP7; class II, variants resistant to phage E79; class III, variants resistant to both PP7 and E79.
Bacterial motility and rhamnolipid production.
All of the variants challenged with different phage treatments exhibited decreased twitching motility (Table 3; see also Fig. S2a and b in the supplemental material). Twitching motility is a flagellum-independent mode of surface translocation mediated by type IV pili (58). Most notably, the twitching diameters decreased by half for the variants challenged by phage PP7, namely, classes I and III (see Fig. S1a in the supplemental material). As shown in Fig. S1b, all of the colony morphotypes in these two classes have lower twitching motilities compared to the same colony morphotypes in class I and in the control. Bacterial swimming diameters in semisolid agar showed a decrease compared to the control for the variants challenged with phage E79, namely, classes II and III, even after 40 h of incubation (Table 3; see also Fig. S2c in the supplemental material). Interestingly, colony morphotypes A and E in class II exhibit a lower swimming diameter compared to the same colony morphotypes in other classes (see Fig. S2d in the supplemental material). Classes II and III also showed significant difference from the swarming diameter of the control population after 20 h of incubation (Table 3; see also Fig. S2e in the supplemental material); all colony groups in class II showed a lower swarming diameter compared to the same colony groups in other classes after this time period (Fig. S2f). However, after 40 h of incubation, the swarming motility of class II increased to the level of the control (see Fig. S2e, inset, in the supplemental material), due to a significant increase in the swarming diameter of colony groups A and E in class II (data not shown). ANOVA showed that the case in which the difference within the different colony groups in a class was statistically significant was that of swimming diameters of class III.
To identify the reason for the decrease in swarming motility for a number of isolates, we quantified the production of rhamnolipids, surfactants known to affect bacterial swarming motility. A significant difference in the production of rhamnolipids (indicated by the diameter of the halo formed on CTAB plates) was not observed when whole populations of various classes were compared to the control population (Table 3; see also Fig. S3a in the supplemental material). However, when the variants in each class were categorized based on their colony morphotypes (see Fig. S3b in the supplemental material), it became clear that (i) colony groups A and B in class I exhibited greater rhamnolipid production compared to the control, something that was not observed in variants with the same colony morphology in other classes, and (ii) colony group E has very little rhamnolipid production in class II, and no rhamnolipid activity was detected for colony group E in class III.
These results were further confirmed by measuring the zone of wetting on the swarm plates around the swarming zone (data not shown). The wetting zone has been described in the literature to be correlated to biosurfactant production. A noteworthy observation on the swarm plates was the extremely diverse swarming patterns; specifically for classes I and III (see Fig. S4 in the supplemental material). These variants formed a range of patterns, all different from the well-known dendrimer (branching) swarming pattern characteristic of P. aeruginosa.
Resistance to oxidative stress and antibiotics.
Class II variants (E79 resistant) showed a significant increase in the MIC level for H2O2 (Table 3; see also Fig. S5a in the supplemental material). All of the colony morphotypes in this class had a higher resistance to oxidative stress compared to the same colony morphotype in other classes with one exception: the H2O2 MIC for colony group E (pyomelanogenic isolates) was equally high in both classes II and III, which was significantly higher than the control and most other colony groups in all classes. The MIC levels for tobramycin (Table 3; see also Fig. S5a in the supplemental material) and streptomycin (Table 3; see also Fig. S5c in the supplemental material) did not show a significant difference when the total populations in different classes were compared to the control population. However, when the individual colony groups in each class were studied separately, it became evident that (i) in class I, colony group D had a lower tobramycin MIC, and colony group C had a lower streptomycin MIC compared to the control, (ii) colony group E had lower MICs for both antibiotics in classes II and III, and (iii) colony group C in class III exhibited a higher MIC compared to the control for streptomycin. Only the streptomycin MIC levels for class III showed a significant variation between different colony groups in the same class according to ANOVA.
Siderophore and pigment production.
Class III variants (resistant to both phages) exhibited heightened levels of pyocyanin, pyoverdin, and pyorubin production, and class II variants (E79 resistant) produced high levels of pyocyanin (Table 3; see also Fig. S6 in the supplemental material). Figure S6b shows that the increase in pyoverdin for class III isolates is mainly due to colony morphotypes A and B in this class. These isolates produced significantly higher levels of pyoverdin compared to isolates with similar colony morphology in other classes and the control group (which contained exclusively colonies of group A). Colony group E in this class produced very low levels of pyoverdin. Colony groups A and B, however, did not show increases in the production of other pigments in class III, but instead groups C and D produced higher pyocyanin and pyorubin levels in this class (see Fig. S6d and f in the supplemental material). It is noteworthy that all colony groups in class II produced higher levels of pyocyanin compared to similar colony morphotypes in other classes.
Although, as a population, class I did not reveal significant changes in the level of secreted pigments compared to the control, when different colony morphotypes in this class were studied separately it was observed that colony group C produced significantly lower levels of pyorubin and colony group D produced lower levels of both pyocyanin and pyorubin. Class I was also the only class to exhibit a variation within the group for pyocyanin production levels for different colony morphotypes according to ANOVA.
Pyomelanin production was limited to some variants in classes II and III (treated with phage E79), namely, those in colony group E, as indicated by the pigment production on furunculosis agar.
Transcription of selected genes and sequencing.
To investigate the reason for the changes in phenotypic traits of variants, mRNA transcription levels were quantified via RT-qPCR for selected genes involved in the biogenesis of flagella, the biogenesis of type IV pili, rhamnolipid production, pyocyanin biogenesis, and iron uptake. One variant from each colony group from class III with phenotypic traits representative of that group was selected for RT-qPCR experiments. The results are presented in Table 4. The transcription of genes involved in flagellar biogenesis was observed to have decreased for almost all of the samples, whereas the transcription of pilA increased for all but groups B and D1 (Table 4). The transcription levels of rhlB showed an increase for groups A and B and a decrease for all of the other groups. The transcription of phzS gene (pyocyanin biogenesis) increased for group B isolates. Iron uptake genes (pchG and pvdA) show an increase in transcription for all samples except for group E, which is in agreement with the data reported in Fig. S6b in the supplemental material. This may appear to be in contradiction to the phenotype of group D1 isolate, which had completely colorless colonies on FeSO4 plates; however, it should be noted that many other genes are involved in the iron uptake process and the transcription of other genes involved in this process, the posttranscription regulation of genes, and the status of export machinery may affect the phenotype observed for group D1 despite the transcription of the two genes in the present study.
Table 4.
Fold change in mRNA transcription levels of selected genes
| Group | Fold changea in mRNA transcription levels of: |
|||||||
|---|---|---|---|---|---|---|---|---|
| fliC | fliD | pilA | rhlA | rhlB | phzS | pchG | pvdA | |
| A | 0.78 (0.1)‡ | 0.19 (0.05) | 46.39 (6) | 0.94 (0.1) | 7.19 (0.1) | 0.72 (0.02) | 6.22 (0.9) | 294.50 (11) |
| B | 0.14 (0.02) | 0.30 (0.02) | 0.02 (0.05) | 1.46 (0.1) | 6.95 (0.1) | 1.07 (0.08) | 10.37 (0.8) | 414.21 (8) |
| C | 1.45 (0.1) | 0.02 (0.001) | 60.33 (5) | 0.43 (0.1) | 0.02 (0.1) | 0.29 (0.02) | 4.33 (0.5) | 83.74 (5) |
| D1 | 0.66 (0.06) | 0.16 (0.02) | 0.02 (0.001) | 1.10 (0.1) | 0.11 (0.1) | 0.61 (0.01) | 6.87 (1) | 758.85 (13) |
| D2 | 0.57 (0.01) | 0.19 (0.01) | 35.69 (3) | 0.63 (0.1) | 0.91 (0.1) | 0.63 (0.03) | 7.94 (1) | 616.85 (9) |
| E | 0.11 (0.01) | 0.12 (0.01) | 362.65 (9) | 0.06 (0.1) | 0.68 (0.1) | 0.04 (0.005) | 0.48 (0.1) | 13.45 (2) |
Values represent the fold increase or reduction relative to the control group. Findings for genes involved in the biogenesis of flagella (fliC, flagellin type B; fliD, flagellar capping protein), the biogenesis of type IV pili (pilA, type 4 fimbrial precursor PilA), rhamnolipid production (rhlA and rhlB, rhamnosyltransferase chains A and B), pyocyanin biogenesis (phzS, flavin-containing monooxygenase), and iron uptake (pchG, pyochelin biosynthetic protein PchG; pvdA, l-ornithine N5-oxygenase) are presented. Data represent averages for three independent RT-qPCR analyses. Values in parentheses represent 95% confidence intervals.
Due to the decrease in twitching motility for all variants, the genes associated with the surface appendage responsible for this mode of motility (bacterial pili) were studied in more detail. All of the genes known to be involved in pili biogenesis and regulation were sequenced (see Table S2 in the supplemental material) for a variant from colony group C (the group exhibiting the highest decrease in twitching motility) chosen from class III. The results revealed that there were no single nucleotide polymorphisms (SNP) in the genes sequenced. Although this does not rule out the possibility of a point mutation in any of the other genes that were not sequenced, it is possible that other mechanisms (e.g., phase variation) could be responsible for the observed change in phenotype.
DISCUSSION
In the present study, a genotypically uniform culture of P. aeruginosa was challenged with various phage environments, which led to the emergence of phage-resistant variants and the diversification of the initial population into five main colony groups. Various fitness determinants and virulence factors were found to have changed significantly for variants that emerged as a result of selective pressure from different phage treatments. A summary of these phenotypic changes is presented in Table 3, and the qualitative trends are presented in Table S4 in the supplemental material. Interestingly, resistance development was observed to be maintained in the bacterial variants in the absence of the phage to which they conferred resistance. Although the variants eventually reverted back to phage susceptibility, their reversion rate was context dependent, and variants resistant to two phages were more stable in maintaining their phage resistance. It would be interesting to verify the fitness and virulence determinants of these reverted variants to understand whether they have reverted back to their original phenotype or whether they have a change in fitness/virulence compared to the wild type. This is a very important concept in phage therapy and should be the subject of further studies.
As expected, the changes in phenotype were different for the variants that developed resistance to E79 or PP7, most likely because of the differences in the mode of infection of these phages. E79 attaches to host lipopolysaccharide (LPS) (59) and uses a classic two-component endolysin/holin system to liberate the newly formed particles from the host cell (60). Bacteriophage PP7 adsorbs to the sides of the polar pili of P. aeruginosa (61) and lyses the host by a short hydrophobic protein that crosses the cytoplasmic membrane and destroys the membrane potential (62). Because these two viruses differ in their host receptor and mode of lysis, it follows that they would impart different kinds of selective pressure on their host, thus leading to phage-resistant variants with different phenotypes. Furthermore, the population that developed in contact with a heterogeneous phage environment (class III) exhibited significantly different phenotypic traits from the ones that were developed in homogeneous phage environments. We hypothesize that there is a strong connection between the phenotypic traits observed for the variants and the type of phage treatment (and thus the phage selective pressure) that they have been subject to. This hypothesis is reinforced by the fact that variants with the same colony morphotype exhibit different phenotypic traits based on their evolutionary context (i.e., phage treatment). Therefore, we have chosen to discuss the results (specifically the qualitative trends) based on the type of the phage environment that resulted in the emergence of the resistant variants.
Phenotypic traits of isolates from a homogeneous phage environment: pilus-associated traits for PP7-resistant variants.
One of the main mechanisms of phage resistance is envelope modification by blocking or modifying the phage receptors on the host cell (63). The receptors that are used by some phages to attack bacteria might be capsules (64, 65) or other virulence determinants, which implies that the development of phage resistance has the potential to impact virulence (66). PP7 is a pilus-specific phage, and thus we hypothesized that variants resistant to this phage may have altered pilus synthesis levels or changed the pilus protein sequence or structure in such a way that the phage cannot bind to it. Type IV pili are responsible for the flagellum-independent mode of surface translocation called twitching motility (58). We observed that class I (and III) variants (exposed to PP7 phage) were severely defective in twitching motility irrespective of their colony morphology, thus supporting our hypothesis that the pili may have been affected in these variants. Pili are also principal adhesins, mediating the adherence to eukaryotic cell surfaces and abiotic surfaces (58), and play an important role in the formation of mature multicellular structures in Pseudomonas biofilms (67). The observation that none of the colony morphotypes in class I was unable to form a stable biofilm (even though they were able to attain growth rates as high as the control) is additional evidence pointing to defective pili. Interestingly, mRNA transcription results show both increased and decreased transcription of pilA in PP7-resistant variants with different colony morphologies. An increase in the transcription of pilA is suggestive of hyperpiliation, which has been linked in some instances to defective pilus function (68, 69). The sequencing data, however, did not reveal point mutations in any of the pilus-related genes that were sequenced and thus did not provide any insight into the mechanism of PP7 resistance. It is possible that mutations occurred in a global regulatory gene. Further research needs to be conducted to verify this hypothesis.
Cost of resistance on E79-resistant variants.
Class II variants (resistant to E79) displayed compromised growth rates, growth yields, and swimming and swarming motilities, among others, for all of the colony morphotypes in this group. Since these effects were not observed for the same colony morphotypes in class I, our results imply that for the system under study, the cost of resistance depends on the phage-host system.
SCVs.
An interesting trait observed for class II variants was the increase of small colony variants (SCVs). Most of the isolates resistant to E79 exhibited small colonies. Slow-growing subpopulations of bacterial pathogens, termed small colony variants, have been observed in various bacterial strains and have been, in some cases, associated with persistent and relapsing infection (70) and heightened antibiotic resistance (71). We did not observe significant increases in the antibiotic susceptibility of the isolated SCVs in class II; however, they did exhibit a slower growth rate and yield. Also, SCVs have been reported to have a higher tendency to form biofilms (9, 72–74). However, we did not observe a significant difference between the biofilm-forming ability of the SCVs in group D (class II) and the control group. The reduced colony diameter size for resistant variants could also suggest that they are impaired in motility and/or chemotaxis. The decreased swimming motility of colony groups D and E in class II provides evidence for this hypothesis.
Production of pyomelanin.
We observed the production of pyomelanin for some of the variants that were challenged with E79 which were classified as colony group E. Variants resistant to this phage have been previously reported to be strong pyorubin producers (75); however, we did not find instances of pyomelanin production reported in the literature for this system. Pyomelanin is a brown pigment that diffuses through agar, and its color varies in intensity from light brown to a deep coffee color. Proposed roles for pyomelanin include the enhancement of bacterial surface attachment, extracellular electron transfer, iron reduction/acquisition, induction of virulence factor expression, heavy metal binding, and protection from environmental stress (76). Pyomelanin-containing supernatants have been demonstrated to protect nonpyomelanogenic strains against oxidative stress (77). This is in agreement with our results, since variants in colony group E were observed to have high resistance to oxidative stress. The pyomelanogenic variants also showed a decreased MIC level to tobramycin and streptomycin, which means they have a higher sensitivity to this antibiotic. They also produced less pyoverdin, and their mRNA transcription levels for the gene that encodes the pyochelin biosynthetic protein PchG were decreased. These variants also exhibited a decrease in rhamnolipid secretion and in the transcription of the rhamnolipid genes rhlA and rhlB and of the flagellar biogenesis genes fliC and fliD.
Effect of heterogeneous phage environment.
The cumulative cost of carrying multiple resistance mutations has been extensively studied for antibiotics (78, 79) and is believed to depend on the cost of each individual mutation, as well as on any interactions between them (80). Combinations of pairs of drugs have been characterized as additive, synergistic, antagonistic, or suppressive (81). By analogy, the right combination of phages could affect the cost of resistance for the host bacterium in such a manner as to constrain its evolution. The evolution of phage-resistant bacteria is expected for many phage applications (66). Our results indicate that although the general fitness determinants (growth rate and motility) decreased for class III variants (resistant to both phages), the production of certain virulence factors, in fact, increased, rendering these variants potentially more virulent.
Decrease in swarming motility.
We observed that although the frequency of development of resistance was lower in the heterogeneous phage environment consisting of both E79 and PP7 (class III), the change in various phenotypic traits was much more pronounced. Accordingly, Koskella et al. found P. syringae populations that were exposed to multiple phage environments to pay a higher cost in terms of population growth (46). We observed the specific traits of classes I (PP7 resistant) and II (E79 resistant) to have been manifested in class III, namely, the significantly lower twitching, lower growth rate and lower yield, the development of SCVs, the production of pyomelanin, and lower swimming and swarming motilities. Although class II variants also had a significantly lower swarming motility after 20 h, they managed to regain their swarming ability with increased incubation time, but overall, the swarming motility of class III variants did not increase much with increased incubation time. Furthermore, class III variants were observed to form a variety of different patterns on the swarm plates which has been reported to result from a change in the relative amounts of the different biosurfactants (e.g., rhamnolipids) produced by these variants (82, 83). Swarming motility is a very complex mode of bacterial motility believed to depend on flagella and pili, as well as on biosurfactant production (84). This trait is strongly affected by the quorum-sensing system (85). There are many genes that are involved directly or indirectly in the swarming motility (86), and a mutation in any of these genes could result in altered swarming ability. Additional research needs to be conducted in order to fully characterize the observed decrease in swarming motility in the present study.
Siderophores and pyocins.
We observed a significant increase in the production of pyoverdin (colony groups A and B) and pyocyanin (colony groups C and D) for class III variants. P. aeruginosa excretes siderophores, e.g., pyoverdin, into the environment which function as powerful iron chelators, solubilizing and transporting iron through the bacterial membranes (87). Pyoverdin is thus essential for the virulence of Pseudomonas (88). Pyocyanin is also a virulence factor and, besides demonstrating antibiotic activity against a wide variety of microorganisms, it has been shown to interfere with mammalian cell respiration, growth, and proliferation (89, 90). Thus, the observed increase in the production of these two compounds has the potential to increase the virulence of phage-resistant variants of Pseudomonas. The increase in the production of pyocyanin and pyoverdin, along with the emergence of variants expressing a number of other virulence determinants (e.g., pyomelanin production and resistance to oxidative stress), raises serious concerns for the application of the tested phage for therapeutic means and highlights the need for more mechanistic studies in this field. Although the use of phage mixtures is believed to be the secret of the apparent success of phage therapy in the East (91), our results indicate that the heterogeneous phage environment under study led to the emergence of variants with heightened levels of a number of important virulence factors. However, this could potentially be tempered by the decreased growth rate of these variants. The true virulence of these variants remains to be investigated in vitro toward mammalian cells and in vivo; the latter in particular would provide a more realistic picture of the extent to which decreased fitness can counteract the increase in some virulence determinants for this class of variants.
Concluding remarks.
It is known that bacteria can develop mechanisms to become resistant to phages and that phages can evolve new ways to attack the resistant bacteria. We demonstrated here that resistance development is associated with multiple changes in bacterial fitness and virulence determinants that are maintained in the absence of the agent to which they confer resistance. This modification of phenotypic traits proved to be associated with changes in gene regulation levels. It was further observed that the phenotypic changes were context dependent and differed for variants exhibiting the same colony morphotype but developed through different evolutionary contexts. Certain traits—e.g., the emergence of SCVs, an increased production of pyomelanin, pyocyanin, and pyoverdin, and heightened resistance to oxidative stress—can potentially increase the virulence of variants, although the true fitness of the variants remains to be investigated in contact with mammalian cells. It is important to acknowledge that repeating this study in vivo may lead to different results in terms of phenotypes that may emerge. We finally emphasize that carefully controlled studies of the genetic interactions in homogeneous and heterogeneous phage environments will not only increase our understanding of the evolutionary biology of the prokaryotes, but are also crucial to pave the way for financial and regulatory agencies' support and approval of phage therapy and use.
ACKNOWLEDGMENTS
This study is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC Strategic Research Network on Bioactive Paper–SENTINEL) and the Canada Research Chairs (CRC) program.
We thank M. Elimelech (Yale University) for providing P. aeruginosa PAO1, H. Lam (McGill University) for designing the primers for reverse transcription-PCR, S. Brunet (Genome Quebec Innovation Center) for performing the sequencing, and G. Hidalgo and C. O'May (McGill University) for technical advice and valuable feedback on the manuscript.
Footnotes
Published ahead of print 22 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03817-12.
REFERENCES
- 1. Breidenstein EB, de la Fuente-Nunez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19:419–426 [DOI] [PubMed] [Google Scholar]
- 2. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GKS, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 3. Kipnis E, Sawa T, Wiener-Kronish J. 2006. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Maladies Infect. 36:78–91 [DOI] [PubMed] [Google Scholar]
- 4. Zierdt CH, Schmidt PJ.1964. Dissociation in Pseudomonas aeruginosa. J. Bacteriol. 87:1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rakhimova E, Munder A, Wiehlmann L, Bredenbruch F, Tümmler B. 2008. Fitness of isogenic colony morphology variants of Pseudomonas aeruginosa in murine airway infection. PLoS One 3:e1685 doi:10.1371/journal.pone.0001685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scott AE, Timms AR, Connerton PL, Loc Carrillo C, Adzfa Radzum K, Connerton IF. 2007. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 3:e119 doi:10.1371/journal.ppat.0030119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ciofu O, Mandsberg LF, Wang H, Høiby N. 2012. Phenotypes selected during chronic lung infection in cystic fibrosis patients: implications for the treatment of Pseudomonas aeruginosa biofilm infections. FEMS Immunol. Med. Microbiol. 65:215–225 [DOI] [PubMed] [Google Scholar]
- 8. Häussler S, Tümmler B, Weissbrodt H, Rohde M, Steinmetz I. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621–625 [DOI] [PubMed] [Google Scholar]
- 9. Häussler S, Ziegler I, Löttel A, Götz FV, Rohde M, Wehmhöhner D, Saravanamuthu S, Tümmler B, Steinmetz I. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52:295–301 [DOI] [PubMed] [Google Scholar]
- 10. Tielen P, Narten M, Rosin N, Biegler I, Haddad I, Hogardt M, Neubauer R, Schobert M, Wiehlmann L, Jahn D. 2011. Genotypic and phenotypic characterization of Pseudomonas aeruginosa isolates from urinary tract infections. Int. J. Med. Microbiol. 301:282–292 [DOI] [PubMed] [Google Scholar]
- 11. Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 35:652–680 [DOI] [PubMed] [Google Scholar]
- 12. Krahn T, Gilmour C, Tilak J, Fraud S, Kerr N, Lau CHF, Poole K. 2012. Determinants of intrinsic aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56:5591–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa: a phenomenon of bacterial resistance. J. Med. Microbiol. 58:1133–1148 [DOI] [PubMed] [Google Scholar]
- 14. Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, Hauser A, McDougald D, Webb JS, Kjelleberg S. 2009. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 3:271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brockhurst MA, Buckling A, Rainey PB. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc. R. Soc. B Biol. Sci. 272:1385–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rohwer F, Edwards R. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213–231 [DOI] [PubMed] [Google Scholar]
- 18. Bohannan BJM, Lenski RE. 1997. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology 78:2303–2315 [Google Scholar]
- 19. Bohannan BJM, Lenski RE. 1999. Effect of prey heterogeneity on the response of a model food chain to resource enrichment. Am. Nat. 153:73–82 [DOI] [PubMed] [Google Scholar]
- 20. Brockhurst MA, Rainey PB, Buckling A. 2004. The effect of spatial heterogeneity and parasites on the evolution of host diversity. Proc. R. Soc. B Biol. Sci. 271:107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buckling A, Rainey PB. 2002. The role of parasites in sympatric and allopatric host diversification. Nature 420:496–499 [DOI] [PubMed] [Google Scholar]
- 22. Chao L, Levin BR, Stewart FM. 1977. A complex community in a simple habitat: an experimental study with bacteria and phage. Ecology 58:369–378 [Google Scholar]
- 23. Lenski RE, Levin BR. 1985. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 125:585–602 [Google Scholar]
- 24. Lythgoe KA, Chao L. 2003. Mechanisms of coexistence of a bacteria and a bacteriophage in a spatially homogeneous environment. Ecol. Lett. 6:326–334 [Google Scholar]
- 25. Schrag SJ, Mittler JE. 1996. Host-parasite coexistence: the role of spatial refuges in stabilizing bacteria-phage interactions. Am. Nat. 148:348–377 [Google Scholar]
- 26. Pirnay JP, Verbeken G, Rose T, Jennes S, Zizi M, Huys I, Lavigne R, Merabishvili M, Vaneechoutte M, Buckling A, De Vos D. 2012. Introducing yesterday's phage therapy in today's medicine. Future Virol. 7:379–390 [Google Scholar]
- 27. Heo YJ, Lee YR, Jung HH, Lee J, Ko G, Cho YH. 2009. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrob. Agents Chemother. 53:2469–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McVay CS, Velásquez M, Fralick JA. 1934. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob. Agents Chemother. 51:2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soothill J, Hawkins C, Anggard E, Harper D. 2004. Therapeutic use of bacteriophages. Lancet Infect. Dis. 4:544–545 [DOI] [PubMed] [Google Scholar]
- 30. Debarbieux L, Leduc D, Maura D, Morello E, Criscuolo A, Grossi O, Balloy V, Touqui L. 2010. Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J. Infect. Dis. 201:1096–1104 [DOI] [PubMed] [Google Scholar]
- 31. Marza JA, Soothill JS, Boydell P, Collyns TA. 2006. Multiplication of therapeutically administered bacteriophages in Pseudomonas aeruginosa-infected patients. Burns. 32:644–646 [DOI] [PubMed] [Google Scholar]
- 32. Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, Glonti T, Krylov V, Mast J, Van Parys L, Lavigne R, Volckaert G, Mattheus W, Verween G, De Corte P, Rose T, Jennes S, Zizi M, De Vos D, Vaneechoutte M. 2009. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One 4:e4944 doi:10.1371/journal.pone.0004944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wright A, Hawkins CH, Änggård EE, Harper DR. 2009. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa: a preliminary report of efficacy. Clin. Otolaryngol. 34:349–357 [DOI] [PubMed] [Google Scholar]
- 34. Comolli JC, Hauser AR, Waite L, Whitchurch CB, Mattick JS, Engel JN. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drake D, Montie TC. 1988. Flagella, motility, and invasive virulence of Pseudomonas aeruginosa. J. Gen. Microbiol. 134:43–52 [DOI] [PubMed] [Google Scholar]
- 36. Barclay ML, Begg EJ, Chambers ST, Thornley PE, Pattemore PK, Grimwood K. 1996. Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J. Antimicrob. Chemother. 37:1155–1164 [DOI] [PubMed] [Google Scholar]
- 37. Boucher JC, Mudd H, Deretic V. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deretic V, Schurr MJ, Boucher JC, Martin DW. 1994. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J. Bacteriol. 176:2773–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gerber AU, Craig WA. 1982. Aminoglycoside-selected subpopulations of Pseudomonas aeruginosa: characterization and virulence in normal and leukopenic mice. J. Lab. Clin. Med. 100:671–681 [PubMed] [Google Scholar]
- 40. Mizoguchi K, Morita M, Fischer CR, Yoichi M, Tanji Y, Unno H. 2003. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl. Environ. Microbiol. 69:170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koskella B, Taylor TB, Bates J, Buckling A. 2011. Using experimental evolution to explore natural patterns between bacterial motility and resistance to bacteriophages. ISME J. 5:1809–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meyer JR, Agrawal AA, Quick RT, Dobias DT, Schneider D, Lenski RE. 2010. Parallel changes in host resistance to viral infection during 45,000 generations of relaxed selection. Evolution 64:3024–3034 [DOI] [PubMed] [Google Scholar]
- 43. Hawkey PM, Munday CJ. 2004. Multiple resistance in Gram-negative bacteria. Rev. Med. Microbiol. 15:51–61 [Google Scholar]
- 44. Li XZ. 2003. Efflux-mediated multiple antibiotic resistance in Pseudomonas aeruginosa. Chin. J. Antibiot. 28:577–596 [Google Scholar]
- 45. Livermore DM. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634–640 [DOI] [PubMed] [Google Scholar]
- 46. Koskella B, Lin DM, Buckling A, Thompson JN. 2012. The costs of evolving resistance in heterogeneous parasite environments. Proc. R. Soc. B Biol. Sci. 279:1896–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bohannan BJM, Travisano M, Lenski RE. 1999. Epistatic interactions can lower the cost of resistance to multiple consumers. Evolution 53:292–295 [DOI] [PubMed] [Google Scholar]
- 48. Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572–581 [DOI] [PubMed] [Google Scholar]
- 49. Spilker T, Coenye T, Vandamme P, LiPuma JJ. 2004. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J. Clin. Microbiol. 42:2074–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carlson K. 2004. Working with bacteriophages, appendix A. In Kutter E, Sulakvelidze A. (ed), Bacteriophages. CRC Press, Inc, Boca Raton, FL [Google Scholar]
- 51. Merritt JH, Kadouri DE, O'Toole GA. 2011. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 22:1B1.1–1B.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 53. Palumbo SA. 1972. Role of iron and sulfur in pigment and slime formation by Pseudomonas aeruginosa. J. Bacteriol. 111:430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ogunnariwo J, Hamilton-Miller MT. 1975. Brown and red pigmented Pseudomonas aeruginosa: differentiation between melanin and pyorubrin. J. Med. Microbiol. 8:199–203 [DOI] [PubMed] [Google Scholar]
- 55. Siegmund I, Wagner F. 1991. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Techniques 5:265–268 [Google Scholar]
- 56. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 57. Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bardy SL, Ng SYM, Jarrell KF. 2003. Prokaryotic motility structures. Microbiology 149:295–304 [DOI] [PubMed] [Google Scholar]
- 59. Jarrell K, Kropinski AM. 1977. Identification of the cell wall receptor for bacteriophage E79 in Pseudomonas aeruginosa strain PAO. J. Virol. 23:461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ceyssens PJ, Lavigne R. 2010. Bacteriophages of Pseudomonas. Future Microbiol. 5:1041–1055 [DOI] [PubMed] [Google Scholar]
- 61. Bradley DE. 1972. Shortening of Pseudomonas aeruginosa pili after RNA-phage adsorption. J. Gen. Microbiol. 72:303–319 [DOI] [PubMed] [Google Scholar]
- 62. Young R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 64. Lindberg AA. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27:205–241 [DOI] [PubMed] [Google Scholar]
- 65. Rakhuba DV, Kolomiets EI, Szwajcer-Dey E, Novik GI. 2010. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Polish J. Microbiol. 59:145–155 [PubMed] [Google Scholar]
- 66. Levin BR, Bull JJ. 2004. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2:166–173 [DOI] [PubMed] [Google Scholar]
- 67. Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, Givskov M, Whitchurch CB, Engel JN, Tolker-Nielsen T. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331–2343 [DOI] [PubMed] [Google Scholar]
- 68. Whitchurch CB, Hobbs M, Livingston SP, Krishnapillai V, Mattick JS. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene 101:33–44 [DOI] [PubMed] [Google Scholar]
- 69. Whitchurch CB, Mattick JS. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13:1079–1091 [DOI] [PubMed] [Google Scholar]
- 70. Proctor RA, van Langevelde P, Kristjansson M, Maslow JN, Arbeit RD. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20:95–102 [DOI] [PubMed] [Google Scholar]
- 71. Balwit JM, Langevelde PV, Vann JM, Proctor RA. 1994. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 170:1033–1037 [DOI] [PubMed] [Google Scholar]
- 72. Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743 [DOI] [PubMed] [Google Scholar]
- 73. Déziel E, Comeau Y, Villemur R. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Häussler S. 2004. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ. Microbiol. 6:546–551 [DOI] [PubMed] [Google Scholar]
- 75. Jarrell KF, Kropinski AM. 1981. Isolation and characterization of a bacteriophage specific for the lipopolysaccharide of rough derivatives of Pseudomonas aeruginosa strain PAO. J. Virol. 38:529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hunter RC, Newman DK. 2010. A putative ABC transporter, hatABCDE, is among molecular determinants of pyomelanin production in Pseudomonas aeruginosa. J. Bacteriol. 192:5962–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rodríguez-Rojas A, Mena A, Martín S, Borrell N, Oliver A, Blázquez J. 2009. Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance, and increased persistence in chronic lung infection. Microbiology 155:1050–1057 [DOI] [PubMed] [Google Scholar]
- 78. Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I. 2009. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5:e1000578 doi:10.1371/journal.pgen.1000578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ward H, Perron GG, Maclean RC. 2009. The cost of multiple drug resistance in Pseudomonas aeruginosa. J. Evol. Biol. 22:997–1003 [DOI] [PubMed] [Google Scholar]
- 80. Yeh P, Tschumi AI, Kishony R. 2006. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 38:489–494 [DOI] [PubMed] [Google Scholar]
- 81. Chait R, Craney A, Kishony R. 2007. Antibiotic interactions that select against resistance. Nature 446:668–671 [DOI] [PubMed] [Google Scholar]
- 82. Caiazza NC, Shanks RMQ, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187:7351–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tremblay J, Richardson A-P, Lépine F, Déziel E. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behavior. Environ. Microbiol. 9:2622–2630 [DOI] [PubMed] [Google Scholar]
- 84. Kearns DB. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tremblay J, Deziel E. 2010. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics 11:587 doi:10.1186/1471-2164-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Overhage J, Lewenza S, Marr AK, Hancock REW. 2007. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 189:2164–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Heinrichs DE, Young L, Poole K. 1991. Pyochelin-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. Infect. Immun. 59:3680–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. McDermott C, Chess-Williams R, Grant GD, Perkins AV, McFarland AJ, Davey AK, Anoopkumar-Dukie S. 2012. Effects of Pseudomonas aeruginosa virulence factor pyocyanin on human urothelial cell function and viability. J. Urol. 187:1087–1093 [DOI] [PubMed] [Google Scholar]
- 90. Muller M, Li Z, Maitz PKM. 2009. Pseudomonas pyocyanin inhibits wound repair by inducing premature cellular senescence: role for p38 mitogen-activated protein kinase. Burns 35:500–508 [DOI] [PubMed] [Google Scholar]
- 91. Brüssow H. 2005. Phage therapy: the Escherichia coli experience. Microbiology 151:2133–2140 [DOI] [PubMed] [Google Scholar]