Abstract
In this study, we constructed and expressed six fusion proteins composed of oligopeptides attached to the N terminus of the alkaline α-amylase (AmyK) from Alkalimonas amylolytica. The oligopeptides had various effects on the functional and structural characteristics of AmyK. AmyK-p1, the fusion protein containing peptide 1 (AEAEAKAKAEAEAKAK), exhibited improved specific activity, catalytic efficiency, alkaline stability, thermal stability, and oxidative stability compared with AmyK. Compared with AmyK, the specific activity and catalytic constant (kcat) of AmyK-p1 were increased by 4.1-fold and 3.5-fold, respectively. The following properties were also improved in AmyK-p1 compared with AmyK: kcat/Km increased from 1.8 liter/(g·min) to 9.7 liter/(g·min), stable pH range was extended from 7.0 to 11.0 to 7.0 to 12.0, optimal temperature increased from 50°C to 55°C, and the half-life at 60°C increased by ∼2-fold. Moreover, AmyK-p1 showed improved resistance to oxidation and retained 54% of its activity after incubation with H2O2, compared with 20% activity retained by AmyK. Finally, AmyK-p1 was more compatible than AmyK with the commercial solid detergents tested. The mechanisms responsible for these changes were analyzed by comparing the three-dimensional (3-D) structural models of AmyK and AmyK-p1. The significantly enhanced catalytic efficiency and stability of AmyK-p1 suggests its potential as a detergent ingredient. In addition, the oligopeptide fusion strategy described here may be useful for improving the catalytic efficiency and stability of other industrial enzymes.
INTRODUCTION
Most industrial enzymes are obtained from the natural environment. However, these enzymes are often used under conditions drastically different from their natural environment, in which the catalytic performance of the enzymes decreases, restricting their applications. Consequently, increasing attention is being paid to improving the catalytic performance of enzymes under extreme but application-relevant conditions such as high temperature, strong acid and alkali, and oxidative stress (1).
Protein engineering has emerged as an important tool to overcome the limitations of natural enzymes (2). Common strategies to alter proteins include site-directed mutagenesis and directed evolution (e.g., error-prone PCR). However, site-directed mutagenesis requires a clear understanding of the relationship between enzyme structure and function, and directed evolution requires a straightforward and efficient high-throughput screening method (3–5).
Alkaline α-amylases have high catalytic efficiency and stability at an alkaline pH range between 9 and 11 (6, 7) and are widely used in the detergent and textile industries for starch hydrolysis under alkaline conditions (8–10). This application requires high catalytic efficiency, high alkaline stability, and high oxidative stability (11). Although site-directed mutagenesis has been used to improve the oxidative stability of α-amylases, this was accompanied by decreased catalytic efficiency, alkaline stability, and thermal stability (4, 5).
Fusion of oligopeptides to the N or C terminus of enzymes promotes the expression of soluble proteins in Escherichia coli and also affects their structure (12, 13). Therefore, we reasoned that it may be possible to improve the stability or catalytic efficiency of enzymes by fusing them with oligopeptides. To the best of our knowledge, there have been no reports of such modifications to enzymes.
In this study, we investigated whether the fusion of oligopeptides to the N terminus of alkaline α-amylase could improve its stability and catalytic efficiency. Specifically, six oligopeptides (Table 1) were synthesized and fused to the N terminus of alkaline α-amylase from Alkalimonas amylolytica. The biochemical properties (e.g., specific activity, catalytic efficiency, alkaline stability, thermal stability, and oxidative stability) of the fusion proteins were characterized and compared with those of the wild-type alkaline α-amylase. Fusions containing peptide 1 were observed to improve both catalytic efficiency and stability. A three-dimensional (3-D) structural model of this fusion protein was obtained using computer simulation and compared with that of wild-type alkaline α-amylase to investigate the structural changes responsible for the improved characteristics.
Table 1.
Sequences, characteristics, and sources of oligopeptides used in this study
| Peptide | Sequence | Characteristic | Source | Reference |
|---|---|---|---|---|
| 1 | AEAEAKAKAEAEAKAK | Form β-sheet structure in aqueous solution; highly hydrophilic | Zuotin protein sequence in Saccharomyces cerevisiae | 14, 15 |
| 2 | VNYGNGVSCSKTKCSVN WGQAFQERYTAGTNSF VSGVSGVASGAGSIGRR | Form a well-defined central amphipathic α-helical structure (residues 18–39); disordered N and C termini forming a coil structure; highly hydrophobic | Synthesized by Soliman et al. (2010) | 16 |
| 3 | DWLKAFYDKVAEKLKEA FKVEPLRADWLKAFYDK VAEKLKEAF | Helix-turn-helix peptide; form α-helical fibril after incubation (37°C, 10 mM sodium phosphate, pH 7.60); amphiphilic | Synthesized by L. Lazar et al. (2005) | 17, 18 |
| 4 | DWLKAFYDKVAEKLKEA FGLLPVLEDWLKAFYDK VAEKLKEAF | Helix-turn-helix peptide; form α-helical fibril after the same incubation with peptide 3; amphiphilic | Synthesized by L. Lazar et al. (2005) | 17, 18 |
| 5 | DWLKAFYDKVAEKLKEA FKVQPYLDDWLKAFYDK VAEKLKEAF | Helix-turn-helix peptide; form α-helical fibril after the same incubation with peptide 3; amphiphilic | Synthesized by L. Lazar et al. (2005) | 17, 18 |
| 6 | DWLKAFYDKVAEKLKEA FNGGARLADWLKAFYD KVAEKLKEAF | Helix-turn-helix peptide; form a small amount of nonfibrillar precipitate after the same incubation with peptide 3; amphiphilic | Synthesized by L. Lazar et al. (2005) | 17, 18 |
MATERIALS AND METHODS
Bacterial strains and plasmids.
Alkaline α-amylase (AmyK) gene (accession no. AY268953) from alkaliphilic A. amylolytica strain N10 was synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) with modifications based on the preferred codon usage of Escherichia coli. Plasmid pET-22b (+) was used for subcloning and expression. E. coli JM109 was used as the host strain for cloning, and E. coli BL21(DE3) was used for protein expression.
Fusion of oligopeptides with alkaline α-amylase.
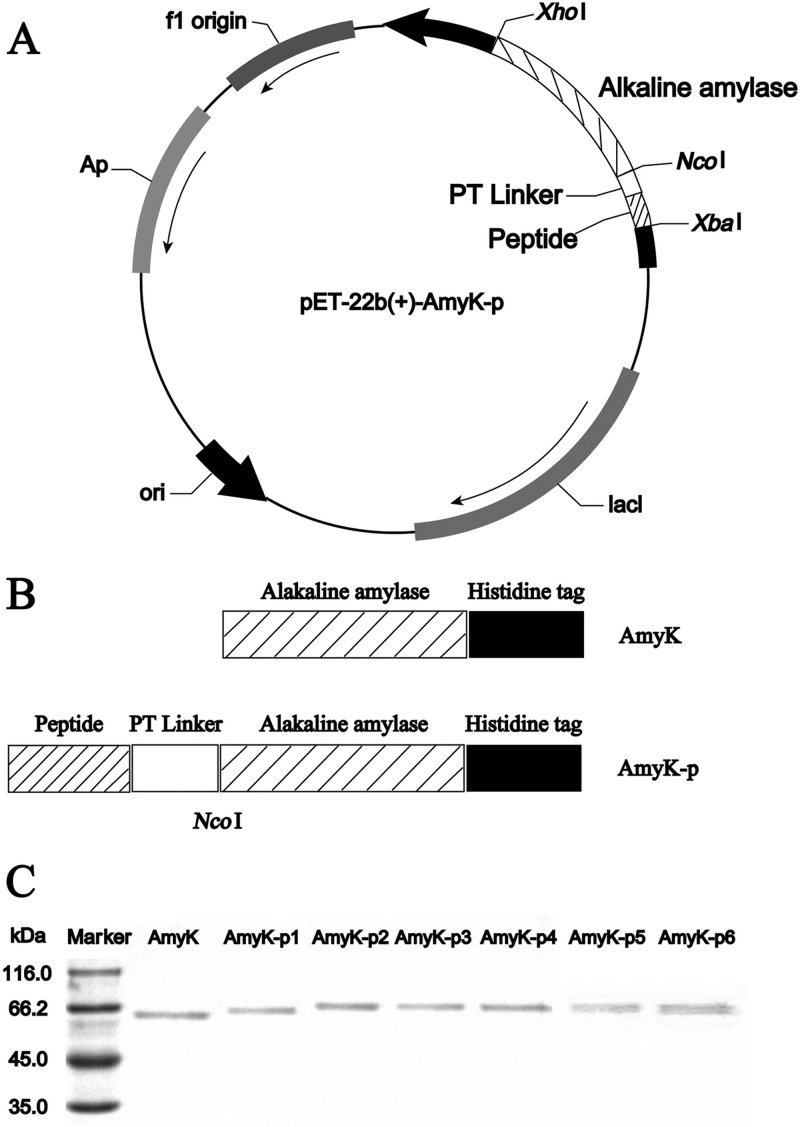
Oligonucleotides encoding six peptides (including a proline-threonine [PT] linker) were codon-optimized for expression in E. coli and ligated into plasmid pET-22b (+) obtained from Sangon Biotech (Fig. 1). The C terminus of the PT linker contained an NcoI restriction site. The AmyK sequence was ligated into the recombinant plasmid between the NcoI and XhoI sites to create an expression construct encoding the N terminus oligopeptide and PT linker, AmyK, and a C terminus His6 tag to enable protein purification. Plasmids were transformed into E. coli BL21(DE3), and proteins were expressed as described below. The fusion proteins lacked signal peptides (Fig. 1B) and were thus retained within the bacteria. Native AmyK was expressed with a functional signal peptide and was recovered from the medium. The peptide sequences and their properties are given in Table 1.
Fig 1.
Genetic constructs of the peptide fusion proteins. Peptide 1, AEAEAKAKAEAEAKAK; peptide 2, VNYGNGVSCSKTKCSVNWGQAFQERYTAGTNSFVSGVSGVASGAGSIGRR; peptide 3, DWLKAFYDKVAEKLKEAFKVEPLRADWLKAFYDKVAEKLKEAF; peptide 4, DWLKAFYDKVAEKLKEAFGLLPVLEDWLKAFYDKVAEKLKEAF; peptide 5, DWLKAFYDKVAEKLKEAFKVQPYLDDWLKAFYDKVAEKLKEAF; peptide 6, DWLKAFYDKVAEKLKEAFNGGARLADWLKAFYDKVAEKLKEAF; and linker, PTPPTTPTPPTTPTPT. (A) Plasmid map. (B) Schematic of inserted fusion sequences. (C) SDS-PAGE analysis of the purified AmyK and fusion proteins. The first lane of gels is the molecular mass marker lane.
Media and culture conditions.
E. coli seed cultures were grown at 37°C for 10 h in Luria-Bertani medium, with shaking at 200 rpm. E. coli transformants were screened on corn starch-agar plates in the presence of 1.0 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) to induce recombinant protein production. After incubation at 37°C for 10 h, the plates were examined for the appearance of a clear halo around individual colonies, which indicated secretion of AmyK. Colonies were inoculated into 25 ml of Terrific broth and incubated at 37°C in 250-ml shaker flasks agitated at 200 rpm. When the optical density at 600 nm reached 0.6, protein production was induced by the addition of 1 mM IPTG and the cultures were incubated at 20°C for 48 h.
Purification of alkaline α-amylase proteins.
Cells were harvested by centrifugation at 6,000 × g for 10 min, resuspended in Tris-HCl buffer (pH 9.0, 20 mM), and lysed by sonication on ice. The supernatants were removed by centrifugation at 15,000 × g for 15 min. For purification of His6-tagged enzymes, the supernatant was filtered and injected onto a Ni2+ column by using an AKTA purifier (GE Healthcare, Houston, TX). The column was washed with buffer A (50 mM phosphate buffer [pH 7.4] containing 0.3 M NaCl and 20 mM imidazole) to remove impurities and then eluted in buffer B (50 mM phosphate buffer [pH 7.4] containing 0.3 M NaCl and 250 mM imidazole). The flow rate was 1.0 ml/min. Proteins were eluted with a linear gradient from 0% to 100% buffer B. Fractions were collected for activity assays and analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The eluted enzymes were stored in Tris-HCl buffer (pH 9.0, 20 mM) at 4°C and diluted to 16 U/ml with glycine-NaOH buffer (pH 9.5, 20 mM) for the activity assays.
Enzyme activity assays.
Alkaline α-amylase activity was measured as the release of reducing sugar during hydrolysis of soluble starch by using a modified dinitrosalicylic acid (DNS) method (19). Soluble starch (1%, wt/vol) dissolved in glycine-NaOH buffer (pH 9.5, 20 mM) was preheated at 50°C for 5 min, a 3-ml aliquot was mixed with 0.4 ml of enzyme solution, and the mixture was incubated at 50°C for an additional 5 min. Then, 1 ml of the reaction mixture was mixed with 1 ml of DNS reagent (5.0 g/liter 3,5-dinitrosalicylic acid, 1.0 g/liter phenol, 0.15 g/liter Na2SO3, 5.0 g/liter NaOH, and 100 g/liter sodium potassium tartrate) and incubated in a boiling water bath for 10 min. The solution was cooled in ice water, and the volume was made to 10 ml with water. DNS reaction solution without enzyme was used as a control. Absorbance of the samples at 540 nm was determined. One unit of AmyK activity was defined as the amount of enzyme that released 1 μmol of reducing sugar as glucose per minute under these assay conditions. Protein concentrations were measured using the Bradford method (8), with bovine albumin (Sangon Biotech) as the standard.
Measurement of kinetic parameters and substrate specificities.
The kinetic parameters (Km, Vmax, kcat, and kcat/Km) of AmyK and the fusion proteins were determined in glycine-NaOH buffer (pH 9.5, 20 mM) at 50°C. Assays were performed using enzyme at a fixed starting level of 16 U/ml, and soluble starch was added to a concentration between 1 and 10 g/liter. Km and Vmax were estimated from Eadie-Hofstee plots (20). Substrate specificities were determined under standard assay conditions by using soluble starch, cornstarch, potato starch, amylopectin, amylose, glycogen, α-cyclodextrin, β-cyclodextrin, γ-cyclodextrin, dextran, pullulan, or cellulose as the substrates.
Measurement of alkaline and thermal stabilities.
The optimal pH for enzyme activity was determined by performing the assay in the following pH buffers: Na2HPO4-NaH2PO4 (pH 7.0 to 8.0, 10 mM), Tris-HCl buffer (pH 8.0 to 9.5, 20 mM), glycine-NaOH buffer (pH 9.5 to 11.0, 20 mM), and Na2HPO4-NaOH buffer (pH 12.0, 10 mM). The activity determined at pH 9.5 (glycine-NaOH buffer, 20 mM) was taken as 100% activity. For analysis of pH stability, the enzymes were incubated in the above-described buffers at 25°C for 24 h, and residual enzyme activity was then measured using the standard assay conditions. For each protein, the highest activity at any pH was taken as 100%, and for the same protein, the percent relative activity at other pH values was calculated relative to the highest activity.
To determine the optimal temperature for activity, the assay was performed at temperatures between 30°C and 70°C in glycine-NaOH buffer (pH 9.5, 20 mM). For each protein, the highest activity at any temperature was taken as 100%, and the percentage of relative activity at other temperatures was calculated accordingly. The thermal stability of the enzymes was determined by incubation for 20 min at 60°C in Tris-HCl buffer (pH 9.0, 20 mM). The activation energy (Ea) was determined from the Arrhenius equation ln (k) = (−Ea/RT) + ln (A) at temperatures ranging from 20°C to 50°C. The reaction rate constant (k) was calculated from the equation ln (c) = −kt + ln (c0), where c is the concentration of soluble starch (g/liter) and c0 is the initial concentration of the substrate (0.1 g/liter). Soluble starch concentrations were measured using the iodometric method. For this purpose, 1 ml of soluble starch was mixed with 5.0 ml iodine solution (0.088 g/liter iodine and 40.0 g/liter potassium iodide), and the absorbance at 660 nm was determined. The concentration was determined from a standard plot ranging from 0.1 to 0.8 g/liter of soluble starch.
Measurement of oxidative stability.
The enzymes (16 U/ml) were incubated with H2O2 (100 to 500 mM) in Tris-HCl buffer (pH 9.0, 20 mM) at 35°C for 30 min. Catalase (final concentration of 2,000 U/ml) was then added to quench H2O2, and residual enzyme activity was measured under standard assay conditions.
Determination of enzyme activity and stability in the presence of commercial detergents.
Enzyme activity was determined in the presence of 4 solid and 4 liquid commercial detergents. The solid detergents used were laundry soap (Nice, Lishui, China), toilet soap (Safeguard, Cincinnati, OH), washing powder 1 (Tide, Cincinnati, OH), and washing powder 2 (Nice, Lishui, China). The liquid detergents used were laundry detergent 1 (Blue Noon, Guangzhou, China), laundry detergent 2 (Liby, Guangzhou, China), liquid detergent 1 (Nice, Lishui, China), and liquid detergent 2 (Liby, Guangzhou, China). Detergents were diluted with tap water to 70 mg/ml and 10% for solid and liquid detergents, respectively. Endogenous proteases were inactivated by incubation of detergents at 65°C for 1 h. The detergents were present in the enzyme assays at final concentrations of 7 mg/ml and 1% for solid and liquid detergents, respectively, to simulate washing conditions (1). The assays were performed under standard assay conditions with 16 U/ml enzyme. The enzyme activity without detergent was taken as 100%, and for the same protein, the percent relative activity in the presence of detergents was calculated relative to the activity without any detergents.
CD and DSC analysis.
Circular dichroism (CD) spectra were measured using a MOS-450/AF-CD-STP-A (Bio-Logic, Grenoble, France) with a 1-cm path-length quartz cuvette at a protein concentration of 0.1 mg/ml in 20 mM Tris-HCl buffer (pH 9.0). The spectropolarimeter and xenon lamp were warmed up for at least 30 min prior to experiments to minimize baseline signal drift. Ellipticity data were collected between 190 and 250 nm, and the spectrum of a buffer blank was subtracted. The lengths and fractions of α-helixes and β-sheets were determined by online analysis of CD data using DichroWeb (http://dichroweb.cryst.bbk.ac.uk/html/process.shtml) according to a previously reported method (21, 22). The melting temperature (Tm) was determined using a Q2000 differential scanning calorimeter (DSC) (TA, New Castle, DE) at a protein concentration of 50 mg/ml in 20 mM Tris-HCl buffer (pH 9.0). The temperature was increased from 20°C to 90°C at 10°C/min.
Construction of a structural model of AmyK enzymes.
Swiss-Model (http://swissmodel.expasy.org/) was used for the identification of structural homologues and for structure prediction. The theoretical structure of AmyK was obtained by homology modeling from the Swiss-Model server, using the crystal structure of AmyB (3bc9) as the template (23). Stereochemical analysis of the structure was performed using PROCHECK (http://nihserver.mbi.ucla.edu/SAVS/). The final model used in this study displayed good geometry, with less than 1% of residues disallowed. Details of the modeling procedure are provided in the supplemental material.
Structural modeling of fusion proteins and molecular dynamics analysis.
The fusion protein amino acid sequences were verified by sequencing the N and C termini. Molecular dynamics simulations were carried out using NAMD software with the charmM force field (http://www.ks.uiuc.edu/Research/namd/). Proteins were solvated in a cubic box consisting of TIP3P water molecules, and the box size was chosen using the criterion that protein atoms must be more than 10.0 Å from the wall. The particle mesh Ewald summation method was used for calculating the total electrostatic energy in a periodic box.
Structure minimization was performed to remove unexpected coordinate collision and to obtain the local minima. The water box and the whole system were minimized using the descent method and the conjugate gradient method. Subsequently, system heating, equilibration, and data sampling were carried out. System heating was performed in an NTV (N particles, constant temperature, constant volume) ensemble, followed by 150-ps simulation for equilibration, and 2-ns or longer simulation for data sampling in an NTP (N particles, constant temperature, constant pressure) ensemble. The temperatures were set at 300 K and 330 K at 1 atm of pressure. A weak coupling algorithm was used for temperature and pressure regulation with a coupling time of 1.0 ps. The SHAKE method was used for constraining all hydrogen bonds.
Statistical analysis.
All experiments were performed at least 3 times, and the results are expressed as the means ± standard deviations (SD). Statistical analyses were performed using Student's t test. P values less than 0.05 were considered statistically significant.
RESULTS AND DISCUSSION
Construction and expression of AmyK and fusion proteins.
Six fusion proteins consisting of an oligopeptide and PT linker fused to the N terminus of A. amylolytica AmyK were constructed, and the proteins were expressed in E. coli BL21 (Fig. 1; Table 1). The nucleotide sequences of all fusion proteins (termed AmyK-p1 to AmyK-p6) were identical to that of AmyK (data not shown). In addition, the C terminus sequence of the fusion protein AmyK-p1 was confirmed to be identical to that of AmyK (GGFHHHHHH). SDS-PAGE analysis showed that the fusion proteins were approximately 66 kDa in size (Fig. 1C). In contrast to AmyK, which contained a signal peptide and was secreted into the culture medium, the fusion proteins were retained intracellularly and were purified as soluble proteins from cell sonicates.
Influence of peptide fusion on the specific activity and kinetic parameters of AmyK.
Table 2 shows the specific activities and catalytic parameters of AmyK and the fusion proteins. The specific activity of AmyK-p1 was significantly higher (∼4.1-fold) than that of AmyK, whereas the specific activities of the other fusion proteins were considerably lower than that of AmyK. The Michaelis constants (Km values) of all fusion proteins were lower than that of AmyK, indicating that peptide fusion had increased substrate-binding ability. The Vmax of AmyK-p1 was higher than that of AmyK (59.5 versus 37.8 μmol/[ml·min]), whereas the Vmax values for the other fusion mutants were significantly lower than that of AmyK. The catalytic constant (kcat) of AmyK-p1 (5.8 × 105 min−1) was higher than that of AmyK (1.7 × 105), whereas those of the other fusion proteins were lower than that of AmyK by 62.3% (AmyK-p2), 79.7% (AmyK-p3), 93.6% (AmyK-p4), 85.5% (AmyK-p5), and 95.9% (AmyK-p6). The kcat/Km of AmyK-p1 (1.8 liter/[g·min]) was higher than that of AmyK (9.7 liter/[g·min]), whereas those of the other fusion proteins were significantly lower. These results indicate that the fusion of peptide 1 to the N terminus of AmyK significantly improved its specific activity and catalytic efficiency.
Table 2.
Enzyme kinetic parameters of AmyK and oligopeptide fusion proteins
| Enzyme kinetic parametera | Value ± SD |
||||||
|---|---|---|---|---|---|---|---|
| AmyK | AmyK-p1 | AmyK-p2 | AmyK-p3 | AmyK-p4 | AmyK-p5 | AmyK-p6 | |
| Km (g/liter) | 9.2 ± 0.2 | 6.0 ± 0.2 | 5.9 ± 0.1 | 5.5 ± 0.4 | 6.2 ± 0.2 | 8.9 ± 0.3 | 6.1 ± 0.5 |
| Vmax (μmol/[ml·min]) | 37.8 ± 2.1 | 59.5 ± 1.1 | 26.3 ± 1.2 | 19.1 ± 0.9 | 27.6 ± 1.5 | 33.7 ± 2.2 | 23.9 ± 1.8 |
| kcat (·103 min−1) | 166.7 ± 2.3 | 582.8 ± 2.9 | 62.9 ± 1.7 | 33.8 ± 2.0 | 10.6 ± 1.5 | 24.1 ± 1.9 | 6.8 ± 1.1 |
| kcat/Km (·103 liter/[g·min]) | 18.1 ± 0.7 | 97.1 ± 3.9 | 10.7 ± 0.5 | 6.2 ± 0.8 | 1.7 ± 0.3 | 2.7 ± 0.3 | 1.1 ± 0.3 |
| Specific activity (·106 U/μmol) | 6.2 ± 0.8 | 25.5 ± 1.1 | 4.0 ± 0.5 | 2.1 ± 0.3 | 0.6 ± 0.1 | 1.3 ± 0.2 | 0.4 ± 0.1 |
Km, substrate dissociation constant; kcat, μmol dextrose equivalents per minute per μmol protein.
There were no significant differences among AmyK and the fusion proteins in their substrate specificities (soluble starch, cornstarch, potato starch, amylopectin, amylose, glycogen, α-cyclodextrin, β-cyclodextrin, γ-cyclodextrin, dextran, pullulan, and cellulose; data not shown). AmyK-p1 showed the highest specific activity on soluble starch, and its specific activity toward amylopectin, amylose, potato starch, cornstarch, and glycogen was 68.5%, 61.3%, 47.8%, 33.6%, and 32.2%, respectively, of that on soluble starch. None of the proteins showed activity toward dextran, pullulan, α-cyclodextrin, β-cyclodextrin, γ-cyclodextrin, or cellulose.
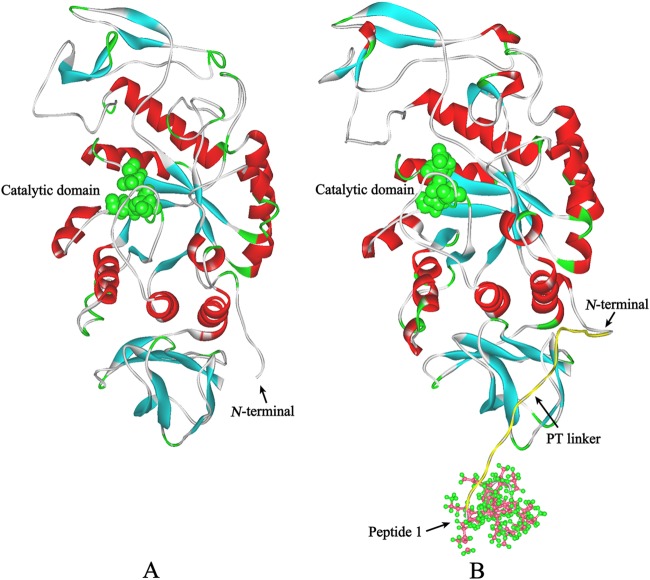
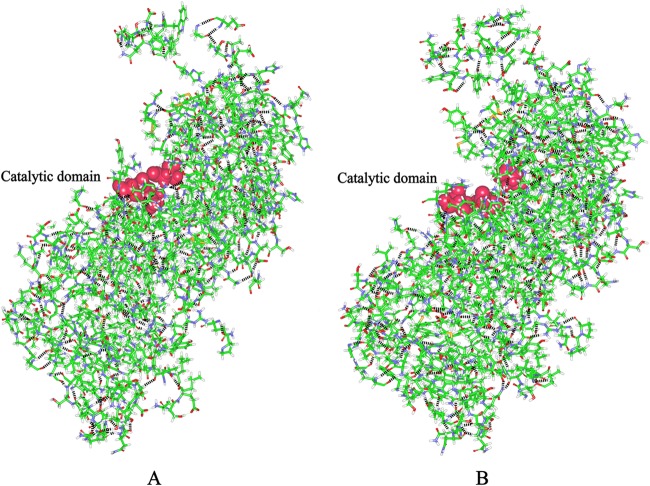
To understand the structural changes responsible for the improved catalytic activity of AmyK-p1, we compared computer-simulated 3-D structural models of AmyK (Fig. 2A) and AmyK-p1 (Fig. 2B). The active site of AmyK includes residues Asp248, Glu278, and Asp340 (23), of which Asp248 is the catalytic nucleophile and Glu278 is the catalytic hydrogen donor (24). The essential amino acid Asp340 is believed to assist in catalysis by hydrogen bonding to the substrate. In the proposed first step of the catalytic mechanism, protonated Glu278 is the proton donor to glycosidic oxygen and the nucleophile Asp248 must be deprotonated (25). In the second step of the reaction, deprotonated Glu278 activates a water molecule. The active site residues must thus be in catalytically competent protonation states for the enzyme to be active (26, 27).
Fig 2.
Structural models of AmyK and AmyK-p1. The α helices and β sheets are shown in red and cyan, respectively. (A) The structural model of AmyK was constructed using the crystal structure of AmyB (3bc9) as a template. The catalytic residues are in green according to the Corey-Pauling-Koltun (CPK) representation scheme. (B) The structural model of AmyK-p1 was constructed using the structural model of AmyK as a template, and molecular dynamics simulations were carried out using the NAMD software with the charmM force field (http://www.ks.uiuc.edu/Research/namd/). The catalytic residues are shown in green according to the CPK representation scheme. The yellow coil is the PT linker. The coil with ball-and-stick residues is peptide 1.
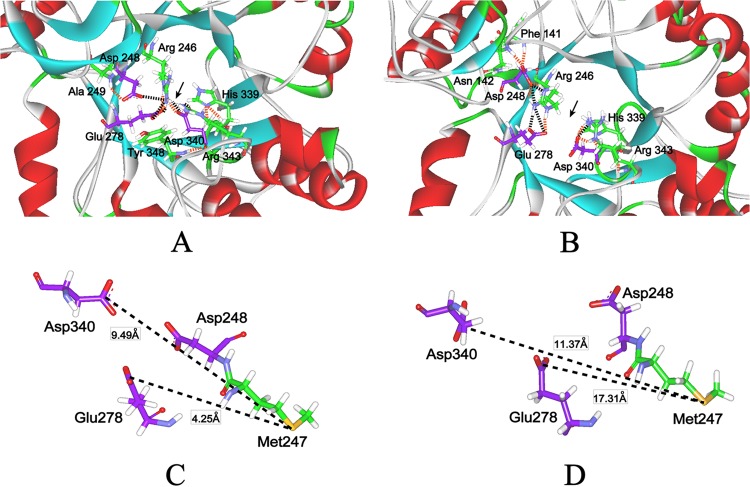
Conformational flexibility plays an important role in enzyme stability (24, 28). High flexibility, particularly around the active site, tends to result in an enzyme with high specific activity and low activation energy (28). Chemical modification has been reported to increase the conformational flexibility of a protease, thereby enhancing the enzyme's catalytic efficiency (28). Flexibility in the active site of AmyK is suggested by the loop between residues 337 and 347 (Fig. 3A). Asp340 and His339 within the loop contribute to substrate distortion and intermediate binding, and the loop assists in these processes by steering the residues in the appropriate direction (24, 29). In AmyK, the catalytic residues Asp248 and Glu278 are connected to Arg246 by salt bridges and hydrogen bonds, and Asp340 and Arg246 are connected by a hydrogen bond and a salt bridge (Fig. 3A). The latter connections may interfere with the ability of Asp340 to assist in catalysis by hydrogen bonding to the substrate. The interaction between peptide 1 and the enzyme may have a slight allosteric effect on the structure because the hydrogen bond and salt bridge between Asp340 and Arg246 are not present in AmyK-p1 (Fig. 3B). However, the flexibility between the catalytic residues (Asp248 and Glu278) was increased in AmyK-p1, which might have improved the protein's ability to bind and hydrolyze the substrate. These observations suggest that greater flexibility around the active site is the main reason for the improved catalytic efficiency of AmyK-p1.
Fig 3.
Changes in structure (e.g., salt bridge, hydrogen bond, and flexibility) around the active sites of AmyK after fusion with peptide 1, and the change in distance between active sites and Met247 for AmyK after fusion with peptide 1. The salt bridge and the hydrogen bond were calculated using Discovery Studio 2.5. The purple sticks are the active sites. (A) The changes in structure (e.g., salt bridge, hydrogen bond, and flexibility) around the active sites of AmyK. The dark dotted line is the salt bridge. The orange dotted line is the hydrogen bond. (B) The changes in structure (e.g., salt bridge, hydrogen bond, and flexibility) around the active sites of AmyK-p1. The dark dotted line is the salt bridge. The orange dotted line is the hydrogen bond. (C) The distance between the active site and Met247 for AmyK. The dotted line is the distance between the active site and Met247 for AmyK. (D) The distance between the active site and Met247 for AmyK-p1. The dotted line is the distance between the active site and Met247 for AmyK-p1.
Peptides 3 to 6 differed only in the linkers between the repeat units, yet the resulting fusion proteins had considerably different catalytic properties. We constructed structural models of AmyK-3, -4, -5, and -6 (see the supplemental material) to investigate this and found that the peptides exhibited different grand average of hydropathicity (Gravy) values and root mean square deviation (RMSD) values. Gravy values of peptides 3, 4, 5, and 6 were −0.505, −0.184, −0.553, and −0.451, respectively, and RMSD values were 1.183, 1.037, 1.394, and 1.647, respectively. Thus, the differences in hydropathicity among the 4 peptides may have affected their catalytic properties.
Effect of peptide fusion on the alkaline stability of AmyK.
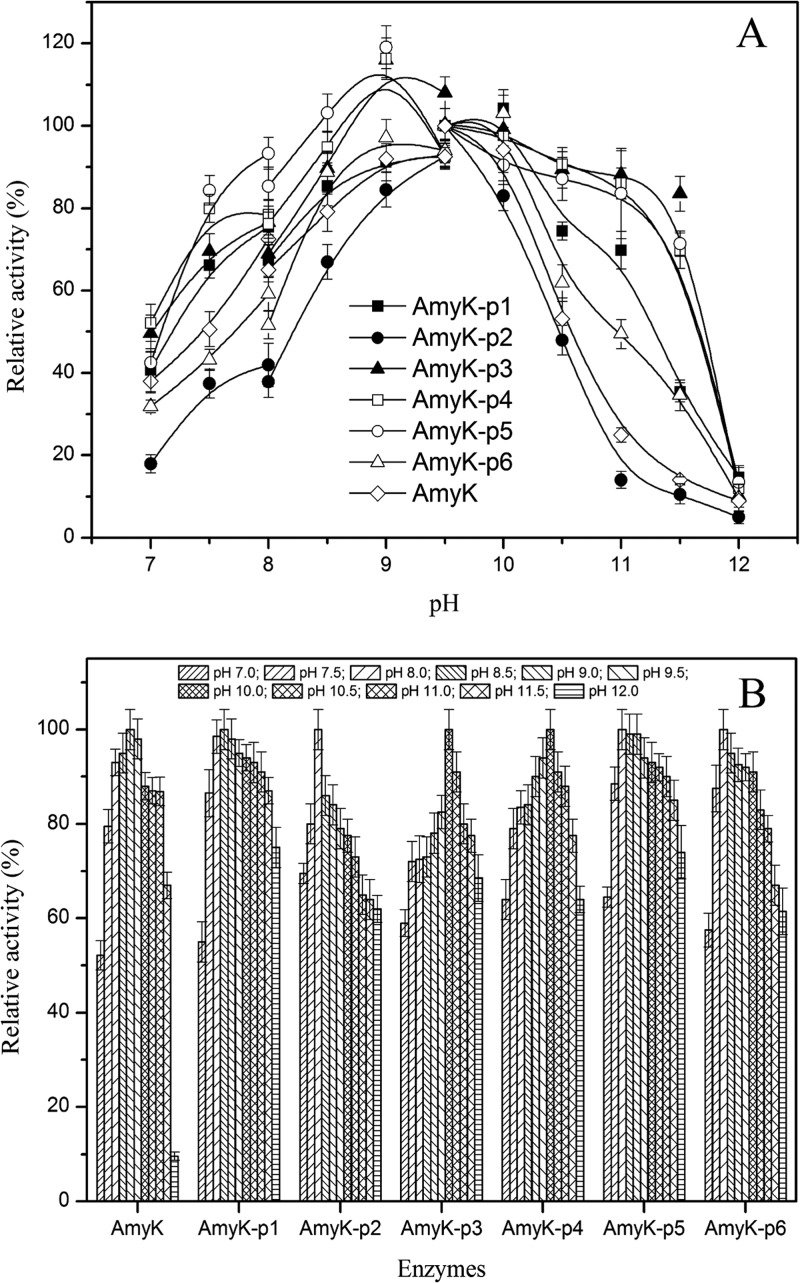
As shown in Fig. 4A, the pH optimum for AmyK and AmyK-p2 activity was 9.5, whereas pH 10.0 was optimal for AmyK-p1 and AmyK-p6, and pH 9.0 was optimal for AmyK-p3, AmyK-p4, and AmyK-p5. The stable pH range of all fusion proteins (7.0 to 12.0) was broader than that of AmyK (7.0 to 11.0) (Fig. 4B).
Fig 4.
Effect of peptides on pH stability of the enzyme. (A) Effect of peptides on optimum pH of enzyme activity. Diamond, AmyK; filled rectangle, AmyK-p1; filled circle, AmyK-p2; filled triangle, AmyK-p3; open rectangle, AmyK-p4; open circle, AmyK-p5; open triangle, AmyK-p6. Activity determined at pH 9.5 (glycine-NaOH buffer, 20 mM) was considered 100%. (B) Effect of peptides on pH stability of enzyme. After incubation at different pH values, the residual activity was measured under standard assay conditions.
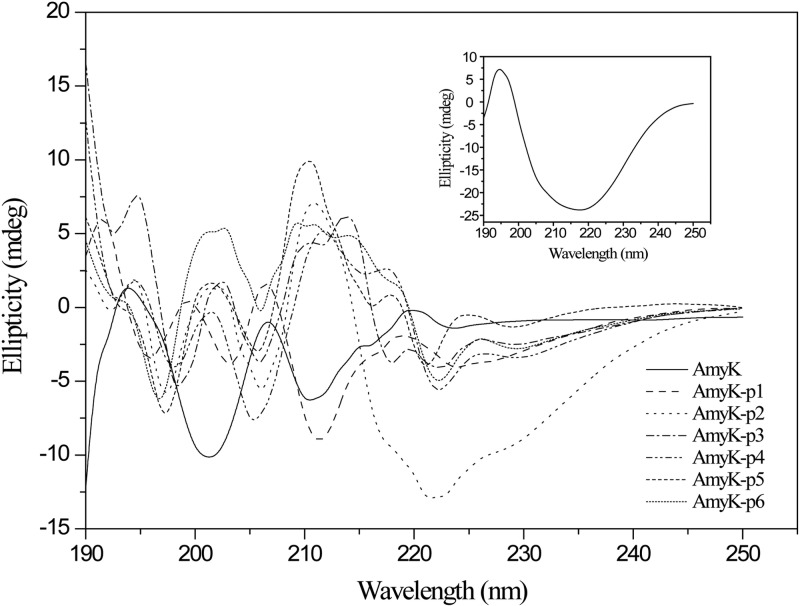
The stability of α-amylase from Bacillus licheniformis has been reported to be enhanced by replacement of Thr353 by Ile, due to an increased propensity for helix formation (30). To understand how the fusion peptides influenced the pH stability of AmyK, we compared their CD spectra (Fig. 5). The CD spectra between 190 and 250 nm at 25°C showed that the secondary structure of AmyK was greatly altered by peptide fusion, and the helical component of AmyK-p1 was significantly improved. During protein denaturation, breaking contacts between neighboring residues in a helix is known to be the most energy-demanding part of the process (30), thus suggesting that the increased helical component of AmyK-p1 contributes to its improved stability at an alkaline pH.
Fig 5.
(A) Conformational changes of AmyK and the fusion enzymes were verified using circular dichroism (CD). The inset shows the far UV-CD spectrum of peptide 1. AmyK: the average length of helices, 5.986 Å; the average length of strands, 6.980 Å. AmyK-p1: the average length of helices, 8.503 Å; the average length of strands, 4.516 Å. AmyK-p2: the average length of helices, 9.226 Å; the average length of strands, 4.811 Å. AmyK-p3: the average length of helices, 9.196 Å; the average length of strands, 4.382 Å. AmyK-p4: the average length of helices, 8.530 Å; the average length of strands, 4.386 Å. AmyK-p5: the average length of helices, 3.897 Å; the average length of strands, 6.189 Å. AmyK-p6: the average length of helices, 4.396 Å; the average length of strands, 5.700 Å.
Effect of oligopeptide fusion on the thermal stability of AmyK.
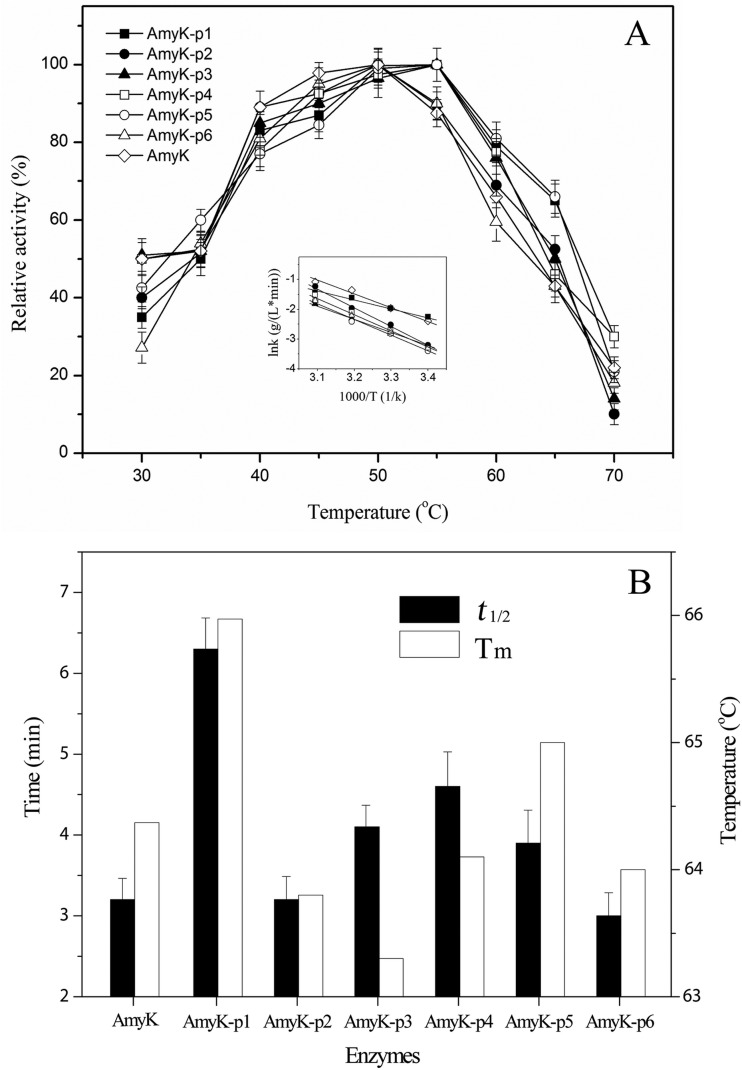
Figure 6 shows the effects of temperature on the activity and stability of AmyK and the fusion proteins. The optimum temperature for activity of AmyK, AmyK-p2, and AmyK-p6 was 50°C, whereas 55°C was optimal for AmyK-p1, -p3, -p4, and -p5 (Fig. 6A). The Ea of AmyK and AmyK-p1, -p2, -p3, -p4, -p5, and -p6 were 36.1, 22.1, 52.1, 40.5, 48.4, 44.2, and 41.8 kJ/mol, respectively. The thermal stability of AmyK was also enhanced after fusion with peptide 1, as shown by the increase in melting temperature (Tm) by 1.6°C (Fig. 6B). The half-life (t1/2) of AmyK-p1 was about 2-fold that of AmyK at 60°C (Fig. 6B). In contrast, the t1/2 and Tm values of the other fusion proteins were almost the same as those of AmyK.
Fig 6.
Effect of peptides on thermal stability of the enzyme. (A) Effect of peptides on optimum temperature for enzyme activity. The inset shows the Arrhenius plot of the logarithm of the k values against the reciprocal of absolute temperature (T). The values shown are activation energies calculated from the plot. Diamond, AmyK; filled rectangle, AmyK-p1; filled circle, AmyK-p2; filled triangle, AmyK-p3; open rectangle, AmyK-p4; open circle, AmyK-p5; open triangle, AmyK-p6. (B) The half-life (t1/2) and the melting temperature (Tm) of different peptide fusions at 60°C. Black, t1/2; white, Tm.
Thermal stabilization of engineered enzymes has been shown to decrease their catalytic activity at moderate temperatures, presumably because of changes in the overall protein rigidity (5). Here, the thermal stability of AmyK-p1 alone was significantly better than that of AmyK. The enhanced thermostability of AmyK-p1 may have been due to changes in AmyK folding after fusion with peptide 1. Indeed, as shown in Fig. 5, the far UV-CD spectrum of peptide 1 showed a 218-nm minimum and a 195-nm maximum, indicating that peptide 1 had a characteristic β-sheet structure. This result was consistent with that of a previous study (14). In addition, the α-helical component of AmyK-p1 was higher than that of AmyK (28.9% versus 20.7%), and the turn component was lower (17.6% versus 19%). These conformational changes are consistent with the structural models of AmyK and AmyK-p1 (Fig. 2). The increased helical component of AmyK-p1 may have helped stabilize the protein structure, as has been previously reported (31). However, the helical component of the other fusion proteins was not significantly different from that of AmyK (Fig. 5).
Hydrogen bonding contributes to enzyme stability at high temperatures, as shown by the inability of enzymes to retain their tightly coiled, thermostable, and catalytically active structures in the absence of proper hydrogen bonding (32). As shown in Fig. 7, fusion of peptide 1 to AmyK increased the number of hydrogen bonds from 383 to 389, which probably contributed to the improved stability of AmyK-p1 at high temperatures.
Fig 7.
Local hydrogen bonding network of the enzyme. The active site is shown in magenta according to the CPK representation scheme. The sticks indicate the residues that could produce hydrogen bonds. The hydrogen bonds are shown with black dotted lines. (A) The local hydrogen bonding network of AmyK. (B) The local hydrogen bonding network of AmyK-p1.
Effect of oligopeptide fusion on the oxidative stability of AmyK.
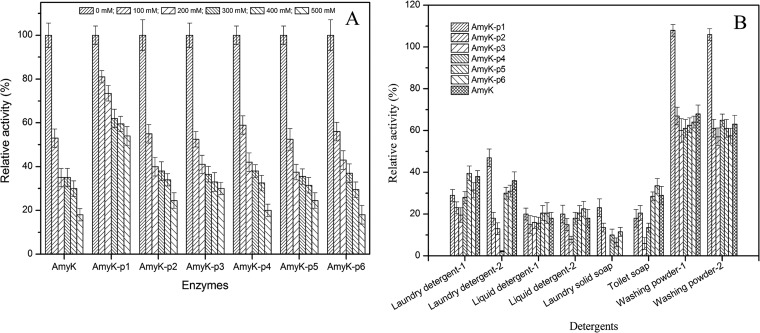
AmyK was found to be susceptible to oxidation, and it retained less than 20% of its activity after incubation with 500 mM H2O2 (Fig. 8A). AmyK-p1 showed greater resistance to oxidation and retained 54% of its activity under the same conditions. However, other fusion proteins showed similar degrees of susceptibility to oxidation as AmyK.
Fig 8.
Effect of peptides on oxidative stability and antidetergents of the enzyme. (A) Effect of peptides on oxidative stability of enzyme. The relative activity was calculated based on a determination of activity without the addition of H2O2, which was considered 100%. (B) Effect of peptides on antidetergents of enzyme.
Oxidation of the methionine residue situated in the cavity of the AmyK active site has been shown to decrease its activity or even inactivate the enzyme (11, 33). The model structure of AmyK from Bacillus sp. strain KSM-K38 shows an equivalent methionine residue (Met197) enclosed in the active site, and this residue is also susceptible to chemical oxidation (34). In our model, Met247 was closest to the catalytic residues and may be susceptible to chemical oxidation. However, the distances between Met247 and Asp278 and between Met247 and Asp340 were greater in AmyK-p1 than in AmyK (17.31 Å versus 4.25 Å and 11.37 Å versus 9.49 Å, respectively) (Fig. 3C and D). Thus, Met247 in AmyK-p1 was located further from the catalytic residues, which may have increased its oxidative stability (Fig. 3C and D).
Effect of oligopeptide fusion on the detergent stability of AmyK.
To evaluate the potential applications of AmyK fusion proteins in the detergent industry, we examined the effects of a variety of solid and liquid detergents on their enzyme activities (detergent compositions are provided in the supplemental material). The fusion proteins were found to be more stable in the presence of the solid detergents (washing powders 1 and 2) than the solid soaps and liquid detergents (Fig. 8B). Interestingly, AmyK-p1 activity increased slightly when incubated with washing powders 1 and 2 (8% and 6%, respectively). However, neither AmyK nor any of the fusion enzymes were stable in the presence of a solid soap or liquid detergent, which reduced their activities to less than 50% of the control activities.
The compatibility of AmyK-p1 with the washing powders may be the result of ionic interactions and hydrophobic or hydrophilic interactions between the detergents and the fusion peptide (11). This might be explained by the fact that for liquid or laundry detergents, the physical isolation of enzymes was difficult and the presence of solvent (water) amplified the detrimental effects of surfactants due to the numerous surfactants in those detergents (11, 35). In addition, AmyK-p1 activity, which is optimal at alkaline pH, was greatly decreased in the presence of liquid detergents with acidic pH (5.5 to 8.0) compared to solid washing powders with pH ranging from 9.0 to 11.0. This observation is in agreement with other studies showing that the activity of AmyUS100ΔIG/M197A from Geobacillus stearothermophilus was increased by 10% to 20% after incubation in the presence of Lav+ and Nadhif detergents (11).
In conclusion, in this study, we described a protein engineering strategy to improve enzyme stability and activity by fusion with oligopeptides, and we verified its effectiveness using AmyK as a model protein. Although fusion of peptide 1 to AmyK improved its specific activity, catalytic efficiency, thermal stability, and oxidative stability, the oligopeptide fusion strategy may be not suitable for all microbial enzymes, and the selection of oligopeptides will need to be tailored to each enzyme. This technique has advantages over other protein engineering strategies such as site-directed mutagenesis and directed evolution, in that it can be implemented without structural information or an efficient high-throughput screening method. Our method therefore appears to have great potential for the molecular engineering of microbial enzymes. We evaluated several fusion peptides, but at present, it is not possible to give general guidelines for the design of peptides that will be effective for a specific enzyme. However, the results of the present study indicate that the oligopeptide should be short and have simple secondary structure. In addition, hydrophilic peptides are preferable to enable expression of soluble fusion proteins. In future studies, we plan to design oligopeptides for engineering other industrial enzymes, which may help identify a general approach for the effective design of peptides for a specific enzyme.
ACKNOWLEDGMENTS
This work was supported by Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 Project (111-2-06), and the National High Technology Research and Development Program of China (863 Program, project 2012AA022202, and 973 Program, project 2012CB720806).
We thank Yi Liu and Zhemin Zhou for help with the structural modeling of fusion proteins.
Footnotes
Published ahead of print 1 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03785-12.
REFERENCES
- 1. Kazlauskas RJ, Bornscheuer UT. 2009. Finding better protein engineering strategies. Nat. Chem. Biol. 5: 526– 529 [DOI] [PubMed] [Google Scholar]
- 2. Böttcher D, Bornscheuer UT. 2010. Protein engineering of microbial enzymes. Curr. Opin. Microbiol. 13: 274– 282 [DOI] [PubMed] [Google Scholar]
- 3. Hong SY, Lee JS, Cho KM, Math RK, Kim YH, Hong SJ, Cho YU, Cho SJ, Kim H, Yun HD. 2007. Construction of the bifunctional enzyme cellulase-β-glucosidase from the hyperthermophilic bacterium Thermotoga maritima. Biotechnol. Lett. 29: 931– 936 [DOI] [PubMed] [Google Scholar]
- 4. Yang HQ, Liu L, Li JH, Du GC, Chen J. 2012. Structure-based replacement of methionine residues at the catalytic domains with serine significantly improves the oxidative stability of alkaline amylase from alkaliphilic Alkalimonas amylolytica. Biotechnol. Prog. 28: 1271– 1277 [DOI] [PubMed] [Google Scholar]
- 5. Yang HQ, Liu L, Wang MX, Li JH, Wang NX, Du GC, Chen J. 2012. Structure-based engineering methionine residues in the catalytic cores of alkaline amylase from Alkalimonas amylolytica for improved oxidative stability. Appl. Environ. Microbiol. 78: 7519– 7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hagihara H, Igarashi K, Hayashi Y, Endo K, Ikawa-Kitayama K, Ozaki K, Kawai S, Ito S. 2001. Novel alpha-amylase that is highly resistant to chelating reagents and chemical oxidants from the alkaliphilic Bacillus isolate KSM-K38. Appl. Environ. Microbiol. 67: 1744– 1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Igarashi K, Hatada Y, Hagihara H, Saeki K, Takaiwa M, Uemura T, Ara K, Ozaki K, Kawai S, Kobayashi T. 1998. Enzymatic properties of a novel liquefying alpha-amylase from an alkaliphilic Bacillus isolate and entire nucleotide and amino acid sequences. Appl. Environ. Microbiol. 64: 3282– 3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim TU, Gu BG, Jeong JY, Byun SM, Shin YC. 1995. Purification and characterization of a maltotetraose-forming alkaline (alpha)-amylase from an alkalophilic Bacillus strain, GM8901. Appl. Environ. Microbiol. 61: 3105– 3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuilderd H, Wu G. 2008. Applied technology-simultaneous desizing and scouring with enzymes-simultaneous fabric desizing and scouring, using alkaline alpha-amylase and an alkaline scouring enzyme, reduces water. Am. Assoc. Text. Chem. Color. 8: 33– 36 [Google Scholar]
- 10. Murakami S, Nagasaki K, Nishimoto H, Shigematu R, Umesaki J, Takenaka S, Kaulpiboon J, Prousoontorn M, Limpaseni T, Pongsawasdi P. 2008. Purification and characterization of five alkaline, thermotolerant, and maltotetraose-producing alpha-amylases from Bacillus halodurans MS-2-5, and production of recombinant enzymes in Escherichia coli. Enzyme Microb. Technol. 43: 321– 328 [Google Scholar]
- 11. Khemakhem B, Ali MB, Aghajari N, Juy M, Haser R, Bejar S. 2009. Engineering of the alpha-amylase from Geobacillus stearothermophilus US100 for detergent incorporation. Biotechnol. Bioeng. 102: 380– 389 [DOI] [PubMed] [Google Scholar]
- 12. Wu W, Xing L, Zhou BH, Lin ZL. 2011. Active protein aggregates induced by terminally attached self-assembling peptide ELK16 in Escherichia coli. Microb. Cell Fact. 10: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xing L, Wu W, Zhou BH, Lin ZL. 2011. Streamlined protein expression and purification using cleavable self-aggregating tags. Microb. Cell Fact. 10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang S, Holmes T, Lockshin C, Rich A. 1993. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc. Nat. Acad. Sci. 90:3334– 3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang S, Lockshin C, Herbert A, Winter E, Rich A. 1992. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO J. 11: 3787– 3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soliman W, Bhattacharjee S, Kaur K. 2010. Adsorption of an antimicrobial peptide on self-assembled monolayers by molecular dynamics simulation. J. Phys. Chem. B. 114: 11292– 11302 [DOI] [PubMed] [Google Scholar]
- 17. Anantharamaiah G, Jones J, Brouillette C, Schmidt C, Chung BH, Hughes T, Bhown A, Segrest J. 1985. Studies of synthetic peptide analogs of the amphipathic helix. Structure of complexes with dimyristoyl phosphatidylcholine. J. Biol. Chem. 260: 10248– 10255 [PubMed] [Google Scholar]
- 18. Lazar KL, Miller-Auer H, Getz GS, Orgel JPRO, Meredith SC. 2005. Helix-turn-helix peptides that form α-helical fibrils: turn sequences drive fibril structure. Biochemistry 44: 12681– 12689 [DOI] [PubMed] [Google Scholar]
- 19. Fuwa H. 1954. A new method for microdetermination of amylase activity by the use of amylose as the substrate. J. Biochem. 41: 583– 603 [Google Scholar]
- 20. Fuhrmann GF, Völker B. 1993. Misuse of graphical analysis in nonlinear sugar transport kinetics by Eadie-Hofstee plots. Biochim. Biophys. Acta 1145: 180– 182 [DOI] [PubMed] [Google Scholar]
- 21. Chen YH, Yang JT, Martinez HM. 1972. Determination of the secondary structure of proteins by circular dichroism and optical rotatory dispersion. Biochemistry 11: 4120– 4131 [DOI] [PubMed] [Google Scholar]
- 22. Yang JT. 1986. Calculation of protein conformation from circular dichroism. Methods Enzymol. 130: 208– 269 [DOI] [PubMed] [Google Scholar]
- 23. Tan TC, Mijts BN, Swaminathan K, Patel BKC, Divne C. 2008. Crystal structure of the polyextremophilic alpha-amylase AmyB from Halothermothrix orenii: details of a productive enzyme-substrate complex and an N-domain with a role in binding raw starch. J. Mol. Biol. 378: 850– 868 [DOI] [PubMed] [Google Scholar]
- 24. Uitdehaag JCM, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat. Struct. Mol. Biol. 6: 432– 436 [DOI] [PubMed] [Google Scholar]
- 25. Davies G, Henrissat B. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3: 853– 859 [DOI] [PubMed] [Google Scholar]
- 26. Jacobson MA, Colman RF. 1984. Distance relationships between the catalytic site labeled with 4-(iodoacetamido)salicylic acid and regulatory sites of glutamate dehydrogenase. Biochemistry 23: 3789– 3799 [DOI] [PubMed] [Google Scholar]
- 27. Wind RD, Uitdehaag J, Buitelaar RM, Dijkstra BW, Dijkhuizen L. 1998. Engineering of cyclodextrin product specificity and pH optima of the thermostable cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1. J. Biol. Chem. 273: 5771– 5779 [DOI] [PubMed] [Google Scholar]
- 28. Siddiqui KS, Parkin DM, Curmi PMG, Francisci DD, Poljak A, Brarrow K, Noble MH, Trewhella J, Cavicchioli R. 2009. A novel approach for enhancing the catalytic efficiency of a protease at low temperature: reduction in substrate inhibition by chemical modification. Biotechnol. Bioeng. 103: 676– 686 [DOI] [PubMed] [Google Scholar]
- 29. Svensson B. 1994. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol. Biol. 25: 141– 157 [DOI] [PubMed] [Google Scholar]
- 30. Liu YH, Hu B, Xu YJ, Bo JX, Fan S, Wang JL, Lu FP. 2012. Improvement of the acid stability of Bacillus licheniformis alpha amylase by error-prone PCR. J. Appl. Microbiol doi:10.1111/j.1365-2672.2012.05359.x [DOI] [PubMed] [Google Scholar]
- 31. Shaw A, Bott R. 1996. Engineering enzymes for stability. Curr. Opin. Struct. Biol. 6: 546– 550 [DOI] [PubMed] [Google Scholar]
- 32. Brosnan MP, Kelly CT, Fogarty WM. 1992. Investigation of the mechanisms of irreversible thermoinactivation of Bacillus stearothermophilus α-amylase. Eur. J. Biochem. 203: 225– 231 [DOI] [PubMed] [Google Scholar]
- 33. Chi MC, Chen YH, Wu TJ, Lo HF, Lin LL. 2010. Engineering of a truncated α-amylase of Bacillus sp. strain TS-23 for the simultaneous improvement of thermal and oxidative stabilities. J. Biosci. Bioeng. 109: 531– 538 [DOI] [PubMed] [Google Scholar]
- 34. Hagihara H, Hayashi Y, Endo K, Igarashi K, Ozawa T, Kawai S, Ozaki K, Ito S. 2001. Deduced amino-acid sequence of a calcium-free α-amylase from a strain of Bacillus. Eur. J. Biochem. 28: 3874– 3982 [DOI] [PubMed] [Google Scholar]
- 35. Stoner MR, Dale DA, Gualfetti PJ, Becker T, Randolph TW. 2005. Ca2+-surfactant interactions affect enzyme stability in detergent solutions. Biotechnol. Prog. 21: 1716– 1723 [DOI] [PubMed] [Google Scholar]