Abstract
Gammaproteobacterial sulfur oxidizers (GSOs), particularly SUP05-related sequences, have been found worldwide in numerous oxygen-deficient marine environments. However, knowledge regarding their abundance, distribution, and ecological role is scarce. In this study, on the basis of phylogenetic analyses of 16S rRNA gene sequences originating from a Baltic Sea pelagic redoxcline, the in situ abundances of different GSO subgroups were quantified by CARD-FISH (catalyzed reporter fluorescence in situ hybridization) with oligonucleotide probes developed specifically for this purpose. Additionally, ribulose bisphosphate carboxylase/oxygenase form II (cbbM) gene transcript clone libraries were used to detect potential active chemolithoautotrophic GSOs in the Baltic Sea. Taken together, the results obtained by these two approaches demonstrated the existence of two major phylogenetic subclusters embedded within the GSO, one of them affiliated with sequences of the previously described SUP05 subgroup. CARD-FISH analyses revealed that only SUP05 occurred in relatively high numbers, reaching 10 to 30% of the total prokaryotes around the oxic-anoxic interface, where oxygen and sulfide concentrations are minimal. The applicability of the oligonucleotide probes was confirmed with samples from the Black Sea redoxcline, in which the SUP05 subgroup accounted for 10 to 13% of the total prokaryotic abundance. The cbbM transcripts presumably originating from SUP05 cells support previous evidence for the chemolithoautotrophic activity of this phylogenetic group. Our findings on the vertical distribution and high abundance of SUP05 suggest that this group plays an important role in marine redoxcline biogeochemistry, probably as anaerobic or aerobic sulfur oxidizers.
INTRODUCTION
Areas of hypoxia, i.e., dissolved oxygen concentrations below 60 μmol kg water−1 (1), occur naturally in a variety of marine systems throughout the world, including in highly productive upwelling zones, inland seas with restricted water exchanges, and periodically stratified fjords. In these hypoxic zones, steep gradients in nutrient and energy availability are accompanied by changes in microbial abundances, community composition, and metabolic activities (1, 2).
Coupled energy-yielding processes in nitrogen cycling (nitrification, denitrification, and anammox) and in sulfur cycling (sulfide oxidation, sulfate reduction) (1, 3, 4), both of which are characteristic for these redox gradients, fuel light-independent carbon dioxide fixation, referred to as chemolithoautotrophy, at depths below photic zones (5, 6).
Along with members of the epsilonproteobacteria, which constitute a major group of chemolithoautotrophs (7), potentially thiotrophic gammaproteobacteria belonging to the GSO (gammaproteobacterial sulfur oxidizer) cluster in the Baltic and Black Seas were identified in RNA-based stable isotope probing analyses (8, 9). These as-yet-unclassified gammaproteobacteria were originally discovered as typical thiotrophic, potentially autotrophic endosymbionts in the gills of vesicomyid clams, such as Calyptogena sp. and Vesicomya sp., in hydrothermal vents and in cold seeps (10). Diversity analyses in a hydrothermal plume of the Suiyo seamount off the coast of Japan led to the discovery of a highly abundant, free-living gammaproteobacterium closely related to these symbiotic bacteria (11). In fact, it was determined that SUP05, named after the Suiyo seamount plume, contributed up to 58% of the total cell number (TCN) and up to 90% of the total bacteria in this hydrothermal system (11). More recent studies demonstrated the significant appearance of this group in seasonally anoxic fjords with changing sulfide conditions (12, 13), where its abundance may be as high as 30% of the total bacteria, and in marine coastal, highly productive upwelling regions (14, 15). In one such study, the maximal cell abundance of SUP05-related cells, as determined by CARD-FISH (catalyzed reporter fluorescence in situ hybridization), was 11% of the total prokaryotic community, and their area of detection coincided with the occurrence of elemental sulfur (15).
Metagenome analyses of fosmid clones originating from the anoxic fjord Saanich Inlet (12) and metatranscriptome analyses of samples from the oxygen minimum zone (OMZ) of the Eastern Tropical South Pacific (16) indicated that SUP05 is capable of chemoautotrophic denitrification, fueled by the oxidation of reduced sulfur compounds. The assignment of certain gene products to SUP05, some of them present in significant amounts and involved in the oxidation of reduced sulfur compounds (e.g., the sox multienzyme complex, sox), the reduction of oxidized nitrogen (e.g., respiratory nitrate reductase, nar), and CO2 fixation (ribulose bisphosphate carboxylase/oxygenase [RubisCO], cbbM) suggested that members of the GSO contribute substantially to major biogeochemical cycles in oxygen-depleted habitats such as marine OMZs and hydrothermal vent plumes (17, 18). Sequences affiliated with the SUP05 cluster were also recovered from the largest sulfidic marine system, the Black Sea (19, 20). Here, RNA stable-isotope probing with 13C-bicarbonate incubation produced direct evidence of the chemolithoautotrophic metabolism of SUP05 around the oxic-anoxic interface (9).
Members of the GSO group were also detected by different approaches in Baltic Sea redoxclines (8, 21–23). However, in contrast to the highly abundant, chemolithoautotrophic epsilonproteobacterial group GD17 (24, 25), there is as yet no detailed information on the diversity, abundance, or function of GSO in Baltic Sea redoxclines. Thus, the major objective of the present study was to gain insights into the identity, abundance, and distribution of the major GSOs in pelagic redoxclines of the central Baltic Sea.
MATERIALS AND METHODS
Sampling.
Sampling from the central Baltic Sea was carried out onboard different research vessels (R/Vs) between 2004 and 2009 with a conductivity, temperature, and depth probe connected with free-flow bottles as a sampling device. Collection dates and stations are provided in Table S1 in the supplemental material. Physicochemical profiles of the water column at the sampling location were obtained as described elsewhere (26). Sampling from the Black Sea was carried out in May 2007 onboard the R/V Meteor (see Table S1). Natural water samples for nucleic acid analyses were filtered onto membrane filters (Millipore; pore size, 0.2 μm) that were immediately shock frozen and stored at −80°C until further analyses. For CARD-FISH, 40- to 60-ml volumes of Formol (2% final concentration)-fixed water samples were filtered onto membrane filters (type GTTP, Millipore, pore size, 0.2 μm, or Nuclepore, Whatman, pore size, 0.2 μm) that were stored at −80°C until further processing.
Cloning of 16S rRNA gene and cbbM transcripts.
To identify the dominant taxa within the gammaproteobacterial sulfur-oxidizing group, 16S rRNA gene transcripts of the whole bacterial community representative of a depth of a Baltic Sea pelagic redoxcline from which GSO sequences were retrieved previously (22) were cloned and sequenced. The basis of the 16S rRNA gene clone library was a water sample obtained at a depth of 119 m (suboxic zone) in the redoxcline of the Gotland Deep in February 2006.
To identify potential chemoautotrophic organisms affiliated with the beta- and gammaproteobacteria, transcripts of ribulose bisphosphate carboxylase/oxygenase (cbbM, RubisCO form II) were cloned with RNA isolated from a water sample collected in July 2006 from the dark CO2 fixation maximum (located at 77 m) of the redoxcline in the Landsort Deep, where chemolithoautotrophic gammaproteobacteria were previously identified (8).
Nucleic acids were extracted according to reference 27, but the procedure was modified as described in reference 8. Coprecipitated DNA was removed by digestion with DNase I (Ambion). DNA-free RNA was reverse transcribed to cDNA with the iScript Select kit (Bio-Rad) and universal reverse primer 1492r (28) for 16S rRNA gene transcripts and cbbM1033r (29). RNA samples without reverse transcriptase were used as the negative control in subsequent PCRs. The cDNA served as the template in the amplification of nearly full-length 16S ribosomal cDNA copies, carried out with the universal bacterial primer set 27f/1492r (28). The PCR conditions and reaction mixture were the same as those described earlier (24). The cbbM cDNA was amplified with primers cbbM663f/cbbM1033r (29) under the same PCR conditions. The purified amplicons were inserted into the pSC-A vector of the Strataclone system (Stratagene) or pGEM-Teasy (Promega) and subsequently transformed into the respective competent cells according to the manufacturer's instructions. Positive clones were selected by blue-white screening. Preliminary grouping into operational taxonomic units (OTUs) was done by restriction fragment length polymorphism (RFLP) analyses as described in reference 9.
Sequencing and phylogenetic analyses.
Inserts of representative clones were sequenced by the Sanger method by AGOVA (Berlin, Germany) and Qiagen (Hilden, Germany) with vector-specific primer sets T3/T7 and SP6/T7, respectively. The phylogenetic affiliations of the quality-checked 16S rRNA gene and cbbM sequences were preliminarily estimated with BLAST (30). Chimeras were identified with the program DECIPHER (31). The ARB software package V5.1 and the SILVA database SSU_102 (nonredundant) were used for the alignment and phylogenetic analyses of the 16S rRNA gene sequences obtained (32). The cbbM sequences were translated into amino acid sequences and aligned with the ClustalW algorithm included in the ARB software package. Reference sequences from different environmental samples and reference strains were obtained from GenBank and included in the alignment.
Sequences for further analysis were reduced to unambiguously alignable positions (1,285 bases for the 16S rRNA gene, 414 amino acids for cbbM) with group-specific filters. For phylogenetic analyses, three different trees were calculated by the neighbor-joining, parsimony, and maximum-likelihood algorithms (RAxML, 100 repeats) on the basis of nearly full-length sequences (16S rRNA gene, >1,350 bp; cbbM, >400 amino acids). Shorter sequences were subsequently added to the basis tree without changing the overall topology.
Probe design.
The ARB software package was used to design specific probes against the SUP05 group and the BBAL-2 (Baltic/Black Sea autotrophic lineage 2) group on the basis of 16S rRNA gene sequences. The specificity of the newly designed probes was checked with the PROBE_MATCH tool of ARB, BLAST, and the Probe Check online tool (33) and tested by hybridization with negative-control bacterial strains at different formamide concentrations. Thiorhodovibrio winogradskyi served as the negative control for GSO826 (two mismatches), and Escherichia coli and Pseudomonas fluorescens served as the negative controls for GSO1032 (one mismatch). In accordance with the results of the formamide curve revealed the necessity of applying a nonlabeled competitor (one base altered) was applied for GSO1032.
All probe sequences and optimal hybridization conditions are listed in Table 1. The probe targeting the whole SUP05 cluster was named according to the nomenclature described in reference 38 as S-*-GSO1032-a-A-18 and abbreviated as GSO1032. The unlabeled competitor excluding nontargeting organisms was named S-*-GSO1032-b-A-18 and abbreviated as GSO1032-comp. The probe targeting the BBAL-2 group was named S-*-GSO0826-a-A-18 and abbreviated as GSO826.
Table 1.
HRP-labeled oligonucleotide probes used in this study
| Probe | Sequence (5′–3′) | Positiona | Specificity | % FAb | Reference |
|---|---|---|---|---|---|
| EUB338 Ic | GCT GCC TCC CGT AGG AGT | 338–355 | Most Bacteria | 55 | 34 |
| EUB338 IIc | GCA GCC ACC CGT AGG TGT | 338–355 | Planctomycetales | 55 | 35 |
| EUB338 IIIc | GCT GCC ACC CGT AGG TGT | 338–355 | Verrucomicrobiales | 55 | 35 |
| NonEUB | ACT CCT ACG GGA GGC AGC | None (negative control) | 55 | 36 | |
| Gam42ad | GCC TTC CCA CAT CGT TT | 1027–1043e | Gammaproteobacteria | 55 | 37 |
| GSO826 | AAA TGC TCC CAA CGG CTA | 826–843 | BBAL-2 within Gammaproteobacteria | 50 | This study |
| GSO1032f | CCT GTA TCA CGG TTC CCG | 1032–1050 | BBAL-1 (SUP05) within Gammaproteobacteria | 60 | This study |
Refers to E. coli numbering.
% FA, final concentration in hybridization buffer at a hybridization temperature of 35°C.
Application of a mixture of all three probes.
The nonlabeled competitor Bet42a-none was added.
Targets the 23S rRNA.
The unlabeled competitor GSO1032-comp (CCT GTC TCA CGG TTC CC) was added.
Stringency was successfully tested by means of a formamide concentration series within a range of 20 to 70%, increasing the formamide concentration in 5% increments. The highest formamide concentration yielding a clearly detectable signal was chosen for further analyses. All hybridizations were conducted at 35°C.
Enumeration of cells by CARD-FISH.
The cells were hybridized and enumerated according to references 39 and 40, with modification as described in reference 24. Fixed samples (2% [final concentration] Formol) were filtered onto polycarbonate filters (0.22-μm diameter) that were stored at −80°C until further analyses. Horseradish peroxidase (HRP)-labeled oligonucleotide probes were synthesized by Biomers (Ulm, Germany). The fluorescent dye used in these experiments was carboxyfluorescein-labeled tyramide.
Hybridized filter pieces were counterstained with a 4′,6-diamidino-2-phenylindole (DAPI)–Citiflour–Vectashield mixture and examined with an epifluorescence microscope (Zeiss Axioskop) with appropriate filter sets. At least 200 cells in 10 randomly chosen microscopic fields were counted. For Gam42a and GSO826, several 5-mm transects across the filter were analyzed because of the low numbers of hybridized cells.
Exemplary hybridization and enumeration of replicate filters revealed standard deviations of triplicate samples of <5% (relative cell counts) and <10% (absolute cell counts).
Statistical analyses.
All statistical analyses were done with the program SPSS 20.0 (IBM). Significant differences in cell abundances at different oxygen and sulfide concentrations were detected by means of nonparametric analyses (Kruskal-Wallis test, pairwise comparison). Correlations between abundance and environmental parameters such as oxygen, sulfide, nitrogen species, and phosphate were determined with the Spearman rank algorithm.
Nucleotide sequence accession numbers.
The 16S rRNA gene and cbbM sequences generated in this study were deposited in the GenBank database under accession numbers JX974825 to JX974832 and KC492833 to KC492892.
RESULTS
Phylogenetic analyses of 16S rRNA gene libraries.
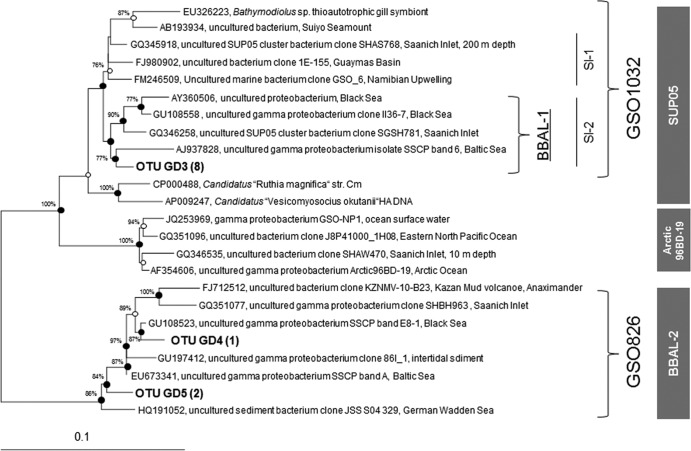
Cloning of the 16S ribosomal cDNA from the lower suboxic zone of the Gotland Deep yielded 196 clones with positive inserts. From these, we obtained 118 nonchimeric sequences that were grouped by subsequent phylogenetic analyses into 59 different OTUs (>97% sequence identity of each OTU), 14 of which were assigned to the gammaproteobacteria. More-detailed phylogenetic analyses showed that three OTUs were phylogenetic members of the GSO and that they belonged to two different subclusters (Fig. 1) designated BBAL-1 and BBAL-2.
Fig 1.
Unrooted maximum-likelihood tree (based on 1,285 bp) of the 16S rRNA gene sequences generated in this study (in bold type) that are phylogenetically affiliated with the sulfur-oxidizing gammaproteobacteria. If available, the isolation sources of the reference sequences are provided. Symbols: ●, validation of the branching point by maximum likelihood, parsimony, and neighbor joining; ○, validation of the subtree by two algorithms. The number in parentheses is the number of clones sequenced. Bootstrap values of >70% are shown. Coverage of newly designed probes is defined by curly brackets.
BBAL-1 was embedded within the SUP05 clade (11) and had <89% identity to BBAL-2. No related cultured representatives are thus far available for members of either subcluster. Eight of the GSO clones were found to be phylogenetically closely related members of the BBAL-1 subcluster united within a single OTU referred to as GD3 (Fig. 1) and embedded within the already described ecotype Sl-2 (12). These sequences were affiliated with clones from the Namibian upwelling (97.2%), Saanich Inlet SUP05 Sl-2 (98.0%), Saanich Inlet SUP05 Sl-1 (97.1%), the Black Sea (97.7%), and the East Tropical South Pacific upwelling (95.5%) and endosymbionts of vesicomyid clams such as Candidatus “Ruthia magnifica” (94.7%). Two different OTUs (GD4, GD5), with an identity of 96.9%, occurred within the BBAL-2 subcluster. GD4 formed a stable cluster with band A, identified by single-strand conformation polymorphism fingerprinting in an earlier study (8). Both OTUs were affiliated with environmental sequences from the Black Sea (92.6 and 93.6%, respectively), from intertidal sediments (97.4 and 96.0%), mud volcanoes (94.2 and 95.9%), and the Saanich Inlet (95.5 and 96.4%). The subcluster ARCTIC19BD-96 (41), presumed to be the sister group of SUP05, was not detected in our clone library.
Identification of SUP05 and BBAL-2 by means of CARD-FISH.
Probe GSO477 (15) failed to generate a significant signal, despite the fact that its sequence fully matched that of the 16S rRNA gene of SUP05-related cells. Hence, on the basis of nearly full-length 16S rRNA gene (>1,300 bp) sequences originating from the Black Sea (9), the Baltic Sea, and other oxygen-depleted marine systems (obtained from GenBank), we designed new phylogenetic probes specifically targeting the two GSO clusters examined in this study.
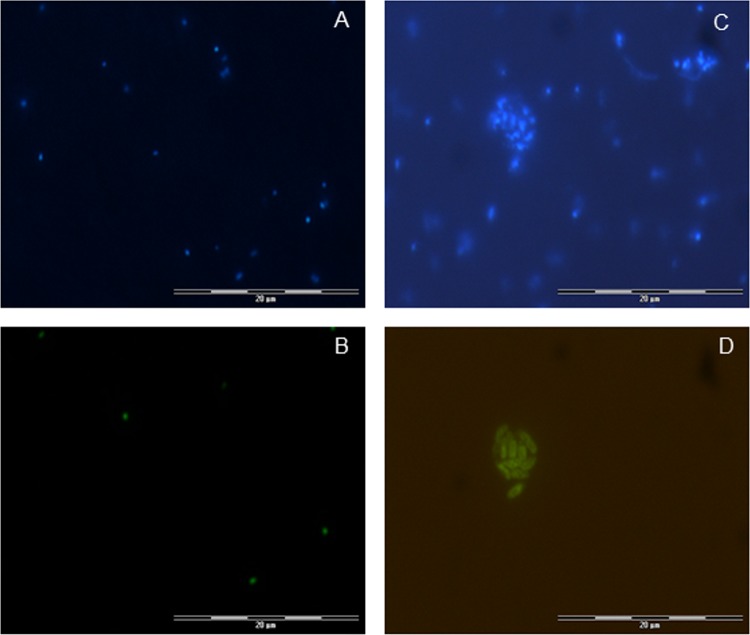
Optimal stringency of gene probe GSO1032, targeting all reported sequences of the SUP05 cluster, but not the ARCTIC96BD-19 cluster, was obtained with a hybridization buffer containing 60% formamide and the inclusion of a nonlabeled competitor with one central mismatch to the original probe sequence (Table 1). GSO1032-positive cells were small, coccoid to slightly rod-shaped cells (Fig. 2B). Probe GSO826, targeting the BBAL-2 subcluster, hybridized with relatively large, rod-shaped cells that were frequently organized in clusters (Fig. 2D). BBAL-2 hybridization was determined to be optimal at a formamide concentration of 50% (Table 1).
Fig 2.
Microscopic views of DAPI-stained environmental samples (A, C), cells hybridized with the GSO1032 probe (B), and cells hybridized with the GSO826 probe (D). (A, B) Landsort Deep, September 2009, 75-m depth. (C, D) Landsort Deep, July 2006, 85-m depth.
Abundance of GSOs in Baltic Sea redoxclines.
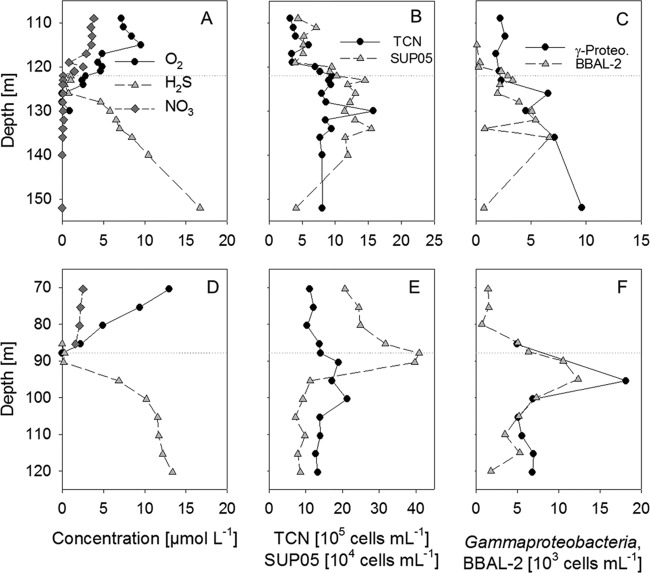
Ten different sample series from pelagic redoxclines of the Baltic Sea, covering two different geographic locations over a period of 5 years, were analyzed (see Table S1 in the supplemental material). In all of the samples investigated, significant numbers of SUP05 and, to a lesser extent, BBAL-2 cells were detected. Representative depth profiles of the nutrient chemistry and distribution of the two GSO clusters are shown in Fig. 3. Maximal SUP05 cell counts were usually detected around the oxic-anoxic interface, where both oxygen and hydrogen sulfide concentrations are close to the detection limit. At these depths, SUP05 cell abundance reached a maximum of 1 × 105 to 4 × 105 cells ml−1, thereby accounting for 10 to 30% of the TCN (Fig. 3B and E). In contrast, BBAL-2 cell counts were about 1 order of magnitude lower, and in the suboxic part of the redoxcline, they were nearly below the detection limit of the method. An increase in the sulfide concentration was paralleled by an increase in BBAL-2 cell counts, which reached an abundance maximum at low sulfide concentrations (<7 μmol liter−1). Nonetheless, cell counts remained below 1% of the TCN in most samples (Fig. 3C and F).
Fig 3.
Depth profiles of a Gotland Deep pelagic redoxcline in February 2006 (top) and a Landsort Deep pelagic redoxcline in September 2009 (bottom). The dotted line marks the chemocline. (A, D) H2S, NO3, and O2 concentrations. (B, E) TCN and SUP05 cell numbers (C, F) Cell counts of gammaproteobacteria and the BBAL-2 subgroup. Note the different scaling in the different panels.
Whereas the Gotland Deep redoxcline was characterized by an extended layer of overlapping oxygen and sulfide (Fig. 3A), at several depths of the Landsort Deep, oxygen and sulfide were below their detection limits (Fig. 3D). In the suboxic zone of the Gotland Deep, SUP05 cell counts of 4.4 × 104 cells ml−1 (corresponding to 8.2% of the TCN) were determined. Cell abundances increased steadily with depth, reaching a maximum of 1.6 × 105 cells ml−1 (13.3% of the TCN) at 134 m (H2S concentration, 6.95 μmol liter−1). At a depth of 152 m, where the H2S concentration was 16.7 μmol liter−1, the cell number declined to 4.1 × 104 ml−1, accounting for 3.5% of the TCN (Fig. 3B). At the Landsort Deep redoxcline, SUP05 cells were highly abundant at its upper, slightly oxygenated part, reaching a maximal abundance of 4.1 × 105 cells ml−1 (29.2% of the TCN) at the oxic-anoxic interface (88 m). The cell number decreased strongly, to 1.1 × 105 cells ml−1, below 95 m, where the H2S concentration exceeded 10 μmol liter−1, but still corresponded to 6.5% of the TCN (Fig. 3E).
The abundance of the BBAL-2 cluster, which hybridized with probe GSO826, was low at most depth profiles (Fig. 3C and F; see Fig. S3 in the supplemental material), with maximal cell counts between 5 × 103 and 1 × 104 cells ml−1, corresponding to <1% of the TCN. In the suboxic water layers of the pelagic redoxclines, the absolute cell numbers were below the detection limit of the method (∼1 × 103 cells ml−1, 0.1% of the TCN) (Fig. 3C and F). Starting from depths at which the oxygen concentration was below 5 μmol liter−1, cell numbers began to increase, with maximal abundances usually reached beneath the chemocline (the shallowest appearance of hydrogen sulfide). Maxima of 6.7 × 103 cells ml−1 (0.8% of the TCN) and 1.2 × 104 cells ml−1 (0.7% of the TCN) were determined in the Gotland Deep and the Landsort Deep, respectively (Fig. 3C and F). In general, the abundance of Gam42a-positive cells was comparable to that of GSO826-positive cells, thus presumably not covering the SUP05 cluster. The cell numbers ranged from 1.8 × 103 to 9.6 × 103 ml−1 (0.3 to 1.2% of the TCN) in the Gotland Deep (Fig. 3C) and from 5.0 × 103 to 1.8 × 104 ml−1 (0.3 to 1.3% of the TCN) in the Landsort Deep (Fig. 3F). Beneath the depth of the first appearance of sulfide, Gam42a-positive cells were closely followed by GSO826-positive cells (Fig. 3C and F).
An exceptional depth profile reflecting a major inflow of oxygenated water into the Baltic proper in 2003 was taken in August 2004 (see Fig. S4 in the supplemental material). The chemocline was located at 232 m (17 m above the sediment), with 16.8 μmol H2S liter−1. At this depth, the abundance of GSO826 increased to 1.5 × 104 cells ml−1, corresponding to a relative cell count of 0.5% of the TCN. The highest cell number, 7.3 × 104 ml−1 (2.5% of the TCN), was detected in the upper sulfidic zone, at 235 m. Additionally, this redoxcline was characterized by an increased total prokaryotic abundance; in absolute terms, the TCN was as high as 3.1 × 106 ml−1.
Statistical analyses.
Taking into account all of the data on SUP05 abundance (n = 70), Spearman rank correlation analyses revealed a significant negative correlation of SUP05 absolute cell abundances with H2S, O2, and nitrate. Relative cell numbers correlated negatively with H2S, ammonia, nitrate, and phosphate and positively with nitrite (Table 2). Box-and-whisker plots of the cell counts in different redox categories showed that the highest SUP05 cell counts occurred around the oxic-anoxic interface and in the lower suboxic zone of the redoxcline (Fig. 4A), as also determined by cumulative plotting of all measurements (see Fig. S3 in the supplemental material). A Kruskal-Wallis test identified significant differences between the categories (P < 0.01), while in subsequent pairwise comparisons, samples originating from depths with H2S concentrations of more than 10 μmol liter−1 were found to differ significantly from those originating at depths with O2 concentrations of less than 10 μmol liter−1 and H2S concentrations of less than 5 μmol liter−1.
Table 2.
Spearman rank correlation analysis of numbers of BBAL-2 and SUP05 cells obtained from Baltic Sea redoxclines
| Parameter | SUP05 |
BBAL-2 |
||||
|---|---|---|---|---|---|---|
| No. of samples | Correlation of parameter with: |
No. of samples | Correlation of parameter with: |
|||
| Absolute cell no. | Relative cell no. | Absolute cell no. | Relative cell no. | |||
| H2S concn | 45 | −0.700a | −0.809a | 50 | 0.140 | 0.091 |
| O2 concn | 34 | −0.344b | −0.146 | 36 | −0.680a | −0.606a |
| NO3 concn | 34 | −0.426b | −0.263 | 41 | −0.626a | −0.508a |
| NH4 concn | 65 | 0.091 | −0.342a | 52 | 0.499a | 0.429a |
| PO4 concn | 69 | 0.048 | −0.333a | 61 | 0.526a | 0.560a |
| NO2 concn | 61 | 0.227 | 0.324b | 57 | −0.042 | −0.211 |
| CO2 fixationc | 40 | 0.122 | 0.196 | 32 | 0.587a | 0.530a |
P < 0.01.
P < 0.05.
μmol liter−1 day−1.
Fig 4.
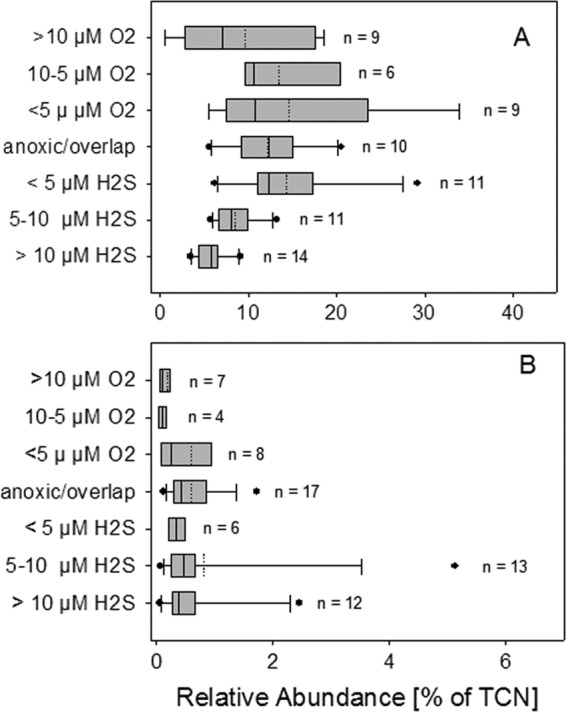
Abundances of SUP05 (A) and BBAL-2 (B) cells versus different concentrations of sulfide and oxygen. The box-and-whisker plots shown include median (solid line) and mean (dashed line) values.
There was a highly significant negative correlation between the cellular abundance of BBAL-2 (n = 67) and oxygen and nitrate, whereas the correlations with ammonia, phosphate, and CO2 fixation were positive (Table 2). Box-and-whisker plots (Fig. 4B) showed that maximal cell numbers occurred at depths with the lowest oxygen and increasing sulfide concentrations. Significant differences between the categories were detected (Kruskal-Wallis test, P < 0.01), whereas subsequent post hoc analyses failed to identify differences between specific groups. Outliers and long whiskers in the box plot of BBAL-2 could be explained by the significantly increased cell numbers of depth profiles sampled 1 year after the major inflow in 2003 (see Fig. S4 in the supplemental material).
Abundance of SUP05-related cells in a pelagic redoxcline of the Black Sea.
The physicochemical parameters of the Black Sea pelagic redoxcline were described in reference 8. Briefly, the chemocline was located at a 135-m depth, with a sulfide concentration of 0.6 μmol liter−1. The H2S concentration increased steadily with increasing depth, reaching 9.9 μmol liter−1 at 160 m (Fig. 5A). SUP05 cells were detectable with the probe GSO1032, with a maximal absolute cell concentration of 7.2 × 104 ml−1 recorded at 145 m (H2S concentration, 4.9 μmol liter−1). A maximal relative cell abundance corresponding to 13.1% of the TCN was determined at a 135-m depth, where the sulfide concentration was 0.6 μmol liter−1. Depths with measurable O2 concentrations were characterized by a reduced abundance of SUP05-related cells, averaging only 0.5 to 4.5% of the TCN. However, at the suboxic depth of 130 m (H2S concentration below the detection limit), the SUP05 abundance already accounted for 10.3% of the TCN (Fig. 5B).
Fig 5.
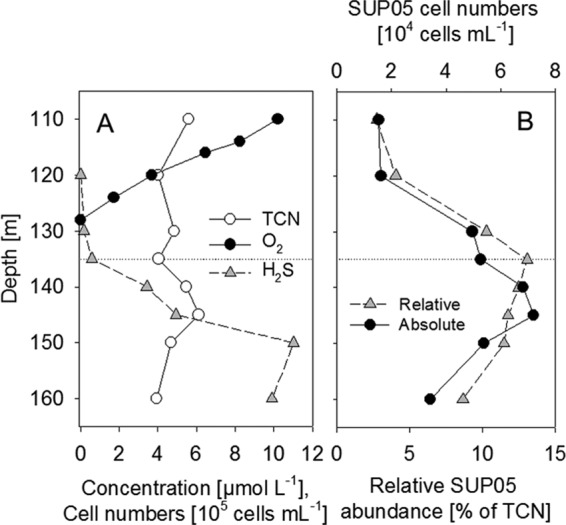
Depth profile of a Black Sea pelagic redoxcline in May 2007. (A) H2S and O2 concentrations and TCN (modified from reference 9). (B) Relative and absolute cell counts of SUP05.
Diversity of the RubisCO form II (cbbM) gene transcript in Baltic Sea redoxclines.
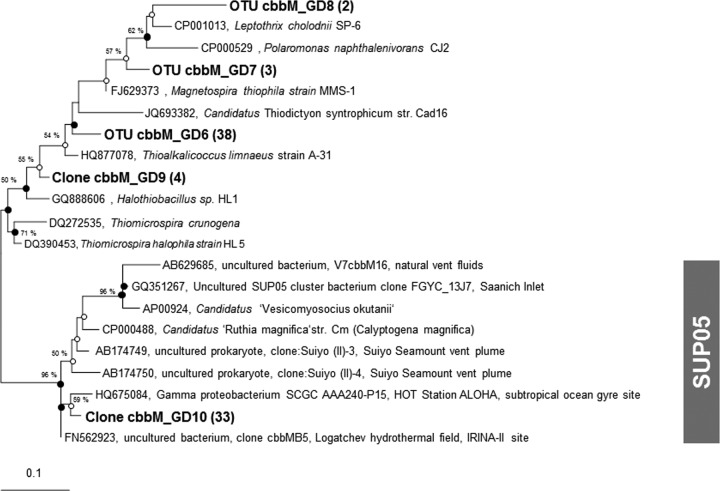
Cloning of the cbbM gene transcripts (RubisCO form II) that originated from the dark CO2 fixation maximum of a pelagic redoxcline of the central Baltic Sea yielded 105 clones with positive inserts. Phylogenetic analyses revealed only five different OTUs (sequence identity, >95%) (Fig. 6). One OTU (cbbM_GD10, 33 clones, 31% of the total clones) was affiliated with SUP05-related metagenome sequences from the Saanich Inlet (90.5%), the Suiyo seamount (87.3%), and vesicomyid mollusk endosymbionts such as Candidatus “Ruthia magnifica” (92.1%) and Candidatus “Vesicomyosocius okutanii” (88.0%).
Fig 6.
Unrooted maximum-likelihood tree of cbbM sequences based on 140 amino acids. Sequences generated in this study are in bold type. If available, the isolation sources of the sequences are provided. Symbols: ●, validation of the branching point by maximum likelihood, parsimony, and neighbor joining; ○, validation of the subtree by two algorithms. The number in parentheses is the number of clones identified by RFLP analyses. Bootstrap values of >50% are shown.
The other OTUs present in this clone library were not as clearly affiliated with distinct phylogenetic groups. The OTU cbbM_GD6, constituting the majority of the clones (38 clones, 36%), affiliated with sequences originating from Thioalkalicoccus limnaeus (gammaproteobacteria, 92.8%) but also with betaproteobacterial sequences from Sulfuricella denitrificans (94.2%) and Thiobacillus thiophilus (90.7%). The OTUs cbbM_GD7 and cbbM_GD8 were related to betaproteobacteria such as Leptothrix cholodnii (82.7 and 85.0%, respectively) and Polaromonas naphthalenivorans (81.0 and 81.6%, respectively). In general, only a few sequences related to this cluster are available; hence, a reliable phylogenetic classification was not possible (Fig. 6).
DISCUSSION
The aim of this study was to obtain more-detailed information on the identity, abundance, and putative ecological function of gammaproteobacterial assemblages, and particularly GSOs, in pelagic redoxclines of the central Baltic Sea and Black Sea. By the full-cycle 16S rRNA gene approach, we identified two different groups, of which the SUP05 cluster was shown to be an abundant member of the microbial community. In contrast, the phylogenetic group BBAL-2, already identified as a chemolithoautotrophic member of the microbial community (8), was negligible in terms of cellular abundance.
This work also describes two newly developed specific oligonucleotide probes that match all reported sequences affiliated with both phylotypes; accordingly, they should be applicable to all of the environments inhabited by these GSOs.
Occurrence of GSO in redoxclines of the Baltic and Black Seas.
By 16S rRNA gene cloning, we identified a single OTU, belonging to the BBAL-1 subcluster, which is embedded within the SUP05 cluster. Subsequent CARD-FISH analyses with a newly developed oligonucleotide probe yielded relatively high cell counts, with abundance maxima ranging from 10 to 30% of the TCN, for the central Baltic Sea and Black Sea pelagic redoxclines. The cellular abundances determined for the Baltic Sea are consistent with previous estimates of SUP05-related 16S rRNA gene abundances based on 454 pyrosequencing, in which up to 20% of the total reads were in waters with oxygen concentrations below 100 μmol liter−1 (42). These abundances are comparable to estimates for the Saanich Inlet (12, 13), based on 16S rRNA gene abundance (approximately 30% of the total bacteria) and to those of a marine OMZ off Namibia, based on direct cell counts (11% of the TCN) (15). Our investigation of the pelagic redoxclines of the Baltic Sea clearly showed that the highest cell counts occurred at depths with a minimal sulfide concentration, pointing to an adaptation to low-sulfidic conditions.
The SUP05 cell counts determined at the abundance maximum were in a range similar to that recently determined for other putative key organisms in Baltic Sea redoxclines, such as one group of epsilonproteobacteria (24) and one cluster of ammonia-oxidizing Archaea (43). The composition of the microbial community, and thus microbe-mediated biogeochemical processes over the pelagic redoxcline, reflects the strong stratification of the water column, which is apparent in terms of temperature, salinity, and nutrient availability (22). At suboxic depths, characterized by detectable nitrification rates (44), the prokaryotic community is dominated by putative ammonia-oxidizing strain GD2, within the phylum Thaumarchaeota (43). This group, in turn, embedded within Marine Group I, constitutes up to 26% of the TCN. In the upper sulfidic part of the redoxcline, usually at depths with overlapping sulfide and nitrate occurrences, the epsilonproteobacterial Sulfurimonas subgroup GD17, identified as a key organism in autotrophic denitrification, constitutes up to 25% of the TCN (7, 24). The high cellular abundance of SUP05 and the location of the abundance maximum between the maxima of GD2 and GD17 suggest that the ecological niche of SUP05 is located around the intersection of the concentration profiles of oxygen and hydrogen sulfide (and probably other reduced sulfur compounds).
It is unclear whether the observed distribution reflects the toxicity of hydrogen sulfide or the lack of potential terminal electron acceptors such as oxygen and nitrate. For the Black Sea redoxcline, maximal absolute cell numbers were detected at depths with higher sulfide concentrations; however, this measurement was based on only one depth profile and is hence not sufficient to allow conclusions to be drawn regarding the distribution of SUP05 in Black Sea redoxclines.
To our knowledge, direct cell counts of SUP05-related cells are available only for the Suiyo seamount (11) and the Namibian upwelling (15); a probe for BBAL-2 has yet to be developed. All other estimates of cellular abundances are based only on nucleic acid abundances, which may be biased because of differences in nucleic acid extraction and PCR efficiencies (12, 13, 16, 20, 45). However, those estimated abundances and distributions across physicochemical gradients are indeed comparable to the cellular abundances determined in this study.
Remarkably, the application of the Gam42a probe (37) led to a large underestimation of the gammaproteobacterial community of Baltic Sea redoxclines, since this phylum-specific probe hybridized with less than 2% of the TCN. Similar gammaproteobacterial cell counts were obtained in a previous quantitative analysis of microbial community composition in Black Sea pelagic redoxclines (46), in which case the probe hybridized with maximally 8% of the TCN in suboxic waters and 6% in the upper sulfidic zone. At depths at which SUP05 cells were detected, the Gam42a-positive cell percentages declined sharply, to as low as 2% of the TCN. The same discrepancies were reported by Fuchsman et al. (20) in a study in which 25% of the 454 pyrosequencing tags were assigned to gammaproteobacteria at depths where maximally 6% of the TCN hybridized with the Gam42a probe according to Lin et al. (46). For the Baltic and Black Seas, there is as yet no 23S rRNA sequence information but the phylogenetic relationships suggest that this probe is insufficient to cover the entire gammaproteobacterial communities in these habitats. In silico analyses of the 23S rRNA of C. “Ruthia magnifica,” the SUP05 metagenome, and related sequences showed that this widely applied oligonucleotide probe contains two central mismatches to the target sequence and thus is not applicable to the detection of SUP05 cells.
Instead, Gam42a-positive cells below the chemocline of Baltic Sea redoxclines are most likely to be represented by the BBAL-2 group, as deduced in this study from cell abundances but also on the basis of its typical morphology. According to our CARD-FISH analyses, the abundance of this gammaproteobacterial group was low but the cells are relatively large (approximately 5 μm). Additionally, previous studies in which rRNA-based fingerprinting was applied to water samples from the Black Sea and Baltic Sea evidenced high relative band intensities for this cluster, pointing to an enhanced cellular RNA pool and thus to highly active cells (8, 9). Although environmental sequences for this group have been frequently reported from sediments, as well as mud volcanoes (47, 48), indicating adaptation to anoxic conditions, the functional and ecological role of this clade is as yet unclear.
Contribution of GSO to the biogeochemical cycling of nitrogen, sulfur, and inorganic carbon.
The most relevant issues that remain to be resolved are the function and ecological niche fulfilled by the GSO, and particularly the SUP05 cluster, in pelagic redoxclines and other oxygen-deficient systems. To date, most of the evidence, i.e., from metagenomic surveys and investigations of functional genes, is indirect. With these approaches, genes frequently involved in denitrification (nirK, narGH) and sulfur oxidation (sox, apr, dsr) affiliated with the SUP05 cluster were identified, predicting sulfur oxidation as the energy-yielding metabolic process, in which NO3 is the terminal electron acceptor (12, 15, 16, 49). Moreover, the cellular abundance of SUP05 in the Namibian upwelling OMZ coincided with peaks in denitrification and the occurrence of elementary sulfur, suggesting the coupling of sulfur oxidation and respiratory nitrate reduction (15). Recently, also evidence of an important role for H2 oxidation in the energy metabolism of a SUP05 cluster in the deep ocean has been obtained (50). For Baltic Sea redoxclines, environmental sequences of narGH, nirK, and genes involved in sulfur oxidation pathways are lacking, such that the genetic potential of SUP05 with respect to nitrogen and sulfur cycles cannot be examined at the moment. However, the abundance of SUP05 cells in redoxclines of the Baltic and Black Seas is maximal at depths characterized by minimal sulfide and oxygen concentrations; this depth distribution resembles that of SUP05 in marine OMZs, where higher concentrations of hydrogen sulfide are not detected (2).
For Baltic Sea redoxclines, the contribution of SUP05 to chemolithoautotrophic production is currently unknown, but our detection of cbbM gene transcripts assignable to SUP05 also indicates chemolithoautotrophic activity. For a Black Sea redoxcline, a recent study provided direct evidence of light-independent CO2 fixation (9). Here, stable isotope probing experiments applying 13C-labeled bicarbonate to water samples from the upper sulfidic zone led to the identification of two different gammaproteobacteria within the GSO, i.e., SUP05 and the BS-GSO2 cluster, as well as one epsilonproteobacterium related to Sulfurimonas denitrificans, as actively bicarbonate-incorporating taxa. Although there is as yet no direct evidence of the chemolithoautotrophic activities of SUP05 in the Baltic Sea redoxcline, both CO2 fixation activity and SUP05-related 16S rRNA gene abundance were stimulated in an earlier experiment in which thiosulfate and manganese oxide were added to water samples from the maximum CO2 fixation zone of a Baltic Sea pelagic redoxcline (21).
By cloning cbbM transcripts, we identified one OTU affiliated with SUP05. To our knowledge, the diversity and expression of key genes involved in CO2-fixing cycles in anoxic basins, such as those of the Baltic Sea, the Black Sea, and the Cariaco Basin, have not been examined so far. The only reference sequences we obtained from GenBank were those from a metagenomic survey in the Saanich Inlet (12) and the Suiyo seamount (51) and originating from endosymbionts of Calyptogena sp. and Vesicomya sp. (52, 53). Nonetheless, the identification of cbbM transcripts affiliated with SUP05 is consistent with the idea that these organisms are active members of the chemolithoautotrophic community of Baltic Sea pelagic redoxclines, a hypothesis that deserves further investigations.
In contrast, BBAL-2 was previously recognized as an inorganic-carbon-assimilating cluster in a Baltic Sea redoxcline (8) and was also reported to be present, albeit without autotrophic activities, in a Black Sea redoxcline (9). However, because of a lack of cultivated representatives or metagenome data from this group or from closely related organisms, sequence data on key enzymes of CO2-fixing mechanisms are not available; hence, OTUs from our cbbM clone library could not be assigned to BBAL-2.
In Baltic Sea pelagic redoxclines, the majority of the chemolithoautotrophic activity was previously shown to be caused by the denitrifying epsilonproteobacterial subgroup GD17, which is closely related to Sulfurimonas denitrificans (24, 25). In contrast, at depths with the highest SUP05 abundances, low chemolithoautotrophic activities are usually recorded, even though substrates for fueling this energy-demanding process should be present because of mixing, lateral intrusion, and diffusion (54). However, since these results were based on incubation-dependent experiments, they may not reflect the actual in situ activity. In a recent study that determined the in situ δ13C values of fatty acid methyl esters, it was shown that directly at the chemocline, i.e., the abundance maximum of SUP05, the 13C content of proteobacterial fatty acids is significantly enriched, which is indicative of chemolithoautotrophy (8). However, assuming that SUP05 and GD17 exhibit comparable metabolisms, as well as comparable, but not similar, depth distributions, the precise ecological niche occupied by these organisms and potential competitive interactions remain to be specified.
ACKNOWLEDGMENTS
We are grateful to the captains and crews of R/V Professor Albrecht Penck, R/V Alkor, R/V Maria S. Merian, R/V Poseidon, and R/V Meteor. We thank Günter Jost for helpful discussions and Steven Hallam for comments on an earlier version of this paper. The excellent technical support of Katja Becker and Bärbel Buuk is greatly appreciated.
This work was funded by the IOW and by grants from the European Science Foundation (09-EuroEEFG-FP-034, MOCA) and the German Science Foundation to M.L. (LA 1466/4-1,4-2) and K.J. (JU 367/12-1).
Footnotes
Published ahead of print 15 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03777-12.
REFERENCES
- 1. Wright JJ, Konwar KM, Hallam SJ. 2012. Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10:381–394 [DOI] [PubMed] [Google Scholar]
- 2. Ulloa O, Canfield DE, DeLong EF, Letelier RM, Stewart FJ. 2012. Microbial oceanography of anoxic oxygen minimum zones. Proc. Natl. Acad. Sci. U. S. A. doi:10.1073/pnas.1205009109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jørgensen BB, Fossing H, Wirsen CO, Jannasch HW. 1991. Sulfide oxidation in the anoxic Black Sea chemocline. Deep Sea Res. A 38:S1083–S1103 [Google Scholar]
- 4. Lam P, Kuypers MMM. 2011. Microbial nitrogen cycling processes in oxygen minimum zones. Annu. Rev. Mar Sci. 3:317–345 [DOI] [PubMed] [Google Scholar]
- 5. Taylor GT, Iabichella M, Ho T-Y, Scranton MI, Thunell RC, Muller-Karger F, Varela R. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol. Oceanogr. 46:148–163 [Google Scholar]
- 6. Jost G, Zubkov MV, Yakushev E, Labrenz M, Jürgens K. 2008. High abundance and dark CO2 fixation of chemolithoautotrophic prokaryotes in anoxic waters of the Baltic Sea. Limnol. Oceanogr. 53:14–22 [Google Scholar]
- 7. Grote J, Schott T, Bruckner CG, Glöckner FO, Jost G, Teeling H, Labrenz M, Jürgens K. 2012. Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc. Natl. Acad. Sci. U. S. A. 109:506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glaubitz S, Lueders T, Abraham W-R, Jost G, Jürgens K, Labrenz M. 2009. 13C-isotope analyses reveal that chemolithoautotrophic Gamma- and Epsilonproteobacteria feed a microbial food web in a pelagic redoxcline of the central Baltic Sea. Environ. Microbiol. 11:326–337 [DOI] [PubMed] [Google Scholar]
- 9. Glaubitz S, Labrenz M, Jost G, Jürgens K. 2010. Diversity of active chemolithoautotrophic prokaryotes in the sulfidic zone of a Black Sea pelagic redoxcline as determined by rRNA-based stable isotope probing. FEMS Microbiol. Ecol. 74:32–41 [DOI] [PubMed] [Google Scholar]
- 10. Dubilier N, Bergin C, Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6:725–740 [DOI] [PubMed] [Google Scholar]
- 11. Sunamura M, Higashi Y, Miyako C, Ishibashi J-I, Maruyama A. 2004. Two bacteria phylotypes are predominant in the Suiyo Seamount hydrothermal plume. Appl. Environ. Microbiol. 70:1190–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, Tortell PD, Hallam SJ. 2009. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326:578–582 [DOI] [PubMed] [Google Scholar]
- 13. Zaikova E, Walsh DA, Stilwell CP, Mohn WW, Tortell PD, Hallam SJ. 2010. Microbial community dynamics in a seasonally anoxic fjord: Saanich Inlet, British Columbia. Environ. Microbiol. 12:172–191 [DOI] [PubMed] [Google Scholar]
- 14. Stevens H, Ulloa O. 2008. Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ. Microbiol. 10:1244–1259 [DOI] [PubMed] [Google Scholar]
- 15. Lavik G, Stührmann T, Brüchert V, Van der Plas A, Mohrholz V, Lam P, Mußmann M, Fuchs BM, Amann R, Lass U, Kuypers MMM. 2009. Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457:581–584 [DOI] [PubMed] [Google Scholar]
- 16. Stewart FJ, Ulloa O, DeLong EF. 2012. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ. Microbiol. 14:23–40 [DOI] [PubMed] [Google Scholar]
- 17. Stewart FJ. 2011. Dissimilatory sulfur cycling in oxygen minimum zones: an emerging metagenomics perspective. Biochem. Soc. Trans. 39:1859–1863 [DOI] [PubMed] [Google Scholar]
- 18. Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ. 2012. The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J. 6:2257–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vetriani C, Tran HV, Kerkhof LJ. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69:6481–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fuchsman CA, Kirkpatrick JB, Brazelton WJ, Murray JW, Staley JT. 2011. Metabolic strategies of free-living and aggregate-associated bacterial communities inferred from biologic and chemical profiles in the Black Sea suboxic zone. FEMS Microbiol. Ecol. 78:586–603 [DOI] [PubMed] [Google Scholar]
- 21. Labrenz M, Jost G, Pohl C, Beckmann S, Martens-Habbena W, Jürgens K. 2005. Impact of different in vitro electron donor/acceptor conditions on potential chemolithoautotrophic communities from marine pelagic redoxclines. Appl. Environ. Microbiol. 71:6664–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Labrenz M, Jost G, Jürgens K. 2007. Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic-anoxic interface. Aquat. Microb. Ecol. 46:177–190 [Google Scholar]
- 23. Brettar I, Christen R, Höfle MG. 2012. Analysis of bacterial core communities in the central Baltic by comparative RNA-DNA-based fingerprinting provides links to structure-function relationships. ISME J. 6:195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grote J, Labrenz M, Pfeiffer B, Jost G, Jürgens K. 2007. Quantitative distributions of Epsilonproteobacteria and a Sulfurimonas subgroup in pelagic redoxclines of the central Baltic Sea. Appl. Environ. Microbiol. 73:7155–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grote J, Jost G, Labrenz M, Herndl GJ, Jürgens K. 2008. Epsilonproteobacteria represent the major portion of chemoautotrophic bacteria in sulfidic waters of pelagic redoxclines of the Baltic and Black Seas. Appl. Environ. Microbiol. 74:7546–7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grashoff K, Erhardt M, Kremling K. 1983. Methods of seawater analysis, 2nd ed Verlag Chemie, Weinheim, Germany [Google Scholar]
- 27. Weinbauer MG, Fritz I, Wenderoth DF, Höfle MG. 2002. Simultaneous extraction from bacterioplankton of total RNA and DNA suitable for quantitative structure and function analyses. Appl. Environ. Microbiol. 68:1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Ltd., Chichester, United Kingdom [Google Scholar]
- 29. Elsaied H, Naganuma T. 2001. Phylogenetic diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from deep-sea microorganisms. Appl. Environ. Microbiol. 67:1751–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wright ES, Yilmaz LS, Noguera DR. 2012. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 78:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. 2008. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ. Microbiol. 10:2894–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 36. Wallner G, Amann R, Beisker W. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136–143 [DOI] [PubMed] [Google Scholar]
- 37. Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. 1992. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593–600 [Google Scholar]
- 38. Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. 2003. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bano N, Hollibaugh JT. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Herlemann DPR, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5:1571–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Labrenz M, Sintes E, Toetzke F, Zumsteg A, Herndl GJ, Seidler M, Jürgens K. 2010. Relevance of a crenarchaeotal subcluster related to Candidatus Nitrosopumilus maritimus to ammonia oxidation in the suboxic zone of the central Baltic Sea. ISME J. 4:1496–1508 [DOI] [PubMed] [Google Scholar]
- 44. Hietanen S, Jäntti H, Buizert C, Jürgens K, Labrenz M, Voss M, Kuparinen J. 2012. Hypoxia and nitrogen processing in the Baltic Sea water column. Limnol. Oceanogr. 57:325–337 [Google Scholar]
- 45. Fuchsman C, Murray AJW, Staley JT. 2012. Stimulation of autotrophic denitrification by intrusions of the Bosporus Plume into the anoxic Black Sea. Front. Microbiol. 3:257 doi:10.3389/fmicb.2012.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin X, Wakeham SG, Putnam IF, Astor YM, Scranton MI, Chistoserdov AY, Taylor GT. 2006. Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl. Environ. Microbiol. 72:2679–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pachiadaki MG, Lykousis V, Stefanou EG, Kormas KA. 2010. Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol. Ecol. 72:429–444 [DOI] [PubMed] [Google Scholar]
- 48. Lenk S, Arnds J, Zerjatke K, Musat N, Amann R, Mußmann M. 2011. Novel groups of Gammaproteobacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment. Environ. Microbiol. 13:758–774 [DOI] [PubMed] [Google Scholar]
- 49. Newton I, Girguis P, Cavanaugh C. 2008. Comparative genomics of vesicomyid clam (Bivalvia: Mollusca) chemosynthetic symbionts. BMC Genomics 9:585 doi:10.1186/1471-2164-9-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anantharaman K, Breier JA, Sheik CS, Dick GJ. 2013. Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc. Natl. Acad. Sci. U. S. A. 110:330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kato S, Nakawake M, Ohkuma M, Yamagishi A. 2012. Distribution and phylogenetic diversity of cbbM genes encoding RubisCO form II in a deep-sea hydrothermal field revealed by newly designed PCR primers. Extremophiles 16:277–283 [DOI] [PubMed] [Google Scholar]
- 52. Newton ILG, Woyke T, Auchtung TA, Dilly GF, Dutton RJ, Fisher MC, Fontanez KM, Lau E, Stewart FJ, Richardson PM, Barry KW, Saunders E, Detter JC, Wu D, Eisen JA, Cavanaugh CM. 2007. The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315:998–1000 [DOI] [PubMed] [Google Scholar]
- 53. Kuwahara H, Takaki Y, Shimamura S, Yoshida T, Maeda T, Kunieda T, Maruyama T. 2011. Loss of genes for DNA recombination and repair in the reductive genome evolution of thioautotrophic symbionts of Calyptogena clams. BMC Evol. Biol. 11:285 doi:10.1186/1471-2148-11-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bruckner CG, Mammitzsch K, Jost G, Wendt J, Labrenz M, Jürgens K. 30 August 2012, posting date Chemolithoautotrophic denitrification of epsilonproteobacteria in marine pelagic redox gradients. Environ. Microbiol. [Epub ahead of print.] doi:10.1111/j.1462-2920.2012.02880.x [DOI] [PubMed] [Google Scholar]