Abstract
Cytochrome P450 monooxygenases (P450s) are known to oxidize hydrocarbons, albeit with limited substrate specificity across classes of these compounds. Here we report a P450 monooxygenase (CYP63A2) from the model ligninolytic white rot fungus Phanerochaete chrysosporium that was found to possess a broad oxidizing capability toward structurally diverse hydrocarbons belonging to mutagenic/carcinogenic fused-ring higher-molecular-weight polycyclic aromatic hydrocarbons (HMW-PAHs), endocrine-disrupting long-chain alkylphenols (APs), and crude oil aliphatic hydrocarbon n-alkanes. A homology-based three-dimensional (3D) model revealed the presence of an extraordinarily large active-site cavity in CYP63A2 compared to the mammalian PAH-oxidizing (CYP3A4, CYP1A2, and CYP1B1) and bacterial aliphatic-hydrocarbon-oxidizing (CYP101D and CYP102A1) P450s. This structural feature in conjunction with ligand docking simulations suggested potential versatility of the enzyme. Experimental characterization using recombinantly expressed CYP63A2 revealed its ability to oxidize HMW-PAHs of various ring sizes, including 4 rings (pyrene and fluoranthene), 5 rings [benzo(a)pyrene], and 6 rings [benzo(ghi)perylene], with the highest enzymatic activity being toward the 5-ring PAH followed by the 4-ring and 6-ring PAHs, in that order. Recombinant CYP63A2 activity yielded monohydroxylated PAH metabolites. The enzyme was found to also act as an alkane ω-hydroxylase that oxidized n-alkanes with various chain lengths (C9 to C12 and C15 to C19), as well as alkyl side chains (C3 to C9) in alkylphenols (APs). CYP63A2 showed preferential oxidation of long-chain APs and alkanes. To our knowledge, this is the first P450 identified from any of the biological kingdoms that possesses such broad substrate specificity toward structurally diverse xenobiotics (PAHs, APs, and alkanes), making it a potent enzyme biocatalyst candidate to handle mixed pollution (e.g., crude oil spills).
INTRODUCTION
Environmental chemicals such as fused-ring polycyclic aromatic hydrocarbons (PAHs) and long-chain alkylphenols (APs) pose a serious threat to human health, as these chemicals are highly mutagenic/carcinogenic (1, 2) and exhibit endocrine-disrupting (ED) activity (3), respectively. As the genotoxicity of PAHs increases with the number of aromatic rings in the molecule, high-molecular-weight PAHs (HMW-PAHs; ≥4 aromatic rings) are a particular human health concern because of their carcinogenic potential. Both HMW-PAHs and APs are generated via anthropogenic and industrial activities and are highly persistent in the environment (3, 4). The hydrophobic nature (low solubility in water) and extended conjugated aromatic structures make these chemicals recalcitrant to biodegradation due to poor bioavailability and high chemical stability (3, 4) and cause their bioaccumulation in the food chain (5, 6). Aliphatic hydrocarbons, particularly alkanes, occur concomitantly with PAHs in crude oils, and their biodegradability decreases with the increase in their carbon chain lengths (because of the resulting hydrophobic nature). Hence, the biodegrading microorganisms in natural pollution scenarios, such as crude oil spills, may preferably require enzymatic capability to metabolize and/or thrive on this major oil fraction (alkanes) in addition to degrading more recalcitrant PAHs (7).
For the past 4 decades much emphasis has been placed on characterization of microorganisms and their enzymes for biodegradation of persistent environmental chemicals, such as HMW-PAHs and endocrine-disrupting chemicals (EDCs). Both the prokaryotes (bacteria) and eukaryotes (fungi and animals) seem to follow similar mechanisms of initial oxidation of these chemicals via oxygenases (8, 9). This usually rate-limiting initial oxidation step in the biodegradation of these chemicals is catalyzed by cytochrome P450 monooxygenases (mainly eukaryotes) or nonheme iron dioxygenases (mainly prokaryotes). Cytochrome P450 monooxygenases (CYPs/P450s) are heme-thiolate proteins distributed across biological kingdoms and phyla and are known to catalyze a wide variety of reactions such as hydroxylation, epoxidation, dealkylation, sulfoxidation, deamination, desulfuration, dehalogenation, and N-oxide reduction (10).
Fungi, particularly the white rot group of basidiomycete fungi, are lower eukaryotic microorganisms which have shown an extraordinary capability to oxidize and degrade or mineralize (to CO2) PAHs and APs via peroxidase- or P450-mediated mechanisms (11, 12). While their peroxidases have been extensively studied, little is known about the specific fungal P450s involved in the oxidation of HMW-PAHs and long-chain APs. Isolation and characterization of these P450s from biodegradative fungi will be important in efforts to develop improved biocatalysts for efficient removal of recalcitrant toxicants from the environment.
Phanerochaete chrysosporium has been the most intensively studied model white rot basidiomycete for understanding the mechanisms of degradation of lignin, the most abundant aromatic polymer on earth (13), and various xenobiotic structures, including PAHs and APs (14). Pregenomic studies from our laboratory led to the first isolation of complete P450 genes, namely, CYP63A1 and CYP63A2 (15) in P. chrysosporium. These genes, though tandemly linked, were found to be inducible to different extents, by a wide range of xenobiotic structures, including aliphatic, aromatic/polyaromatic, and alkyl-substituted aromatic compounds (16). Subsequent whole-genome sequencing of P. chrysosporium (17) revealed the presence of an extraordinarily large contingent of P450 genes (149 complete P450 genes and 12 pseudogenes) in this organism (14, 17). Although xenobiotic induction data on several individual P450s implied their functional potential, experimental analysis to understand their specific role in catalysis was hampered due to the difficulty in active heterologous expression of these enzymes (18, 19). In this direction, we have recently developed a coexpression strategy that allowed successful active expression of P. chrysosporium P450s along with their homologous cytochrome P450 reductase (Pc-CPR) in the yeast Pichia pastoris (20, 21). Using this coexpression strategy, the current study focused on functional characterization of CYP63A2, one of the first cloned Pc-P450 enzymes, and unveiled its unusual and versatile catalytic characteristics. Computational analysis of the modeled three-dimensional (3D) structure in conjunction with experimental catalytic analysis showed that CYP63A2 possesses a broad and unique substrate specificity, oxidizing HMW-PAHs as well as APs and alkanes of various chain lengths. This study constitutes the first characterization of a microbial native P450 capable of oxidizing the recalcitrant HMW-PAHs with up to 6 fused aromatic rings, APs with various alkyl side chain lengths, including the recalcitrant (C8 and C9) forms, and long-chain n-alkanes (C9 to C12 and C15 to C19). Furthermore, this study marks the beginning of systematic functional characterization of the members in the first cloned P450 family, CYP63, in the white rot fungus.
MATERIALS AND METHODS
Chemicals.
All PAH compounds and related authentic standards [1-hydroxypyrene and 3-hydroxybenzo(a) pyrene], the alkylphenols, including technical grade nonylphenol (tNP), 4-n-nonylphenol (4-n-NP), and 4-n-octylphenol (4-n-OP), and the n-alkane mixture containing 40 mg/liter each of the C8 to C20 n-alkanes in hexane were obtained from Sigma-Aldrich (St. Louis, MO). 4-n-Heptylphenol (4-n-HTP), 4-n-pentylphenol (4-n-PTP), 4-n-butylphenol (4-n-BP), and 4-n-propylphenol (4-n-PP) were purchased from Fisher Scientific (Pittsburgh, PA).
Coexpression of CYP63A2 and its homologous redox partner in P. pastoris.
In order to assess the oxidation capability of CYP63A2 toward PAHs and APs, two P. pastoris clones generated in our laboratory by transforming with a binary vector construct containing Pc-CYP63A2 and Pc-CPR (named PC2) (21) and empty vector pPICZB (named PP C) (20) were used. Media and growth conditions for culturing P. pastoris were as described in the Pichia expression manual supplied by the manufacturer (Invitrogen, Life Technologies, USA).
Coexpression of CYP63A2 and its homologous redox partner Pc-CPR in recombinant P. pastoris clones was assessed using microsomes prepared as described previously (21). Briefly, the test clone (PC2 or PP C) was cultured in buffered minimal glycerol (BMG) medium till it reached an absorbance (A600) of 2.0. Pelleted cells were resuspended in buffered minimal (BM) medium (21 ml) supplemented with aminolevulinic acid (2 mM) and methanol (0.5% [vol/vol]). Following incubation at 30°C for 24 h (220 rpm), the cells were pelleted and resuspended in a lysis buffer containing 50 mM potassium phosphate buffer, pH 7.4, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Sigma; catalog no. P-8465). Cells were lysed by vortexing with an equal volume of acid-washed glass beads using 10 cycles of alternate pulsing and cooling for 30 s each. The cell lysate was centrifuged at 5,000 × g for 10 min at 4°C to remove cell debris. The resulting supernatant was centrifuged (20,000 × g for 30 min at 4°C) to pellet the mitochondria. The refined supernatant was collected and recentrifuged at 100,000 × g for 3 h to isolate microsomes. The pellet (microsomes) was resuspended in the lysis buffer and aliquoted and stored at −80°C until used for analyzing the expression levels. P450 and CPR expression levels in microsomes were determined on the basis of the reduced CO difference spectrum and NADPH-dependent cytochrome c reducing activity, respectively, as described elsewhere (22). In addition, expression of the CPR and P450 proteins in the microsomes was analyzed by SDS-PAGE and Western blotting using anti-His antibodies.
Heterologous expression of CYP63A2 in Escherichia coli.
In order to assess the oxidation capability of CYP63A2 toward alkanes, it was heterologously expressed in E. coli. Briefly, the CYP63A2 cDNA, truncated at the 5′ end by 255 nucleotides (encoding 85 amino acids) for optimal expression, was cloned in the pCWOri vector (kindly provided by M. J. Paine, Liverpool School of Tropical Medicine, Liverpool, United Kingdom). The expressing E. coli clone was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) as described elsewhere (23). The induced cells were resuspended in 100 mM Tris-HCl buffer containing 20% glycerol, 5 mM dithiothreitol (DTT), and 1 mM PMSF and lysed using a sonicator. Cell debris was removed by centrifugation (3,000 × g for 5 min), and the resulting crude cell extract was used for analyzing the CYP63A2 expression via detection of a characteristic P450 reduced CO difference spectrum (22).
Yeast whole-cell oxidation assays.
For whole-cell oxidation of PAHs and APs (tNP and linear APs with alkyl chains ranging from C3 to C9), a single colony from each of the recombinant P. pastoris clones (PC2 and PP C) was cultured in BMG medium until the absorbance (A600) reached 2.0. Cells were pelleted and resuspended in BM medium (21 ml) containing aminolevulinic acid (2 mM). The cell suspension was subdivided equally (7 ml each) into three conical flasks. Each flask was spiked with the test xenobiotic compound dissolved in methanol. All compounds were tested at a 20-ppm final concentration, and the concentration of the vehicle (methanol), which was also the carbon source in the medium, did not exceed 0.5% (vol/vol). An uninoculated control meant for estimation of the initial level of the compound and for assessing the degree of any abiotic degradation was prepared using the same medium and run alongside the test flasks. All treatments were performed in triplicate. The cultures were incubated (30°C, 250 rpm) for 24 h. Subsequently, 5-ml culture aliquots were removed aseptically and extracted (3 times) with an equal volume of methylene chloride. The solvent extracts were dried on anhydrous sodium sulfate and resuspended in acetonitrile (ACN) or methanol. Simultaneously, after incubation, 1 ml of the culture was centrifuged and the cell pellet was dried to estimate dry biomass weight, as described elsewhere (20).
In vitro reconstituted P450 enzyme reaction-based oxidation assay.
CYP63A2 oxidation activity toward alkanes was tested in an in vitro reconstituted P450 enzyme reaction. The reaction mixture (1 ml) contained 100 mM Tris-HCl buffer, 200 mM NAPDH, 0.001% alkane mixture, and the enzymes. Equal volumes of the E. coli cell extract containing CYP63A2 enzyme (prepared as described above) and a cell extract containing homologous P450 reductase (Pc-CPR) prepared from a recombinant E. coli clone generated in our previous study (19) were mixed and used as the source of enzymes. The reaction mixture was incubated at room temperature for 1 h under constant shaking. The completed reaction mixtures and the no-enzyme controls (prepared by incubating alkanes with the control host E. coli cell extracts) were extracted with methylene chloride (3 times). The pooled extract from an individual reaction was evaporated to dryness and resuspended in a final volume of 1 ml using methylene chloride, and the resulting extract was used for gas chromatography-mass spectrometry (GC-MS) analysis of parent alkanes. For reliable identification of the oxidation products and the position of hydroxylation in the products, methylene chloride extracts were derivatized using N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) (catalog no. 33027; Sigma-Aldrich). Briefly, each reaction extract residue was resuspended in 100 μl of the same solvent. An equal volume of BSTFA (100 μl) was added, and the vial was tightly sealed and incubated at 65°C for 20 min. The resulting derivatized samples were analyzed by GC-MS.
Analytical methods. (i) HPLC.
The oxidation status of the PAHs [pyrene, phenanthrene, benzo(a)pyrene] and AP compounds (tNP and linear APs with alkyl chains ranging from C3 to C9) was analyzed using high-pressure liquid chromatography (HPLC). The analysis was done on Prostar 210/215 HPLC system (Varian, Inc.) equipped with a C18 reverse-phase column (4.6 mm by 250 mm) and a UV detector. The separation was achieved using a 20-min linear gradient of acetonitrile in water [60% to 100% acetonitrile for fluorene, anthracene, acenaphthene, phenanthrene, benzo(a)pyrene, and benzo(ghi)perylene and 70% to 100% for pyrene and fluoranthene] as the mobile phase at a flow rate of 2 ml/min. HPLC separation of APs was achieved using a 20-min linear gradient of ACN in water (50% to 100% for tNP, 4-n-NP, and 4-n-OP) and a 25-min linear gradient of ACN in water (50% to 90% for 4-n-HTP, 50% to 80% for 4-n-PTP, 50% to 60% for 4-n-BP and 4-n-PP) at a flow rate of 2 ml/min. PAHs and their metabolites were detected at 254 nm, whereas APs and their metabolites were detected at 277 nm. These compounds were quantified based on corresponding standard curves generated using their defined concentrations.
(ii) LC-ESI/MS.
Hydroxylated metabolites of PAHs and APs generated by the action of CYP63A2 were analyzed by liquid chromatography-electrospray ionization/mass spectrometry (LC-ESI/MS) using the LTQ-Orbitrap coupled with a surveyor MS Pump Plus (Thermo Finnigan) as described in our previous studies (20, 24). The solvent system comprised water and methanol, with 10 mM ammonium acetate included in both solvents. The LC gradient for PAH metabolites was as follows: pyrene and benzo(a)pyrene metabolites, 65% methanol for 2 min, 65 to 100% methanol in 25 min, followed by a hold at 100% methanol for 3 min; fluoranthene and benzo(ghi)perylene metabolites, 65% methanol for 5 min, 65 to 100% methanol in 50 min, followed by a hold at 100% methanol for 5 min. The LC gradient for AP metabolites was as follows: 50% methanol for 5 min, 45-min linear gradient of methanol in water (50% to 90%), followed by a hold at 100% methanol for 6 min and 50% methanol for 4 min. Full-scan mass spectra (m/z 100 to 500 [for PAHs] and m/z 50 to 500 [for APs]) were acquired at a resolution of 30,000 (full width at half-maximum intensity [FWHM]) throughout the chromatographic runs. The results were analyzed, and extracted ion chromatograms were constructed with XCalibur (Thermo). Experimental masses of metabolites were obtained by averaging 25 scans across the full width at the half maximum of the peak.
(iii) GC-MS.
The alkane oxidation rate and oxidation products were analyzed by GC-MS. Analysis was performed using an Agilent 7890A GC/5975C MSD, equipped with an Rxi-5sil MS (Restek Corp.) column (30 m in length with a 0.25-mm inside diameter [ID] and 0.25-μm film). The injector was run in splitless mode at 275°C, with a 1-μl sample injection. The carrier gas (helium) flow rate was set to 1 ml/min. The GC oven was programmed at 60°C initially with a 1-min hold and then ramped to 300°C at 10°C/min; this was followed by a hold for 3 min. The MS detector was set to scan over the m/z range of 40 to 400, with an emission current set to 35 μA and electron energy set to −70 eV. The MS source was set to 230°C, and the quadrupole was set to 150°C.
Alkanes were verified using retention time and MS matching to the stock solution of the C8 to C20 alkane mixture, coupled with National Institute of Standards and Technology (NIST) MS library verification. Alcohol metabolites, in the form of trimethylsilyl (TMS) derivatives, were verified using the NIST MS library, with mass spectra of TMS ethers of primary alcohols downloaded from NIST standard reference database number 69 (available at http://webbook.nist.gov/chemistry/).
The GC-MS profiles of CYP63A2 reaction extracts were compared with those of the control reaction extracts. Rates of oxidation of different alkanes by CYP63A2 were expressed as percentages of oxidation of the alkanes compared to that for the control reaction. Control reactions showed the same amount of alkanes as the buffer control. The P450-specific metabolites were identified based on the mass spectral fragmentation profile and the GC-MS NIST library.
Homology modeling and ligand docking simulations.
3D models of CYP63A2 were obtained using the Phyre server (25). Structure annotation and comparative structure analysis were performed using POLYVIEW-2D (26). Docking simulations of the substrates were conducted with AutoDock 4 (27). Analysis of ligand docking results was performed using POLYVIEW-MM (28). Volumes and surface areas of the cavities were calculated with CASTp (29) using a 1.4-Å probe. The final 3D model of CYP63A2 and active-site cavities were rendered using PyMol (http://www.pymol.org/).
RESULTS
Structural characteristics and substrate-binding potential of CYP63A2.
A 3D model of CYP63A2 was built using the Phyre server (25). Three top-ranking homology models of CYP63A2 based on the CYP3A4, CYP102A1 (BM3), and CYP1A2 structural templates (Protein Data Bank [PDB] identifiers 1tqn, 2ij2, and 2hi4, respectively) were evaluated by the following model assessors: DFire (30), QMEAN (31), and Verify3D (32). As can be seen from Table 1, DFire favors the model based on the 1tqnA template, whereas QMEAN ranks the 2ij2A-based model top. Verify3D was unable to distinguish the evaluated models. The 1tqnA-based model encompasses more residues of CYP63A2, while keeping the same level of sequence identity, than the 2ij2A-based model. Considering the quality of the model, level of sequence homology, taxonomy distance to the template, and CYP63A2 sequence coverage by the model, the CYP3A4 (1tqn)-based model was selected for further analysis reported in this work.
Table 1.
Comparison of the top-ranked 3D models of CYP63A2 generated by Phyre
| Templatea | Sequence identity (%)b | Lengthc | DFire resultd | QMEAN6 scoree | Avg Verify3D score ± SD |
|---|---|---|---|---|---|
| 1tqnA CYP3A4 | 22 | 495 | −629.46 | 0.484 | 0.33 ± 0.21 |
| 2ij2A CYP102A1 (BM3) | 23 | 479 | −607.73 | 0.606 | 0.31 ± 0.22 |
| 2hi4A CYP1A2 | 18 | 471 | −607.73 | 0.518 | 0.33 ± 0.18 |
Template used for 3D modeling (PDB identifier and chain label).
Sequence identity between CYP63A2 and the template used.
Number of CYP63A2 amino acids modeled.
DFire pseudoenergy (lower values signify a better model).
QMEAN6 composite score (higher values signify a better model).
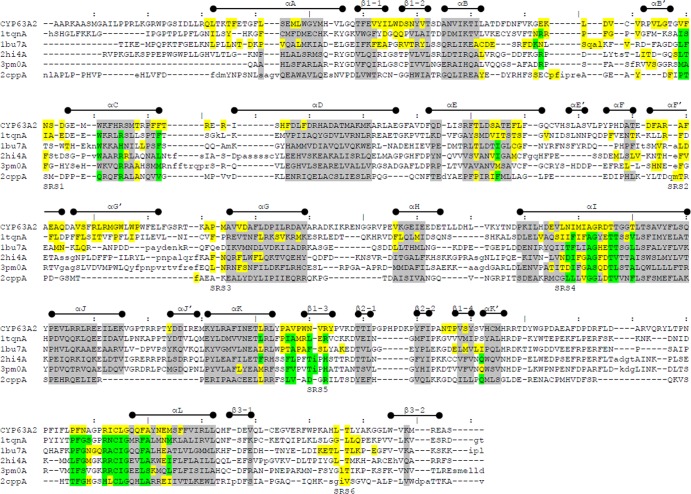
Pairwise structure alignment of selected P450s (known to oxidize PAHs and with crystal structures available) to the CYP63A2 model was conducted using the DaliLite server (33). Figure 1 provides a summary of the structural data, including major secondary structure elements (α-helices and β-strands) and substrate recognition sites (SRS), mapped into the corresponding sequences. Residues within conserved structural regions (CSR) are highlighted gray. Residues highlighted yellow appear in the active-site cavity found using CASTp (29) with the 1.4-Å probe. Residues found in contact with heme in the corresponding crystal structure are highlighted green. Table 2 presents a comparison of the CYP63A2 model with other P450s used in Fig. 1. In these comparisons, CYP63A2 turned out to be the largest protein in terms of sequence length and the one possessing the largest active-site cavity (Fig. 2). Ligand docking simulations conducted for selected substrates using AutoDock 4 (27) showed that the estimated binding affinities in terms of inhibition constant for benzo(a)pyrene and benzo(ghi)perylene were at the nanomolar level (653.99 nM and 620.41 nM, respectively), whereas for pyrene, nonylphenol, and n-nonane, they were in the micromolar range (6.72 μM, 3.93 μM, and 620.71 μM, respectively).
Fig 1.
Structural alignment of the CYP63A2 3D model with P450s CYP3A4 (1tqnA), CYP102A1 (1bu7A), CYP1A2 (2hi4A), CYP1B1 (3pm0A), and CYP101D (2cppA) using DaliLite (33). Residues in lowercase indicate insertions relative to CYP63A2. Overall boundaries of secondary structure (SS) states indicated above the sequences are based on the CYP3A4 (1tqnA) structure. Substrate recognition sites (SRS) are labeled under the sequences. Residues highlighted gray share the same SS state (α-helix or β-strand) across all aligned proteins. Residues highlighted yellow appear in the active-site cavity found using CASTp (29). Residues highlighted green were found in contact with heme using POLYVIEW-2D (26). In case of overlap, yellow supersedes gray and green supersedes yellow.
Table 2.
Comparative analysis of the homology-based 3D structure of CYP63A2 and the 3D crystal structures of selected cytochrome P450s
| Property | Value for: |
|||||
|---|---|---|---|---|---|---|
| CYP63A2 | CYP3A4 (1tqn) | CYP1A2 (2hi4) | CYP1B1 (3pm0) | CYP101D (2cpp) | CYP102A1 (1bu7) | |
| La | 601/495 | 503/468 | 515/480 | 543/459 | 415/405 | 472/455 |
| Sb (Å2) | 3,254 | 2,363 | 1,288 | 1,177 | 1,138 | 2,609 |
| Vc (Å3) | 4,719 | 3,275 | 1,613 | 1,519 | 1,496 | 4,065 |
Sequence length of the protein. The first number represents total length, and the second indicates the number of amino acids in the corresponding crystal structure (or model for CYP63A2). In case of the fused P450 CYP102A1, length refers to the cytochrome P450 part only.
Surface area of the active-site cavity.
Volume of the active-site cavity.
Fig 2.
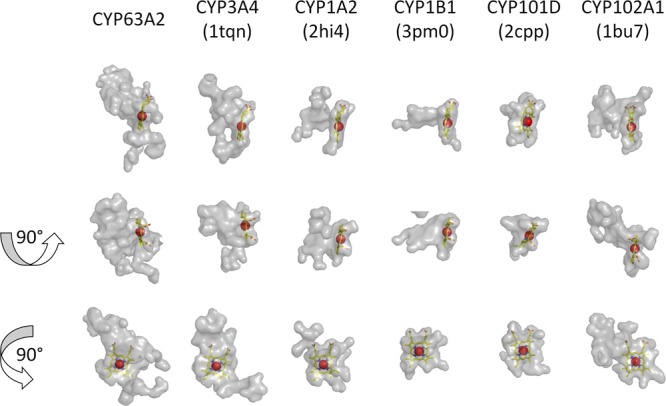
Active-site cavities of selected cytochrome P450s. Cavity identification and the image rendering were performed using PyMol. Cavities are rendered as semitransparent gray surfaces. Heme is shown using the stick representation. Iron is rendered as a red sphere. Second and third rows are results of rotation of 90 degrees around x and y axes, respectively, from the initial projection in the first row. Resolved protein structures were taken from PDB, whereas CYP63A2 was modeled using Phyre (see Results for details).
Expression analysis of CYP63A2 and Pc-CPR in P. pastoris.
The P. pastoris clone expressing CYP63A2, its homologous reductase partner Pc-CPR (designated the PC2 clone), and a control P. pastoris transformant carrying the empty vector pPICZB (designated the PP C clone), the last two generated in our recent efforts to investigate the comparative role of P450 redox proteins (21), were utilized in this study to investigate the catalytic properties of CYP63A2.
Before proceeding to check the CYP63A2 substrate specificity, heterologous expression levels of the cloned P450 and the redox protein Pc-CPR were analyzed in the microsomal fractions prepared from the recombinant P. pastoris clone PC2. Microsomes from the PP C culture were used as a negative control. SDS-PAGE analysis of the microsomal preparations showed a clear band coinciding with the Pc-CPR protein compared to the control microsomes (Fig. 3A). Western blot analysis using anti-His antibody showed expression of full-length CYP63A2 (Fig. 3B). A typical P450 reduced CO difference spectrum was observed in the microsomal fractions prepared from PC2, whereas no such spectrum was observed in the microsomes prepared from PP C cells (Fig. 3C). This indicated that the cloned CYP63A2 was expressed in an active form. The expressed CYP63A2 showed a maximum absorption peak (Soret peak) at 448 nm. PC2 microsomes showed a P450 content of 60 pmol/mg microsomal protein (Fig. 3E) and a higher level of CPR activity (1,616 nmol/min/mg) than the PP C microsomes, which showed negligible native host CPR activity (Fig. 3D).
Fig 3.
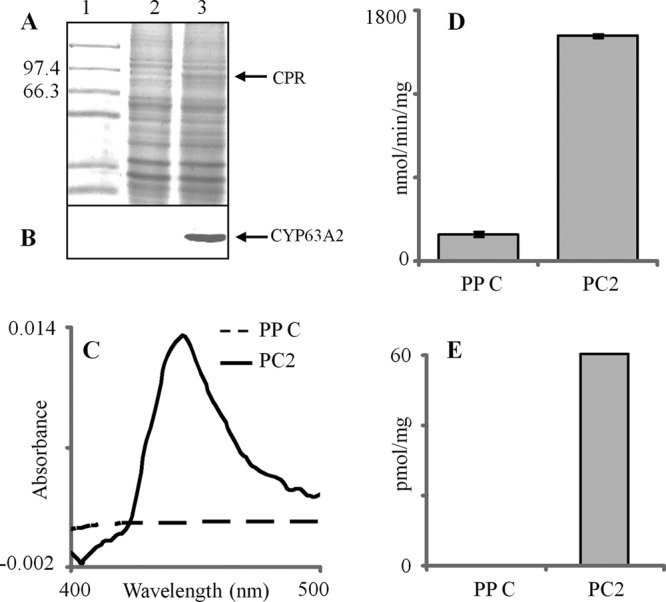
Recombinant coexpression and activity analysis for CYP63A2 and the homologous P450 reductase (Pc-CPR) partner in P. pastoris. The expressed recombinant enzymes were assessed in yeast microsomes. (A) SDS-PAGE analysis of the microsomal protein preparations from PP C (lane 2) and PC2 (lane 3) clones. Lane 1 indicates protein molecular weight markers. (B) Western blot analysis of CYP63A2 protein using anti-His antibody. (C) Characteristic reduced CO difference spectra for the microsomes prepared form PP C and PC2 clones. (D) Cytochrome P450 oxidoreductase (CPR) activity. The differences in CPR activities in microsomes between the recombinant clones (PP C and PC2) were statistically significant (P ≤ 0.05). (E) P450 enzyme concentrations derived from reduced CO difference spectra.
Substrate specificity and catalytic activity of CYP63A2 toward xenobiotics.
Whole-cell oxidation assays using PC2 and PP C clones were carried out to assess catalytic activity and specificity. The data on CYP63A2 activity toward different xenobiotic compounds, including PAHs with different ring sizes (3 to 6 rings), APs with various alkyl chain lengths (ranging from C3 to C9), technical grade nonylphenol (tNP) containing a mixture of nonylphenol p isomers (mixture of compounds with branched side chain), and n-alkanes with various chain lengths (C9 to C12 and C15 to C19) are shown in Table 3.
Table 3.
Xenobiotic substrate specificity and oxidation activity of CYP63A2
| Xenobiotic (class and compound) | Structural variable |
CYP63A2 activity | Mean oxidation (%) (±SD)a | ||
|---|---|---|---|---|---|
| No. of rings | Alkyl chain length (mixture of p isomers) | No. of carbon atoms | |||
| Aromatic/polycyclic aromatic hydrocarbons | |||||
| Benzo(ghi)perylene | 6 | Yes | 18.5 (±3.7) | ||
| Benzo(a)pyrene | 5 | Yes | 35.7 (±2.3) | ||
| Fluoranthene | 4 | Yes | 24.5 (±4.7) | ||
| Pyrene | 4 | Yes | 21.0 (±3.6) | ||
| Fluorene | 3 | NDc | |||
| Anthracene | 3 | ND | |||
| Phenanthrene | 3 | ND | |||
| Acenaphthene | 3 | ND | |||
| Alkylphenols | |||||
| tNP | Yes | 43.3 (±3.4) | |||
| 4-n-Nonylphenol | C9 | Yes | 88.2 (±4.5) | ||
| 4-n-Octylphenol | C8 | Yes | 97.1 (±3.6) | ||
| 4-n-Heptylphenol | C7 | Yes | 90.2 (±5.0) | ||
| 4-n-Pentylphenol | C5 | Yes | 87.5 (±3.6) | ||
| 4-n-Butylphenol | C4 | Yes | 85.1 (±3.7) | ||
| 4-n-Propylphenol | C3 | Yes | 60.1 (±5.7) | ||
| Aliphatic hydrocarbons: n-alkanes | 9–12, 15–19 | Yes | 9.1–31.1b | ||
Values are for three biological replicates. Each culture was spiked with an individual PAH or an alkylphenol, and the oxidation activity was assessed at 24 h of incubation.
For details on oxidation of individual alkanes, see Table 5.
ND, not detected.
(i) Oxidation of PAHs.
PC2 oxidized HMW-PAHs ranging from 4 to 6 rings (Table 3); the activity varied with the ring size (Table 3). Among the tested 4- to 6-ring HMW-PAHs, PC2 showed higher levels of oxidation of benzo(a)pyrene (35.7 ± 2.3%) than of benzo(ghi)perylene (18.5% ± 3.7%). In addition, the oxidation levels of pyrene (21.0% ± 3.6%) and fluoranthene (24.5% ± 4.7%) were close to those for benzo(ghi)perylene (Table 3). No oxidation activity in the empty-vector-containing yeast cells (negative control) suggested that the host yeast native P450s did not contribute to the PAH oxidation activity and that the activity was exclusively due to the recombinantly expressed CYP63A2.
Initial HPLC screening of the reaction extracts for the metabolite peak(s) and subsequent LC-ESI/MS analyses allowed identification of the metabolites (see Fig. S1 in the supplemental material) for pyrene and benzo(a)pyrene. An inspection of the extracted ion chromatograms of the possible hydroxylated metabolites indicated that the patterns were in agreement with those observed in the HPLC UV profile. Both reaction extracts showed a single monohydroxylated metabolite peak each (see Fig. S1A in the supplemental material). The retention times and monoisotopic masses of these metabolites (see Fig. S1B in the supplemental material) matched those of 1-hydroxypyrene and 3-hydroxybenzo(a)pyrene, respectively (Table 4). In the case of fluoranthene and benzo(ghi)perylene, the hydroxylated metabolite(s) could not be conclusively identified by LC-ESI/MS because of lack of commercially available standards.
Table 4.
LC-ESI/MS detection and identification of P450 oxidation metabolites from different PAHsa
| PAH compound | Oxidation product peak | Rtb (min) | [M-H]− |
ppmc | Oxidation product ID | |
|---|---|---|---|---|---|---|
| Measured | Theoretical | |||||
| Pyrene | I | 11.9 | 217.0641 | 217.0659 | −8.2 | 1-Hydroxypyrene |
| Benzo(a)pyrene | I | 16.2 | 267.0789 | 267.0815 | −9.9 | 3-Hydroxybenzo(a)pyrene |
| Standards | ||||||
| 1-Hydroxypyrene | 12.1 | 217.0634 | ||||
| 3-Hydroxybenzo(a)pyrene | 16.2 | 267.0788 | ||||
Further details are provided in the “LC-ESI/MS” section of Materials and Methods.
Rt, retention time.
Values are derived based on the formula parts per million of the theoretical value (Δppm) = [(a − b)/b] × 1,000,000, where a is the measured m/z value of the target metabolite and b is the theoretical m/z value of the authentic standard.
(ii) Oxidation of APs.
HPLC analysis of the AP reaction extracts prepared from PC2 cultures showed substantial oxidation (43.3% ± 3.4%) of tNP (Table 3), whereas no change in tNP levels was observed for the negative control (PP C), compared to the uninoculated control. Analysis of the regioselectivity of CYP63A2 toward tNP was difficult, as tNP contains a mixture of >100 p isomers (34), which generated an array of oxidized metabolites showing overlapping separation peaks and variable hydroxylation patterns. Hence, individual linear AP compounds (analytical grade) were employed to determine the CYP63A2 substrate specificity and regioselectivity. APs with alkyl chain lengths ranging from C3 to C9 were used (Table 3). As shown in Table 3, PC2 cells expressing CYP63A2 oxidized all tested APs, albeit to various extents. The C4 to C9 alkyl chain length APs were oxidized to a greater extent (85 to 97%) than the C3 alkyl chain length AP (4-n-propylphenol [4-n-PP]) (60.1% ± 5.6%) (Table 3). Differences in oxidation rates among the C4 to C9 APs were not statistically significant (Table 3).
HPLC screening of the reaction extracts for the individual linear AP congeners showed a single metabolite peak compared to the negative control (Fig. 4A). The metabolite peaks observed in PC2 AP extracts showed the same retention times (Fig. 4A) as those observed for the corresponding AP aldehydes in our preceding study (24). In order to confirm the aldehyde identity of the metabolites, representative PC2 AP extracts (4-n-nonylphenol [4-n-NP] and 4-n-octylphenol [4-n-OP]) were subjected to LC-ESI/MS analysis. As shown in Fig. 4B, the corresponding extracted ion chromatogram showed molecular masses equivalent to those of hydroxybenzaldehydes. Based on the HPLC UV and MS analyses and comparison with the results from the CYP5136A3 study (24), it is suggested that CYP63A2 oxidizes APs (C3 to C9) at the terminal carbon atom of the alkyl chain.
Fig 4.
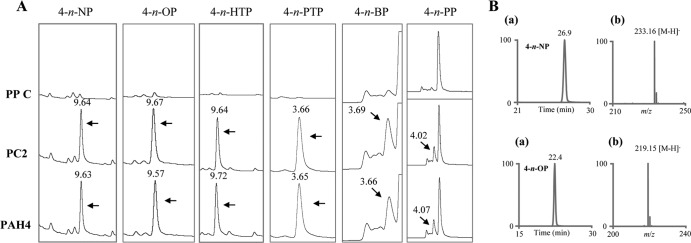
Detection and identification of CYP63A2 oxidation metabolites from different alkylphenols (APs). (A) HPLC separation profiles of the oxidation products of APs from whole-cell assays using P. pastoris clones PP C (control), PC2 (expressing CYP63A2), and PAH4 (expressing CYP5136A3) (24). The HPLC chromatographic peaks corresponding to the AP metabolites formed by the action of CYP63A2 and CYP5136A3 are shown, along with retention times in min. Abbreviations: 4-n-NP, 4-n-nonylphenol; 4-n-OP, 4-n-octylphenol; 4-n-HTP, 4-n-heptylphenol; 4-n-PTP, 4-n-pentylphenol; 4-n-BP, 4-n-butylphenol; 4-n-PP, 4-n-propylphenol. (B) LC-ESI/MS analysis of alkylphenols 4-n-NP and 4-n-OP. (a) Extracted ion chromatograms for APs constructed with mass windows (±0.5 amu) centered at m/z 233.2 (for the 4-n-NP metabolite) and at m/z 219.2 (for the 4-n-OP metabolite); (b) mass spectra for the respective AP metabolites. Accurate mass measurements were carried out by averaging 25 scans across the full-width half maximum of the extracted ion profile.
(iii) Oxidation of alkanes.
In order to assess CYP63A2 capability to oxidize aliphatic hydrocarbons, in vitro oxidation reactions were performed using E. coli cell extracts containing the coexpressed CYP63A2 and its reductase partner (Pc-CPR). A standard mixture containing C8 to C20 n-alkanes was used as a substrate. As shown in Table 5, CYP63A2 was found to oxidize several alkanes, ranging from carbon lengths of 9 to 12 to 15 to 19. We were unable to assess CYP63A2 activity toward octane (C8), possibly due to the evaporation of octane from the reaction mixture. GC-MS analysis of the organic extracts showed the presence of novel metabolite peaks in the CYP63A2 sample, compared to the control. Based on NIST mass spectral library data, the alkane metabolites were tentatively identified as monohydroxylated alkanes, suggesting CYP63A2 hydroxylation of alkanes to aliphatic alcohols. To identify the specific alcohols and the positions of hydroxylation, extracts were derivatized with BSTFA before being subjected to GC-MS analysis. As shown in Fig. S2 in the supplemental material, the hydroxyl group was found to be present at the terminal carbon atom of the alkane chain based on NIST mass spectral matching. For C16 and C18 alkanes, the corresponding alcohol metabolite was detected but the position of hydroxylation could not be confirmed due to the low abundance of the derivatized metabolite (Table 5).
Table 5.
GC-MS analysis of alkane oxidation by CYP63A2a
| Alkane | No. of carbon atoms | Alkane oxidation (%) | Oxidation metabolite (BSTFA derivative) |
||
|---|---|---|---|---|---|
| MWb | Ion fragmentsc | Metabolited | |||
| n-Nonane | 9 | 24.7 | 216.4 | 201, 171, 143, 115, 103, 83, 75, 55, 41 | 1-Nonanol |
| n-Decane | 10 | 15.1 | 230.4 | 215, 171, 139, 115, 103, 83, 75 | 1-Decanol |
| n-Undecane | 11 | 16.5 | 244.4 | 229, 143, 83, 75, 55 | 1-Undecanol |
| n-Dodecane | 12 | 17.0 | 258.5 | 243, 215, 185, 155, 129, 103, 75, 43 | 1-Dodecanol |
| n-Pentadecane | 15 | 9.1 | 300.6 | 285, 257, 199, 159, 103, 75, 43 | 1-Pentadecanol |
| n-Hexadecane | 16 | 19.8 | —e | NDf | — |
| n-Heptadecane | 17 | 30.7 | 328.6 | 313, 238, 199, 167, 103, 75, 43 | 1-Heptadecanol |
| n-Octadecane | 18 | 31.1 | — | ND | — |
| n-Nonadecane | 19 | 19.9 | 356.7 | 341, 255, 213, 167, 103, 75, 43 | 1-Nonadecanol |
See Fig. S2 in the supplemental material for the GC-MS profiles of metabolites.
MW, molecular weight.
Numbers are m/z values.
Identification of metabolites was performed using a standard NIST traceable MS database, with TMS-derivatized alcohols downloaded from NIST standard reference database number 69 (available at http://webbook.nist.gov/chemistry/).
—, not applicable.
ND, not detected.
DISCUSSION
Bioremediation of PAHs, which often exist as a part of mixed pollution such as in petroleum/crude oil spills, would ideally require a P450 biocatalyst with both an aromatic ring hydroxylation activity and an ability to oxidize the co-occurring alkanes and alkane substructures. Human P450s possess PAH-oxidizing capability (35) but may offer poor prospects for use as bioremediation agents for such mixed pollution considering that these enzymes lack alkane hydroxylation activity. In the microbial world, while the otherwise well-characterized model bacterial P450s, CYP101 (P450cam) from Pseudomonas putida (36) and CYP102 (P450BM-3) from Bacillus megaterium (37), have been genetically modified to confer PAH oxidation ability (38–41), the engineered enzymes could oxidize only three- and four-ring PAHs and had negligible activity toward five-ring PAHs; their ability to oxidize six-ring PAHs is not known. To date, there are no reports on oxidation of APs by prokaryotic P450s, native or engineered. A microbial P450 that can oxidize different classes of xenobiotic chemicals could be an ideal candidate for bioremediation applications. Here we report such a P450 (CYP63A2), from the model white rot fungus P. chrysosporium, which showed the extraordinary capability to oxidize HMW-PAHs (up to 6 aromatic rings) as well as environmental chemicals with aliphatic structures as in APs (C3 to C9) and alkanes (C9 to C19) (Fig. 5).
Fig 5.
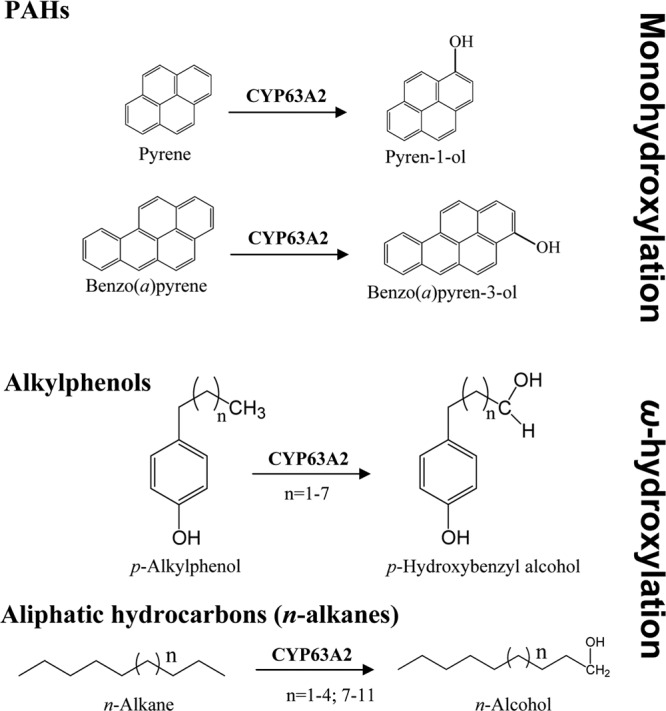
CYP63A2-catalyzed oxygenation reactions for structurally diverse xenobiotics, including polycyclic aromatic hydrocarbons (PAHs), alkylphenols (APs), and linear alkanes. In the case of PAHs, monohydroxylation was observed, whereas APs and alkanes were hydroxylated at the terminal carbon of the aliphatic chain (ω-hydroxylation).
As a part of our initial attempts to understand the functional significance of P450s in P. chrysosporium (16), CYP63A2 was found to be inducible in response to various classes of xenobiotics. To make this follow-up experimental functional characterization more efficient, we first utilized a computational modeling and docking strategy to estimate the substrate range and catalytic potential of CYP63A2. Comparison of the CYP63A2 3D model with the crystal structures of P450s from prokaryotes (42, 43) and higher eukaryotes (44–46) indicated that CYP63A2 is the largest among the analyzed P450s (Fig. 2 and Table 2). The CYP63A2 active-site cavity is about 1.5 times larger than that of CYP3A4 (Table 2). As can be seen from Fig. 6, the modeled structure of CYP63A2 shares the same conserved structural regions with other PAH-oxidizing P450s. With the overall longer amino acid sequence, the structure displays elongated unstructured loops, specifically between αG′ and αG, αH and αI, and β2-1 and β2-2. This may provide additional flexibility to the structure to increase the volume of the active-site cavity and to accommodate larger ligands, such as benzo(a)pyrene and benzo(ghi)perylene. As anticipated, CYP63A2 preserves the residues responsible for heme binding (Fig. 1; alignments with other enzymes where the heme-binding residues are highlighted green). Interestingly, its larger cavity does not extend substrate recognition sites. Only SRS2 appears to be enlarged (Fig. 1; note the residues highlighted yellow in PC2 [CYP63A2] located within the SRS regions, which are labeled under the alignments). This may imply that, while the overall cavity is increased and more residues are found lining the cavity, the amino acids critical in substrate recognition and regioselectivity may still be confined to these short sequence stretches representing the SRS regions. Furthermore, ligand docking studies based on selected PAHs [pyrene, benzo(a)pyrene, and benzo(ghi)perylene], alkylphenols (4-n-nonylphenol), and alkanes (n-nonane) confirmed that these compounds can be potential substrates for CYP63A2, with estimated binding affinities ranging from the nanomolar to the micromolar level.
Fig 6.
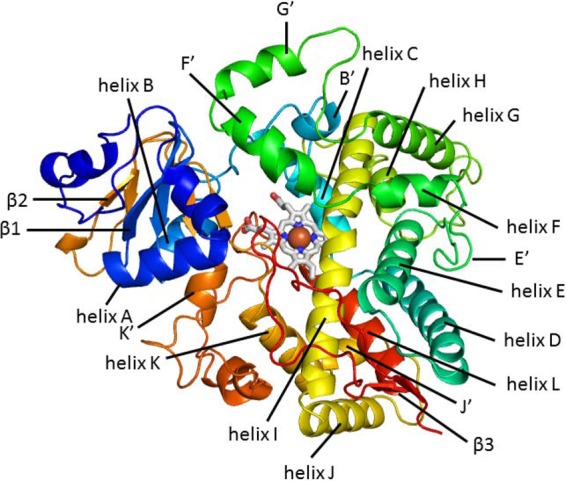
A 3D model of CYP63A2 with all the structured regions labeled, shown by a cartoon representation of the structure colored using the rainbow gradient, where blue corresponds to the N terminus and red to the C terminus.
Validation of the in silico results using experimental approaches (whole-cell-based and in vitro enzymatic studies) confirmed the catalytic versatility and revealed an unusual combination of activities toward compounds of different classes, namely, PAHs, APs, and alkanes.
CYP63A2 oxidized the 4-ring PAHs pyrene and fluoranthene. While pyrene oxidation is commonplace, to our knowledge no specific P450(s) oxidizing fluoranthene has yet been identified in eukaryotes, making this study the first report on a specific eukaryotic P450 capable of oxidizing fluoranthene. Similar to human CYPs (47), pyrene was oxidized to 1-hydroxypyrene (Fig. 5), a metabolite that has been used as a biomarker for biomonitoring human occupational exposures to PAHs (48). In this context, CYP63A2 differed from the other PAH-oxidizing P450s in P. chrysosporium, which yielded two monohydroxylated metabolites (20).
Benzo(a)pyrene (5 rings), a proven carcinogen in living organisms, was oxidized by CYP63A2 into 3-hydroxybenzo(a)pyrene (Fig. 5), which is consistent with what has been observed in microsomal preparations in whole-fungus studies on P. chrysosporium (49). This suggested the in vivo catalytic role of this P450 in the native organism.
One of the unique catalytic capabilities of CYP63A2 is oxidation of the 6-ring PAH benzo(ghi)perylene. This compound represents the group of PAHs lacking a “classic” bay region and is known to exhibit mutagenicity (50) via a synergistic response wherein benzo(a)pyrene-induced CYP1A1 gene transcription via aryl hydrocarbon receptor (AhR) activation gets enhanced (51). To our knowledge, a specific P450 from any phylum (prokaryotic or eukaryotic) causing oxidation of benzo(ghi)perylene has not yet been reported. While P450 involvement in benzo(ghi)perylene oxidation was suggested based on whole-microsome studies (rat liver microsomes) and the formation of characteristic P450 metabolites (50), no specific P450 enzyme catalyzing this oxidation was identified. The current study on CYP63A2 therefore constitutes the first report on a specific eukaryotic P450 capable of oxidizing benzo(ghi)perylene.
Among the tested PAHs with various numbers of fused rings, CYP63A2 showed the highest enzymatic activity toward the 5-ring PAH, followed by the 4-ring and 6-ring PAHs, in that order (Table 3). No observed activity toward the 3-ring PAHs suggested that this fungal P450 has a preference for HMW-PAHs. This contrasts with the other recently characterized set of PAH-oxidizing P450s (Pc-Pah1 through Pc-Pah6) from this organism, which showed greater specificity for lower-molecular-weight PAHs in general in our recent study (20). Interestingly, CYP63A2 showed a greater rate of oxidation of PAHs [pyrene and benzo(a)pyrene], as it oxidized the same amount of PAHs in 24 h as other P. chrysosporium P450s did in the 36-h time period (20) despite the smaller amount of expressed P450 content (170 to 285 pmol/mg earlier versus 60 pmol/mg in this study) under otherwise identical culturing and oxidation conditions. Furthermore, none of the other PAH-oxidizing P450s (Pc-Pah1 through Pc-Pah6) oxidized benzo(ghi)perylene (data not shown).
In the class of endocrine-disrupting alkylphenols, CYP63A2 oxidized linear APs with alkyl chain lengths ranging from C3 to C9, both as individual congeners and as a mixture (tNP) (Table 3), suggesting that CYP63A2 has a broad substrate specificity. CYP63A2 hydroxylated the terminal carbon (ω-carbon) in the AP alkyl chain, while host yeast enzymes subsequently converted the hydroxylated metabolite into the corresponding aldehyde metabolite. This suggested that CYP63A2 is an AP alkyl chain ω-hydroxylase. Considering the known pathway in yeasts/fungi for degradation of an alkylphenol moiety that begins via terminal oxidation of the alkyl side chain followed by removal of the terminal carbons via the β-oxidation pathway (52), we can assume that CYP63A2 plays a key role in the initial oxidation of alkylphenols (C3 to C9) in P. chrysosporium. Compared to CYP5136A3, another AP-oxidizing P450 from P. chrysosporium identified in our recent study (24), CYP63A2 preferred APs with longer alkyl chains and exhibited a higher rate of oxidation of APs. For instance, the extent of oxidation of individual linear APs by CYP63A2 in the 24-h period was the same as that by CYP5136A3 in the 48-h period (24) despite the fact that the P450 content was higher in the latter. Taken together, these results suggest that CYP63A2 is catalytically more efficient than CYP5136A3 and thus could potentially serve as a superior biocatalyst candidate for bioremediation of APs, particularly for the environmentally recalcitrant APs with longer alkyl chain lengths.
Considering that CYP63A2 showed inducibility by aliphatic hydrocarbons in our past study (16) and an oxidizing activity at the terminal carbon (ω-carbon) of the linear alkyl side chain in APs (this work), we chose to assess this P450's capability to oxidize alkanes. In this respect, whole-cell oxidation studies using recombinant P. pastoris clones were not successful, as the host yeast P. pastoris showed an inherent alkane oxidation pathway (data not shown). Hence, we expressed this P450 in E. coli (a host not capable of oxidizing alkanes) and performed in vitro oxidation assays using whole-cell lysates containing the solubly expressed P450 enzyme and alkanes of various chain lengths (C8 to C20) in a mixture form. As we anticipated, CYP63A2 oxidized a range of alkanes (Table 5; see Fig. S2 in the supplemental material). The oxidation occurred at the terminal carbon atom (ω-carbon), and monohydroxylated alcohols were detected as the reaction products. This suggested that CYP63A2 is an n-alkane ω-hydroxylase. To date, P450s belonging to the CYP52 family from yeasts (53), CYP153 family from bacteria (54), and the mammalian P450 CYP4B1 (rabbit) (55) have been shown to oxidize alkanes. Among the characterized bacterial ω-hydroxylases, only the CYP153 family oxidizes alkanes (C5 to C16) at the terminal carbon (54). CYP102A1 (BM3) oxidizes fatty acids (C12 to C20) at internal-chain carbons (ω-1, ω-2, and ω-3; 36:30:34) with no ω-hydroxylation (56). CYP125A1 from Rhodococcus jostii RAH1 (57) and CYP124, CYP125, and CYP142 from Mycobacterium tuberculosis ω-hydroxylate the sterol side chain of cholesterol and its 3-keto-4-ene derivative (58). Furthermore, CYP124 also ω-hydroxylates fatty acids and other long-chain lipids (59). However, none of these P450s have been reported to possess additional catalytic activity toward PAHs and APs. Hence CYP63A2 is unique among the aliphatic hydroxylase P450s identified to date, considering its comprehensive ability to oxidize both the thermodynamically disfavored ω-hydrocarbon (60) and difficult-to-oxidize aromatic rings in the fused-ring HMW-PAHs. Collectively, the experimental studies demonstrating growth of P. chrysosporium on alkanes (61), induction of CYP63A2 in response to alkanes (16), and the ability of CYP63A2 to oxidize alkanes into alcohols (this study) suggest that CYP63A2 plays a key role in utilization of aliphatic hydrocarbons by this fungus.
In conclusion, the study revealed that CYP63A2 has a unique ability to oxidize high-molecular-weight PAHs of up to 6 rings and is catalytically versatile, considering its additional abilities to oxidize endocrine-disrupting alkylphenols with alkyl side chains of various lengths (C3 to C9) and lower- to higher-molecular-weight alkanes with various numbers of carbons (ranging from 9 to 12 to 15 to 19). CYP63A2 performs the thermodynamically disfavored reaction of ω-hydroxylation of the aliphatic chains (60) present in alkylphenols and alkanes. CYP63A2 possesses one of the largest active-site cavities among known P450s, as deduced by the homology model, which may explain the observed extraordinary oxidation activity toward larger and diverse substrates. The versatile and uncommon catalytic properties of CYP63A2 indicate that this fungal P450 is a strong candidate for developing new potent bioremediation agents targeting mixed pollution comprising environmentally recalcitrant compounds with polyaromatic, substituted aromatic, and aliphatic substructures, such as the crude oil spills.
ACKNOWLEDGMENTS
The work was primarily supported by National Institute of Environmental Health Sciences (NIEHS) grants R01ES10210 and R01ES015543 to J.S.Y. The efforts of A.P. were supported in part by NIEHS Center for Environmental Genetics funding (P30-ES006096). LC-ESI/MS analysis by Y.W.L. was supported in part by the Vermont Genetics Network Proteomics Facility through grant number 8P20GM103449 from the INBRE Program of the National Institute of General Medical Sciences and the National Center for Research Resources, components of the National Institutes of Health.
The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency.
Footnotes
Published ahead of print 15 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03767-12
REFERENCES
- 1. Xue W, Warshawsky D. 2006. Molecular mechanisms of action of selected organic carcinogens, p. 45–77 In Warshawsky D, Landolph JR. (ed), Molecular carcinogenesis and molecular biology of human cancer. Taylor and Francis Group, CRC Press, Boca Raton, FL [Google Scholar]
- 2. Haritash AK, Kaushik CP. 2009. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J. Hazard. Mater. 169:1–15 [DOI] [PubMed] [Google Scholar]
- 3. Soares A, Guieysse B, Jefferson B, Cartmell E, Lester JN. 2008. Nonylphenol in the environment: a critical review on occurrence, fate, toxicity and treatment in wastewaters. Environ. Int. 34:1033–1049 [DOI] [PubMed] [Google Scholar]
- 4. Shuttleworth KL, Cerniglia CE. 1995. Environmental aspects of PAH biodegradation. Appl. Biochem. Biotechnol. 54:291–302 [DOI] [PubMed] [Google Scholar]
- 5. Ekelund R, Bergman A, Granmo A, Berggren M. 1990. Bioaccumulation of 4-nonylphenol in marine animals—a reevaluation. Environ. Pollut. 64:107–120 [DOI] [PubMed] [Google Scholar]
- 6. Ademollo N, Ferrara F, Delise M, Fabietti F, Funari E. 2008. Nonylphenol and octylphenol in human breast milk. Environ. Int. 43:984–987 [DOI] [PubMed] [Google Scholar]
- 7. Zahed MA, Aziz HA, Isa MH, Mohajeri L. 2010. Enhancement biodegradation of n-alkanes from crude oil contaminated seawater. Int. J. Environ. Res. 4:655–664 [DOI] [PubMed] [Google Scholar]
- 8. Corvini PFX, Schaffer A, Schlosser D. 2006. Microbial degradation of nonylphenol and other alkylphenols—our evolving view. Appl. Microbiol. Biotechnol. 72:223–243 [DOI] [PubMed] [Google Scholar]
- 9. Peng RH, Xiong AS, Xue Y, Fu XY, Gao F, Zhao W, Tian YS, Yao QH. 2008. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 32:927–955 [DOI] [PubMed] [Google Scholar]
- 10. Sono M, Roach MP, Coulter ED, Dawson JH. 1996. 1996. Heme-containing oxygenases. Chem. Rev. 96:2841–2888 [DOI] [PubMed] [Google Scholar]
- 11. Pszczynski A, Crawford RL. 1995. Potential for bioremediation of xenobiotic compounds by the white rot fungus Phanerochaete chrysosporium. Biotechnol. Prog. 11:368–379 [Google Scholar]
- 12. Gold MH, Wariishi H, Valli K. 1987. Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium. ACS Symp. Ser. 389:127–140 [Google Scholar]
- 13. Kirk TK, Farrell RL. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465–505 [DOI] [PubMed] [Google Scholar]
- 14. Syed K, Yadav JS. 2012. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit. Rev. Microbiol. 38:339–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yadav JS, Soellner MB, Loper JC, Mishra PK. 2003. Tandem cytochrome P450 monooxygenase genes and splice variants in the white rot fungus Phanerochaete chrysosporium: cloning, sequence analysis, and regulation of differential expression. Fungal Genet. Biol. 38:10–21 [DOI] [PubMed] [Google Scholar]
- 16. Doddapaneni H, Yadav JS. 2004. Differential regulation and xenobiotic induction of tandem P450 monooxygenase genes pc-1 (CYP63A1) and pc-2 (CYP63A2) in the white rot fungus Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 65:559–565 [DOI] [PubMed] [Google Scholar]
- 17. Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695–700 [DOI] [PubMed] [Google Scholar]
- 18. Doddapaneni H, Subramanian V, Yadav JS. 2005. Physiological regulation, xenobiotic induction, and heterologous expression of P450 monooxygenase gene pc-3 (CYP63A3), a new member of CYP63 gene cluster in the white rot fungus Phanerochaete chrysosporium. Curr. Microbiol. 50:292–298 [DOI] [PubMed] [Google Scholar]
- 19. Subramanian V, Yadav JS. 2008. Regulation and heterologous expression of P450 enzyme system components of the white rot fungus Phanerochaete chrysosporium. Enzyme Microb. Technol. 43:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Syed K, Doddapaneni H, Subramanian V, Lam YW, Yadav JS. 2010. Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbons (PAHs). Biochem. Biophys. Res. Commun. 399:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Syed K, Kattamuri C, Thompson TB, Yadav JS. 2011. Cytochrome b5 reductase-cytochrome b5 as an active P450 redox enzyme system in Phanerochaete chrysosporium: atypical properties and in vivo evidence of electron transfer capability to CYP63A2. Arch. Biochem. Biophys. 509:26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guengerich PF, Martin MV, Sohl CD, Cheng Q. 2009. Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat. Protoc. 4:1245–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barnes HJ. 1996. Maximizing expression of eukaryotic cytochrome P450s in Escherichia coli. Methods Enzymol. 272:3–14 [DOI] [PubMed] [Google Scholar]
- 24. Syed K, Porollo A, Lam YW, Yadav JS. 2011. A fungal P450 (CYP5136A3) capable of oxidizing polycyclic aromatic hydrocarbons and endocrine disrupting alkylphenols: role of Trp129 and Leu324. PLoS One 6:e28286 doi:10.1371/journal.pone.0028286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the web: a case study using the phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 26. Porollo A, Adamczak R, Meller J. 2004. POLYVIEW: a flexible visualization tool for structural and functional annotations of proteins. Bioinformatics 20:2460–2462 [DOI] [PubMed] [Google Scholar]
- 27. Goodsell DS, Morris GM, Olson AJ. 1996. Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 9:1–5 [DOI] [PubMed] [Google Scholar]
- 28. Porollo A, Meller J. 2010. POLYVIEW-MM: web-based platform for animation and analysis of molecular simulations. Nucleic Acids Res. 38:662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. 2006. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34:116–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou H, Zhou Y. 2002. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 11:2714–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benkert P, Biasini M, Schwede T. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lüthy R, Bowie JU, Eisenberg DR. 1992. Assessment of protein models with three-dimensional profiles. Nature 356:83–85 [DOI] [PubMed] [Google Scholar]
- 33. Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ieda T, Horii Y, Petrick G, Yamashita N, Ochiai N, Kannan K. 2005. Analysis of nonylphenol isomers in a technical mixture and in water by comprehensive two-dimensional gas chromatography-mass spectrometry. Environ. Sci. Technol. 39:7202–7207 [DOI] [PubMed] [Google Scholar]
- 35. Shimada T. 2006. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab. Pharmacokinet. 21:257–276 [DOI] [PubMed] [Google Scholar]
- 36. Katagiri M, Ganguli BN, Gunsalus IC. 1968. A soluble cytochrome P-450 functional in methylene hydroxylation. J. Biol. Chem. 243:3543–3546 [PubMed] [Google Scholar]
- 37. Narhi LO, Fulco AJ. 1986. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 261:7160–7169 [PubMed] [Google Scholar]
- 38. England PA, Harford-Cross CF, Stevenson JA, Rouch DA, Wong LL. 1998. The oxidation of naphthalene and pyrene by cytochrome P450cam. FEBS Lett. 424:271–274 [DOI] [PubMed] [Google Scholar]
- 39. Harford-Cross CF, Carmichael AB, Allan FK, England PA, Rouch DA, Wong LL. 2000. Protein engineering of cytochrome P450cam (CYP101) for the oxidation of polycyclic aromatic hydrocarbons. Protein Eng. 13:121–128 [DOI] [PubMed] [Google Scholar]
- 40. Carmichael AB, Wong LL. 2001. Protein engineering of Bacillus megaterium CYP102. The oxidation of polycyclic aromatic hydrocarbons. Eur. J. Biochem. 268:3117–3125 [DOI] [PubMed] [Google Scholar]
- 41. Li QS, Ogawa J, Schmid RD, Shimizu S. 2001. Engineering cytochrome P450 BM-3 for oxidation of polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 67:5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poulos TL, Finzel BC, Howard AJ. 1987. High-resolution crystal structure of cytochrome P450cam. J. Mol. Biol. 195:687–700 [DOI] [PubMed] [Google Scholar]
- 43. Ravichandran KG, Boddupalli SS, Hasemann CA, Peterson JA, Deisenhofer J. 1993. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450's. Science 261:731–736 [DOI] [PubMed] [Google Scholar]
- 44. Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. 2004. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-Å resolution. J. Biol. Chem. 279:38091–38094 [DOI] [PubMed] [Google Scholar]
- 45. Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. 2007. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J. Biol. Chem. 282:14348–14355 [DOI] [PubMed] [Google Scholar]
- 46. Wang A, Savas U, Stout CD, Johnson EF. 2011. Structural characterization of the complex between α-naphthoflavone and human cytochrome P450 1B1. J. Biol. Chem. 286:5736–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim YD, Todoroki H, Oyama T, Isse T, Matsumoto A, Yamaguchi T, Kim H, Uchiyama I, Kawamoto T. 2004. Identification of cytochrome P450 isoforms involved in 1-hydroxylation of pyrene. Environ. Res. 94:262–266 [DOI] [PubMed] [Google Scholar]
- 48. Jongeneelen FJ, Anzion RB, Scheepers PT, Bos RP, Henderson PT, Nijenhuis EH, Veenstra SJ, Brouns RM, Winkes A. 1988. 1-Hydroxypyrene in urine as a biological indicator of exposure to polycyclic aromatic hydrocarbons in several work environments. Ann. Occup. Hyg. 32:35–43 [DOI] [PubMed] [Google Scholar]
- 49. Masaphy S, Levanon D, Henis Y, Venkateswarlu K, Kelly SL. 1996. Evidence for cytochrome P-450 and P-450-mediated benzo(a)pyrene hydroxylation in the white rot fungus Phanerochaete chrysosporium. FEMS Microbiol. Lett. 135:51–55 [DOI] [PubMed] [Google Scholar]
- 50. Platt KL, Grupe S. 2005. Microsomal biotransformation of benzo(ghi)perylene, a mutagenic polycyclic aromatic hydrocarbon without a “classic” bay region. Chem. Res. Toxicol. 18:700–710 [DOI] [PubMed] [Google Scholar]
- 51. Cherng SH, Lin P, Yang JL, Hsu SL, Lee H. 2001. Benzo[g,h,i]perylene synergistically transactivates benzo[a]pyrene induced CYP1A1 gene expression by aryl hydrocarbon receptor pathway. Toxicol. Appl. Pharmacol. 170:63–68 [DOI] [PubMed] [Google Scholar]
- 52. Rozalska S, Szewczyk R, Dlugonski J. 2010. Biodegradation of 4-n-nonylphenol by the non-ligninolytic filamentous fungus Gliocephalotrichum simplex: a proposal of a metabolic pathway. J. Hazard. Mater. 180:323–331 [DOI] [PubMed] [Google Scholar]
- 53. Iida T, Sumita T, Ohta A, Takagi M. 2000. The cytochrome P450ALK multigene family of an n-alkane-assimilating yeast, Yarrowia lipolytica: cloning and characterization of genes coding for new CYP52 family members. Yeast 16:1077–1087 [DOI] [PubMed] [Google Scholar]
- 54. van Beilen JB, Funhoff EG, van Loon A, Just A, Kaysser L, Bouza M, Holtackers R, Röthlisberger M, Li Z, Witholt B. 2006. Cytochrome P450 alkane hydroxylase of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl. Environ. Microbiol. 72:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fisher MB, Zheng YM, Rettie AE. 1998. Positional specificity of rabbit CYP4B1 for omega-hydroxylation1 of short-medium chain fatty acids and hydrocarbons. Biochem. Biophys. Res. Commun. 248:352–355 [DOI] [PubMed] [Google Scholar]
- 56. Miura Y, Fulco FJ. 1975. ω-1, ω-2 and ω-3 hydroxylation of long-chain fatty acids, amides and alcohols by a soluble enzyme system from Bacillus megatyerium. Biochim. Biophys. Acta 388:305–317 [DOI] [PubMed] [Google Scholar]
- 57. Rostoniec KZ, Wilbrink MH, Capyk JK, Mohn WW, Ostendorf M, van der Geize R, Dijkhuizen L, Eltis LD. 2009. Cytochrome P450 125 (CYP125) catalyzes C26-hydroxylation to initiate sterol side chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 74:1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Johnston JB, Ouellet H, Ortiz de Montellano PR. 2010. Functional redundancy of sterol C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analysis. J. Biol. Chem. 285:36352–36360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Johnston JB, Kells PM, Podust LM, Oritz de Montellano PR. 2009. Biochemical and structural characterization of CYP124: a methyl-branched lipid ω-hydroxylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 106:20687–20692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnston JB, Ouellet H, Podust LM, Oritz de Montellano PR. 2011. Structural control of cytochrome P450-catalyzed ω-hydroxylation. Arch. Biochem. Biophys. 507:86–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanaly RA, Hur HG. 2006. Growth of Phanerochaete chrysosporium on diesel fuel hydrocarbons at neutral pH. Chemosphere 63:202–211 [DOI] [PubMed] [Google Scholar]