Abstract
The consumption of fresh tomatoes has been linked to numerous food-borne outbreaks involving various serovars of Salmonella enterica. Recent advances in our understanding of plant-microbe interactions have shown that human enteric pathogenic bacteria, including S. enterica, are adapted to survive in the plant environment. In this study, tomato plants (Solanum lycopersicum cv. Micro-Tom) grown in sandy loam soil from Virginia's eastern shore (VES) were inoculated with S. enterica serovars to evaluate plausible internalization routes and to determine if there is any niche fitness for certain serovars. Both infested soil and contaminated blossoms can lead to low internal levels of fruit contamination with Salmonella. Salmonella serovars demonstrated a great ability to survive in environments under tomato cultivation, not only in soil but also on different parts of the tomato plant. Of the five serovars investigated, Salmonella enterica serovars Newport and Javiana were dominant in sandy loam soil, while Salmonella enterica serovars Montevideo and Newport were more prevalent on leaves and blossoms. It was also observed that Salmonella enterica serovar Typhimurium had a poor rate of survival in all the plant parts examined here, suggesting that postharvest contamination routes are more likely in S. Typhimurium contamination of tomato fruit. Conversely, S. Newport was the most prevalent serovar recovered in both the tomato rhizosphere and phyllosphere. Plants that were recently transplanted (within 3 days) had an increase in observable internalized bacteria, suggesting that plants were more susceptible to internalization right after transplant. These findings suggest that the particular Salmonella serovar and the growth stage of the plant were important factors for internalization through the root system.
INTRODUCTION
Nontyphoidal Salmonella spp. are some of the leading causes of hospitalization due to food-borne illnesses in the United States (1). The incidence of Salmonella infection has not declined significantly in more than a decade (2). On the other hand, fruits and vine stalk vegetables are increasingly implicated as vehicles of Salmonella spp. in food-borne outbreaks (3). In particular, the consumption of fresh tomatoes has been linked to numerous food-borne outbreaks involving various serovars of Salmonella enterica.
Contamination of produce might occur during preharvest field production or in the postharvest processing facility. At the preharvest stage, several potential routes for S. enterica colonization and internalization to contaminate tomato fruits have been examined previously (4–8). Some of the findings point to irrigation with contaminated water as a potential source of fruit contamination (4, 7); however, evidence that S. enterica is able to enter tomato plants through contaminated irrigation water remains inconsistent. Hintz et al. (7) reported that repeated application of Salmonella enterica serovar Newport to the root zone via irrigation water can result in the contamination of various tomato plant tissues (Solanum lycopersicum cv. Solar Fire), when sampled throughout different plant growth stages. Yet, Jablasone et al. (9) recovered no Salmonella enterica serovar Enteritidis from plant tissue after applying contaminated water directly onto the soil of pots containing tomatoes (S. lycopersicum cv. Cherry Gold). In addition, another study found no evidence of Salmonella enterica serovar Montevideo survival in the stems, leaves, or fruit of tomato plants inoculated via irrigation water (S. lycopersicum L. cv. Trust) (10).
The tomato blossom represents another potential route for Salmonella contamination. When “Better Boy” tomato flowers were brushed with a five-strain cocktail of S. enterica serovars Enteritidis, Hartford, Michigan, Montevideo, and Poona, 25% of the ripened fruit was found to be contaminated by at least one of the five serovars (5). Most recently, Barak et al. (11) demonstrated that populations of S. enterica present in the phyllosphere resulted in contaminated tomato fruit (S. lycopersicum cv. Micro-Tom). Evidence was also presented by Gu et al. (6) that S. Typhimurium can be internalized into tomato plants via the leaves when inoculated in a suspension of the surfactant Silwet L-77, leading to high levels of colonization in fruits, without inducing any symptoms in the tomato plant (S. lycopersicum cv. Florida Lanai).
Survival of bacterial populations in the plant environment is often directly associated with both plant and microbial factors. Varied contamination rates of S. enterica have been observed between tomato cultivars. Barak et al. (11) found S. enterica population levels on tomato leaves to be cultivar-dependent. In that study, type 1 trichomes were identified as the preferred colonization site on tomato leaves. However, the ability of S. enterica to colonize and survive on the tomato plant is unlikely to be only cultivar-dependent, but might also be Salmonella serovar-specific. That is, a certain serovar(s) of S. enterica might be more adapted to survive in the tomato plant microenvironment than others. In this study, tomato plants (S. lycopersicum cv. Micro-Tom) were inoculated with S. enterica serovars Javiana, Montevideo, Newport, Saintpaul, and Typhimurium to evaluate plausible internalization routes and the relative fitness levels of tomato-associated S. enterica serovars.
MATERIALS AND METHODS
Bacterial cultures.
Five S. enterica serovars were obtained from the stock culture collection of the Division of Microbiology, Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, College Park, MD: S. Newport (serogroup C2), S. Saintpaul (serogroup B), S. Javiana (serogroup D), S. Montevideo (serogroup C1), and S. Typhimurium (serogroup B). These strains were all isolated from tomato- or other-produce-associated outbreaks.
Inoculum preparation.
Stock cultures were stored in brain heart infusion (BHI) broth containing 25% glycerol at −80°C. Cultures were streaked onto tryptic soy agar (TSA) plates and incubated at 35°C for 18 h. Subsequently, a single colony was transferred to 5 ml of tryptic soy broth (TSB) and underwent shaking at 35°C for 18 h. Each culture was harvested by centrifugation at 7,000 × g for 10 min followed by washing with 0.01 M phosphate-buffered saline (PBS) (pH 7.2). This centrifugation and washing procedure was performed three times. Bacterial cultures were resuspended in 5 ml of PBS to an optical density at 600 nm of 1.0, which approximates 109 CFU/ml. An equal volume of cell suspension of each serovar was combined as the inocula for tomato plants. The five-strain cocktail was further diluted in PBS at a 1:4 ratio as the inoculum for the soil inoculation studies. The inoculum was not adapted to any cold or stress conditions prior to its application into the soil. The concentration of each serovar in the cocktail was determined by plate count immediately before inoculation.
Plant preparation.
Tomato seeds (S. lycopersicum Micro-Tom) were surface-sterilized with bleach (NaOCl). Briefly, seeds were disinfected with 70% ethyl alcohol (EtOH) for 3 min, rinsed in sterile double-distilled deionized water (ddH2O), and soaked in Clorox commercial bleach (0.525% sodium hypochlorite) for 15 min. Seeds were then rinsed in sterilized ddH2O three times (5 min each rinse) and germinated in the dark in sterile petri dishes containing sterile filter paper moistened with sterile ddH2O. Germinated seeds were then planted in a commercially available potting mix (Sun Gro Metro-Mix) in a greenhouse at the USDA-Beltsville Agricultural Research Center (BARC) West (Beltsville, MD). In order to eliminate weed seeds and plant pathogens or pests, seedlings were transplanted at 2 weeks postseeding to steam-sterilized soil (∼300 g) from Virginia's eastern shore (VES) in 4-in.-diameter plastic Azalea pots (ITML Horticultural Products, Middlefield, OH) that were placed in a plastic saucer to serve as a water reservoir for indirect irrigation. Sandy loam soil was collected from intensively managed research plots used for tomato and cucumber production at the Eastern Shore Agricultural Research and Extension Center (AREC) of Virginia Tech (Painter, VA). The fields were managed with typical fertilizer, fungicide, insecticide, and herbicide application schedules. The amounts of N, P, and K in the soil were 320, 136, and 103 mg/kg, respectively, and the pH of the soil was 6.2. Drip irrigation with well water was applied to the plots at 2-day intervals to maintain soil moisture at around 11%. Plants were transferred to Conviron E7/2 climate-controlled growth chambers (Winnipeg, Canada) for the duration of the study. Growth chamber temperatures were maintained at 25°C (daytime) and 23°C (nighttime) with a day-and-night cycle of 12 h and a constant relative humidity of 65%. The saucer was refilled with ca. 10 ml water at 2-day intervals. Additionally, plants received tomato and blossom-set spray treatments, per the manufacturer's instructions (Bonide Products, Inc., Oriskany, NY), to speed up harvest time and increase yields. Neptune's Harvest organic fish and seaweed fertilizer (Ocean Crest Seafoods, Inc., Gloucester, MA) was applied according to the manufacturer's instructions to maintain plant growth. Pots were randomized throughout the growth chamber.
Soil inoculation with S. enterica.
A total of 22 seedlings at 1 week posttransplant were used in the soil experiment. Plants were divided into two treatment groups: a negative control (PBS only, n = 4) and an experimental group (inoculated with Salmonella inoculum, n =18). Four milliliters of Salmonella inoculum (five-strain cocktail to PBS in a 1:4 volume ratio, with a cell density of ∼1 × 108 CFU/ml) or PBS was directly injected into soil at two loci using a pipette tip. One core sample of rhizosphere (10 g) from each plant was taken with a sterile cork borer (15 mm diameter) at 4, 8, and 16 days postinoculation (dpi) to determine the survival rates of Salmonella. Between 6 and 10 colonies were randomly selected from each Salmonella-positive soil sample, for a total of approximately 100 colonies, and were subjected to molecular serotyping for serological surveillance at days 8 and 23 following inoculation, respectively. Stems from all 22 plants were used for the recovery of endophytically colonized Salmonella at 23 dpi using the method described below.
Leaf inoculation with S. enterica.
A total of 22 seedlings were divided into two treatment groups at 14 days posttransplant (dpt): a negative control group (inoculated with PBS, n =4) and an experimental group (inoculated with the five-strain cocktail, n =18). Leaflets (n = 6 to 9) on each plant were lightly dusted with 400-mesh carborundum to abrade the surfaces and create the wounds necessary for the entry of bacteria. A total of 2 drops of inoculum (5 μl/drop, with cell density of ∼5 × 108 CFU/ml) was spread over the upper surface of the leaflet. Three inoculated leaflets from each plant were sampled with a sterile scalpel at 0, 8, and 16 dpi to determine the survival rates of Salmonella. Eight to 10 colonies were randomly selected from each Salmonella-positive leaf sample, for a total of approximately 100 colonies, and were subjected to molecular serotyping for serological surveillance at 8 and 23 dpi, respectively.
Blossom inoculation with S. enterica.
A total of 38 plants at the blossom stage were divided into two treatment groups: a negative control group (inoculated with PBS, n =4) and an experimental group (inoculated with five-strain cocktail, n =34). More than 170 blossoms were painted inside with cotton swabs containing the five-strain cocktail inoculum (approximately 10 μl) and 30 blossoms with control PBS. Inoculated blossoms were marked individually. One blossom from each plant was sampled with a sterile scalpel at 0 and 7 dpi to determine the survival rate of Salmonella. Approximately 100 Salmonella colonies, with 10 colonies from each Salmonella-positive blossom sample, were randomly picked for serological surveillance at 7 dpi.
Inoculation of soil with S. enterica for the internalization experiment.
Two-tiered experiments were conducted to investigate the translocation of S. enterica into tomato plants (S. lycopersicum cv. Micro-Tom) from soil. The first experiment was conducted with 18 inoculated and 4 control plants as described previously in the soil experiment, except that all 22 seedlings in this experiment were used within 3 days after transplant. In the second experiment, a total of 24 seedlings were transplanted into pots and placed in individual saucers before being distributed evenly between two Conviron growth chambers operated under the conditions described above. One chamber contained the set of plants inoculated immediately after transplant, and the other chamber contained the set of plants inoculated 1 week after transplant. Plants in both chambers were divided into two treatment groups: a negative control group (inoculated with PBS, n =2) and an experimental group (inoculated with Salmonella inoculum, n =10). Four milliliters of Salmonella inoculum (five-strain cocktail to PBS in a 1:4 volume ratio, with a cell density of ∼1 × 108 CFU/ml) or PBS was directly injected into the soil at two loci, at approximately a 40-mm depth using a sterile pipette tip. Plants were watered in the saucers as described above to avoid splashes from watering the soil surface, and to ensure that no contact was made between the treatments during application to the surface and the rest of the plant to avoid cross-contamination throughout both experiments. Stems from six inoculated plants in the first experiment and all 24 plants in the second experiment were subjected to isolation of Salmonella at 7 dpi. Middle and top leaves, as well as fruit samples, were collected from the remaining 16 plants in the first experiment (12 inoculated plants and 4 control plants) at the early fruit stage.
Soil, leaf, and blossom sample testing.
Each sample was aseptically transferred into an individual sterile Whirl-Pak filter bag (Fort Atkinson, WI). Modified buffered peptone water (mBPW) (Becton, Dickinson, and Company, Sparks, MD) (12) was added to each sample bag according to the sample type: soil (20 ml), leaflets (10 ml), or blossoms (10 ml). Each sample bag was hand-massaged for 2 min, and then the homogenate was diluted 10-fold in PBS, and 0.1-ml aliquots of the appropriate dilutions were spread onto xylose-lysine-Tergitol 4 (XLT-4) agar (Becton, Dickinson, and Company, Sparks, MD). After 20 to 24 h of incubation at 37°C, typical S. enterica colonies, i.e., pink-red with a black center and yellow periphery, were considered presumptive positive. Those colonies were transferred to triple sugar iron agar (TSI) and lysine iron agar (LIA) slants and incubated for 24 h at 35°C. Growth from presumptive-positive TSI slants was confirmed as Salmonella with somatic group antisera (Statens Serum Institut, Copenhagen, Denmark) and Salmonella molecular serotyping using the Luminex/Bio-Plex system (13, 14).
Recovery of endophytically colonized Salmonella from stems and leaves.
At 7 or 23 dpi, each stem from 1 cm above the soil surface was aseptically removed from a plant with sterile scissors. After removing all side branches aseptically, the main stem remained for analysis. Sterile distilled water was used immediately to wipe the outside of the main stem. After aseptic transport back to the lab in plastic Ziploc bags, stem samples were immersed in 70% EtOH for 1 min, 5% Clorox for 1 min, 70% EtOH for 1 min, and 1% silver nitrate (AgNO3) for 20 min for surface wetting and sterilization (15). Stem samples were then washed with sterile ddH2O for 1 min to remove the silver nitrate. The stem was divided into 0.5-cm long pieces with a sterile scalpel from the top (apical) to bottom (basal), and the last basal piece was discarded. Each piece of stem tissue was placed immediately after sectioning onto the surface of XLT-4 agar medium in a positional order from top to bottom. The appearance of typical Salmonella colonies on XLT-4 was observed daily for 3 days at room temperature (RT), and further isolation and confirmation were performed as described above.
Middle and top leaves were aseptically removed from the plant using sterile scissors. Leaves were surface sterilized with a 70% EtOH spray and allowed to dry under a flow hood until no visible solution remained (7, 10). The excised leaves were aseptically combined in one stomacher bag for each plant and treated as a single sample. Both surface decontamination methods (i.e., silver nitrate and 70% EtOH) were verified in preliminary trials by treating both stem and leaf samples that were previously surface-inoculated with Salmonella with silver nitrate and 70% EtOH, respectively. Verification included plating stem tissue directly onto XLT-4 agar and rinsing leaf surfaces with mBPW followed by plating onto XLT-4.
Fruit sampling and testing procedures.
For each experiment, both green and red ripe tomato fruits were harvested from the plants and divided into experimental and control groups. For the soil and leaf inoculation experiments, one or two fruits were randomly sampled from each plant to test for the presence of S. enterica. For blossom inoculation experiments, a total of 90 tomatoes, 71 of which were produced from inoculated blossoms and 19 from emergent (i.e., postinoculation) uninoculated blossoms, were harvested in the experimental group. Tomatoes were aseptically picked using sterile scalpels and immediately placed individually into sterile Whirl-Pak filter bags for transport to the laboratory. Ten milliliters of mBPW was added at RT to each tomato sample bag. Sample bags were then hand-rubbed for 2 min to aid in the suspension of potential surface populations of microbes on the fruit. Each tomato was removed from the mBPW wash suspension and immersed in 70% alcohol for 2 min for surface disinfection and then allowed to dry under a laminar flow hood until no visible EtOH remained. Tomatoes from the control groups were always treated last using the same EtOH reservoir to confirm EtOH disinfection efficiency. After aseptically removing the pedicle and calyx, each fruit was then placed in an individual sterile Whirl-Pak filter bag containing 10 ml of mBPW and stomached for 60 s at 230 rpm with a Stomacher 400 circulator (Seward, London, United Kingdom). The mBPW wash suspensions and fruit homogenates were incubated for 24 h at 35°C. Aliquots of 0.1 ml from the incubated pre-enrichments were subcultured in 10 ml of tetrathionate (TT) broth with incubation for 24 h at 35°C. Incubated TT selective enrichment broth was streaked (10 μl) for isolation of salmonellae onto XLT-4 agar plates. After 24 h of incubation at 35°C, colonies typical of S. enterica on XLT-4 agar were considered to be presumptive positive and were transferred to TSI and LIA slants for 24 h at 35°C. Growth from presumptive-positive TSI slants was confirmed as Salmonella as described above. Tomato fruit that was shown to retain any confirmed Salmonella colony was regarded as a Salmonella-positive tomato.
Molecular serotyping.
The standard protocol for the molecular determination of serotype in Salmonella from the Centers for Disease Control and Prevention (CDC) was followed (16). Briefly, the O-grp-1 assay (a six-plex PCR specific for the O group 1 antigen) (13) and H-ag assay (a 20-primer multiplex PCR specific for the H antigen) (14) were performed in a thermal cycler (Bio-Rad, Hercules, CA) with the following parameters: initial denaturation at 15 min, then 30 cycles of 94°C for 30 s, 48°C for 90 s, and 72°C for 90 s, followed by a 72°C incubation for 10 min. PCR amplicons were then used directly with coupled beads (Radix Biosolutions, Georgetown, TX) in a hybridization reaction. The reaction mixture involved adding 33 μl of corresponding bead mix to 5 μl of PCR product from the O-grp-1 assay or H-ag assay and 12 μl of Tris-EDTA (TE) buffer in a single well of a low-profile 96-well microtiter plate (Bio-Rad, Hercules, CA). The reaction mixture was incubated first for 5 min at 94°C and then for 30 min at 52°C to denature the DNA and allow for hybridization of the probes to the PCR amplicons. Microspheres were then suspended in 75 μl of detection buffer (R-phycoerythrin-conjugated streptavidin; Life Technologies, Grand Island, NY) and diluted to 4 μg/ml in 1× tetramethylammonium chloride (TMAC) hybridization buffer. Samples were incubated for an additional 10 min at 52°C and then analyzed on the Bio-Plex (Bio-Rad, Hercules, CA) per the manufacturer's instructions. The median fluorescence intensity (MFI) for each bead set was calculated automatically by the Bio-Plex software. A positive signal was defined as an MFI yielding 6× the background fluorescence intensity for each bead-probe set.
Statistical analysis.
For serological surveillance of Salmonella colonies isolated from leaf, blossom, and soil samples, an estimated percentage and a confidence interval (CI) were calculated for each of the five serovars used in the study. The estimated percent equals the number of samples of the specified serovar divided by the total number of samples (∼100 colonies per each sample type). In the case of one serovar (S. Typhimurium) that did not show up in the surveillance data, its upper confidence bound was the highest proportion such that observing none had a 5% chance of occurring. Otherwise, all possible outcomes were ordered by the number of serovars of the specified type. The method of Clopper and Pearson (17) was used to calculate the confidence interval results with the lower confidence bound being a minimum P value such that the probability Pr (all possible outcomes were less than or equal to the actual outcome) was 0.025. By this same approach, the upper confidence bound was the maximum P such that Pr (all possible outcomes were less than or equal to the actual outcome) was 0.025. In both cases, P denotes the probability that a serovar was the type specified.
RESULTS
Survival of S. enterica on tomato plants.
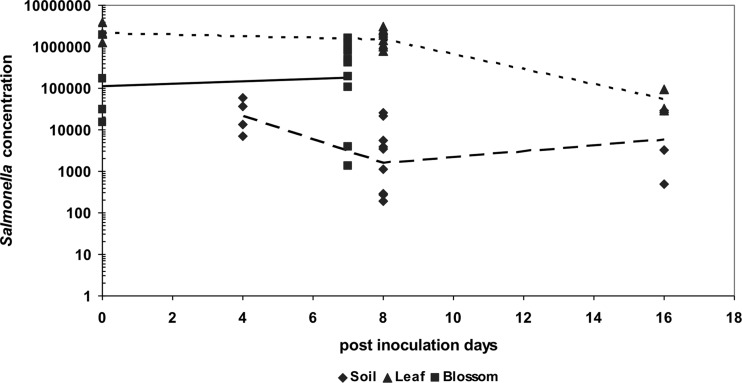
The ability of S. enterica to persist on tomato plants was indicated by the detection of Salmonella CFU in soil rhizosphere, leaflet, and blossom samples (Fig. 1). The trend of Salmonella growth varied across inoculation sites. The concentration of Salmonella almost doubled in blossoms (from 1.12 × 105 to 1.77 × 105 CFU/blossom) at 7 dpi. Salmonella concentrations, while showing no significant differences on leaflets within the first 8 days postinoculation, demonstrated a >13× decrease in soil (from 2.13 × 104 to 1.62 × 103 CFU per gram [dry weight] of soil) during this same time course. Thereafter, the average concentration of Salmonella dropped to 5.38 × 104 CFU per leaflet. Conversely, a >3× increase was observed in soil (from 1.62 × 103 to 6.00 × 103 CFU per g soil) at 16 dpi.
Fig 1.
Salmonella enterica populations in soil (CFU/g), on leaves (CFU/leaflet), and blossoms (CFU/blossom) of tomato plants after inoculation. Average S. enterica populations are shown as lines: soil (– –), leaves (- - -) and blossoms (––).
S. enterica serovar-specific niche colonization of tomato plants.
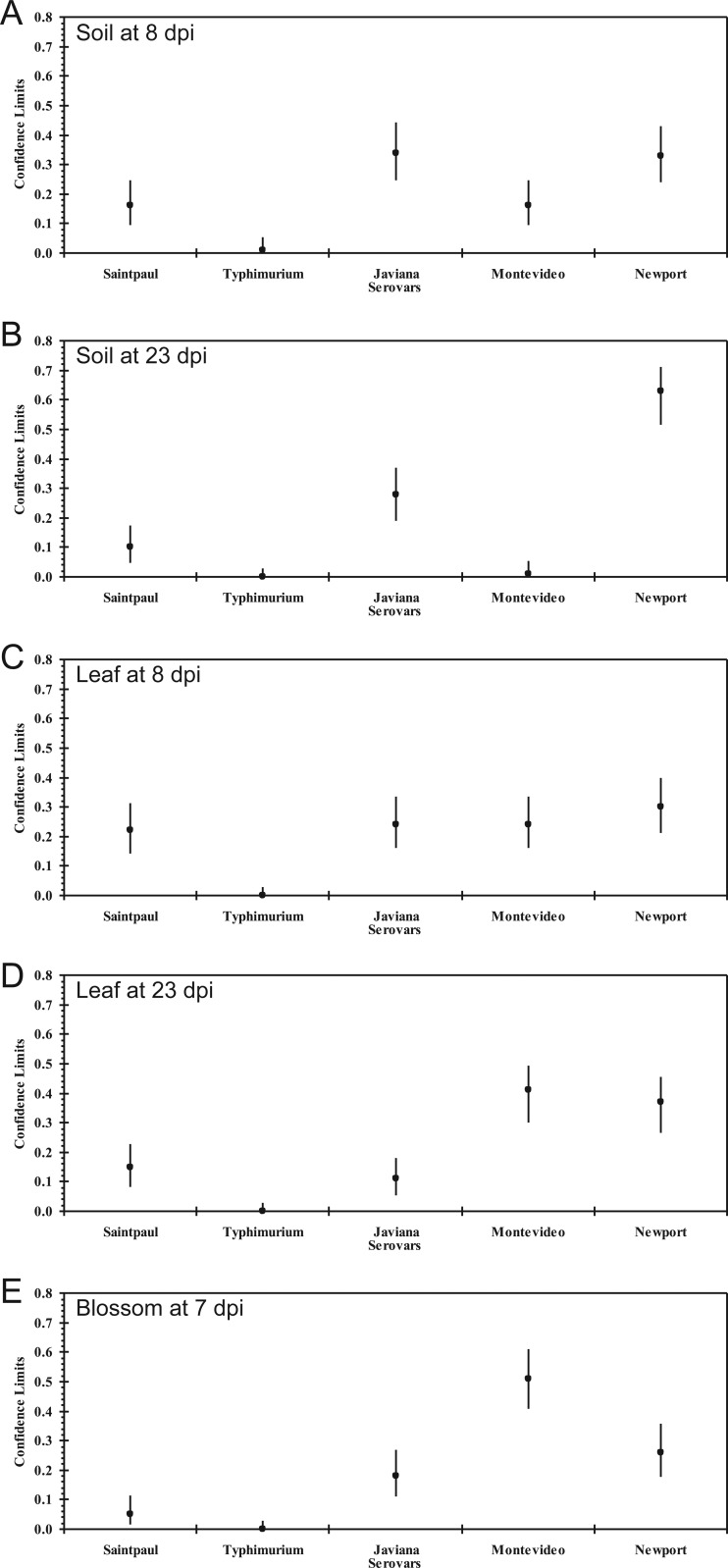
The number of Salmonella-colonized plants was significantly affected by the S. enterica serovar added to soil (χ2 = 57.61, P < 0.0001 at 23 dpi), leaf (χ2 = 38.89, P < 0.0001 at 23 dpi), and blossom (χ2 = 36.98, P < 0.0001 at 7 dpi) samples. Surprisingly, a different colonization pattern by the five S. enterica serovars comprising the Salmonella cocktail was observed based on molecular serology screening (Fig. 2). In particular, S. Newport and S. Javiana were recovered from rhizosphere samples at a higher probability than other serovars at both 8 dpi (S. Newport, 33% [95% confidence interval, 24% to 43%]; S. Javiana, 34% [95% confidence interval, 25% to 44%]) and 23 dpi (S. Newport, 62% [95% confidence interval, 52% to 71%]; S. Javiana, 27% [95% confidence interval, 19% to 37%]) (Fig. 2A and B, respectively). On the other hand, S. Montevideo and S. Newport showed greater fitness on leaves over time (Fig. 2D). Moreover, the colonization of leaves was observed to be similar among all serovars, save for S. Typhimurium, at 8 dpi (Fig. 2C). In blossom samples, 51 colonies were identified as S. Montevideo (95% confidence interval, 41% to 61%) out of the 100 Salmonella-positive colonies sampled, suggesting its superior fitness for this plant organ (Fig. 2E). It is interesting to note that almost no S. Typhimurium was recovered in samples from either of the plant organs studied here.
Fig 2.
Molecular serology prevalence per Salmonella enterica serovars Saintpaul, Typhimurium, Javiana, Montevideo, and Newport in the rhizosphere, on leaves, and on blossoms of tomato plants after inoculation. A five-strain cocktail was inoculated into soil and onto leaves and flowers of tomato plants in the corresponding experimental groups. At day 8 (A) and day 23 (B) after soil inoculation, day 8 (C) and day 23 (D) after leaf inoculation, or day 7 after blossom inoculation (E), around 100 Salmonella colonies, with 6 to 10 colonies from each Salmonella-positive sample, were randomly picked for serological surveillance. Each dot represents the estimated fraction for each of the five serovars in around 100 Salmonella colonies isolated from each sampling, and each line represents the lower and upper values of the 95% confidence interval (CI).
S. enterica contamination of fruit via blossoms.
In total, 112 red or green cherry tomatoes were harvested and analyzed for the presence of Salmonella (Table 1). Twenty-two of these tomatoes originated from control plants that were not inoculated with Salmonella, and 90 from plants inoculated by blossom brushing, of which 19 were from emergent blossoms that were not inoculated with Salmonella. Salmonella was not recovered from tomatoes harvested from uninoculated control plants, which was performed by plating either enriched samples of mBPW wash water or tomato pulp homogenates. However, Salmonella was detected on or in tomatoes that developed from experimentally inoculated blossoms as well as from emergent uninoculated blossoms (Table 1).
Table 1.
Salmonella enterica contamination of tomato fruits via blossomsa
| Group | No. of tomatoes | S. enterica-positive rate (%) | No. of S. enterica-positive samples from: |
||
|---|---|---|---|---|---|
| Surface only | Inside only | Both surface and inside | |||
| Exptl | |||||
| Inoculated blossoms | 71 | 70.4 | 21 | 1 | 28 |
| Noninoculated blossoms | 19 | 15.8 | 2 | 0 | 1 |
| Control | 22 | 0 | 0 | 0 | 0 |
A five-strain cocktail was inoculated onto individually labeled blossoms of tomato plants. All the tomato fruits derived from the inoculated and emergent flowers in the experimental and control groups were harvested and screened for surface and internal populations of Salmonella.
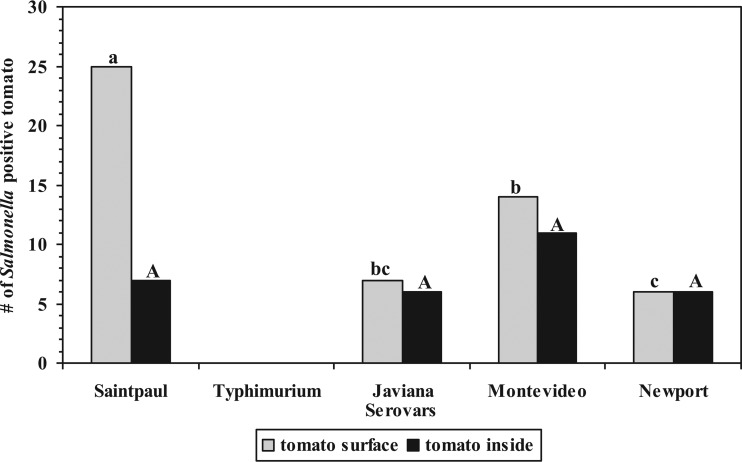
Fifty of 71 tomato fruits (70%) harvested from inoculated blossoms were positive for Salmonella. Of the 50 Salmonella-positive tomatoes, 21 contained the pathogen on the fruit surfaces only, as Salmonella was detected only in mBPW wash water but not in the whole tomato fruit homogenate (Table 1). Twenty-eight tomato fruits, however, were found to have Salmonella both on the surface and internally, and one tomato was found to harbor the pathogen only internally (Table 1). Interestingly, three out of 19 tomato fruits harvested from adjacent emergent uninoculated blossoms on the same plant were positive for Salmonella as well, with two retaining Salmonella on the surface only, and one with Salmonella both on the surface and internally. Furthermore, 17 tomatoes were found to harbor more than one serotype. Consistent with previous observations described above, S. Typhimurium was not found in or on tomato fruits. While S. enterica serovar Saintpaul was the most prevalent serovar isolated on tomato fruit surfaces (25/52, 48%), S. Montevideo was the serovar most frequently isolated internally in tomato fruits (11/30, 37%) (Fig. 3). Also, even though no significant differences between breaker and red ripe tomatoes were noted in the rates of Salmonella recovery, a significant interaction was indicated between fruit ripeness (green versus red or breaker) (P < 0.05) (Table 2).
Fig 3.
Recovery of each Salmonella serovar on and within tomato fruit derived from inoculated blossoms. A five-strain cocktail was inoculated onto individually labeled blossom of tomato plants. All the tomato fruits derived from inoculated and emergent flowers in the experimental group were harvested and screened for surface and internal populations of Salmonella. Molecular serotyping was used to determine the serovar of isolated Salmonella colonies. Each bar represents the serovar-associated number of Salmonella-positive tomatoes. Serovars with different letters denote significant differences (P < 0.05) as determined by Tukey-Kramer honestly significant difference (HSD) testing on two-way analysis of variance (ANOVA) results. Uppercase letters refer to differences on tomato surfaces; lowercase letters refer to differences inside tomato fruit.
Table 2.
Salmonella enterica contamination of tomato fruits with respect to fruit ripeness
| Tomato ripeness | Total no. of tomatoes |
Salmonella-positive samples from: |
|||
|---|---|---|---|---|---|
| On/within tomatoes (%) | On tomatoes only (%) | Within tomatoes only (%) | Total no. (%)a | ||
| Green | 43 | 11 | 6 | 0 | 17 (39.5) B |
| Breaker | 21 | 8 | 8 | 0 | 16 (76.2) A |
| Red | 26 | 10 | 8 | 1 | 19 (73.1) A |
Different uppercase letters represent the number of positives of the total that are significantly different (P < 0.05).
Internalization and migration of S. enterica in tomato plants via soil.
In the first internalization experiment, a total of 48 plant samples were collected from inoculated plants, with 6 stems collected at 7 days posttransplant (dpt) as described previously, and 12 top and middle leaves and 30 fruits collected at the early fruit stage for recovering endophytically colonized Salmonella. Eighteen plant samples were collected at the early fruit stage from four control (i.e., noninoculated) plants (4 stems, 4 leaves, and 10 fruits). All leaves or tomato fruits sampled were composited for each plant after surface disinfection. No samples from control plants were positive for the presence of Salmonella. Of the tomato plants grown with Salmonella-infested soil, 22% (4 out of 18) contained endophytically colonized Salmonella based on direct plating or enrichment procedures, including two stem samples (11.1%), one leaf sample (5.5%), and one fruit sample (5.5%). S. enterica serovar Saintpaul was isolated from the single positive leaf sample and S. Newport was found on the surface and inside the single positive tomato sample (5.5%). More interestingly, Salmonella was recovered from inside the stem up to 10 cm from the soil line within a week after inoculation, and multiple serovars, including S. Newport, S. Montevideo, and S. Saintpaul, were recovered from different stem segments.
In the second experiment, two of 10 stem samples (20%) were positive for endophytically colonized Salmonella when the plant was inoculated ≤3 dpt. Conversely, no Salmonella was recovered from the plants inoculated 7 dpt (Table 3). The interaction between the inoculation time posttransplant and the recovery of endophytic Salmonella was significant (χ2 = 7.7, P < 0.01).
Table 3.
Recovery of endophytically colonized S. enterica from tomato plants (Solanum lycopersicum cv. Micro-Tom)a
| Inoculation time posttransplant (days) | No. of plants analyzed in each expt |
Salmonella-positive plants in: |
|
|---|---|---|---|
| Each expt (no. [%]) | Total (no./total no.)e | ||
| 1–3 | 18b | 4 (22) | 6/28 A |
| 10c | 2 (20) | 6/28 A | |
| 7 | 18d | 0 (0) | 0/28 B |
| 10c | 0 (0) | 0/28 B | |
A five-strain cocktail was inoculated into the soil of tomato plants' root zones. The presence of S. enterica inside the plant tissues was evaluated by sanitizing the exterior of the sample before direct plating or enrichment.
Among 18 plants, only stems were sampled from 6 plants at 7 days postinoculation (dpi); top and middle leaves and tomato fruits were sampled from the remaining 12 plants at the early fruit stage (plants had one or more green fruits). Leaves or fruits were composited for each plant after surface disinfection.
Only stems were sampled from plants at 7 dpi.
Only stems were sampled from plants at 23 dpi.
Different uppercase letters represent the number of positives out of the total that are significantly different (P < 0.05).
DISCUSSION
Previous studies have shown the persistence of Salmonella in various ecological niches of Virginia's eastern shore agricultural region (8, 18–20). Most recently, ecological surveillance data on VES tomato farms by Bell et al. (21) and Micallef et al. (22) provide further evidence that Salmonella persists in the VES tomato-growing environment, particularly in pond water often used for irrigation, or in creek water that flowed downstream of the pond, pond or creek sediment, and soil. The recent isolation of an S. Newport strain from an irrigation pond that matched an outbreak strain (23) suggested that it is still possible for pond water to contribute to Salmonella contamination of tomatoes even when drip irrigation and plasticulture are used for tomato production. Studies have been conducted experimentally to examine possible routes for S. enterica to contaminate preharvest tomatoes. Salmonella has been shown to be capable of internalizing tomato plants through the roots (7), leaves (6), and blossoms (5, 24), provided there are favorable conditions for this to occur. Here, our findings provide further evidence of root and blossom uptake of Salmonella in S. lycopersicum Micro-Tom (11) grown in VES sandy loam soil, leading to the contamination of developing tomato fruits. It is noteworthy that the fruit contamination rate was much higher with Salmonella introduction through flowers (70.4%) than through the rhizosphere (5.5%). Equally remarkable was the observation that after Salmonella colonized the blossom, it not only proliferated, but also persisted, for at least 35 additional days of fruit development, accentuating the potentially high risk to the safety of fresh tomatoes. The possible routes of Salmonella transmission to tomato blossoms still require further investigation.
Contradictory opinions abound regarding whether Salmonella enterica can internalize tomato plants through the root system (4, 7, 10). However, our findings revealed that, aside from cultivar type, serovar type and the introduction time posttransplant are two key factors affecting Salmonella internalization through the root system. Bernstein et al. (25) reported that S. Newport is capable of persisting in potting medium for 4.7 to 10 weeks. In our study, S. Newport and S. Javiana appeared to colonize sandy loam soil more efficiently than other serovars, including S. Montevideo, S. Saintpaul, and S. Typhimurium. Specifically, S. Newport was recovered from 100% of plant rhizosphere samples that were inoculated and assayed, and it comprised 62% of the salmonellae isolated at 23 dpi. Through the recovery of endophytic Salmonella from stems, this study illustrated clearly that time-dependent factors are also important for Salmonella internalization via the root system. Specifically, inoculation within 3 days of transplanting yielded a significantly higher recovery of endophytically colonized Salmonella (average of 20%) than did inoculations 1 week after transplantation (0%). Plant wounding or stress induced by abiotic factors (26) during transplantation probably underscores this bias that was observed for Salmonella entrance. In practice, tomato crops are grown from plants started in greenhouses, hotbeds, or cold frames. Seedlings are then transplanted when they are about 2 to 5 in. tall. Transplanting tends to promote the natural taproot system to become a more fibrous root system, permitting earlier ripening of fruit and a longer season for growth (27). However, interior root colonization might occur passively through wounds in roots that are damaged during transplantation (26). Moreover, methyl bromide has had a long history of use in tomato cultivation as a soil fumigant in the eastern United States, and recent metagenomic studies have shown that such practices have diminished overall soil microbial diversity (28), perhaps increasing the potential for Salmonella colonization and persistence in the soil. Taken together, these findings support a hypothesis whereby Salmonella might be introduced in the soil via potentially contaminated irrigation water. In VES, certain serovars, such as S. Newport, seem well adapted to survival in soil, surface water, and tomato crops, the last niche of which is supported in this report. During the transplantation stage, a tomato plant is more susceptible to internalization, thereby increasing the occurrence of Salmonella internalization in the plant, and, subsequently, causing an increased risk of Salmonella contamination of preharvest tomato fruits.
Not surprisingly, our study failed to recover Salmonella from tomato fruits via inoculated leaf samples. A lack of recovery of Salmonella from other noninoculated but adjacent leaf surfaces or tomato fruits might have been due to the usage of saucers as water reservoirs, which avoided splashing from direct watering and cross-contamination, throughout the experiment. Evidence of Salmonella internalization through leaf surfaces, even when they are modified by the presence of carborundum or surfactants, indicates that this being a route for internal fruit contamination (presumably due to limited or no internal translocation) is significantly unlikely (6). In this study, the various possible mechanisms by which Salmonella-inoculated leaves resulted in the contamination of tomato fruit were not investigated.
Multiple Salmonella serovars have been reported concurrently in the same freshwater (19, 29) and sediment (19, 21, 22, 29) samples. However, only a few serovars of Salmonella have been repeatedly linked to outbreaks associated with tomato fruit, leading to the thesis that certain serovars (i.e., S. Newport) are more adapted for survival and persistence in the tomato plant environment. To further address this question, strains from different serovars that were associated previously with tomato- or other-produce-linked outbreaks were selected, and a five-strain cocktail was introduced in this study, as opposed to in previous studies that inoculated different serovars individually (24). As is documented here, S. Newport is the most persistent and dominant serovar over time in the rhizosphere. Conversely, no differences were noted among serovars recovered from leaves in terms of prevalence, with the exception of S. Typhimurium at 7 dpi. It is noteworthy, however, that over time, S. Montevideo and S. Newport appeared to be more adapted to survival in leaf tissues than were other serovars. Guo et al. (5) and Shi et al. (24) both reported that S. Montevideo was the most persistent serovar recovered within tomato fruits when introduced via blossoms. In line with those findings, our study supports that S. Montevideo is the most highly adapted for survival in tomato blossoms, followed by serovars S. Newport and S. Javiana, which also appeared to be well adapted for survival in tomatoes. All Salmonella serovars introduced onto blossoms, except S. Typhimurium, were recovered within developing tomato fruits at similar levels (P > 0.05), even though S. Montevideo was most frequently isolated within tomato fruits. However, accounting for tomato ripeness, both S. Montevideo and S. Newport were more frequently recovered (P < 0.05) than other serovars. Consistent with a report by Garcia et al. (30), the results from the current study show that S. Typhimurium survived poorly in all plant parts examined, suggesting that tomato contamination by S. Typhimurium might be more likely associated with postharvest handling.
It has been postulated that competition between various salmonellae might influence which serovars dominate within specific tomato niches (24), albeit this suggestion stemmed from earlier work involving alfalfa seeds (31) and not tomato plants. Nonetheless, given our application of a five-serovar cocktail, the role of competition among serovars merits further discussion. Although it was noted that in vitro growth curves of these Salmonella strains were essentially indistinguishable (data not shown), it is important to note that such growth rates might not be readily extrapolated to growth on and in tomato plants. That is, a competitive advantage achieved through either robust persistence in tomato plants or an ecological inhibition among distinct serovars might have been enjoyed by one or more cocktail members when applied to actual soil-tomato plant environments. Indeed, competition among these tomato-associated serovars might, in part, explain the differences observed here in tomato microniche adaptation and colonization. However, a role for competition would not be unexpected. Competition among closely related species is a central driver of evolutionary fitness among microbial species (32, 33), and in this case, would likely further augment the specialization of certain serovars for particular tomato niches over time (e.g., tomato fruits, blossoms, leaves, or the rhizosphere). It is interesting to note that the Salmonella cocktail used in this study comprised five serovars known to be associated with the contamination of tomatoes or related produce commodities (e.g., peppers) with several of these already known to reside in tomato farm environments.
It is important to note that the Salmonella inoculation levels used here (highest concentration was ∼5 × 108 CFU/ml) most likely did not emulate the Salmonella levels found among naturally contaminated tomato plants. Had this study focused on the persistence of specific serovars on tomatoes over time, such an aggravating factor might have warranted concern. However, despite this caveat, the purpose of this study was met largely through an empirical demonstration of tomato plant microenvironment fitness among different Salmonella serovars. Thus, bacterial inoculum levels seemingly had little to no effect on the specific outcomes related to serovar fitness on and in tomato plants.
As mentioned above, a significant difference was noted in the recovery of Salmonella from tomatoes related to fruit ripeness. The practice of vine-ripening tomatoes to the red or middle-to-late breaker stage of development prior to harvest might present a greater chance of Salmonella contamination than does harvesting at the green stage. Red ripened tomato fruits have decreased acid levels and increased levels of sugars and lycopene (34). These changes might favor the survival of increased numbers of Salmonella within the tomato (35) and thereby contribute to an increased risk of preharvest contamination of vine-ripened field-grown fresh market tomato fruits.
In summary, this study sheds light on Salmonella internalization by root uptake into healthy tomato plants. Our results strongly suggest that internalization through the root system is a function of at least serovar and plant growth stage. These data also demonstrate categorically that both infested soil and contaminated blossoms can lead to internal fruit contamination under the circumstances investigated. Certain salmonellae already appear to be far more adapted to tomato cultivation than other serovars. Akin to these findings, it is imperative to better understand how tomatoes become contaminated with Salmonella so that more effective and targeted agricultural practices can improve the microbiological safety of the fresh tomato supply.
ACKNOWLEDGMENTS
We thank Michael Mahovic and James Gorny for their helpful critique of the manuscript.
Footnotes
Published ahead of print 1 February 2013
We dedicate this paper to Thomas Hill (retired), who tirelessly championed tomato safety for all of us.
REFERENCES
- 1. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. CDC 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 1996–2010. MMWR Morb. Mortal. Wkly. Rep. 60:749–755 [PubMed] [Google Scholar]
- 3. CDC 2011. Surveillance for foodborne disease outbreaks—United States, 2008. MMWR Morb. Mortal. Wkly. Rep. 60:1197–1202 [PubMed] [Google Scholar]
- 4. Guo X, van Iersel MW, Chen J, Brackett RE, Beuchat LR. 2002. Evidence of association of salmonellae with tomato plants grown hydroponically in inoculated nutrient solution. Appl. Environ. Microbiol. 68:3639–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo X, Chen J, Brackett RE, Beuchat LR. 2001. Survival of salmonellae on and in tomato plants from the time of inoculation at flowering and early stages of fruit development through fruit ripening. Appl. Environ. Microbiol. 67:4760–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gu G, Hu J, Cevallos-Cevallos JM, Richardson SM, Bartz JA, van Bruggen AH. 2011. Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS One 6:e27340 doi:10.1371/journal.pone.0027340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hintz LD, Boyer RR, Ponder MA, Williams RC, Rideout SL. 2010. Recovery of Salmonella enterica Newport introduced through irrigation water from tomato (Lycopersicum esculentum) fruit, roots, stems, and leaves. HortScience 45:675–678 [Google Scholar]
- 8. Barak JD, Liang AS. 2008. Role of soil, crop debris, and a plant pathogen in Salmonella enterica contamination of tomato plants. PLoS One 3:e1657 doi:10.1371/journal.pone.0001657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jablasone J, Brovko LY, Griffiths MW. 2004. A research note: the potential for transfer of Salmonella from irrigation water to tomatoes. J. Sci. Food Agric. 84:287–289 [Google Scholar]
- 10. Miles JM, Sumner SS, Boyer RR, Williams RC, Latimer JG, McKinney JM. 2009. Internalization of Salmonella enterica serovar Montevideo into greenhouse tomato plants through contaminated irrigation water or seed stock. J. Food Prot. 72:849–852 [DOI] [PubMed] [Google Scholar]
- 11. Barak JD, Kramer LC, Hao LY. 2011. Colonization of tomato plants by Salmonella enterica is cultivar dependent, and type 1 trichomes are preferred colonization sites. Appl. Environ. Microbiol. 77:498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng CM, Van Khanh T, Lin W, Ruby RM. 2009. Interlaboratory validation of a real-time PCR 24-hour rapid method for detection of Salmonella in foods. J. Food Prot. 72:945–951 [DOI] [PubMed] [Google Scholar]
- 13. Fitzgerald C, Collins M, van Duyne S, Mikoleit M, Brown T, Fields P. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 45:3323–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McQuiston JR, Waters RJ, Dinsmore BA, Mikoleit ML, Fields PI. 2011. Molecular determination of H antigens of Salmonella by use of a microsphere-based liquid array. J. Clin. Microbiol. 49:565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franz E, Visser AA, Van Diepeningen AD, Klerks MM, Termorshuizen AJ, van Bruggen AH. 2007. Quantification of contamination of lettuce by GFP-expressing Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium. Food Microbiol. 24:106–112 [DOI] [PubMed] [Google Scholar]
- 16. CDC 2009. Standard protocol molecular determination of serotype in Salmonella, workshop on molecular determination of serotype of Salmonella. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 17. Clopper CJ, Pearson ES. 1934. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–413 [Google Scholar]
- 18. Hanning IB, Nutt JD, Ricke SC. 2009. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathog. Dis. 6:635–648 [DOI] [PubMed] [Google Scholar]
- 19. Patchanee P, Molla B, White N, Line DE, Gebreyes WA. 2010. Tracking Salmonella contamination in various watersheds and phenotypic and genotypic diversity. Foodborne Pathog. Dis. 7:1113–1120 [DOI] [PubMed] [Google Scholar]
- 20. Gorski L, Parker CT, Liang A, Cooley MB, Jay-Russell MT, Gordus AG, Atwill ER, Mandrell RE. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bell RL, Cao G, Meng J, Allard MW, Keys C, Hill T, Ottesen A, Brown EW. 2012. Salmonella Newport contamination of produce: ecological, genetic, and epidemiological aspects. In Monte AS, Santos PED. (ed), Salmonella: classification, genetics and disease outbreaks. Nova Science Publishers, Inc., Hauppauge, NY [Google Scholar]
- 22. Micallef SA, Rosenberg Goldstein RE, George A, Kleinfelter L, Boyer MS, McLaughlin CR, Estrin A, Ewing L, Jean-Giles Beaubrun J, Hanes DE, Kothary MH, Tall BD, Razeq JH, Joseph SW, Sapkota AR. 2012. Occurrence and antibiotic resistance of multiple Salmonella serotypes recovered from water, sediment and soil on mid-Atlantic tomato farms. Environ. Res. 114:31–39 [DOI] [PubMed] [Google Scholar]
- 23. Greene SK, Daly ER, Talbot EA, Demma LJ, Holzbauer S, Patel NJ, Hill TA, Walderhaug MO, Hoekstra RM, Lynch MF, Painter JA. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol. Infect. 136:157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi X, Namvar A, Kostrzynska M, Hora R, Warriner K. 2007. Persistence and growth of different Salmonella serovars on pre- and postharvest tomatoes. J. Food Prot. 70:2725–2731 [DOI] [PubMed] [Google Scholar]
- 25. Bernstein N, Sela S, Neder-Lavon S. 2007. Effect of irrigation regimes on persistence of Salmonella enterica serovar Newport in small experimental pots designed for plant cultivation. Irrig. Sci. 26:1–8 [Google Scholar]
- 26. Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. 1997. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43:895–914 [Google Scholar]
- 27. Weaver JE, Bruner WE. 1927. Chapter XXVI: tomato. In Root development of vegetable crops, 1st ed McGraw-Hill Book Company, Inc., New York, NY [Google Scholar]
- 28. Ibekwe AM, Papiernik SK, Gan J, Yates SR, Yang CH, Crowley DE. 2001. Impact of fumigants on soil microbial communities. Appl. Environ. Microbiol. 67:3245–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haley BJ, Cole DJ, Lipp EK. 2009. Distribution, diversity, and seasonality of waterborne salmonellae in a rural watershed. Appl. Environ. Microbiol. 75:1248–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garcia R, Bælum J, Fredslund L, Santorum P, Jacobsen CS. 2010. Influence of temperature and predation on survival of Salmonella enterica serovar Typhimurium and expression of invA in soil and manure-amended soil. Appl. Environ. Microbiol. 76:5025–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Howard MB, Hutcheson SW. 2003. Growth dynamics of Salmonella enterica strains on alfalfa sprouts and in waste seed irrigation water. Appl. Environ. Microbiol. 69:548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lenski RE, Mongold JA, Sniegowski PD, Travisano M, Vasi F, Gerrish PJ, Schmidt TM. 1998. Evolution of competitive fitness in experimental populations of E. coli: what makes one genotype a better competitor than another? Antonie Van Leeuwenhoek 73:35–47 [DOI] [PubMed] [Google Scholar]
- 33. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anthon GE, LeStrange M, Barrett DM. 2011. Changes in pH, acids, sugars and other quality parameters during extended vine holding of ripe processing tomatoes. J. Sci. Food Agric. 91:1175–1181 [DOI] [PubMed] [Google Scholar]
- 35. Chung KC, Goepfert JM. 1970. Growth of Salmonella at low pH. J. Food Sci. 35:326–328 [Google Scholar]