Abstract
Recent reports in North America and Europe of Clostridium difficile being isolated from livestock and retail meats of bovine origin have raised concerns about the risk to public health. To assess the situation in Australia, we investigated the prevalence and genetic diversity of C. difficile in adult cattle and calves at slaughter. Carcass washings, gastrointestinal contents, and feces were collected from abattoirs across five Australian states. Selective culture, toxin profiling, and PCR ribotyping were performed. The prevalence of C. difficile was 56% (203/360 samples) in feces from <7-day-old calves, 3.8% (1/26) in 2- to 6-month-old calves, and 1.8% (5/280) in adult cattle. Three PCR ribotypes (RTs), RT127, RT033, and RT126, predominated in <7-day-old calves and comprised 77.8% (158/203 samples) of isolates. RT056, which has not been reported in cattle before, was found in 16 <7-day-old calves (7.7%). Surprisingly, RT078 strains, which dominate production animal carriage studies in the Northern Hemisphere, were not isolated.
INTRODUCTION
Clostridium difficile is the leading cause of antimicrobial- and health care-associated diarrhea in humans (1). C. difficile infection (CDI) is a significant economic burden to global health care systems (1, 2). Individuals infected with C. difficile present with a wide spectrum of clinical symptoms, ranging from asymptomatic carriage in the mildest form to severe pseudomembranous colitis and, rarely, fulminant colitis with intestinal perforation and megacolon (3). In the past decade, CDI in humans has become more common and more severe (1). Much of the increased C. difficile burden has been driven by a rapid change in the global epidemiology of CDI with the emergence of an epidemic strain of C. difficile, RT027 (BI/NAP1), initially in North America and then in Europe (1, 2). In addition, RT078, which is commonly isolated from livestock in the Northern Hemisphere (4, 5), is now the 3rd most common strain of C. difficile isolated from humans in Europe (6) and is increasingly being isolated from humans in the United States (7).
C. difficile is often isolated from animals, particularly neonatal pigs (8, 9), foals (10), and cattle (5, 11, 12). Detection of C. difficile in production animals has raised concerns that contaminated meat products could be a potential source of CDI in humans. Reports from North America and, to a lesser extent, Europe describe C. difficile as being isolated from cattle and meat destined for human consumption (13–15). Nothing is known about the prevalence of C. difficile in Australian production animals. Given Australia's remoteness and strict quarantine regulations, it is possible that different strains of C. difficile infect both production and native animals in this country. In this study, we investigated the prevalence of C. difficile in Australian adult cattle and calves at slaughter and characterized the isolated strains by genotyping and PCR ribotyping.
(Preliminary results of this investigation were presented at the 4th International Clostridium difficile Symposium [ICDS], Bled, Slovenia, September 2012 [16].)
MATERIALS AND METHODS
Study samples.
A total of 975 samples were collected and analyzed (Table 1). Samples of adult cattle gastrointestinal contents (approximately 50 g; n = 158) and carcass washings (approximately 50 ml; n = 151) were collected on six occasions from November 2007 to January 2008 from an abattoir in Western Australia (WA) (designated W1). Sampling of animal carcasses took place within the processing area of the slaughter line after hosing of the whole carcass was complete. A sterile container was used to collect washings directly from the carcass. Gastrointestinal contents were collected aseptically, directly from the large intestine, within the viscera processing area of the slaughter line.
Table 1.
Isolation of C. difficile from Australian adult cattle and calves at slaughter
| Source | State | Abattoir code | No. of specimens | Age group | No. (%) of C. difficile isolates | No. (%) of isolates obtained by direct culturea |
|---|---|---|---|---|---|---|
| Gastrointestinal contents | WA | W1 | 158 | Adult | 0 (0.0) | NT |
| Carcass washings | WA | W1 | 151 | Adult | 0 (0.0) | NT |
| Feces | NSW | N1-N5 | 85 | Adult | 2 (2.4) | NT |
| Feces | QLD | Q1-Q11 | 130 | Adult | 2 (1.5) | NT |
| Feces | SA | S1-S2 | 15 | Adult | 0 (0.0) | NT |
| Feces | VIC | V1-V4 | 35 | Adult | 1 (2.8) | NT |
| Feces | WA | W2-W3 | 15 | Adult | 0 (0.0) | NT |
| Feces | VIC | V5 | 50 | Calf (<7 days) | 36 (72.0) | NT |
| Feces | VIC | V6 | 222 | Calf (<7 days) | 116 (52.3) | 87 (75.0) |
| Feces | VIC | V6α | 50 | Calf (<7 days) | 24 (48.0) | 16 (66.7) |
| Feces | QLD | Q12 | 38 | Calf (<7 days) | 27 (71.1) | 12 (44.4) |
| Feces | QLD | Q12 | 5 | Calf (2 mo) | 1 (20.0) | 1 (20.0) |
| Feces | QLD | Q12 | 4 | Calf (4 mo) | 0 (0.0) | 0 (0.0) |
| Feces | QLD | Q12 | 17 | Calf (6 mo) | 0 (0.0) | 0 (0.0) |
| Total | 29 abattoirs | 975 | 209 (22.7)b | 116 (69.0)b |
NT, not tested.
Values shown in parentheses are mean %.
Samples of feces from individual adult cattle (approximately 50 g; n = 280) were collected from 25 abattoirs, in New South Wales (NSW) (n = 5 [N1 to N5]), Queensland (QLD) (n = 11 [Q1 to Q11]), South Australia (SA) (n = 2 [S1 and S2]), Victoria (VIC) (n = 4 [V1 to V4]), and WA (n = 3 [W2 to W4]), over the period October 2008 to May 2009. Samples of feces from calves aged <7 days at slaughter (n = 360) were collected over five 2-day periods, from March to September 2012, from three more abattoirs, in VIC (V5 and V6) and QLD (Q12). Calf sampling comprised numerous farms or “lots”: 4 farms and 50 calves in March, 13 farms and 38 calves in April, and 128 farms and 272 calves in August and September. At the same time, some older calves were also sampled, at 2 months of age (n = 5), 4 months of age (n = 4), and 6 months of age (n = 17). All samples were transported to The University of Western Australia, stored at 4°C, and processed within 24 h.
Animals at slaughter in abattoir V6 included a subset of animals from 24 lots in NSW that were transported across the state border (n = 50 [abattoir designated V6α]). The yearly production (per annum [pa]) of the slaughterhouses varies greatly, from ∼5,000 calves pa (Q12) to 26,000 calves pa (V6) or up to ∼80,000 calves pa (V5). The calves sampled were predominantly male dairy calves (Friesan cross and Jersey).
Isolation, culture, and identification of C. difficile.
Isolation of C. difficile was based on previously described methods (17), with some modifications. Intestinal contents were cultured both directly on cycloserine-cefoxitin-fructose agar (CCFA) containing sodium taurocholate to enhance spore germination and in an enrichment broth containing gentamicin, cycloserine, and cefoxitin (GCC broth) (18), while carcass washings were centrifuged and the deposits inoculated into GCC broth.
Fecal samples were plated directly onto C. difficile ChromID (bioMérieux, Marcy l'Etoile, France) and added to GCC broth. After 48 h of incubation, 1 ml of enrichment broth was alcohol shocked with an equal volume of absolute ethanol for 1 h and then plated onto CCFA plates as described above. All plates were incubated in an anaerobic chamber (Don Whitley Scientific Ltd., Shipley, West Yorkshire, United Kingdom) at 37°C, in an atmosphere containing 80% nitrogen, 10% hydrogen, and 10% carbon dioxide.
Putative colonies of C. difficile were identified on the basis of their characteristic colony morphology (yellow, ground-glass appearance), odor (horse dung smell), and chartreuse (yellow-green) fluorescence under long-wave UV light (∼360 nm). The identity of doubtful isolates was confirmed by Gram staining and the presence of l-proline aminopeptidase activity (Remel Inc., Lenexa, KS).
PCR ribotyping and toxin gene profiling.
Crude bacterial template DNA for toxin profiling was prepared by resuspension of cells in a 5% (wt/vol) solution of Chelex-100 resin (Sigma-Aldrich, Castle Hill, NSW, Australia). All isolates were screened by PCR for the presence of the toxin A (tcdA) and toxin B (tcdB) genes (19) and the binary toxin (cdtA and cdtB) genes (20) and for changes in the repeating region of tcdA (21). Confirmation of true toxin A- and toxin B-negative (A− B−) isolates was achieved by amplification of the pathogenicity locus (PaLoc) integration region (22). PCR ribotyping was performed as previously described (23). PCR ribotyping reaction products were concentrated using a Qiagen MinElute PCR purification kit (Ambion Inc., Austin, TX) and run on a QIAxcel capillary electrophoresis platform (Ambion Inc., Austin, TX). Visualization of PCR products was performed with QIAxcel ScreenGel software (v1.0.2.0; Ambion Inc., Austin, TX).
PCR ribotyping banding patterns were identified by comparison of banding patterns with a reference library consisting of a collection of 15 reference strains from the European Centre for Disease Prevention and Control (ECDC), a collection of the most prevalent PCR ribotypes (RTs) currently circulating in Australia (B. Elliott, unpublished data), and a selection of binary toxin-positive strains. Interpretation of the capillary electrophoresis data (PCR ribotyping banding patterns) was performed by dendrogram and cluster analysis using the Dice coefficient within the BioNumerics software package v.6.5 (Applied Maths, Saint-Martens-Latem, Belgium). Isolates that could not be identified with the available reference library were designated with internal nomenclature.
Statistical analysis.
Fisher's exact test and the chi-square test were used where appropriate to compare the prevalences of C. difficile among the sampled abattoirs and to analyze the effects of age and geographic distribution on the number and types of ribotypes identified.
RESULTS
C. difficile prevalence in cattle and calves.
The prevalences of C. difficile in adult cattle and veal calves are presented in Table 1. C. difficile was not isolated from any of the 158 samples of gastrointestinal contents and 151 carcass washings from adult cattle in WA. Of the further 280 adult fecal samples from other regions of Australia, C. difficile was isolated by enrichment from 5 (1.8%) samples. The overall C. difficile recovery from the feces of veal calves (aged <7 days of age) was 56%, with a recovery from Victorian abattoirs of 55% (36/50 samples [72%] from V5, 116/222 [52%] from V6, and 24/50 [48%] from V6α) and from the Queensland abattoir (Q12) of 71% (27/38 samples). A total of 336 calf fecal samples were cultured by both direct culture and enrichment culture. From these samples, 168 isolates of C. difficile were recovered. The mean percent recovery by direct culture was 69.0% (range, 20.0 to 75.0%) (Table 1). C. difficile was isolated from 1 (3.8%) of 26 older calves. This isolate, from a 2-month-old calf at abattoir V5, was recovered by direct culture (Table 1).
The age-related differences in prevalence were statistically significant for both comparisons of <7-day-old calves versus 2- to 6-month-old calves (P = 0.0002) and <7-day-old calves versus adult cattle (P < 0.0001). The overall prevalences of C. difficile in calf fecal samples from abattoirs in Victoria and Queensland were not significantly different, nor was there any difference in recovery of C. difficile from Victorian and NSW calves at abattoir V6.
Toxin gene profiles of C. difficile isolates.
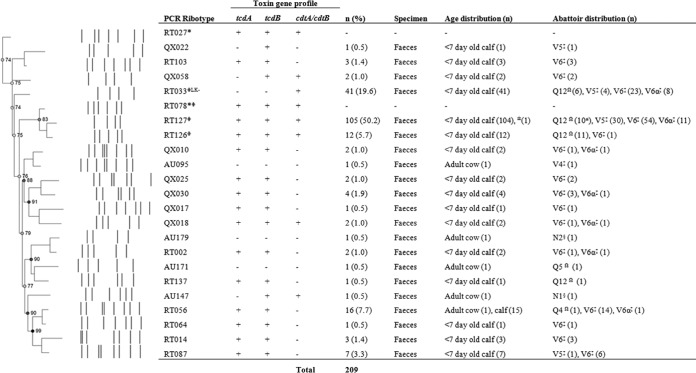
Of the 209 isolates of C. difficile recovered from adult cattle and calves, 76.6% (160/209) were positive for tcdA and tcdB (A+ B+), among which 54.5% (114/160) were also positive for the binary toxin genes (cdtA and cdtB) (CDT+). Forty-one isolates (19.6%) were negative for both tcdA and tcdB but were CDT+ (A− B− CDT+). The remaining 8 isolates yielded the following toxin profiles: A− B+ CDT+ (4/209 [1.9%]), A− B− CDT− (3/209 [1.4%]), and A− B+ CDT− (1/209 [0.5%]). Toxin gene profiles for all isolates, along with their distributions between abattoirs, age groups, and PCR ribotypes, are summarized in Fig. 1.
Fig 1.
Summary of C. difficile PCR ribotypes, toxin gene profiles, and isolate demographics from representative Australian veal calf and adult cow isolates at slaughter. PCR ribotyping pattern analysis was performed by creating a neighbor-joining tree, using the Dice coefficient (optimization, 1.00%; tolerance, 0.5%). *, reference strain; Ω, abattoir(s) in Queensland; ‡, abattoir(s) in Victoria; §, abattoir(s) in New South Wales; ±, includes a single isolate from a 2-month-old calf; Φ, multilocus sequence type clade 5 strains; LK−, PaLoc-negative strains.
PCR ribotyping of C. difficile isolates.
In both the adult cattle and calf samples, multiple PCR ribotypes were identified: 91.4% (191/209) of isolates were assigned 1 of 10 internationally recognized RTs (RT002, RT014, RT033, RT056, RT064, RT087, RT103, RT126, RT127, and RT137) (Fig. 1). The remaining 18 isolates (including 4 of the 5 from adult cattle) gave 11 distinct banding patterns but were unable to be assigned PCR ribotypes based on our reference library at the time. These isolates were classified with internal nomenclature (QX010, QX017, QX018, QX022, QX025, QX030, QX058, AU095, AU147, AU171, and AU179) (Fig. 1).
The most common RT found was RT127 (A+ B+ CDT+), which comprised 50.2% (105/209) of isolates, followed by RT033 (A− B− CDT+) (41/209 [19.6%]), RT056 (A+ B+ CDT−) (16/209 [7.7%]), RT126 (A+ B+ CDT+) (12/209 [5.7%]), and RT087 (A+ B+ CDT−) (7/209 [3.3%]). RT126 was more prevalent in Queensland (11/38 isolates [29.0%] from Q12) than in Victoria (1/322 [0.3%] from V5, V6, and V6α) (P < 0.0001). RT127 had similar prevalences in both states (95/322 isolates [30.0%] from V5, V6, and V6α and 10/38 [26.3%] from Q12), as did RT033 (35/322 [10.9%] from V5, V6, and V6α and 6/38 [15.8%] from Q12). No significant difference was seen for the distribution of RT056 (15/322 isolates [4.7%] from V5, V6, and V6α and 0/38 [0.0%] from Q12) (P = 0.3839). For abattoir V6, no difference was seen in the most prevalent PCR ribotypes between those animals originating from Victoria and those originating from NSW (V6α) (P = 0.3819).
DISCUSSION
To date, there have been no reported studies on carriage of C. difficile by pigs and cattle in the Southern Hemisphere. The prevalence of C. difficile in veal calves reported here (53.4%; range, 44.4% to 75.0%) is significantly higher than those reported for similar studies in Canada (11.2%) (11), the United States (9%) (14), Slovenia (9%) (9), and Switzerland (0.5%) (13) but similar to that in another Canadian study looking at long-term surveillance of C. difficile and tetracycline resistance on a veal farm (32% to 51%) (24). We also found a slightly larger proportion of C. difficile isolates from veal calves with at least one toxin gene present (100%) than that reported elsewhere (80%) (14). Differences in slaughter age between countries may explain the contrasting levels of C. difficile obtained prior to slaughter in this study and studies conducted overseas. In North America, calves may be up to 21 weeks of age when they are slaughtered, whereas the calves sampled in this study were slaughtered at <7 days of age. Slaughter at this early age with such a high prevalence of C. difficile carriage is a possible risk factor for carcass contamination.
The observed decline in prevalence with increasing age supports the results of studies reported elsewhere. Costa et al. (24) noted not only a high prevalence of C. difficile but also a significant rise in prevalence (32% to 51%) 1 week after arrival at a veal farm (calves were aged 2 to 10 days on arrival) and then a decline to 2% 17 weeks and 21 weeks after arrival, at which point the calves were slaughtered. This age-related effect, where prevalence decreases with age, has also been reported for pigs (8) and is true for C. difficile colonization of humans (1). In neonatal piglets, the decrease in prevalence with age is most likely a result of an increase in the gut microflora responsible for colonization resistance. Neonatal animals have an underdeveloped intestinal microflora, and C. difficile is better able to colonize, proliferate, and produce toxins in these younger animals (11).
C. difficile RT078 (A+ B+ CDT+) is the most common production animal ribotype reported worldwide (5, 25), with virulence attributes comparable to those of epidemic RT027 isolates but with a much stronger association with animals, particularly livestock (3, 5). RT078 has been found in 67% (102/152) of isolates from veal calves in Canada (24) and 94% (31/33) of isolates from veal calves in the United States (14). This strain also appears to be an emerging cause of human infections in the United States (7), while it is the 3rd most common human isolate in European hospitals (6).
Of the ribotypes detected in this study, three of the four most prevalent ribotypes, i.e., RT127 (A+ B+ CDT+), RT126 (A+ B+ CDT+), and RT033 (A− B− CDT+), belong to multilocus sequence type (MLST) 11, as does RT078. All strains of this sequence type (ST) are grouped into clade 5 (26). Stabler et al. (27) demonstrated that clade 5 strains, including strains of ribotypes 078, 126, 127, 033, and 237 (A− B+ CDT+), are common in human clinical, animal, and food sources worldwide and are highly divergent from other known ribotypes. He et al. (28) dated the last common ancestor of these strains to between 1.1 and 6.4 million years ago, and it has been suggested that this common ancestor was a nontoxigenic strain that acquired the PaLoc (26). Furthermore, strains belonging to ribotypes 033, 126, and 127 have all been isolated from humans with CDI in Australia in the last decade (T. V. Riley et al., unpublished data).
The absence of RT078 in Australian cattle is significant, albeit not totally unexpected. RT078 has not been found in any of our other livestock surveillance programs (Riley et al., unpublished data). This picture is very different from reports from overseas, where RT078 is the predominant strain in most livestock animals, including pigs, chickens, and cattle (5, 8, 9, 11). In contrast, RT127, which is rare in cattle studies overseas, appears to have emerged as the predominant strain in Australia. The emergence of clade 5 ribotypes as significant causes of disease is a recent event (27). Given the genetic similarity of these strains, their clear epidemiological links to animals, and Australia's geographic isolation, it is conceivable that RT127 has occupied the same niche that is occupied by RT078 overseas. More studies are warranted in this area, specifically to investigate the evolutionary history and phenotypes of these two strains.
In this study, the overall prevalence of CDT-producing isolates was 76.1%. As with epidemic strains RT078 and RT027, RT126, RT127, and RT033 all produce binary toxin. The exact role of CDT is still being debated; however, CDT-positive strains are often found in large numbers in animals (4, 5, 9) and are associated with an increased severity of disease in humans (29). In addition, RT078 is increasingly associated with community-acquired CDI (CA-CDI) (25, 30, 31). Recent data suggest that the incidence of CA-CDI is increasing (32, 33) and accounts for up to one-quarter of all diagnosed CDI cases (33). In the Netherlands, strains of RT078 infecting both humans and animals are genetically related by multiple-locus variable-number tandem-repeat analysis (MLVA) (34). This suggests a common source (34) and implies that C. difficile is part of a zoonosis (25, 30, 31).
Seasonality was reported as a factor affecting C. difficile prevalence in retail veal meat, with the highest prevalence occurring in winter (15). Sampling of veal calves in this study took place in late summer (March and April) and in winter (August and September). No significant increase in C. difficile prevalence was seen between abattoirs over this period. However, without sampling the same abattoir at a second time point, it is not possible to rule out a possible seasonal effect.
The finding of low levels (5/209 isolates [2.4%]) of variant toxin strains (A− B+) in livestock is consistent with other studies (29). Although initially thought not to cause disease, these variants are now described as increasing in number and causing clinically significant disease in human (35) and animal (36) infections.
Several PCR ribotypes (RT002, RT014, RT056, RT064, RT087, RT103, and RT137) that are known to cause disease in humans but are not typically associated with animals (5, 9) were identified in this study. Of these strains, RT014 is the most common ribotype in many countries, including the Netherlands (6) and Australia (T. V. Riley and B. Elliott, submitted for publication). PCR ribotypes RT002 and RT014 have been found in very small numbers in older cattle and poultry in Belgium (37) and in horses, domestic pets, and livestock in the Netherlands (38). These numbers are consistent with those reported in the current study (1 to 2% prevalence). In addition to its presence in livestock, RT014 has also been found in two samples of retail meat in Canada (15).
The presence of RT056 (A+ B+ CDT−) in Australian calves at slaughter was unexpected. This RT has been identified in a single poultry isolate in a Dutch study (38), but to our knowledge, this is the first report of RT056 in cattle. Bauer et al. (6) identified RT056 among the most frequent types of toxigenic isolates found in a recent European hospital survey. In that study, RT056 was significantly associated with a complicated disease outcome in cases of nosocomial CDI.
There are some limitations of this study. A more discriminatory typing method, such as MLVA or whole-genome sequencing, needs to be applied to this isolate set, given the large proportion of strains that grouped into two or three RTs. In addition, follow-up investigations of the same abattoirs are also warranted. In a recent study by Zidaric et al. (39), who looked at C. difficile on a veal farm over a long period, two key findings were made. First, ST 11 strains (RT078 and RT126) became more predominant as the ages of calves increased, and second, strains of a single predominant ribotype could be differentiated further by pulsed-field gel electrophoresis types, sporulation properties, and antibiotic susceptibilities. Studies overseas have shown that C. difficile is associated with disease in cattle, including calf enteritis (11) and neonatal calf diarrhea (12). In the current study, it was not possible to determine whether the veal calves presented any indications of illness or disease, although if they had been clinically ill, it is unlikely that they would have been slaughtered. Finally, future studies should include assessments of C. difficile concentrations in the fecal samples examined. Many of our samples gave a heavy growth of C. difficile on direct culture, suggesting that large numbers of C. difficile cells were present; however, this needs to be confirmed with viable counts.
Continued colonization of veal calves by C. difficile may well depend on exposure to antimicrobials. Analogous with the situation in humans, where C. difficile amplification is driven by exposure to antibiotics (1), C. difficile expansion in animals is likely driven by antimicrobial use in these populations. The extensive use of cephalosporins in Australian swine has been reported by Jordan et al. (40), who found that 25% of 197 large Australian pig herds routinely used the agent ceftiofur. Studies of a similar nature are needed for cattle. C. difficile is intrinsically resistant to cephalosporins, providing an impetus for amplification in animals and contributing to the transmission cycle in intensive animal production facilities. The use of cephalosporins in Australian livestock could be driving the amplification of C. difficile, providing reservoirs of toxigenic strains known to cause clinical disease in humans (41).
In conclusion, RT127 is the dominant strain of C. difficile in Australian cattle, and this ribotype appears to be behaving like RT078, the major production animal ribotype in the Northern Hemisphere. The high rate of carriage/colonization found in this study clearly suggests that veal calves (unlike older cattle) are potentially sources/reservoirs of C. difficile, including toxigenic strains known to cause clinical disease in humans. There is a risk of contamination of retail veal products during the slaughter process, both directly and indirectly, but the extent of this risk is currently unquantified. Further investigations are needed to determine the prevalence and concentration of C. difficile in downstream parts of the supply chain (e.g., carcasses and retail meat), as well as the dose needed to cause CDI in humans. The early slaughter age of Australian veal calves combined with their high rates of carriage of C. difficile is a risk factor for possible food-borne transmission.
ACKNOWLEDGMENTS
We are grateful to Chris Sentance (Food Safety Services, Lyndoch, SA, Australia) and Sharon Koch (Ace Laboratories, Bendigo, VIC, Australia) for assistance with collection of samples. We thank Niki Foster, Briony Elliott, and Kerry Carson for study and technical advice and Kirsty Maher and Michele Squire for assistance with manuscript preparation. Finally, we are indebted to Julie Balan and Ian Jenson, from Meat & Livestock Australia, for their continued support throughout this project.
This study was supported by grants from Meat and Livestock Australia, Sydney, NSW, Australia.
Footnotes
Published ahead of print 8 February 2013
REFERENCES
- 1. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 2. Wiegand PN, Nathwani D, Wilcox MH, Stephens J, Shelbaya A, Haider S. 2012. Clinical and economic burden of Clostridium difficile infection in Europe: a systematic review of healthcare-facility-acquired infection. J. Hosp. Infect. 81:1–14 [DOI] [PubMed] [Google Scholar]
- 3. Keessen EC, Gaastra W, Lipman LJA. 2011. Clostridium difficile infection in humans and animals, differences and similarities. Vet. Microbiol. 153:205–217 [DOI] [PubMed] [Google Scholar]
- 4. Rupnik M, Widmer A, Zimmermann O, Eckert C, Barbut F. 2008. Clostridium difficile toxinotype V, ribotype 078, in animals and humans. J. Clin. Microbiol. 46:1963–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keel K, Brazier JS, Post KW, Weese JS, Songer JG. 2007. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves and other species. J. Clin. Microbiol. 45:1963–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bauer MP, Notermans DW, van Benthem BHB, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73 [DOI] [PubMed] [Google Scholar]
- 7. Jhung MA, Thompson AD, Killgore GE, Zukowski WE, Songer JG, Warny M, Johnson S, Gerding DN, McDonald LC, Limbago BM. 2008. Toxinotype V Clostridium difficile in humans and food animals. Emerg. Infect. Dis. 14:1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hopman NEM, Keessen E, Harmanus C, Sanders IMJG, van Leegoed LAMG, Kuijper EJ, Lipman LJA. 2011. Acquisition of Clostridium difficile by piglets. Vet. Microbiol. 149:186–192 [DOI] [PubMed] [Google Scholar]
- 9. Avbersek J, Janezic S, Pate M, Rupnik M, Zidaric V, Logar K, Vengust M, Zemljic M, Pirs T, Ocepek M. 2009. Diversity of Clostridium difficile in pigs and other animals in Slovenia. Anaerobe 15:252–255 [DOI] [PubMed] [Google Scholar]
- 10. Thean S, Elliott B, Riley TV. 2011. Clostridium difficile in horses in Australia—a preliminary study. J. Med. Microbiol. 60:88–92 [DOI] [PubMed] [Google Scholar]
- 11. Rodriguez-Palacios A, Stämpfli HR, Duffield T, Peregrine AS, Trotz-Williams LA, Arroyo LG, Brazier JS, Weese JS. 2006. Clostridium difficile PCR ribotypes in calves, Canada. Emerg. Infect. Dis. 12:1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammitt MC, Bueschel DM, Keel MK, Glock RD, Cuneo P, DeYoung DW, Reggiardo C, Trinh HT, Songer JG. 2008. A possible role for Clostridium difficile in the etiology of calf enteritis. Vet. Microbiol. 2008:343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffer E, Haechler H, Frei R, Stephan R. 2010. Low occurrence of Clostridium difficile in fecal samples of healthy calves and pigs at slaughter and in minced meat in Switzerland. J. Food Prot. 73:973–975 [DOI] [PubMed] [Google Scholar]
- 14. Houser BA, Soehnlen MK, Wolfgang DR, Lysczek HR, Burns CM, Jayarao BM. 2012. Prevalence of Clostridium difficile toxin genes in the feces of veal calves and incidence of ground veal contamination. Foodborne Pathog. Dis. 9:32–36 [DOI] [PubMed] [Google Scholar]
- 15. Rodriguez-Palacios A, Reid-Smith RJ, Staempfli HR, Daignault D, Janecko N, Avery BP, Martin H, Thomspon AD, McDonald LC, Limbago BM, Weese JS. 2009. Possible seasonality of Clostridium difficile in retail meat, Canada. Emerg. Infect. Dis. 15:802–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knight DR, Thean S, Putsathit P, Fenwick S, Riley TV. 2012. Clostridium difficile from Australian cattle: all will be re(veal)ed!, abstr O25. Abstr. 4th Int. Clostridium difficile Symp. (ICDS), Bled, Slovenia, September 2012 [Google Scholar]
- 17. Bowman RA, Riley TV. 1988. The laboratory diagnosis of Clostridium difficile-associated diarrhoea. Eur. J. Clin. Microbiol. Infect. Dis. 7:476–484 [DOI] [PubMed] [Google Scholar]
- 18. Carroll SM, Bowman RA, Riley TV. 1983. A selective broth for Clostridium difficile. Pathology 15:165–167 [DOI] [PubMed] [Google Scholar]
- 19. Kato N, Ou C, Kato H, Bartley SL, Brown VK, Dowell VR, Jr, Ueno K. 1991. Identification of toxigenic Clostridium difficile by the polymerase chain reaction. J. Clin. Microbiol. 29:33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stubbs SLJ, Rupnik M, Gibert M, Brazier JS, Duerden B, Popoff M. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307–312 [DOI] [PubMed] [Google Scholar]
- 21. Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim SM, Chong Y, Wasito EB. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braun V, Hundsberger T, Leukel P, Sauerborn M, van Eichel-Streiber C. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29–38 [DOI] [PubMed] [Google Scholar]
- 23. Stubbs SLJ, Brazier JS, O'Neill GS, Duerden BI. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Costa MC, Stämpfli HR, Arroyo LG, Pearl DL, Weese JS. 2011. Epidemiology of Clostridium difficile on a veal farm: prevalence, molecular characterization and tetracycline resistance. Vet. Microbiol. 152:379–384 [DOI] [PubMed] [Google Scholar]
- 25. Weese JS. 2010. Clostridium difficile in food—innocent bystander or serious threat? Clin. Microbiol. Infect. 16:3–10 [DOI] [PubMed] [Google Scholar]
- 26. Dingle KE, Griffiths D, Didelot X, Evans J, Vaughan A, Krimanidou M, Stoesser N, Jolley KA, Golubchik T, Harding RM, Peto TE, Fawley W, Walker AS, Wilcox MH, Crook DW. 2011. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One 6:e19993 doi:10.1371/journal.pone.0019993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stabler RA, Dawson LF, Valiente E, Cairns MD, Martin MJ, Donahue EH, Riley TV, Songer JG, Kuijper EJ, Dingle KE, Wren BW. 2012. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS One 7:e31599 doi:10.1371/journal.pone.0031599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He M, Sebaihiaa M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HBH, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U. S. A. 107:7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barbut F, Decré D, Lalande V, Burghoffer B, Noussair L, Gigandon A, Espinasse F, Raskine L, Robert J, Mangeol A, Branger C, Petit JC. 2005. Clinical features of Clostridium difficile-associated diarrhoea due to binary toxin (actin-specific ADP-ribosyltransferase)-producing strains. J. Med. Microbiol. 54:181–185 [DOI] [PubMed] [Google Scholar]
- 30. Hensgens MPM, Keessen EC, Squire MM, Riley TV, Koene MGJ, de Boer E, Lipman LJA, Kuijper EJ. 2012. Clostridium difficile infection in the community: a zoonotic disease? Clin. Microbiol. Infect. 18:635–645 [DOI] [PubMed] [Google Scholar]
- 31. Rupnik M. 2010. Clostridium difficile: (re)emergence of zoonotic potential. Clin. Infect. Dis. 51:584. [DOI] [PubMed] [Google Scholar]
- 32. Kuntz JL, Chrischilles EA, Pendergast JF, Herwaldt LA, Polgreen PM. 2011. Incidence of and risk factors for community-associated Clostridium difficile infection: a nested case-control study. BMC Infect. Dis. 11:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, Harmsen WS, Zinsmeister AR. 2012. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am. J. Gastroenterol. 107:89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bakker D, Corver J, Harmanus C, Goorhuis A, Keessen EC, Fawley WN, Wilcox MH, Kuijper EJ. 2010. Relatedness of human and animal Clostridium difficile PCR ribotype 078 isolates determined on the basis of multilocus variable-number tandem repeat analysis and tetracycline resistance. J. Clin. Microbiol. 48:3744–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elliott B, Squire MM, Thean S, Chang BJ, Brazier JS, Rupnik M, Riley TV. 2011. New types of toxin A-negative, toxin B-positive strains among clinical isolates of Clostridium difficile in Australia. J. Med. Microbiol. 60:1108–1111 [DOI] [PubMed] [Google Scholar]
- 36. Thakur S, Putnam M, Fry PR, Abley M, Gebreyes WA. 2010. Prevalence of antimicrobial resistance and association with toxin genes in Clostridium difficile in commercial swine. Am. J. Vet. Res. 71:1189–1194 [DOI] [PubMed] [Google Scholar]
- 37. Rodriguez C, Taminiau B, van Broeck J, Avesani V, Delmée M, Daube G. 3 October 2012. Clostridium difficile in young farm animals and slaughter animals in Belgium. Anaerobe [Epub ahead of print.] doi:10.1016/j.anaerobe.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 38. Koene MGJ, Mevius D, Wagenaar A, Harmanus JC, Hensgens MPM, Meetsma AM, Putirulan FF, van Bergen MAP, Kuijper EJ. 2012. Clostridium difficile in Dutch animals: their presence, characteristics and similarities with human isolates. Clin. Microbiol. Infect. 18:778–784 [DOI] [PubMed] [Google Scholar]
- 39. Zidaric V, Pardon B, dos Vultos T, Deprez P, Brouwer MSM, Roberts AP, Henriques AO, Rupnik M. 21 September 2012. Multiclonal presence of Clostridium difficile PCR ribotypes 078, 126 and 033 within a single calf farm is associated with differences in antibiotic resistance and sporulation properties. Appl. Environ. Microbiol. doi:10.1128/AEM.02185-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jordan D, Chin JJC, Fahy VA, Barton MD, Smith MG, Trott DJ. 2009. Antimicrobial use in the Australian pig industry: results of a national survey. Aust. Vet. J. 87:222–229 [DOI] [PubMed] [Google Scholar]
- 41. Riley TV. 2009. Is Clostridium difficile a threat to Australia's biosecurity? Med. J. Aust. 190:661–662 [DOI] [PubMed] [Google Scholar]