Abstract
Knowledge of the diversity of magnetotactic bacteria in natural environments is crucial for understanding their contribution to various biological and geological processes. Here we report a high diversity of magnetotactic bacteria in a freshwater site. Ten out of 18 operational taxonomic units (OTUs) were affiliated with the Deltaproteobacteria. Some rod-shaped bacteria simultaneously synthesized greigite and magnetite magnetosomes.
TEXT
Magnetotactic bacteria (MTB) in the class Deltaproteobacteria have been shown to produce magnetite (Fe3O4) or greigite (Fe3S4) magnetosomes, or both within the same cell (1–3). They have been widely found in marine sediments (4), river estuaries (5), coastal salt ponds (6), lagoons (7), and alkaline environments (8) but only occasionally in freshwater lakes (3, 9). Because of their unique ability to biomineralize both magnetite and greigite, the Deltaproteobacteria MTB have attracted great interest as possible tools to aid in deciphering the mechanism of magnetosome biomineralization and the evolution of bacteria magnetotaxis (3, 10).
The Deltaproteobacteria MTB may play an important role in biogeochemical cycling of iron and sulfur elements. Recently, a cultivable strain, BW-1, which was isolated from a brackish spring, was found to mineralize either magnetite or greigite magnetosomes depending on the concentration of environmental hydrogen sulfide (3). In nature, most reported Deltaproteobacteria MTB have been found in saline environments. However, the overall diversity and distribution of these MTB in freshwater environments are still poorly understood.
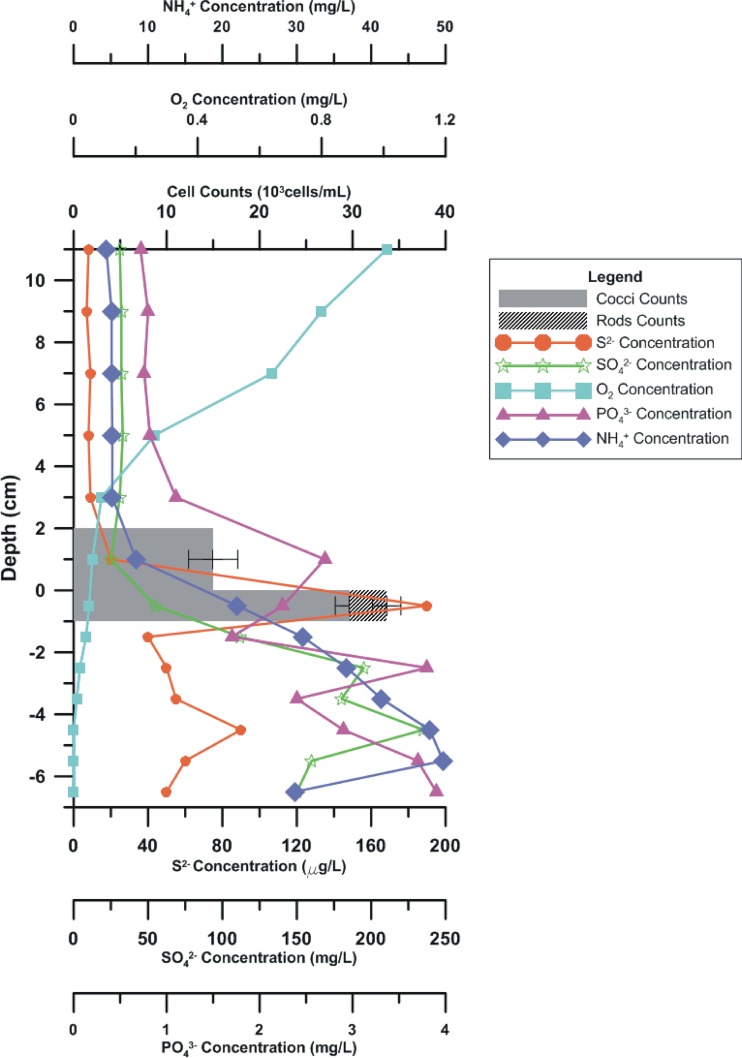
Surface sediment samples (∼10-cm depth) and two in situ vertical cores were collected from a site (34°15′10.00″N, 108°55′13.41″E) in the city moat of Xi'an, China. The water pH ranged from 7.1 to 7.5, and salinity was less than 0.34 ppt. The two vertical cores (about 1 m away from each other) were sampled using a gravity sampler. Geochemical analyses of S2−, SO42−, PO43−, and NH4+ indicated that the Xi'an moat was slightly or moderately polluted (11). The variations of MTB abundance and the concentrations of S2−, SO42−, O2, PO43−, and NH4+ with depth are shown in Fig. 1 and in Table S1 in the supplemental material. Live MTB were magnetically enriched using the MTB trap method described previously (12, 13). We observed that the MTB lived in a narrow layer in the uppermost sediment (0 to 2 cm) and in the water column no more than 2 cm above the sediment in core water that correlated to the critical oxic-anoxic transition zone (OATZ) of the site, where chemical parameters dramatically changed (Fig. 1). The composition of the MTB community was examined using a Bacteriodrome (14), light microscopy, transmission electron microscopy (TEM), and 16S rRNA genes. Magnetococci were the most dominant MTB, occurring 2 cm both above and beneath the water-sediment interface. The rod-shaped MTB were abundant at 1 cm beneath the interface. Spirilla and vibrios were also occasionally found in the same layer. The occurrence of MTB around the OATZ and correlations to geochemical variation were in line with previous studies (15, 16, 17). Detailed information on the sampling site and methods are presented in the supplemental material.
Fig 1.
Vertical distribution of MTB cells in relation to the concentrations of S2−, SO42−, O2, PO43−, and NH4+ in the vertical core (core 1) from Xi'an city moat. The depth was measured relative to the water-sediment boundary.
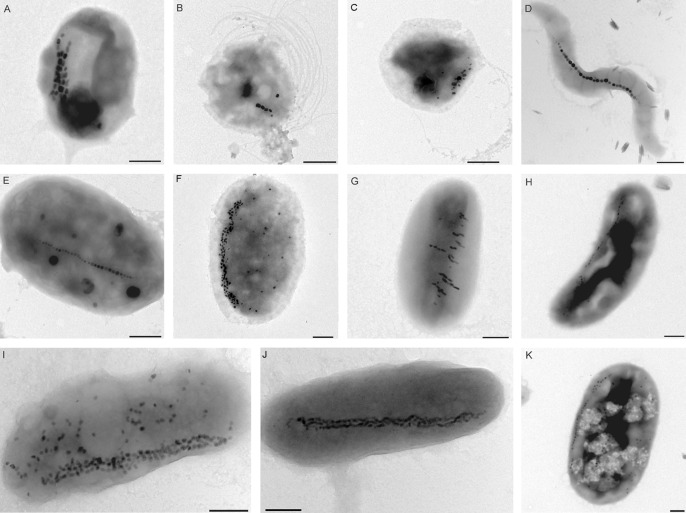
Morphologies of the enriched MTB cells are shown in Fig. 2. These MTB include cocci, spirilla, vibrios, and large rod-shaped bacteria. Within these MTB cells, different morphologies of magnetosome were found: for example, elongated prismatic magnetosomes in magnetotactic cocci (Fig. 2A to C), cuboidal magnetosomes in magnetotactic spirilla (Fig. 2D), and bullet-shaped and irregular magnetosomes in vibrioids and rod-shaped to oval MTB (Fig. 2G to K). The magnetosomes were arranged in single chains (Fig. 2D and E), multiple chains (Fig. 2A, F, and J), clusters of randomly oriented grains concentrated on one side of the cell (Fig. 2C), or multiple short chains parallel to the short axis of the cells (Fig. 2G).
Fig 2.
Representative TEM micrographs of the MTB cells collected from the freshwater Xi'an city moat. These MTB include cocci (A to C), spirilla (D), and vibrios (rod shaped to oval) (E to K). Bars = 500 nm.
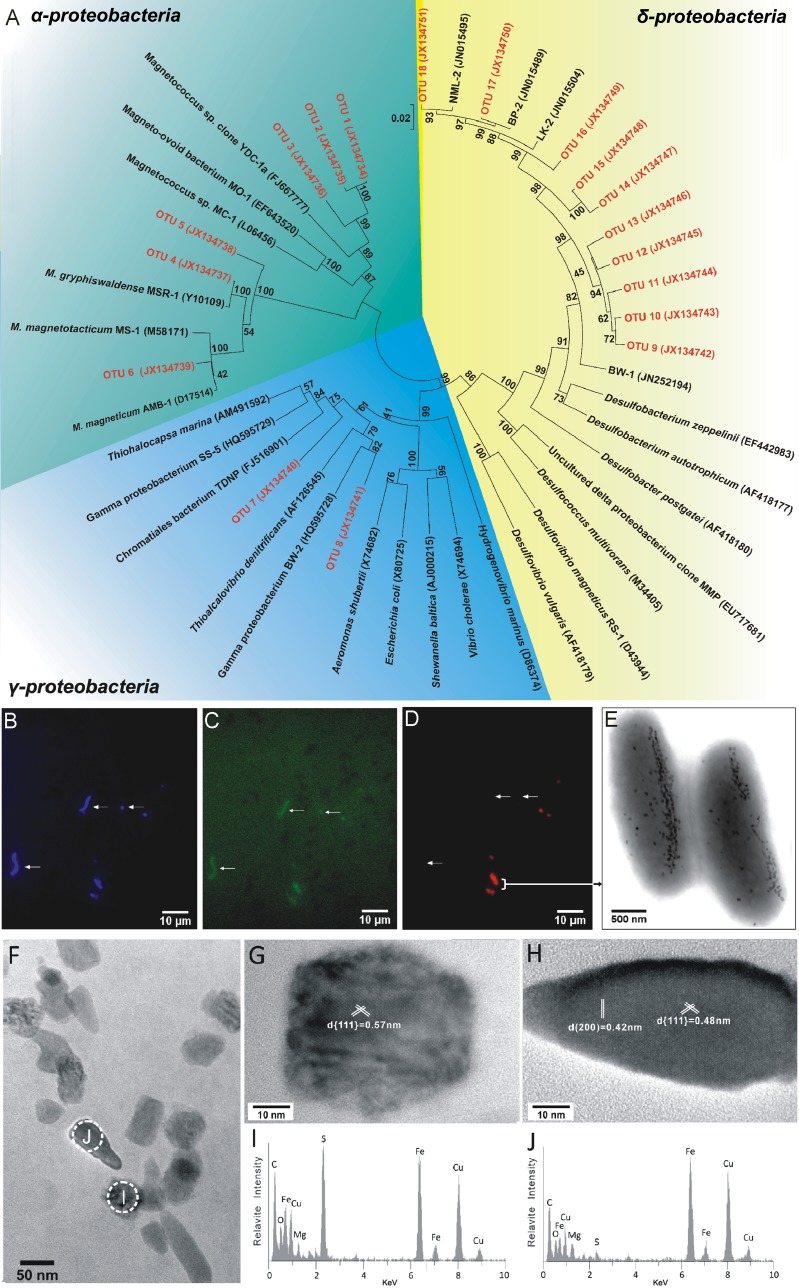
Phylogenetic analysis based on 16S rRNA gene sequences was performed to determine the community structure of MTB. All sequences (50 sequences) were composed of 18 operational taxonomic units (OTUs), which were defined at the 98% similarity level (Fig. 3A; also, see Table S2 in the supplemental material). Ten OTUs (OTUs 9 to 18) were identified as belonging to sulfate-reducing bacteria (SRB) within the Deltaproteobacteria, which accounted for more than 50% of all retrieved sequences. Although three OTUs (OTUs 16 to 18) were closely related to previously described MTB (3), sequences belonging to OTUs 9 to 15 were highly divergent from known MTB species (Fig. 3A) and might therefore represent novel branches. Furthermore, phylogenetic analysis has demonstrated that OTU 8 is 92% identical to the cultured Gammaproteobacteria MTB BW-2, belonging to the family Thiotrichales (18), whereas OTU 7, which has relatively low similarity (90%) to the other cultured strain, SS-5, was affiliated with the family Chromatiales. Both the families Thiotrichales and Chromatiales possess the ability to oxidize sulfur and are known as sulfur-oxidizing bacteria (SOB) (18, 19, 20). Additionally, six OTUs (OTUs 1 to 6) belong to the Alphaproteobacteria, three of which were most similar to three different known magnetotactic cocci, whereas the others had high similarities to the cultured strain Magnetospirillum gryphiswaldense MSR-1 (96% to 99%).
Fig 3.
(A) Phylogenetic tree of 16S rRNA gene sequences, constructed based on the neighbor-joining method. Bootstrap values at each node are based on 1,000 replicates. (B to D) The same microscopic field after staining with DAPI (B), after hybridization with 5′-6-carboxyfluorescein (FAM)-labeled bacterial universal probe EUB338 (C), and after hybridization with a 5′-Cy3-labeled probe SRB385Db (D). Arrows indicate MTB vibrios and MTB cocci as negative controls. (E) TEM images of magnetite and greigite magnetosome-mineralizing rod-shaped bacteria from the Xi'an city moat. (F) The rod-shaped bacteria synthesize both irregular rectangular and bullet-shaped magnetosomes. (G and H) HRTEM analyses of an irregular rectangular magnetosome and a bullet-shaped magnetosome, respectively. (I and J) EDX analyses of the irregular rectangular magnetosome and the bullet-shaped magnetosome shown in panels G and H, respectively. EDX analyses and HRTEM imaging on individual particles indicate that the mineral phase of irregular rectangular and bullet-shaped magnetosomes is greigite and magnetite, respectively.
Fluorescence in situ hybridization (FISH) analysis was performed to confirm that the Deltaproteobacteria 16S rRNA gene sequences originated from the MTB enrichment. Probe SRB385Db (21), specific for SRB in the Deltaproteobacteria, was found to match all the Deltaproteobacteria sequences retrieved in this study and was therefore selected for FISH analysis. The enriched MTB sample was stained with DAPI (4′,6′-diamidino-2-phenylindole) and hybridized with the universal bacterial probe EUB338 and the specific probe SRB385Db. As shown in Fig. 3B to D, large rod-shaped bacteria (2.5 to 5.7 μm in length) and small cocci (1 to 2 μm in diameter) robustly hybridized with the specific probe. TEM analysis of the rod-shaped bacteria revealed that they contain both bullet-shaped and irregular rectangular magnetosomes in the same cell (Fig. 3E and F). High-resolution transmission electron microscopy (HRTEM) imaging (Fig. 3G and H), fast Fourier transform (FFT) patterns (see Fig. S1 in the supplemental material), and energy-dispersive X-ray spectroscopy (EDX) analyses (Fig. 3I and J) indicate that the mineral phase of bullet-shaped magnetosomes was magnetite, while the irregular rectangular magnetosomes were greigite. MTB which exclusively produce either greigite or magnetite and which have the same cell shape and size were also observed in the same microcosm.
MTB cells simultaneously producing magnetite and greigite magnetosomes are of great interest for studies of microbiology, environmental magnetism, and biomineralization (3, 5, 22). These bacteria were first identified in the Pettaquamscutt River estuary in Rhode Island, a saline environment (5). Those authors discovered that these rod-shaped bacteria form bullet-shaped magnetite magnetosomes when they are found within the upper sediment layers, but when they are found in the deeper, hydrogen sulfide-rich layers, most rod-shaped bacteria synthesize greigite magnetosomes (23). The present study has shown that diverse, large rod-shaped MTB of the class Deltaproteobacteria that can produce either magnetite or greigite magnetosomes, or both, can be found in the surface sediments of the freshwater moat of Xi'an (Fig. 3). Microbial community analysis based on 16S rRNA genes and FISH in this study determined that these MTB belong to the sulfate-reducing Deltaproteobacteria, which have similarity to the pure cultured strain BW-1 (90% to 92%). Recently, Lefèvre and coworkers found that the mineralization of greigite and magnetite magnetosomes by BW-1 depends on the concentration of hydrogen sulfide (3). Two candidate gene clusters may control the biomineralization of greigite or magnetite magnetosomes (3). Further investigations, such as metagenomics or single-cell analysis, of these newly isolated Deltaproteobacteria MTB are needed in future to elucidate the detailed function of the magnetosome genes and their regulation networks.
It was interesting to find a high phylogenetic diversity of MTB, especially in the Deltaproteobacteria, in the Xi'an city moat (Fig. 3). These MTB mainly occupied the top layer (less than 2 cm) of the sediments, where chemical gradients were steep, as indicated by the concentrations of S2−, SO42−, NH4+, PO43−, and O2 (Fig. 1; also, see Table S1 in the supplemental material). The sampling site contained large amounts of nutrients and can be classified as a eutrophic environment (11). Therefore, a possible explanation for the highly diversified Deltaproteobacteria MTB in this sampling site could be that the high nutrient loading, steep vertical chemical gradient, and fast changes associated with sewage pollution provide diverse micro-ecological niches for different bacterial lineages and help to stimulate their growth, in contrast to what has been found in other studies of freshwater MTB (15, 24, 25). The availability of nutrients, and hence energy supply, and sharply vertical redox environments have been shown to be important drivers of microbial diversity (6, 26, 27, 28, 29). The distribution of Deltaproteobacteria MTB has been documented in saline environments (3, 4, 5, 7, 30) and freshwater niches (3, 9). Altogether, our results may suggest that the Deltaproteobacteria MTB, which include greigite-producing varieties, may widely exist in both saline and freshwater environments.
Nucleotide sequence accession numbers.
The sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers JX134734 to JX134751.
ACKNOWLEDGMENTS
We thank Haitao Chen, Qinyang Wang, and Liming Wang for help with field sampling. We also thank Greig A. Paterson for improving the English, Mo Huang and Wenfang Wu for useful discussions, and Xin'an Yang and Jingnan Liang for TEM analyses. We also thank the three anonymous referees for valuable comments on an earlier version of the paper.
This work was supported by the CAS/SAFEA International Partnership Program for Creative Research Teams (KZCX2-YW-T10), the CAS project, and NSFC grants 40821091 and 41104041.
Footnotes
Published ahead of print 1 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03635-12.
REFERENCES
- 1. Bazylinski DA, Frankel RB. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217–230 [DOI] [PubMed] [Google Scholar]
- 2. Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875–4898 [DOI] [PubMed] [Google Scholar]
- 3. Lefèvre CT, Menguy N, Abreu F, Lins U, Pósfai M, Prozorov T, Pignol D, Frankel RB, Bazylinski DA. 2011. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science 334:1720–1723 [DOI] [PubMed] [Google Scholar]
- 4. Wenter R, Wanner G, Schüler D, Overmann J. 2009. Ultrastructure, tactic behaviour and potential for sulfate reduction of a novel multicellular magnetotactic prokaryote from North Sea sediments. Environ. Microbiol. 11:1493–1505 [DOI] [PubMed] [Google Scholar]
- 5. Bazylinski DA, Heywood BR, Mann S, Frankel RB. 1993. Fe3O4 and Fe3S4 in a bacterium. Nature 366:218 [Google Scholar]
- 6. Simmons SL, Edwards KJ. 2007. Unexpected diversity in populations of the many-celled magnetotactic prokaryote. Environ. Microbiol. 9:206–215 [DOI] [PubMed] [Google Scholar]
- 7. Lins U, Keim CN, Evans FF, Farina M, Buseck PR. 2007. Magnetite (Fe3O4) and greigite (Fe3S4) crystals in multicellular magnetotactic prokaryotes. Geomicrobiol. J. 24:43–50 [Google Scholar]
- 8. Lefèvre CT, Frankel RB, Pósfai M, Prozorov T, Bazylinski DA. 2011. Isolation of obligately alkaliphilic magnetotactic bacteria from extremely alkaline environments. Environ. Microbiol. 13:2342–2350 [DOI] [PubMed] [Google Scholar]
- 9. Kawaguchi R, Burgess JG, Sakaguchi T, Takeyama H, Thornhill RH, Matsunaga T. 1995. Phylogenetic analysis of a novel sulfate-reducing magnetic bacterium, RS-1, demonstrates its membership of the δ-Proteobacteria. FEMS Microbiol. Lett. 126:277–282 [DOI] [PubMed] [Google Scholar]
- 10. Abreu F, Cantão ME, Nicolás MF, Barcellos FG, Morillo V, Almeida LGP, do Nascimento FF, Lefèvre CT, Bazylinski DA, de Vasconcelos ATR. 2011. Common ancestry of iron oxide-and iron-sulfide-based biomineralization in magnetotactic bacteria. ISME J. 5:1634–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Gao S, Bai K. 2011. Water quality analysis and ecological restoration scheme for moat of Xi'an City. J. Water Resour. Water Eng. 22:63–65 [Google Scholar]
- 12. Jogler C, Lin W, Meyerdierks A, Kube M, Katzmann E, Flies C, Pan Y, Amann R, Reinhardt R, Schüler D. 2009. Toward cloning of the magnetotactic metagenome: identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl. Environ. Microbiol. 75:3972–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin W, Wang Y, Li B, Pan Y. 2012. A biogeographic distribution of magnetotactic bacteria influenced by salinity. ISME J. 6:475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pan Y, Lin W, Tian L, Zhu R, Petersen N. 2009. Combined approaches for characterization of an uncultivated magnetotactic coccus from Lake Miyun near Beijing. Geomicrobiol. J. 26:313–320 [Google Scholar]
- 15. Flies CB, Jonkers HM, Beer D, Bosselmann K, Böttcher ME, Schüler D. 2005. Diversity and vertical distribution of magnetotactic bacteria along chemical gradients in freshwater microcosms. FEMS Microbiol. Ecol. 52:185–195 [DOI] [PubMed] [Google Scholar]
- 16. Jogler C, Niebler M, Lin W, Kube M, Wanner G, Kolinko S, Stief P, Beck A, De Beer D, Petersen N. 2010. Cultivation-independent characterization of ‘Candidatus Magnetobacterium bavaricum’ via ultrastructural, geochemical, ecological and metagenomic methods. Environ. Microbiol. 12:2466–2478 [DOI] [PubMed] [Google Scholar]
- 17. Simmons SL, Sievert SM, Frankel RB, Bazylinski DA, Edwards KJ. 2004. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl. Environ. Microbiol. 70:6230–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lefèvre CT, Viloria N, Schmidt ML, Pósfai M, Frankel RB, Bazylinski DA. 2012. Novel magnetite-producing magnetotactic bacteria belonging to the Gammaproteobacteria. ISME J. 6:440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schauer R, Røy H, Augustin N, Gennerich HH, Peters M, Wenzhoefer F, Amann R, Meyerdierks A. 2011. Bacterial sulfur cycling shapes microbial communities in surface sediments of an ultramafic hydrothermal vent field. Environ. Microbiol. 13:2633–2648 [DOI] [PubMed] [Google Scholar]
- 20. Sylvan JB, Toner BM, Edwards KJ. 2012. Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio 3:e00279-12 doi:10.1128/mBio.00279-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rabus R, Fukui M, Wilkes H, Widdle F. 1996. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl. Environ. Microbiol. 62:3605–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pósfai M, Buseck PR, Bazylinski DA, Frankel RB. 1998. Iron sulfides from magnetotactic bacteria; structure, composition, and phase transitions. Am. Mineral. 83:1469–1481 [Google Scholar]
- 23. Bazylinski DA, Frankel RB, Heywood BR, Mann S, King JW, Donaghay PL, Hanson AK. 1995. Controlled biomineralization of magnetite (Fe3O4) and greigite (Fe3S4) in a magnetotactic bacterium. Appl. Environ. Microbiol. 61:3232–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin W, Li J, Schüler D, Jogler C, Pan Y. 2009. Diversity analysis of magnetotactic bacteria in Lake Miyun, northern China, by restriction fragment length polymorphism. Syst. Appl. Microbiol. 32:342–350 [DOI] [PubMed] [Google Scholar]
- 25. Lin W, Pan Y. 2010. Temporal variation of magnetotactic bacterial communities in two freshwater sediment microcosms. FEMS Microbiol. Lett. 302:85–92 [DOI] [PubMed] [Google Scholar]
- 26. Jørgensen BB. 1982. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 296:643–645 [Google Scholar]
- 27. Zhou J, Xia B, Treves DS, Wu LY, Marsh TL, O'Neill RV, Palumbo AV, Tiedje JM. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mußmann M, Ishii K, Rabus R, Amann R. 2005. Diversity and vertical distribution of cultured and uncultured Deltaproteobacteria in an intertidal mud flat of the Wadden Sea. Environ. Microbiol. 7:405–418 [DOI] [PubMed] [Google Scholar]
- 29. Bienhold C, Boetius A, Ramette A. 2012. The energy-diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 6:724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou K, Zhang W, Yu-Zhang K, Pan H, Zhang S, Zhang W, Yue H, Li Y, Xiao T, Wu L. 2012. A novel genus of multicellular magnetotactic prokaryotes from the Yellow Sea. Environ. Microbiol. 14:405–413 [DOI] [PubMed] [Google Scholar]