Abstract
Marine ecosystems are significant sources of the powerful greenhouse gas nitrous oxide (N2O). A by-product of nitrification and an intermediate in the denitrification pathway, N2O is formed primarily in oxygen-deficient waters and sediments. We describe the isolation of a group of alphaproteobacteria from the suboxic waters of the Arabian Sea that are phylogenetically affiliated with Labrenzia spp. and other denitrifiers. Quantitative PCR assays revealed that these organisms were very broadly distributed in this semienclosed ocean basin. Their biogeographical range extended from the productive, upwelling region off the Omani shelf to the clear, oligotrophic waters that are found much further south and also included the mesotrophic waters overlying the oxygen minimum zone (OMZ) in the northeastern sector of the Arabian Sea. These organisms actively expressed NosZ (N2O reductase, the terminal step in the denitrification pathway) within the OMZ, an established region of pelagic denitrification. They were found in greatest numbers outside the OMZ, however, and nosZ mRNAs were also readily detected near the base of the upper mixed layer in nutrient-poor, oxic regions. Our findings provide firm molecular evidence of a potential sink for N2O within well-ventilated, oceanic surface waters in this biogeochemically important region. We show that the Labrenzia-like denitrifiers and their close relatives are habitual colonizers of the pseudobenthic environment provided by Trichodesmium spp. We develop the conjecture that the O2-depleted microzones that occur within the colonies of these filamentous, diazotrophic cyanobacteria might provide unexpected niches for the reduction of nitrogen oxides in tropical and subtropical surface waters.
INTRODUCTION
Emissions of the greenhouse gas nitrous oxide (N2O) have increased steadily since the early part of the 19th century. Atmospheric N2O concentrations are higher today than at any time during the past 650,000 years (1, 2). Apart from its significant warming potential (∼300-fold that of CO2 over a 100-year period), rising N2O is of further environmental concern because it is presently the single most destructive source of emissions contributing to stratospheric ozone depletion (3). The growing inventory of atmospheric N2O has occurred primarily as a result of an increase in emissions from the terrestrial environment owing to changes in agricultural practices, the combustion of fossil fuels, and other anthropogenically driven perturbations of the nitrogen cycle (2). The marine environment is also an important net source of N2O to the atmosphere, however, and the unperturbed (nonanthropogenic) rates of emissions from coastal margins, shelf, and open waters are of the same order as those from land (1, 4, 5).
Significant feedbacks on marine N2O emissions are anticipated over the coming decades as a result of increasing ocean acidification (5) and the expansion of hypoxic waters owing to surface warming and an acceleration in the prevailing rates of eutrophication from anthropogenic nutrient loading (1, 6). In oxic waters, N2O is produced primarily as a by-product of nitrification, the oxidation of ammonium (NH4+) to nitrite (NO2−) and nitrate (NO3−) carried out by ammonium-oxidizing bacteria (AOB) and archaea (AOA). Recent work has shown that ammonium oxidation rates (and hence N2O production by AOB/AOA) are exquisitely sensitive to comparatively modest (∼0.1-unit) declines in pH (5) and show a mean reduction of ∼20% in response to near-future (20 to 30 years hence) ocean acidification. The pH of oceanic surface waters is set to decline by 0.3 to 0.4 units by the end of the present century (7) with the potential corollary of large negative feedbacks on marine nitrification rates and N2O emissions in the coming decades (5).
The production of N2O by nitrifiers is oxygen sensitive, however, and its emissions increase very substantially under hypoxic conditions (8). Over the past 50 years, the areal extent of hypoxic waters in coastal regions and at intermediate depths in the North Pacific and tropical oceans has expanded and shoaled significantly (9, 10), a trend that will only intensify as the global ocean continues to warm and lose oxygen over the next few decades and beyond (11). Approximately 10% of the contemporary ocean is either hypoxic (O2, <30% saturation) or suboxic (O2, <1% saturation), and even a modest expansion in the present volume of deoxygenated waters is likely to increase N2O production by nitrifiers significantly (1, 6). The net impacts of increasing atmospheric CO2 on marine N2O emissions in the years to come, therefore, will depend on the overall balance of both positive and negative feedbacks on ammonium oxidation rates and other climate-sensitive sources of N2O.
The predominant source of N2O emissions under suboxic conditions is not from ammonium oxidizers but from a taxonomically diverse group of mainly heterotrophic microbes known as denitrifiers (6). Denitrifiers use NO3− as an alternative respiratory electron acceptor to oxidize an electron donor (usually organic carbon) to generate metabolic energy during anaerobic growth. Denitrification leads ultimately to the liberation of dinitrogen gas (N2) in a four-step reductive pathway from NO3− that proceeds via NO2− and two obligate gaseous intermediates, nitric oxide (NO) and N2O. Not all denitrifiers are capable of the final reduction of N2O to N2, however, while for others a sustained lag occurs during the induction of the necessary cellular machinery to carry out this terminal step in the process (12). Some denitrifiers produce N2 only in the complete absence of free oxygen but, nonetheless, are capable of partial denitrification and liberate N2O under hypoxic conditions (13). Denitrification is both a sink and a potential source of emissions, therefore, and another key process in the marine nitrogen cycle known to be sensitive to the future expansion in the volume of deoxygenated waters in the ocean (1, 6).
The only known metabolic pathway for the consumption of N2O is that which requires the copper-containing enzyme nitrous oxide reductase (NosZ). NosZ is found in all denitrifiers that are capable of reducing NO3− as far as N2 and also in a few nondenitrifying bacteria, such as Wolinella (Vibrio) succinogenes, that can use N2O as a terminal electron acceptor (14). The latter organisms are not themselves N2O producers, however, because they lack the NO-producing nitrite reductase (NirK or NirS) that is the definitive biochemical feature of denitrifiers (15), the primary sink for this nitrogen oxide in the oceans (16).
Uncertainties over the future scale and climatic impact of marine N2O emissions and, in particular, their sensitivities to acidification and deoxygenation (17) have led to calls for a better understanding of the biological sources and sinks of this trace gas in the oceans (1, 18). Key to constraining the marine N2O budget is the development of a robust model of the distributions and activities of the organisms that produce and/or consume N2O and their responses to stress induced by global environmental change. Approximately half of the annual emissions of N2O in the contemporary ocean come from the three major oxygen minimum zones (OMZs) that are located in the eastern tropical North Pacific (ETNP), the eastern tropical South Pacific (ETSP), and the Arabian Sea (19, 20). The net contributions of different biological sources of N2O in these regions are debated (see reference 18), but there is general agreement that the OMZs are “hot spots” within the ocean that are especially vulnerable to warming and deoxygenation over the coming decades (1, 17).
In the present study, we describe the isolation into culture of a novel group of pelagic denitrifying alphaproteobacteria from the suboxic waters of the Arabian Sea, one of the most intense regions of N2O production in the world ocean (20). Employing a quantitative PCR protocol targeting the nosZ gene from these organisms, we show that they have a broad biogeographical distribution in this ocean basin, ranging from the upwelling region along the Omani shelf to the highly oligotrophic equatorial waters to the south. We further show that these organisms were expressing the cognate mRNA for nosZ in the suboxic intermediate waters of the OMZ located in the northeastern sector of the Arabian Sea and, quite unexpectedly, were also active at shallower depths within the upper mixed layer of waters well outside the region of the OMZ.
MATERIALS AND METHODS
Study site, sample collection, and nucleic acid purification.
Observations were made in September 2001 aboard RRS Charles Darwin during the Natural Environment Research Council (NERC)-funded AMBITION cruise (CD132) in the Arabian Sea. Eleven stations were occupied along the length of a 5,150-km transect between Victoria, Seychelles, and Muscat, Oman (21). Hydrographical data were collected at each station with a Sea-Bird 911plus conductivity-temperature-depth (CTD) profiler (Sea-Bird Electronics, Inc., Bellevue, WA) configured with a Chelsea Aquatracker III fluorometer (Chelsea Instruments, West Molesey, United Kingdom) to measure chlorophyll fluorescence and auxiliary sensors for dissolved oxygen and photosynthetically active radiation (PAR).
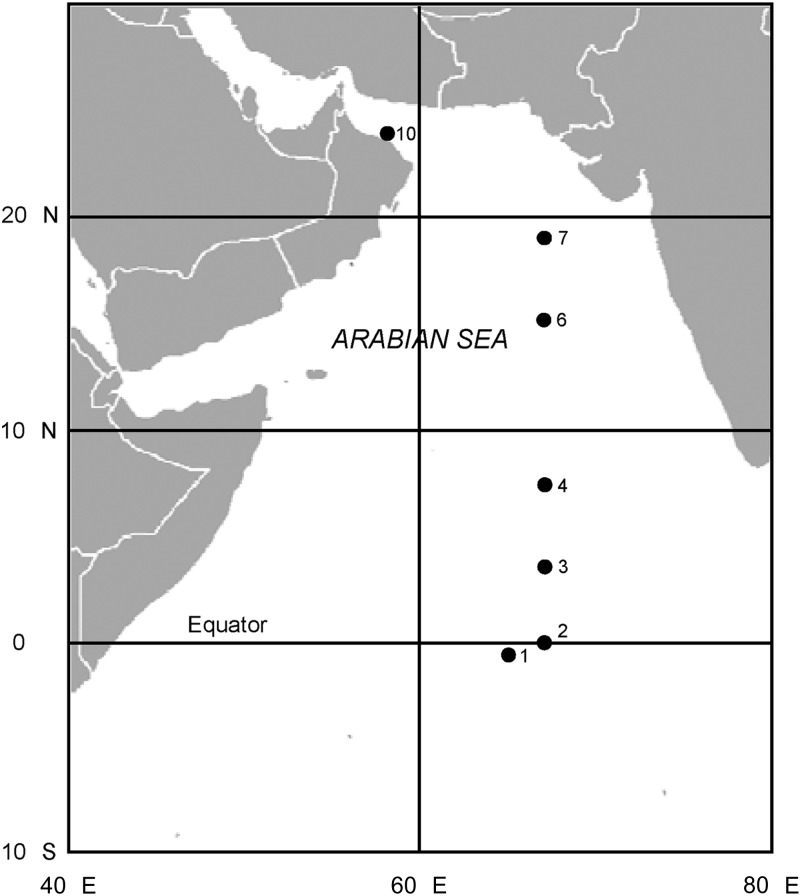
Seawater samples were collected from discrete depths at selected stations (Fig. 1) with a rosette of 24 30-liter volume Niskin bottles mounted on the CTD profiler. Plankton samples were obtained by filtering 4 to 5 liters of seawater (unscreened) through 90-mm-diameter, 0.2-μm-pore-size polycarbonate filters (Osmonics Inc., Minnetonka, MN) at a negative pressure of <20 mm Hg. Cell material collected on the filters was taken up in DNA isolation buffer (250 mM NaCl, 100 mM EGTA, 100 mM Tris-HCl [pH 8.0], and 1% [wt/vol] lithium dodecyl sulfate) and stored frozen at −70°C or, for RNA samples, preserved in RNAlater (Applied Biosystems, Warrington, United Kingdom) and refrigerated at 4°C. At the end of the cruise, the preserved nucleic acid samples were shipped by air to the United Kingdom on dry ice and subsequently stored at −80°C prior to the extraction and purification of DNA and RNA as described by Bird et al. (21).
Fig 1.
Study area in the Arabian Sea showing the locations and designated station numbers of the sites sampled during the south-to-north AMBITION cruise transect in September 2001.
Plankton net hauls for the collection of Trichodesmium colonies and extraction of DNA.
Plankton samples were also collected from depths of 5 or 10 m at each station during short (10- to 15-min) horizontal hauls using a standard WP2 conical net (200-μm mesh size) towed at a ship speed (through the water) of 1 knot. Trichodesmium colonies were sorted into sterile, filtered (0.2-μm-pore-size polycarbonate membranes) surface seawater using a sterile, disposable inoculating loop, rinsed in two changes of sterile seawater, and transferred to 0.5 ml RNAlater prior to storage at 4°C. DNA was extracted from the sorted Trichodesmium colonies (∼5 colonies per extraction) after rinsing twice in sterile artificial seawater (ASW) medium (22) to remove RNAlater. The washed colonies were collected on 0.2-μm-pore-size polycarbonate membranes and then lysed in TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) amended to 0.1% Triton X-100 and 0.2 mg ml−1 lysozyme (Sigma catalog no. L7651) at 37°C for 15 min. The lysates were brought to 50 μg ml−1 proteinase K (Roche) and 0.2% SDS and incubated at 56°C for a further 10 min before purification of DNA using the Qiagen DNeasy kit according to the supplier's abridged protocol for the purification of DNA lysates.
Enrichment culture for the isolation of nitrate-respiring (denitrifying) bacteria from the Arabian Sea.
Seawater was collected from a depth of 120 m in suboxic waters at station 10 (24°19′N, 58°10′E; depth, 2,863 m) (Fig. 1) and amended with 1.5 g liter−1 tryptic soy broth (Thermo Fisher Scientific, Basingstoke, United Kingdom). The amended seawater was further supplemented with 1× Guillard's (F/2) nutrient and vitamin solution (Sigma, Poole, United Kingdom) and incubated in filled, airtight, sterile bottles at 25°C. Following the visible establishment of bacterial growth after 3 to 5 days, subcultures were transferred aseptically to degassed (boiled), filter-sterilized seawater from the same station amended with 0.5 g liter−1 NaNO3, 1.0 g liter−1 NH4Cl, 0.006 g liter−1 FeCl2, 0.001 g liter−1 EDTA, 0.003 g liter−1 K2HPO4, and 1 ml liter−1 A5 trace metal mixture (23) and 1 g liter−1 sodium acetate. The subcultures were incubated in filled, airtight, sterile bottles at 25°C until the medium turbidity increased (7 to 14 days). After two further rounds of subculture, the bacterial suspensions were transferred to streak plates of the same medium solidified with 10 g liter−1 Difco Bacto agar (Becton, Dickinson and Co., Oxford, United Kingdom). The inoculated plates were incubated at 25°C in sealed, anaerobic jars in an oxygen-free nitrogen atmosphere. Individual colonies were isolated by repeated subculture on agar streak plates and maintained thereafter at 25°C under these same incubation conditions.
PCR amplification of nosZ, nirS, and 16S rRNA genes from bacteria isolated from the Arabian Sea.
Independent isolates of putative, nitrate-respiring bacteria were grown aerobically at 25°C for 24 to 48 h in 3 ml ASW medium (22) amended with 10% (vol/vol) Luria-Bertani broth (10 g liter−1 tryptone, 5 g liter−1 yeast extract, 10 g liter−1 NaCl) in an orbital incubator at 180 rpm. The cultures were harvested by centrifugation at 16,000 × g for 2 min, and DNA was isolated from the pelleted cell material using a DNeasy tissue kit according to the supplier's (Qiagen Ltd., Crawley, United Kingdom) recommended protocol for bacteria. The DNA samples were screened for the presence of nosZ by PCR using Thermoprime DNA polymerase master mix (Fisher Scientific, Loughborough, United Kingdom) in reaction volumes of 25 μl containing 2 mM MgCl2, 10 ng DNA, and 50 pmol each of the primers nosZF1 and nosZR (Table 1).
Table 1.
Oligonucleotide primers used in this study
| Oligonucleotide | DNA sequence (5′–3′) | Product size (bp) | Reference |
|---|---|---|---|
| nosZF1 | GAYGTNCANTAYCARCCNGGNCA | 612 | This study |
| nosZR | CATYTCCAKRTGNADNGCRTGRCA | 612 | This study |
| StanosF | GCTGAAACCGGAGAATGAAC | 252 | This study |
| StanosR2 | TGCATGTAGACACGCACCTT | 252 | This study |
| QnosF | GAAGTTCACGGTAAAGCAGG | 98 | This study |
| QnosR | CATAGTTGGACAGGCAGAAG | 98 | This study |
| Probe | ACGAAGTCACCATCTACGTCACCAACA | 98 | This study |
| nirS1F | CCTAYTGGCCGCCRCART | ∼890 | 24 |
| nirS6R | CGTTGARCTTRCCGGT | ∼890 | 24 |
The degenerate primers nosZF1 and nosZR were designed to target the conserved motifs DV(H/Q)YQPGH and CHA(M/I/L)H(L/M)EM identified in the majority of complete NosZ sequences available from the GenBank database at the start of this study and correspond to nucleotide positions 1276 to 1298 and 1864 to 1887 of nosZ from Pseudomonas stutzeri (ZoBell strain) ATCC 14405 (GenBank accession no. CAA37714.2), respectively. After denaturation at 95°C for 2 min, the cycling conditions were 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min for 25 cycles followed by a final extension at 72°C for 10 min.
The PCR products obtained were resolved in 1% (wt/vol) agarose gels and purified using the Wizard SV gel and PCR cleanup system (Promega Ltd., Southampton, United Kingdom) before TA cloning in the plasmid vector pCR-TOPO as recommended by the supplier (Invitrogen, Paisley, United Kingdom). The cloned PCR products from four independent isolates (designated 1N, 4N, 5N, and 8N) were DNA sequenced on both strands using M13 forward and reverse primers in parallel reactions performed with a DYEnamic ET terminator cycle sequencing kit (GE Healthcare Life Sciences, Little Chalfont, United Kingdom) and an ABI Prism 377 automated sequencer.
An ∼890-bp fragment of the gene encoding NirS (nitrite reductase) was amplified from genomic DNA purified from the Arabian Sea isolate designated 4N using the primer pair nirS1F and nirS6R (24) (Table 1). The reactions were carried out as described above for nosZ except that the annealing temperature was increased to 58°C. Genomic DNA from this isolate was also used to amplify a fragment of the 16S rRNA gene with the universal primer pair 27F and 1492R (25) under similar reaction conditions but with a lower annealing temperature of 48°C and an extension step of 90 s. The nirS and 16S rRNA PCR products obtained were gel purified, TA cloned, and sequenced either in-house (16S rRNA) as described for nosZ above or, in the case of nirS, by a commercial provider (Source BioScience Lifesciences, Nottingham, United Kingdom) using M13 primers.
Amplification of nosZ from Trichodesmium consortia by the PCR.
Aliquots of DNA (1 μl from 100 μl of extract) isolated from Trichodesmium colonies collected at three stations (stations 2, 3, and 4 [Fig. 1]) were interrogated for the presence of nosZ closely related to that of the Arabian Sea alphaproteobacteria described in this study (including the isolate designated 4N) using a targeted PCR protocol. Each 25-μl reaction volume contained 1× Thermoprime DNA polymerase master mix (Thermo Fisher Scientific, Loughborough, United Kingdom) amended to 2 mM MgCl2, 0.1 mg ml−1 bovine serum albumin (Promega Corp., Southampton, United Kingdom), and 50 pmol each of the primers StanosF and StanosR2 (Table 1). The primers target a 252-bp internal region of nosZ corresponding to nucleotides 97 to 348 of the trimmed sequence of the Arabian Sea isolate 4N (GenBank accession no. JN850958). The PCR cycling conditions consisted of an initial denaturation at 95°C for 2 min followed by 30 cycles at 94°C for 30 s, 52°C for 30 s, and 72°C for 45 s and a final extension at 72°C for 10 min.
The PCR products were resolved by electrophoresis through 1.5% (wt/vol) NuSieve 3:1 agarose gels (Lonza, Stratech Scientific, Newmarket, United Kingdom), and a fragment of the expected size from the station 3 sample was excised from the gel, purified, and TA cloned as described above. The inserts from 10 of the resulting recombinant clones were screened by digestion with the endonuclease SphI (which has a predicted recognition sequence centered at nucleotide position 105 within the nosZ fragment targeted by the primers), and all 10 clones were found to produce digestion products of the predicted size when resolved on 2.0% (wt/vol) NuSieve 3:1 agarose gels. One of these clones was sequenced on both strands by a commercial provider (Source BioScience Lifesciences, Nottingham, United Kingdom) to confirm its identity as described above.
To examine the wider distribution of nosZ in Trichodesmium consortia, DNA samples purified from colonies collected in surface waters near the Bahama Islands in 1991 and from a cultured isolate (strain IMS101) were also analyzed by PCR, and the products obtained were cloned and sequenced as described above.
Detection and enumeration of nosZ DNA in natural samples of plankton from the Arabian Sea.
The depth and latitudinal distribution of bacteria in the Arabian Sea that harbored a nosZ gene identical to or highly similar in sequence to that amplified from the cultured isolates were investigated by quantitative PCR (QPCR). DNA samples obtained from four stations (designated 1, 4, 6, and 10 [Fig. 1]) occupied during the AMBITION cruise were assayed using a double dye-labeled oligonucleotide probe and the primer pair QnosF and QnosR (Table 1). The primers amplify a product of 98 bp and bind at nucleotide positions 376 to 395 and 454 to 473, respectively, within the original (trimmed) nosZ PCR products amplified from the cultured isolates. The probe, which was coupled with fluorescein at the 5′ end and Elipse dark quencher at the 3′ end (Eurofins MWG Operon, Ebersberg, Germany), binds nucleotide positions 399 to 425 bp starting 4 bp downstream of the 3′ end of the forward primer QnosF.
QPCR assays were performed with a Stratagene Mx3000p thermocycler and Brilliant II reagents (Agilent Technologies Ltd., Wokingham, United Kingdom) in 25-μl volumes containing 1.0 μl of DNA, 25 pmol of each primer, and a 150 nM concentration of probe DNA. Following activation at 95°C for 10 min, reaction mixtures were cycled at 95°C for 30 s, 60°C for 30 s, and 72°C for 45 s for 40 cycles and data were collected at the end of the extension step. The threshold cycle (CT) was determined automatically by the instrument software, and the initial template quantity (nosZ copies) was calculated with reference to a standard curve constructed using a 10-fold dilution series of linear fragments of the cloned nosZ amplicon from the isolate 4N.
Standard DNA was amplified from the original plasmid clone (pnos4N) using the primer pair nosZF1 and nosZR as described above, and the concentration of the gel-purified 612-bp product was determined using a Picodrop 100 microspectrophotometer (Cambridge Bioscience, Cambridge, United Kingdom). Copy number was estimated from the DNA concentration of the standard by assuming that 1 bp has an average molecular mass of 650 Da and using the following expression: copy number = (N × 6.022 × 1023)/(L × 1 × 109 × 650), where N is the DNA amount in ng and L is the length of the DNA fragment.
The lower detection limit of nosZ standard DNA was routinely between 10 and 100 copies or better, and PCR efficiency was 98.3% (r2 = 0.998). The absence of PCR inhibitors in the natural sample DNA preparations was verified by spiking parallel reaction mixtures with 105 copies of the target DNA fragment. Those samples in which the CT of the spiked controls was >1.5 cycles greater than that predicted were excluded from further analysis.
Quantitative reverse transcriptase PCR (QRT-PCR).
RNA samples (0.5 μg) were treated with Ambion Turbo DNase (Applied Biosystems, Warrington, United Kingdom) and reverse transcribed using a Quantitect reverse transcriptase (RT) kit (Qiagen Ltd., Crawley, United Kingdom) and random hexamer primers following treatment with Genomic Wipeout reagent as recommended by the suppliers. cDNAs originating from nosZ mRNA were quantified by QPCR as described above using 1-in-50 dilutions of the RT reaction mixtures and run in parallel with minus-RT controls and no-template blanks. PCR efficiency was 97.7% (r2 = 0.999).
To verify the specificity of the assays, the RT-PCR products amplified from the 50-m depth sample from station 2 were resolved through a 2.5% (wt/vol) NuSieve 3:1 agarose gel and the 98-bp product was purified using the Wizard SV gel and PCR cleanup system. The purified product was screened with the endonucleases EcoRV and NcoI (which have recognition sequences centered at nucleotide positions 58 and 75, respectively, within the 98-bp amplicon) and found to produce digestion products of the expected sizes. The RT-PCR amplicons were TA cloned as described above, and three randomly selected clones were DNA sequenced. The cloned inserts were found to contain the internal region of nosZ targeted by the QPCR primers and to share 98 to 100% identity with nosZ from the Arabian Sea isolate 4N.
Experiments (n = 3) were also conducted to examine the effect of pO2 on nosZ mRNA abundance in cultures of the Arabian Sea isolate 4N grown at 25°C in the acetate-containing medium described above under aerobic conditions (O2 saturation, ≥95%) or after transfer to a suboxic (O2 saturation, ≤1%) atmosphere for 30 min prior to sampling. RNA was extracted, reverse transcribed, and assayed for nosZ by QPCR as described for the natural samples, and the products were run out on 2.5% (wt/vol) NuSieve 3:1 agarose gels. Dissolved oxygen concentrations were determined using a calibrated oxygen electrode (Hansatech Instruments Ltd., Kings Lynn, United Kingdom).
Phylogenetic reconstruction.
The DNA sequences of nosZ and nirS from the Arabian Sea isolate 4N were trimmed of primer sequences and translated in silico. Peptide sequences closely related to each target gene were identified using the Basic Local Alignment Search Tool (BLAST) at NCBI (26) and aligned in MUSCLE with those from the isolate 4N using the unweighted-pair group method using arithmetic (UPGMA) clustering method (27). Evolutionary analyses were performed in MEGA5 (28) using the maximum likelihood method based on the Dayhoff matrix model. The bootstrap consensus trees produced in each case were based on 500 replicate samples.
Nucleotide sequence accession numbers.
The DNA sequences of nirS, nosZ, and the 16S rRNA gene from the Arabian Sea isolate designated 4N have been deposited in GenBank under the accession numbers JX827176 (nirS), JN850958 (nosZ), and JN850957 (16S rRNA). The DNA sequence of the nosZ fragment amplified from Trichodesmium colonies collected at station 3 is deposited in GenBank under accession number JX827177, and that from the cultured isolate strain IMS101 is deposited under accession number KC205088.
RESULTS AND DISCUSSION
Isolation of putative denitrifying bacteria from the Arabian Sea and characterization of the representative isolate, strain 4N.
Liquid enrichment cultures of bacteria from the Arabian Sea that were capable of utilizing acetate as their sole carbon source and nitrate as an alternative electron acceptor were successfully established under suboxic growth conditions. This enrichment culture medium was designed to select for the growth of nitrate-respiring bacteria (and denitrifiers, in particular). With the notable exception of methanogens (29), acetate is a nonfermentable carbon substrate for other bacteria. After transfer to a solid medium under an oxygen-free atmosphere, the growth of individual colonies of putative, nitrate-respiring (denitrifying) bacteria was observed after 7 to 10 days. Four of these were selected for further experimentation and brought into axenic, clonal culture using the same medium and growth conditions.
Following their establishment, all isolates produced very similarly sized, round, smooth, shiny colonies that developed a pink/reddish-brown coloration at their center as they matured. All of the isolates were found to be Gram negative and displayed an elevated requirement for Na+ and Cl− but not for either Mg2+ or Ca2+, i.e., the isolates were halophytic rather than obligately marine (30). In broth culture, the isolates grew primarily as individual, actively motile cells that produced gas (nitrogen) when grown under anaerobic conditions in the acetate-nitrate enrichment medium described above. The ability of the isolate designated 4N to denitrify was subsequently confirmed by the acetylene block technique (15) using the same medium and incubation conditions.
PCR primers were designed to screen DNA extracted from all four isolates for the presence of nosZ (and nirS, see below). The forward nosZF1 primer targets a motif within region H4 of nitrous oxide reductase that includes a highly conserved histidine residue, while the reverse primer, nosZR, targets a region close to the C terminus that includes several ligands for the CuA center of the enzyme (31) and overlaps the coding region targeted by the nondegenerate primer Nos2230R designed by Scala and Kerkhof (32). The primers should amplify DNA from the majority of nosZ sequences deposited in the GenBank database to date, but they are not completely universal. In particular, they are unlikely to recognize nosZ from those bacteria in which a serine rather than a histidine codon occurs adjacent to the 3′ end of the reverse primer target. This substitution occurs most frequently in a subset of NosZ proteins that carry a C-terminal extension of about 200 residues and which also show variability in the conserved motif targeted by the forward primer (33, 34). In this instance, however, a PCR product of the expected size (∼612 bp) was amplified from DNA extracted from all four axenic Arabian Sea isolates screened.
The DNA sequences of the cloned amplicons from all four isolates were virtually identical (>99%), and a BLASTN search of the GenBank database confirmed that they contained the specific region of nosZ targeted by the primers. The most closely related DNA sequence (∼95% nucleotide identity) to that from all four Arabian Sea clones was from the cultured isolate Labrenzia aggregata IAM 12614, a rosette-forming, aerobic, denitrifying, marine heterotroph isolated from a sediment sample from the western Baltic Sea (35). All of the nucleotide differences (29/565 bases) that occur between nosZ in this species and that from the isolate designated 4N (which we studied in the most detail) were located in the third codon position, however (data not shown), and their derived peptide sequences were identical.
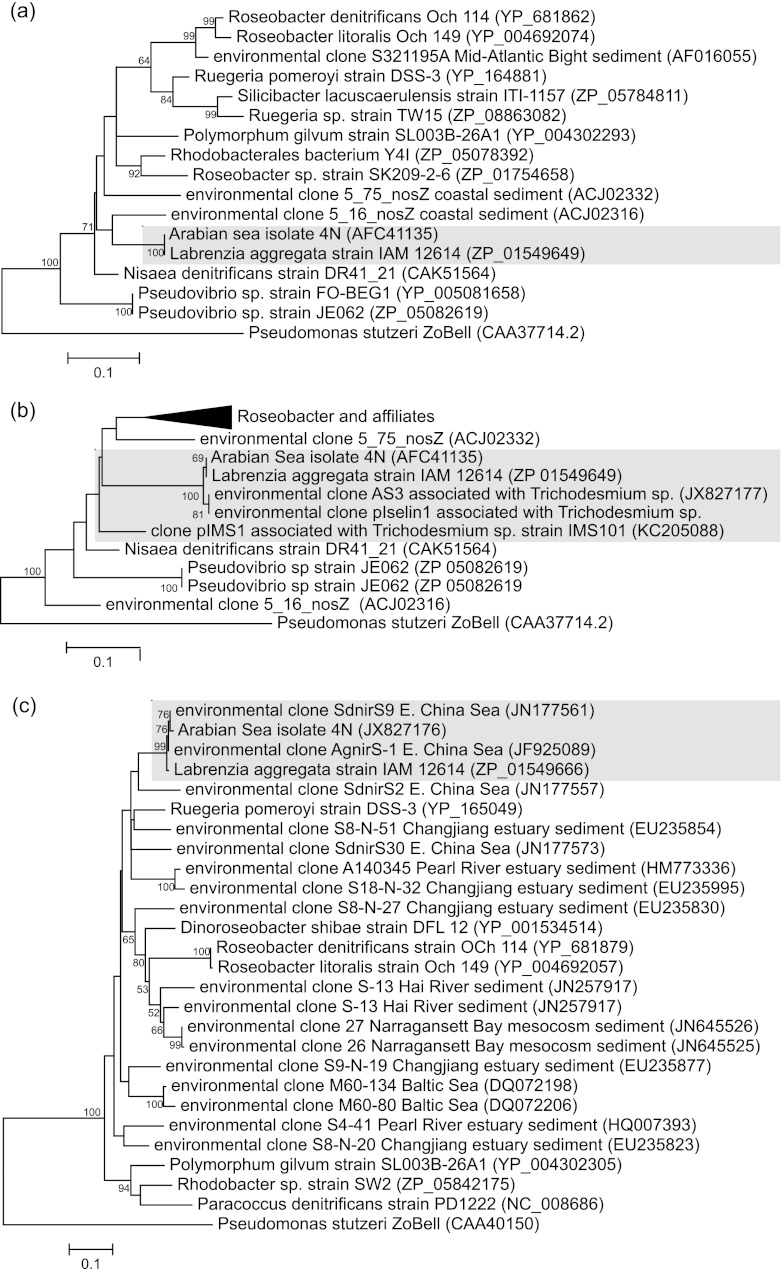
Besides L. aggregata IAM 12614, phylogenetic analysis of the peptide sequence from the Arabian Sea isolate 4N showed that it also clustered with NosZ from several other known marine, denitrifying alphaproteobacteria (Fig. 2a). These included Nisaea denitrificans, originally isolated from the Mediterranean Sea (36), and also Ruegeria pomeroyi, Roseobacter sp. strain SK209-2-6, and Roseobacter denitrificans among members of the widely distributed Roseobacter clade (37, 38). The two most closely related environmental sequences (clones 5_16 and 5_75) originated from the same coastal sediment in the East China Sea (Fig. 2a) and shared a similar level of amino acid identity (between 86 and 90%) with L. aggregata IAM 12614 as they did with Polymorphum gilvum, a hydrocarbon-utilizing halophile that shares significant genome synteny with members of the genus Labrenzia (39).
Fig 2.
Phylograms for nosZ (a and b) and nirS (c) based on partial peptide sequences (188, 83, and 283 residues, respectively) from the Arabian Sea isolate 4N and closely related alphaproteobacterial genes. The maximum likelihood trees were inferred from 500 bootstrap replicates in MEGA5 (28) and rooted with the appropriate orthologue from the ZoBell strain of Pseudomonas stutzeri (ATCC 14405). Those partitions that received greater than 50% support in the bootstrap are indicated at their corresponding nodes. The GenBank accession numbers of the sequences are indicated after the sequence names, and the distance scale is shown to the bottom left of each phylogram. The shaded regions highlight the clusters within which genes from the Arabian Sea isolate 4N are found in each phylogram. Note that the highlighted sequence designated pIselin1 in panel b was obtained from Trichodesmium thiebautii DNA collected in 1991 from near-surface waters in the Bahama Islands aboard RV Columbus Iselin during cruise 91-11 (84) and was identical to that of clone AS3 obtained from station 3 in the Arabian Sea (GenBank accession number JX827177). The template used to amplify the nosZ clone pIMS1 from Trichodesmium strain IMS101 was from a DNA sample originating from the laboratory of Jon Zehr (University of California, Santa Cruz) and was supplied by Jon G. Kramer of the Center of Marine Biotechnology, University of Maryland, in late 1997.
Phylogenetic analysis of NirS peptide sequences showed that the closest known sequence to that obtained from the isolate 4N was also from L. aggregata IAM 2614 (Fig. 2c), with which it shares 93.2% and 98% nucleotide and amino acid identity, respectively. Both the L. aggregata IAM 2614 and the isolate 4N NirS sequences were very closely related to two environmental clones (designated SdnirS9 and AgnirS-1) originally retrieved from shallow water (20-m depth) sponges in the East China Sea and (somewhat more distantly) to numerous benthic environmental sequences that clustered with R. pomeroyi and other members of the Roseobacter clade (Fig. 2c). Two clones of pelagic origin (clones M60-80 and M60-134) that were obtained from the Baltic Sea (35) were also paired within this wider cluster of sequences but shared only 90 to 91% amino acid similarity with NirS from isolate 4N.
To confirm the most likely taxonomic affiliation of the Arabian Sea isolate 4N, a BLASTN search of the GenBank database was also conducted with the 16S rRNA gene (1,405 bp) from this isolate. This search revealed that it was >99% identical to 16S rRNA gene sequences from several Labrenzia strains isolated from a range of saline habitats and also to some unclassified symbiotic or epibiotic bacteria associated with marine invertebrates or phytoplankton (Table 2). Among the top-scoring matches was the 16S rRNA gene from L. aggregata strain 2PR58-2, which its GenBank entry (accession no. EU440961) descriptor reveals was isolated from the deep water column of the southwest Indian Ocean. The remaining high-scoring Labrenzia 16S rRNA gene sequences were mostly from isolates obtained from benthic habitats, as was the case for the source of the majority of the most closely related environmental NirS sequences also (see above).
Table 2.
Taxonomic affiliations, environmental sources, and percent identities of 16S rRNA genes from cultivated and environmental sources closest in sequence to that of the Arabian Sea isolate clone 4N
| Organism | Source | Identity, no. of identical nucleotides/total no. of nucleotides (%) | Accession no. |
|---|---|---|---|
| Labrenzia sp. strain PEB02 | Microbial mat, Ebro delta, Spain | 1,401/1,405 (99.72) | GU213158 |
| Labrenzia sp. strain w-2-2 | Karlodinium micrum (Dinophyceae) | 1,401/1,405 (99.72) | GQ495021 |
| L. aggregata strain 2PR58-2 | Southwest Indian Ocean deep-sea water | 1,401/1,405 (99.72) | EU440961 |
| Bacterium K2-26 | Lake Kauhako, Hawaii | 1,401/1,405 (99.72) | AY345441 |
| Labrenzia sp. strain PEB05 | Microbial mat, Ebro delta, Spain | 1,400/1,405 (99.64) | GU213161 |
| Symbiont CV812-530 | Eastern oyster (Crassostrea virginica) | 1,390/1,394 (99.71) | AF246614 |
| L. aggregata IAM 12614 | Baltic Sea, sediment | 1,397/1,405 (99.43) | D88520 |
| Labrenzia sp. strain MK16 | Salt marsh, southwest South Korea | 1,386/1,395 (99.35) | AY690722 |
| Epibiont alphaproteobacterium PM05 | Pseudo-nitzschia multiseries (Bacillariophyceae) | 1,365/1,377 (99.12) | AY548766 |
The consensus view of the combined molecular data, therefore, is that the Arabian Sea isolate 4N (and, in all probability, isolates 1N, 5N, and 8N) is most likely to be a new member of the genus Labrenzia (40), although this proposed taxonomic affiliation awaits further detailed systematic investigation.
Spatial distribution of Labrenzia-like denitrifying bacteria in the Arabian Sea.
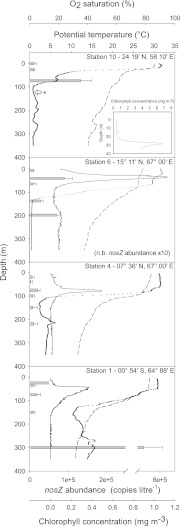
The depth distribution of bacteria closely related to the new Arabian Sea isolates was assayed by QPCR at four stations along the AMBITION cruise track (Fig. 1). The stations were located in waters of markedly contrasting biogeochemical regimes that included highly oligotrophic waters at the southern end of the transect (stations 1 and 4), where primary production was dominated by Prochlorococcus spp.; a more mesotrophic transition zone north of 8°N (station 6), in which Synechococcus spp. predominated (41); and the nutrient-rich shelf waters along the southeastern coast of Oman (station 10), where the biota was impacted by seasonal upwelling. Monsoonal winds that blow from the southwest between June and September each year lead to a pronounced shoaling of the nutricline in these more northerly waters. At station 10, the nutricline was located at a depth of 25 to 30 m, whereas in the waters further to the south it was typically found at depths of 50 to 70 m (42). Mirroring the latitudinal gradient in nutrient inventories, peak chlorophyll concentrations increased from south to north and were in excess of 6 mg m−3 at station 10 (Fig. 3).
Fig 3.
Depth distributions of chlorophyll (solid lines), potential temperature (dashed lines), oxygen saturation (dotted lines), and the concentration (mean ± standard error, n = 3) of the Labrenzia-like nosZ gene (bars) at four stations in the Arabian Sea. The abundance of nosZ at station 6 is shown at 10-fold the actual concentration, and the depth distribution of chlorophyll at station 10 is shown in the inset. The arrow shows the depth (120 m) from which the seawater was obtained for the original enrichment cultures at station 10. *, PCR inhibitors detected.
Oxygen-depleted waters were encountered below the surface mixed layer at all stations (Fig. 3), and the OMZ (<5 μM O2) was located between 11 and 24°N at depths below ∼100 m (43). Nitrite maxima coincident with the depths of the chlorophyll maximum were found at all stations surveyed, whereas a secondary nitrite maximum (SNM) (>3 μM), located at ∼150 m and below, was present only in the central region between 11 and 19°N (M. Woodward, personal communication). The occurrence of a secondary nitrite maximum is often considered to be indicative of active denitrification (44). A recent report has challenged this orthodoxy, however, and its authors (45) measured only low, sporadic N losses within the SNM of the Arabian Sea OMZ at a time of year (September/October 2007) similar to that in the present study.
The Labrenzia-like nosZ gene was detected at all stations sampled along the length of the transect, but unexpectedly, the highest mean concentration (4.9 × 105 copies liter−1) was found at a depth of 300 m at station 1, well outside the OMZ (Fig. 3). More typically, nosZ from these organisms was present at mean concentrations of ∼1 × 105 copies liter−1 or fewer at this and the other three stations sampled. Best estimates suggest, therefore, that these denitrifiers were only moderately abundant at the time of sampling. Nonetheless, their very broad biogeographical distribution is indicative of a lifestyle that is highly adaptable given the extremes in physical forcing that are the signature feature of the Arabian Sea region (46).
Paradoxically, the overall abundance of the Labrenzia-like nosZ gene detected within the OMZ region at station 6 was lower than that encountered elsewhere. Two maxima (∼1 × 104 copies liter−1) were present: the first located in the oxygenated, surface mixed layer at ∼45 m and coincident with the chlorophyll maximum and a second that occurred within the SNM at a depth of 200 m (Fig. 3). The total numbers of heterotrophic denitrifiers within the SNM of the Arabian Sea OMZ can exceed 1 × 108 cells liter−1 (47), although more modest estimates of their concentrations (∼1.5 × 104 liter−1) have been reported recently (45). Such wide variability in population size is most probably related to the episodic nature of the supply of organic carbon to support denitrification (45, 47), and it might be that the Labrenzia-like organisms are more numerous and dynamic contributors to the denitrifying assemblage within the OMZ at other times of the year.
Transcription of nosZ by Labrenzia-like denitrifying bacteria within the OMZ of the Arabian Sea.
The distribution of these organisms at station 6 (and the others sampled outside the OMZ) suggests that they do not operate exclusively as denitrifiers, since they were clearly not confined to the suboxic intermediate waters of the OMZ where active denitrification has been demonstrated experimentally (47). Not surprisingly, therefore, nosZ mRNAs from these organisms were below the limit of detection by QRT-PCR at three depths (4, 15, and 35 m) within the upper mixed layer at this station (data not shown). Deeper RNA samples from within the core of the OMZ were not available from this particular station, but nosZ transcripts were readily detected (9.844 × 103 ± 1.56 × 103 copies liter−1; n = 3) in a 220-m-depth sample from a site (station 7) with a similar oxygen deficit (O2 at 220-m depth = 3.1 μmol liter−1; 1.28% saturation) located immediately to the north (Fig. 1). At least in this mesotrophic region of the Arabian Sea, therefore, the evidence to hand is that the active consumption of N2O by the Labrenzia-like alphaproteobacteria was most probably confined to the deeper, suboxic waters.
Comparable studies of nosZ expression in natural populations of denitrifying bacteria in open waters are sparse. A recent investigation, however, did find nosZ mRNAs within the metatranscriptome of organisms obtained from suboxic depths within the OMZ off the coast of northern Chile but not in samples from shallower waters (see Table S6 in reference 48). Like the Labrenzia-like bacteria in the present study, organisms harboring nosZ were present at these shallower, oxic depths, but their metabolism was probably fueled by aerobic respiration outside the OMZ core because transcripts originating from nirS/K were also close to the limits of detection (48). Indeed, it is thought that many of the organisms that inhabit OMZs are metabolically versatile and that they are involved in other biologically mediated transformations of nitrogen and various other elemental cycles (49). Labrenzia species, for example, have been shown to be capable of CO oxidation and mixotrophic CO2 fixation (50) as well as the production of dimethylsulfide from dimethylsulfoniopropionate (51).
Depth distribution of Labrenzia-like denitrifying bacteria transcribing nosZ outside the Arabian Sea OMZ.
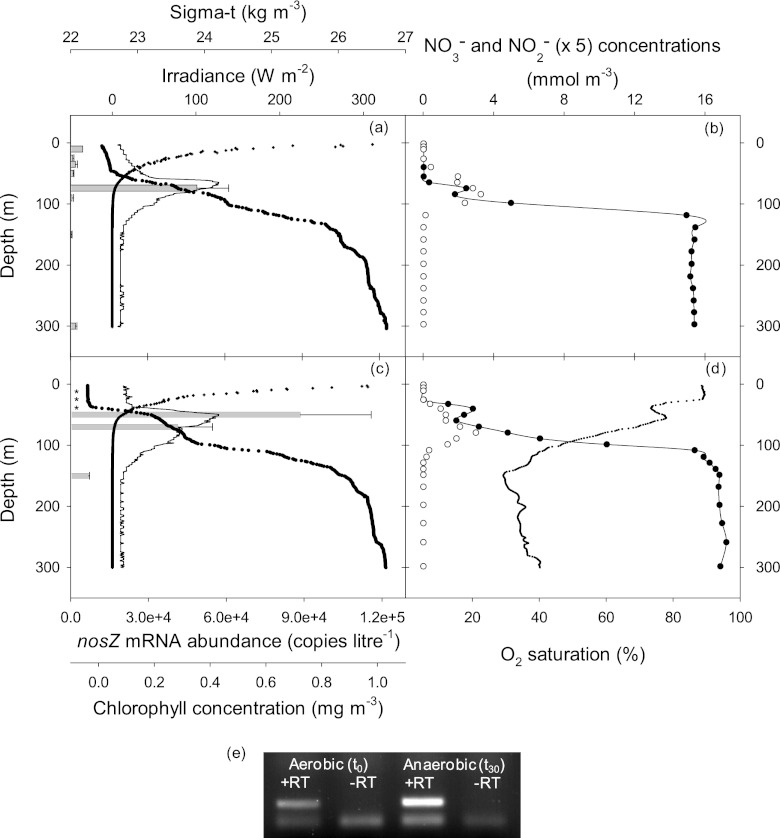
Nevertheless, some denitrifiers are able to reduce nitrogen oxides to N2 and/or NOx under elevated O2 concentrations (52, 53), and it is thought that some water column denitrification must occur outside the major OMZs (54, 55). Indeed, firm evidence for aerobic denitrification in the wider marine environment is accumulating (56). Accordingly, RNA samples that were obtained from the oligotrophic stations 1 and 2 were also analyzed by QRT-PCR for the Labrenzia-like nosZ mRNA even though the water column at these equatorial latitudes was not especially oxygen depleted, i.e., ≥30% saturation throughout the upper 300 m (Fig. 3 and 4). Most unexpectedly, transcripts originating from the Labrenzia-like nosZ gene were readily detected in the upper water column at both stations (Fig. 4). Transcript abundance was greatest, however, within the chlorophyll maximum located toward the base of the euphotic zone at both stations.
Fig 4.
(a to d) Depth distribution of chlorophyll (solid line), PAR (crosses), density (closed circles), and the concentration (mean ± standard error, n = 3) of Labrenzia-like nosZ mRNA (bars; *, not detected) at stations 1 (a) and 2 (c). The corresponding depth distributions of nitrate (closed circles) and nitrite (×5, open circles) concentrations at stations 1 (b) and 2 (d) are shown in the right-hand panels. The additional dotted line shown in panel d is the oxygen saturation profile for station 2. (e) Electrophoretogram of nosZ RT-PCR products from the Arabian Sea isolate 4N grown in denitrification medium under an aerobic atmosphere (t0) and following transfer to suboxic conditions for 30 min (t30). Equal quantities of total RNA were used for the reverse transcriptase (+RT) reactions. The absence of contaminating DNA was verified in parallel reactions in which the enzyme was omitted (−RT). The mean concentration (n = 3) of the nosZ PCR product at t30 determined by QPCR was 4.23-fold that at t0. The lower band in each lane is a mixture of QPCR primers and the fluorescein-labeled nosZ probe.
The highest nosZ mRNA concentrations ([0.49 ± 0.12] × 105 and [0.89 ± 0.27] × 105 copies liter−1 at 74-m and 50-m depths at stations 1 and 2, respectively) were centered on the 23.5 isopycnal (kg m−3) and were also coincident with the primary nitrite maximum (PNM) at each station. The nitrite concentrations (>0.5 μM) within the PNM at these stations were somewhat above the range (0.01 to 0.4 μM) commonly encountered elsewhere (57). The source of nitrite within the PNM is thought to derive from the incomplete assimilation of nitrate by phytoplankton and/or the activities of nitrifying bacteria (57, 58, 59). The predominance of Prochlorococcus spp. at these stations (41), which, in general, do not utilize nitrate, would suggest that nitrifiers, rather than phytoplankton, were the most significant nitrite producers in these waters. The latter organisms, rather than denitrifiers, are also thought to be the most likely sources of N2O found within the oxycline associated with the PNM (18).
The occurrence of nosZ mRNAs in oxic waters is perplexing since the activity of the gene product, nitrous oxide reductase, is considered to be the most oxygen-sensitive step in the denitrification pathway (60). Constitutive expression of nosZ (and the preferential reduction of N2O over NO2−) has been shown under aerobic conditions in the denitrifier Pseudomonas stutzeri strain TR2 (53). In the Arabian Sea isolate 4N, by contrast, the abundance of nosZ mRNA is close to the limit of detection when grown in well-aerated cultures but is upregulated rapidly (within 30 min) in denitrifying medium following the removal of oxygen (Fig. 4). When placed in the field context of the low abundances of Labrenzia-like nosZ mRNAs detected in near-surface waters and below the upper mixed layer at stations 1 and 2, these experimental findings are consistent with the specific induction of nosZ within the localized environment of the PNM.
It has been recognized for more than 25 years that particulate materials such as marine snow and fecal pellets are potential microsites within the oxygenated water column that might provide the reduced niches required to sustain suboxic processes such as denitrification and methanogenesis (61, 62, 63). These materials, which are readily trapped on density surfaces within the pycnocline (64, 65), support an active, sessile, microbial community that includes Roseobacter spp. (66), with which the genus Labrenzia is closely affiliated and with which some species share a particle-associated lifestyle (50). Labrenzia spp. are also known to form close (symbiotic) associations with a range of other organisms that include large diatoms and dinoflagellates (67, 68) and other algae (69). It may not be entirely coincidental, therefore, that the greatest concentrations of Labrenzia-like nosZ transcripts that were detected at stations 1 and 2 were located within the chlorophyll maximum close to the pycnocline.
Trichodesmium spp. as potential hosts for Labrenzia-like denitrifying bacteria in oxic waters of the Arabian Sea.
The phytoplankton present at these oligotrophic stations were dominated by the small-size classes (Prochlorococcus spp. and, to a lesser extent, picoeukaryotic algae [41, 42, 70]). The very low sinking velocities of these organisms would militate against the production of significant numbers of potentially suboxic aggregates within the water column (71). Perhaps tellingly, however, the diazotrophic cyanobacterium Trichodesmium was also present at these southern stations but not in the nutrient-enriched waters to the north of station 4 (21). Trichodesmium spp. form macroscopic colonies that are known to provide a so-called pseudobenthic habitat for many other organisms. These include metazoa such as small copepods, diatoms, protists and other eukaryotic microbes; viruses; and also autotrophic and heterotrophic prokaryotes (72, 73, 74, 75, 76).
As part of a parallel study examining diazotroph diversity in this region, we collected Trichodesmium colonies at the southern stations 2, 3, and 4 and, by serendipity, were able to interrogate DNA isolated from the colonies for the presence of the nosZ gene from the Labrenzia-like Arabian Sea isolates. For this purpose, a different primer pair (StanosF and StanosR2 [Table 1]) from that used for the QPCR experiments was utilized in order to amplify a larger (252-bp), more phylogenetically informative fragment of nosZ from the target organisms.
PCR products of the expected size were readily obtained from DNA samples purified from the Trichodesmium colonies collected at all three southern stations. The amplicon obtained from station 3 was selected for further investigation and found to contain the fragment of nosZ targeted by the primers. Compared to nosZ from the isolate 4N, its DNA sequence (GenBank accession number JX827177) was found to contain a nucleotide substitution (a G/A transition) at position 204 within the 252-bp region amplified, leading to a single amino acid change (Y to C) at amino acid position 68 within the translated peptide. The peptide sequence was otherwise ∼99% identical to those of the Arabian Sea isolate 4N and L. aggregata IAM 12614 and, unsurprisingly, formed part of the same tight cluster when analyzed phylogenetically (Fig. 2b).
While their wider distribution in more northerly waters (and in the deeper waters at station 1) does not suggest an obligate relationship, at least some component of the Labrenzia-like population sampled at the southern stations was part of the epibiotic community associated with Trichodesmium spp. Given the particle-associated and epibiotic lifestyle of both cultured close relatives and the range of host organisms from which so many of the phylogenetically related environmental clone sequences originated (Fig. 2; Table 2), this finding was not unexpected. It acquires some significance, however, because oxygen-depleted microzones are known to occur within the interior of Trichodesmium colonies (77). Owing to their high endogenous respiratory rates, they may become virtually anoxic in subdued light and, in particular, at night (78). The low-oxygen environment that can develop within Trichodesmium colonies, therefore, might provide a potentially novel niche for N2O reduction by colony-associated, Labrenzia-like bacteria and, perhaps, also for earlier steps in the denitrification pathway from NO3− and NO2−.
It should not be forgotten, though, that the Trichodesmium colonies sampled in the present study were from well-illuminated waters where NO3− and NO2− concentrations were in the low-nanomolar range (Fig. 4) (42). Nevertheless, with this caveat in mind a rationale can be developed to explain why nosZ transcripts from the Labrenzia-like bacteria were found to be most abundant close to the chlorophyll maximum within dimly lit waters near the pycnocline. Trichodesmium colonies can undergo rapid (>10-m hour−1) vertical migrations through the water column (79) and may spend extended periods of the day in the lower reaches of the upper mixed layer (80, 81). At these depths, they are physically closer to the sources of nitrogen oxides (NO2− and N2O) produced in situ within the PNM (18) and also to the reservoir of NO3− advected from deeper waters.
Even during the peak daylight hours, endogenous photosynthetic oxygen evolution is likely to be well below the compensation point in Trichodesmium colonies residing at these poorly illuminated but more nitrogen-replete depths (78). The foci of suboxia that develop within the colonies under these conditions are not only within easy reach of a ready supply of nitrogen oxides from surrounding waters but also are bathed in the locally produced fixed carbon (and nitrogen) that is exuded from within the Trichodesmium colony (e.g., see references 82 and 83). This beneficial combination of circumstances should provide the ideal conditions for the rapid induction of nosZ by epibiotic Labrenzia-like organisms associated with Trichodesmium, as was observed for the cultured isolate 4N under suboxia in the laboratory (Fig. 4).
Our observations provide a plausible mechanistic explanation for the expression of one of the signature genes (nosZ) of the denitrification pathway in an oxygenated water column and have pinpointed a potential biological sink for the N2O produced by nitrifiers (and denitrifiers?) in low-latitude near-surface waters. Much further work is required to establish how widespread such close associations between Trichodesmium spp. and epibiotic denitrifiers are, but during the present study a closely related (99% nucleotide identity) nosZ sequence was also amplified from Trichodesmium thiebautii colonies collected off the Bahama Islands in 1991 (84), and a further nosZ gene fragment was recovered from DNA purified from the cultured isolate Trichodesmium erythraeum strain IMS101 (Fig. 2b). We note, furthermore, that 16S rRNA sequences belonging to alphaproteobacteria that are very similar to those of Labrenzia spp. (e.g., clone BATS14_155) and Nisaea denitrificans (e.g., clone BATS14_163), a closely related denitrifier (Fig. 2), were among those recovered from the epibiotic community of Trichodesmium spp. collected from the Sargasso Sea (see Table S1 in reference 76).
Close biogeographical coupling between pelagic nitrogen fixation and denitrification in nitrogen-deficient ocean waters has been proposed before (85) but not, to date, at the microscopic spatial scales implied by the findings reported here.
ACKNOWLEDGMENTS
This research was supported by a project grant (NER/T/S/2000/00633) from the Natural Environment Research Council (NERC) of the United Kingdom under the Marine and Freshwater Microbial Diversity (M&FMB) program.
We thank Principal Scientist Peter Burkill, Captain Keith Avery, and the ship's company of RRS Charles Darwin for their excellent scientific and shipboard support during the AMBITION cruise. We are also most grateful to Malcolm Woodward of Plymouth Marine Laboratory, United Kingdom, for access to the nutrient data.
Footnotes
Published ahead of print 8 February 2013
REFERENCES
- 1. Codispoti LA. 2010. Interesting times for marine N2O. Science 327:1339–1340 [DOI] [PubMed] [Google Scholar]
- 2. Gruber N, Galloway JN. 2008. An Earth-system view of the global nitrogen cycle. Nature 451:293–296 [DOI] [PubMed] [Google Scholar]
- 3. Ravishankara AR, Daniel JS, Portmann RW. 2009. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326:123–125 [DOI] [PubMed] [Google Scholar]
- 4. Dore JE, Popp BN, Karl DM, Sansone FJ. 1998. A large source of atmospheric nitrous oxide from subtropical North Pacific surface waters. Nature 396:63–66 [Google Scholar]
- 5. Beman JM, Chow C-E, King AL, Feng Y, Fuhrman JA, Andersson A, Bates NR, Poppa BN, Hutchins DA. 2011. Global declines in oceanic nitrification rates as a consequence of ocean acidification. Proc. Natl. Acad. Sci. U. S. A. 108:208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Canfield DE, Glazer AN, Falkowski PG. 2010. The evolution and future of the Earth's nitrogen cycle. Science 330:192–196 [DOI] [PubMed] [Google Scholar]
- 7. The Royal Society 2005. Ocean acidification due to increasing atmospheric carbon dioxide. Policy document 12/05. Royal Society, London, United Kingdom [Google Scholar]
- 8. Suntharalingham P, Sarmiento JL, Toggweiler JR. 2000. Global significance of nitrous oxide production and transport from oceanic low-oxygen zones: a modeling study. Glob. Biogeochem. Cycles 14:1353–1370 [Google Scholar]
- 9. Diaz RJ, Rosenberg R. 2008. Spreading dead zones and consequences for marine ecosystems. Science 321:926–929 [DOI] [PubMed] [Google Scholar]
- 10. Stramma L, Johnson GC, Sprintall J, Mohrholz V. 2008. Expanding oxygen-minimum zones in the tropical oceans. Science 320:655–658 [DOI] [PubMed] [Google Scholar]
- 11. Keeling RF, Körtzinger A, Gruber N. 2010. Ocean deoxygenation in a warming world. Annu. Rev. Mar. Sci. 2:199–229 [DOI] [PubMed] [Google Scholar]
- 12. Shapleigh JP. 2006. The denitrifying prokaryotes, p 769–792 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed, vol 2 Springer, New York, NY [Google Scholar]
- 13. Takaya N, Catalan-Sakairi MAB, Sakaguchi Y, Kato I, Zhou Z, Shoun H. 2003. Aerobic denitrifying bacteria that produce low levels of nitrous oxide. Appl. Environ. Microbiol. 69:3152–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshinari T. 1980. N2O reduction by Vibrio succinogenes. Appl. Environ. Microbiol. 39:81–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishii S, Ashida N, Otiska S, Senoo K. 2011. Isolation of denitrifiers carrying previously uncharacterized functional gene sequences. Appl. Environ. Microbiol. 77:338–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Codispoti LA, Brandes JA, Christensen JP, Devol AH, Naqvi SWA, Paerl HW, Yoshinari T. 2001. The oceanic fixed nitrogen and nitrous oxide budgets: moving targets as we enter the anthropocene? Sci. Mar. 65(Suppl 2):85–105 [Google Scholar]
- 17. Gruber N. 2011. Warming up, turning sour, losing breath: ocean biogeochemistry under global change. Philos. Trans. R. Soc. A 369:1980–1996 [DOI] [PubMed] [Google Scholar]
- 18. Freing A, Wallace DWR, Bange HW. 2012. Global oceanic production of nitrous oxide. Philos. Trans. R. Soc. B 367:1245–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bange HW, Naqvi SWA, Codispoti LA. 2005. The nitrogen cycle in the Arabian Sea. Prog. Oceanogr. 65:145–158 [Google Scholar]
- 20. Naqvi SWA, Bange HW, Gibb SW, Goyet C, Hatton AD, Upstill-Goddard RC. 2005. Biogeochemical ocean-atmosphere transfers in the Arabian Sea. Prog. Oceanogr. 65:116–144 [Google Scholar]
- 21. Bird C, Martínez Martínez J, O'Donnell AG, Wyman M. 2005. Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotrophic γ-proteobacteria in the Arabian Sea. Appl. Environ. Microbiol. 71:2079–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wyman M, Gregory RPF, Carr NG. 1985. A novel role for phycoerythrin in a marine cyanobacterium, Synechococcus strain DC2. Science 230:818–820 [DOI] [PubMed] [Google Scholar]
- 23. Stanier RY, Kunisawar R, Mandel M, Cohen-Bazire G. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35:171–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Braker G, Fesefeldt A, Witzel K-P. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY [Google Scholar]
- 26. Altschul SF, Madden TL, Schaeffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edgar RG. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pine MJ, Barker HA. 1956. Studies on the methane fermentation. XII. The pathway of hydrogen in the acetate fermentation. J. Bacteriol. 71:644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Waterbury JB, Willey JM, Franks DG, Valois FW, Watson SW. 1985. A cyanobacterium capable of swimming motility. Science 230:74–76 [DOI] [PubMed] [Google Scholar]
- 31. Charnock JM, Dreusch A, Körner H, Neese F, Nelson J, Kannt A, Michel H, Garner CD, Kroneck PM, Zumft WG. 2000. Structural investigations of the CuA centre of nitrous oxide reductase from Pseudomonas stutzeri by site-directed mutagenesis and X-ray absorption spectroscopy. Eur. J. Biochem. 267:1368–1381 [DOI] [PubMed] [Google Scholar]
- 32. Scala DJ, Kerkhof LJ. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Ecol. 162:61–68 [DOI] [PubMed] [Google Scholar]
- 33. Simon J, Einsle O, Kroneck PMH, Zumft WG. 2004. The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett. 569:7–12 [DOI] [PubMed] [Google Scholar]
- 34. Sievert SM, Scott KM, Klotz MG, Chain PS, Hauser LJ, Hemp J, Hügler M, Land M, Lapidus A, Larimer FW, Lucas S, Malfatti SA, Meyer F, Paulsen IT, Ren Q, Simon J, USF Genomics Class 2008. Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 74:1145–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ahrens R. 1968. Taxonomische Untersuchungen an sternbildenden Agrobacterium-Arten aus der westlichen Ostsee. Kiel Meeresforsch. 24:147–173 [Google Scholar]
- 36. Urios L, Michotey V, Intertaglia L, Lesongeur F, Lebaron P. 2008. Nisaea denitrificans gen. nov., sp. nov. and Nisaea nitritireducens sp. nov., two novel members of the class Alphaproteobacteria from the Mediterranean Sea. Int. J. Syst. Evol. Microbiol. 58:2336–2341 [DOI] [PubMed] [Google Scholar]
- 37. Allgaier M, Uphoff H, Felske A, Wagner-Döbler I. 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 69:5051–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buchan A, González JM, Moran MA. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nie Y, Tang Y-Q, Li Y, Chi C-Q, Cai M, Wu X-L. 2012. The genome sequence of Polymorphum gilvum SL003B-26A1T reveals its genetic basis for crude oil degradation and adaptation to the saline soil. PLoS One 7:e31261 doi:10.1371/journal.pone.0031261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Biebl H, Pukall R, Lünsdorf H, Schulz S, Allgaier M, Tindall BJ, Wagner-Döbler I. 2007. Description of Labrenzia alexandrii gen. nov., sp. nov., a novel alphaproteobacterium containing bacteriochlorophyll a, and a proposal for reclassification of Stappia aggregata as Labrenzia aggregata comb. nov., of Stappia marina as Labrenzia marina comb. nov. and of Stappia alba as Labrenzia alba comb. nov., and emended descriptions of the genera Pannonibacter, Stappia and Roseibium, and of the species Roseibium denhamense and Roseibium hamelinense. Int. J. Syst. Evol. Microbiol. 57:1095–1107 [DOI] [PubMed] [Google Scholar]
- 41. Zubkov MV, Fuchs BM, Tarran GA, Burkhill PH, Amann R. 2003. High rate of uptake of organic compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Appl. Environ. Microbiol. 69:1299–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fuller NJ, Tarran GA, Cummings DG, Woodward EMS, Orcutt KM, Yallop M, Le Gall F, Scanlan DJ. 2006. Molecular analysis of photosynthetic picoeukaryote community structure along an Arabian Sea transect. Limnol. Oceanogr. 51:2502–2514 [Google Scholar]
- 43. Zubkov MV, Tarran GA, Burkhill PH. 2006. Bacterioplankton of low and high DNA content in the suboxic waters of the Arabian Sea and the Gulf of Oman: abundance and amino acid uptake. Aquat. Microb. Ecol. 43:23–32 [Google Scholar]
- 44. Jayakumar DA, O'Mullan G, Naqvi SWA, Ward BB. 2009. Denitrifying bacterial community composition changes associated with stages of denitrification in oxygen minimum zones. Microb. Ecol. 58:350–362 [DOI] [PubMed] [Google Scholar]
- 45. Lam P, Jensen MM, Kock A, Lettmann KA, Plancherel Y, Lavik G, Bange HW, Kuypers MMM. 2011. Origin and fate of the secondary nitrite maximum in the Arabian Sea. Biogeosciences 8:1565–1577 [Google Scholar]
- 46. Ducklow HW, Smith DC, Campbell L, Landry MR, Quinby HL, Steward GF, Azam F. 2001. Heterotrophic bacterioplankton in the Arabian Sea: basinwide response to year-round high primary productivity. Deep Sea Res. II 48:1303–1323 [Google Scholar]
- 47. Ward BB, Devol AH, Rich JJ, Chang BX, Bulow SE, Naik H, Pratihary A, Jayakumar A. 2009. Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature 461:78–82 [DOI] [PubMed] [Google Scholar]
- 48. Stewart FJ, Ulloa O, DeLong EF. 2012. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ. Microbiol. 14:23–40 [DOI] [PubMed] [Google Scholar]
- 49. Lam P, Kuypers MMM. 2011. Microbial nitrogen cycling processes in oxygen minimum zones. Annu. Rev. Mar. Sci. 3:317–345 [DOI] [PubMed] [Google Scholar]
- 50. Weber CF, King GM. 2007. Physiological, ecological, and phylogenetic characterization of Stappia, a marine CO-oxidizing bacterial genus. Appl. Environ. Microbiol. 73:1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Curson ARJ, Rogers R, Todd JD, Brearley CA, Johnston AWB. 2008. Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine α-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10:757–767 [DOI] [PubMed] [Google Scholar]
- 52. Lloyd D, Boddy L, Davies KJP. 1987. Persistence of bacterial denitrification capacity under aerobic conditions: the rule rather than the exception. FEMS Microbiol. Lett. 45:185–190 [Google Scholar]
- 53. Miyahara M, Kim S-W, Fushinobu S, Takaki K, Yamada T, Watanabe A, Miyauchi K, Endo G, Wakagi T, Shoun H. 2010. Potential of aerobic denitrification by Pseudomonas stutzeri TR2 to reduce nitrous oxide emissions from wastewater treatment plants. Appl. Environ. Microbiol. 76:4619–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Codispoti LA, Yoshinari T, Devol AH. 2005. Suboxic respiration in the oceanic water column, p 225–247 In del Giorgio PA, Williams PJLB. (ed), Respiration in aquatic ecosystems. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 55. Codispoti LA. 2007. An oceanic fixed nitrogen sink exceeding 400 Tg N a−1 vs the concept of homeostasis in the fixed-nitrogen inventory. Biogeosciences 4:233–253 [Google Scholar]
- 56. Gao H, Schreiber F, Collins G, Jensen MM, Kostka JE, Lavik G, de Beer D, Zhou HY, Kuypers MMM. 2010. Aerobic denitrification in permeable Wadden Sea sediments. ISME J. 4:417–426 [DOI] [PubMed] [Google Scholar]
- 57. Lomas MW, Lipschultz F. 2006. Forming the primary nitrite maximum: nitrifiers or phytoplankton? Limnol. Oceanogr. 51:2453–2467 [Google Scholar]
- 58. Dore JE, Karl DM. 1996. Nitrite distributions and dynamics at Station ALOHA. Deep Sea Res. II 43:385–402 [Google Scholar]
- 59. Mackey KRM, Bristow L, Parks DR, Altabet MA, Post AF, Paytan A. 2011. The influence of light on nitrogen cycling and the primary nitrite maximum in a seasonally stratified sea. Prog. Oceanogr. 91:545–560 [Google Scholar]
- 60. Bonin P, Gilewicz M, Bertrand JC. 1989. Effects of oxygen on each step of denitrification on Pseudomonas nautica. Can. J. Microbiol. 35:1061–1064 [Google Scholar]
- 61. Alldredge AL, Cohen Y. 1987. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, faecal pellets. Science 235:689–691 [DOI] [PubMed] [Google Scholar]
- 62. Scranton MI, Brewer PG. 1977. Occurrence of methane in the near-surface waters of the western subtropical North Atlantic. Deep Sea Res. II 24:127–138 [Google Scholar]
- 63. Sieburth JM. 1987. Contrary habitats for redox-specific processes: methanogenesis in oxic waters and oxidation in anoxic waters, p 11–38 In Sleigh MA. (ed), Microbes in the sea. Ellis Horwood Ltd, Chichester, United Kingdom [Google Scholar]
- 64. Alldredge AL, Cowles TJ, MacIntyre S, Rines JEB, Donaghay PL, Greenlaw CF, Holliday DV, Dekshenieks MM, Sullivan JM, Zaneveld JRV. 2002. Occurrence and mechanisms of formation of a dramatic thin layer of marine snow in a shallow Pacific fjord. Mar. Ecol. Prog. Ser. 233:1–12 [Google Scholar]
- 65. Herndl GJ. 1992. Marine snow in the Northern Adriatic Sea: possible causes and consequences for a shallow ecosystem. Mar. Microb. Food Webs 6:149–172 [Google Scholar]
- 66. Gram L, Grossart H-P, Schlingloff A, Kiørboe T. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Green DH, Llewellyn LE, Negri AP, Blackburn SI, Bolch CJS. 2004. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 47:345–357 [DOI] [PubMed] [Google Scholar]
- 68. Kaczmarska I, Ehrman JM, Bates SS, Green DH, Leger C, Harris J. 2005. Diversity and distribution of epibiotic bacteria on Pseudonitzschia multiseries (Bacillariophyceae) in culture, and comparison with those on diatoms in native seawater. Harmful Algae 4:725–741 [Google Scholar]
- 69. Hollants J, Leroux O, Leliaert F, Decleyre H, De Clerck O, Willems A. 2011. Who is in there? Exploration of endophytic bacteria within the siphonous green seaweed Bryopsis (Bryopsidales, Chlorophyta). PLoS One 6(10):e26458 doi:10.1371/journal.pone.0026458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fuller NJ, Tarran GA, Yallop M, Orcutt KM, Scanlan DJ. 2006. Molecular analysis of picocyanobacterial community structure along an Arabian Sea transect reveals distinct spatial separation of lineages. Limnol. Oceanogr. 51:2515–2526 [Google Scholar]
- 71. Dunne JP, Armstrong RA, Gnanadesikan A, Sarmiento JL. 2005. Empirical and mechanistic models for the particle export ratio. Glob. Biogeochem. Cycles 19:GB4026 doi:10.1029/2004GB002390 [Google Scholar]
- 72. Paerl HW, Bebout BM, Prufert LE. 1989. Bacterial associations with marine Oscillatoria sp (Trichodesmium sp.) populations: ecological implications. J. Phycol. 25:773–784 [Google Scholar]
- 73. O'Neil JM, Roman MR. 1992. Grazers and associated organisms of Trichodesmium, p 61–73 In Carpenter EJ, Capone DG, Rueter JG. (ed), Marine pelagic cyanobacteria: Trichodesmium and other diazotrophs. Kluwer Academic Press, Dordrecht, Netherlands [Google Scholar]
- 74. Sheridan CC, Steinberg DK, Kling GW. 2002. The microbial and metazoan community associated with colonies of Trichodesmium spp.: a quantitative survey. J. Plankton Res. 24:913–922 [Google Scholar]
- 75. Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ, Montoys JP, Zehr JP. 2009. Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J. 3:1286–1300 [DOI] [PubMed] [Google Scholar]
- 76. Hmelo LR, Van Mooy BAS, Mincer TJ. 2012. Characterization of bacterial epibionts on the cyanobacterium Trichodesmium. Aquat. Microbiol. Ecol. 67:1–14 [Google Scholar]
- 77. Paerl HW, Bebout BM. 1988. Direct measurement of O2-depleted microzones in marine Oscillatoria: relation to N2 fixation. Science 241:442–445 [DOI] [PubMed] [Google Scholar]
- 78. Kana TM. 1993. Rapid oxygen cycling in Trichodesmium thiebautii. Limnol. Oceanogr. 38:18–24 [Google Scholar]
- 79. Walsby AE. 1978. The properties and buoyancy-providing role of gas vacuoles in Trichodesmium Ehrenberg. Br. Phycol. J. 13:103–116 [Google Scholar]
- 80. Villareal TA, Carpenter EJ. 1990. Diel buoyancy regulation in the marine diazotrophic cyanobacterium Trichodesmium thiebautii. Limnol. Oceanogr. 35:1832–1837 [Google Scholar]
- 81. Letelier RM, Karl DM. 1998. Trichodesmium spp. physiology and nutrient fluxes in the North Pacific subtropical gyre. Aquat. Microb. Ecol. 15:265–276 [Google Scholar]
- 82. Sellner KG. 1992. Trophodynamics of marine cyanobacterial blooms, p 75–94 In Carpenter EJ, Capone DG, Rueter JG. (ed), Marine pelagic cyanobacteria: Trichodesmium and other diazotrophs. Kluwer Academic Press, Dordrecht, Netherlands [Google Scholar]
- 83. Mulholland MR, Bronk DA, Capone DG. 2004. Dinitrogen fixation and release of ammonium and dissolved organic nitrogen by Trichodesmium IMS101. Aquat. Microb. Ecol. 37:85–94 [Google Scholar]
- 84. Kramer JG, Wyman M, Zehr JP, Capone DG. 1996. Diel variability in transcription of the structural gene for glutamine synthetase (glnA) in natural populations of the marine diazotrophic cyanobacterium Trichodesmium thiebautii. FEMS Microbiol. Ecol. 21:187–196 [Google Scholar]
- 85. Deutsch C, Sarmiento JL, Sigman DM, Gruber N, Dunne JP. 2007. Spatial coupling of nitrogen inputs and losses in the ocean. Nature 445:163–167 [DOI] [PubMed] [Google Scholar]