Abstract
The methanotrophic potential in sewage treatment sludge was investigated. We detected a diverse aerobic methanotrophic community that potentially plays a significant role in mitigating methane emission in this environment. The results suggest that community structure was determined by conditions specific to the processes in a sewage treatment plant.
TEXT
Next to water vapor and carbon dioxide, methane is one of the most potent greenhouse gases that contribute to radiative forcing (1). Sewage treatment accounts for around 4% of the total methane produced (500 to 600 Tg methane) globally every year (2) and will likely increase with the growing need for clean water and sanitation to sustain the world's growing population (3). Although methane emitted from sewage treatment plants (STPs) may be captured for energy generation, methane slippage occurs and dissolved methane in the effluent can be discharged without recovery (4). This raises the question of whether methanotrophs play a role in attenuating methane emission from these environments. Indeed, biological methane oxidation is an important process, consuming around 80% of the methane entering an activated-sludge reactor (4).
Taxonomically, aerobic proteobacterial methanotrophs have been grouped into type I (Gammaproteobacteria) and type II (Alphaproteobacteria). Type I methanotrophs have been further divided into type Ia and type Ib on the basis of pmoA gene phylogeny. Additionally, Methylocella and Methylocapsa, belonging to Beijerinckiaceae, fall within the class Alphaproteobacteria. These methanotrophs possess a well-known physiology (5, 6). Besides, Crenothrix polyspora and Clonothrix fusca are known proteobacterial methane oxidizers found mainly in groundwater and wells (7, 8). Only recently have nitrite-driven anaerobic methanotrophs belonging to the phylum NC10 been discovered and now include a cultured representative, “Candidatus Methylomirabilis oxyfera” (9). “C. Methylomirabilis oxyfera” is unique in the way it generates molecular oxygen intercellularly and uses it subsequently for methane oxidation (9, 10). Both aerobic proteobacterial methanotrophs and “C. Methylomirabilis oxyfera” possess a particulate methane monooxygenase (pMMO), the key enzyme in methane oxidation. The pmoA gene encodes the β subunit of the particulate form of MMO and is present in the vast majority of methanotrophs, making it a suitable marker for culture-independent studies (11, 12). Methylocella and Methyloferula, however, possess only the soluble form of the MMO (13, 14). In addition to the class Proteobacteria and the phylum NC10, acidophilic and thermophilic verrucomicrobial methanotrophs were discovered, but they have so far been detected only in extreme environments (15, 16).
Previous interest in methanotrophs in groundwater or sewage treatment systems stems from their ability to cometabolically degrade chlorinated aliphatic hydrocarbons (e.g., trichloroethylene, 1,1,1-trichloroethane) (6, 17). However, little attention was given to their role in methane oxidation, and to our knowledge, no other study has yet described the diversity and abundance of aerobic methanotrophs in STPs. This necessitates qualitative and quantitative assessment of the methanotrophic communities in wastewater sludge. Sewage treatment systems are characterized by a high nutrient turnover rate, high shear force, and alternating oxic and anoxic periods or processes. Hence, they provide a dynamic yet homogenized model system in which to study microbial community structure and population dynamics (18, 19). Overall, we aimed to (i) determine the diversity and abundance of the bacterial methane-oxidizing communities in activated and anaerobic digester sludges and biomass from oxygen-limited autotrophic nitrification/denitrification (OLAND) rotating biological contactors and (ii) determine the potential for methane oxidation in these sludges under oxic conditions.
Wastewater sludges were sampled from compartments of different STPs that represent the different processes in domestic wastewater treatment (see Fig. S1 in the supplemental material). The characteristics of the wastewater sludge used are given in Table 1. The OLAND rotating biological contactors used are well characterized and were used to treat wastewater with high nitrogen concentrations (20). Typically, these reactors contain a community of aerobic and anaerobic ammonium oxidizers (21). Triplicate batch incubations (working volume, 8 ml) were performed with 120-ml serum bottles under approximately 25% methane by volume in air on a shaker (120 rpm) in the dark at 28°C for each sludge. DNA was extracted from the starting material and after incubation (12 days) with the QBIOgene soil extraction kit with minor modifications (MP Biomedicals) (22, 23) and stored at −20°C for further molecular analyses. For details of the methodology used and the subsequent molecular analyses, see the supplemental material.
Table 1.
STP wastewater sludge characteristics and total methane consumed over 12 days of incubationa
| Sample description | City, country | pH | DOb (mg O2 liter−1) | ECec (mS cm−1) | VSSd (mg ml−1) | Nutrient contents (mM)f |
CH4 uptake measurementg |
Temp range (°C) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PO43− | SO42− | NO3− | NH4+ | Lag (h) | Total CH4 consumed (mmol g VSS−1) | |||||||
| Anoxic activated sludge | Ghent, Belgium | 7.37 | 0 | 0.88 | 0.64 | BDLe | 0.66 | 0.58 | 1.13 ± 0.09 | <94 | 160.32 ± 17.79 | 9–16h |
| Returned activated sludge | Ghent, Belgium | 7.35 | 0–5.0 | 0.99 | 2.90 | BDL | 1.06 | 0.62 | 0.18 ± 0.03 | <94 | 34.66 ± 0.94 | 9–16h |
| Anaerobic digester sludge | Ghent, Belgium | 7.70 | 0 | 5.27 | 21.56 | BDL | 0.30 | 0.77 | 53.41 ± 1.12 | <94 | 22.09 ± 1.02 | 34 |
| OLAND (lab scale) | Ghent, Belgium | 7.57 | 1.0 | 8.92 | 2.04 | 1.34 | 37.29 | 17.94 | 1.98 ± 0.02 | ≤117 | 85.16 ± 2.69 | 28 |
| OLAND (industrial scale) | Sneek, The Netherlands | 5.78 | 0.6 | 4.73 | 1.74 | 5.63 | 3.41 | 58.38 | 15.68 ± 0.29 | ≤168 | 35.19 ± 5.71 | 27 |
The means ± standard deviations shown are for three samples each. Samples were normalized per gram of VSS (21).
DO, dissolved oxygen.
ECe, electrical conductivity.
VSS are particulate organic solids, excluding soluble solids and inorganic solids.
BDL, below detection limit.
Endpoint nutrient concentrations (after 12 days of incubation) are shown in Fig. S4 in the supplemental material.
Temporal changes in methane uptake are shown in Fig. S3 in the supplemental material.
Sampled in March 2012; subject to seasonal temperature fluctuations.
To capture aerobic methanotroph diversity, a pmoA-based diagnostic microarray analysis was performed as described before (22, 23), with the A189f/T7_A682r primer combination. The amplicons were derived from three DNA extractions of the starting material and from three batch incubations of each sludge sample after 12 days. Analysis of the standardized microarray data was done in R ver. 2.10.0 (24) using heatmap.2 as implemented in gplots ver. 2.7.4. The pmoA gene is present in virtually all methanotrophs, allowing a wider coverage of the methanotroph inventory than mmoX (a gene encoding the soluble form of MMO), which is confined to only some methanotrophs. Methylocella-like mmoX sequences have been retrieved from diverse environments (25), but their ecological relevance at circumneutral pH remains uncertain (26). Hence, we focused on the pmoA gene diversity of the potentially active community after 12 days of in vitro incubation. Although we detected a broadly similar methanotrophic composition comprising both types I and II, a cluster analysis of standardized microarray data revealed grouping of the activated and anaerobic digester sludges and OLAND sludges, respectively (Fig. 1). The methanotrophic communities in the returned activated and anaerobic digester sludges were largely similar but differed in their relative distribution in these sludges (Fig. 1). Similar clusters were observed in the starting material (see Fig. S2 in the supplemental material). Although the microarray covers Crenothrix-like and verrucomicrobial methanotrophs, the corresponding probes did not show a hybridization signal (Fig. 1; see Fig. S2). Despite the different working scales and pHs, it appears that in the OLAND reactor biomass, a specific methanotrophic community was enriched after incubation, largely made up of Methylocaldum-affiliated type Ib (probes Mcl404, Mcl408, and P_MclE302) and a unique type II community (probes peat264, Msi232, and P_Msi423). Interestingly, this community seems to be depauperate in pmoA2 (probe P_NMsiT.271). pmoA2 encodes an isozyme of pMMO that enables methane oxidation and growth at low methane concentrations (27) and is so far confined to type II methanotrophs (28). The dominant type Ib methanotrophs in the OLAND reactor biomass were Methylocaldum-affiliated microorganisms, whereas all other samples were dominated by environmental pmoA lineages without closely related cultivated representatives (freshwater lineages 1 and 2) (29), suggesting that the different processes favored distinct members of a subgroup.
Fig 1.
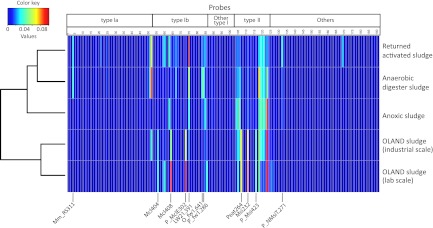
Cluster analysis of standardized microarray data showing the diversity of the aerobic methanotrophic community in wastewater sludge after incubation. Probe names and their intended specificities are shown (see Table S1 in the supplemental material). The results shown are averages of triplicate analyses of each sludge sample. Red and blue indicate the highest and no hybridization signal, respectively. These probes represent type I and II methanotrophs. Probes targeting amoA (a gene encoding ammonia monooxygenase), pmoA2, verrucomicrobial methanotrophs, and environmental sequences clustered between amoA and pmoA are grouped as “Others.”
The OLAND reactor may provide a niche for the aerobic methanotrophs, given oxic or micro-oxic areas in the structured biofilms (21). The OLAND reactor biomass, however, is not known to produce methane, with methanogenesis probably being suppressed by the continuous presence of nitrate. However, methane is supplied via the influent. The biomass in the OLAND reactors experiences continuously high ammonium concentrations, up to 1.0 to 1.1 g N liter−1 in the influent. Type I methanotrophs, particularly Methylomicrobium- and Methylocaldum-like methanotrophs, shown to be selectively stimulated by ammonium amendment (30), may have been favored under this condition. Moreover, intermediates and products of nitrification may have discriminated against methanotrophs, e.g., susceptible to hydroxylamine or nitrite (31, 32). Hence, a large ammonium load may be a strong selecting force for the methanotrophic community composition in these reactors. On the contrary, type II methanotrophs are thought to be present mainly as resting cells in other environments (26, 33), which is also indicated by the qPCR analysis in this study (Fig. 2).
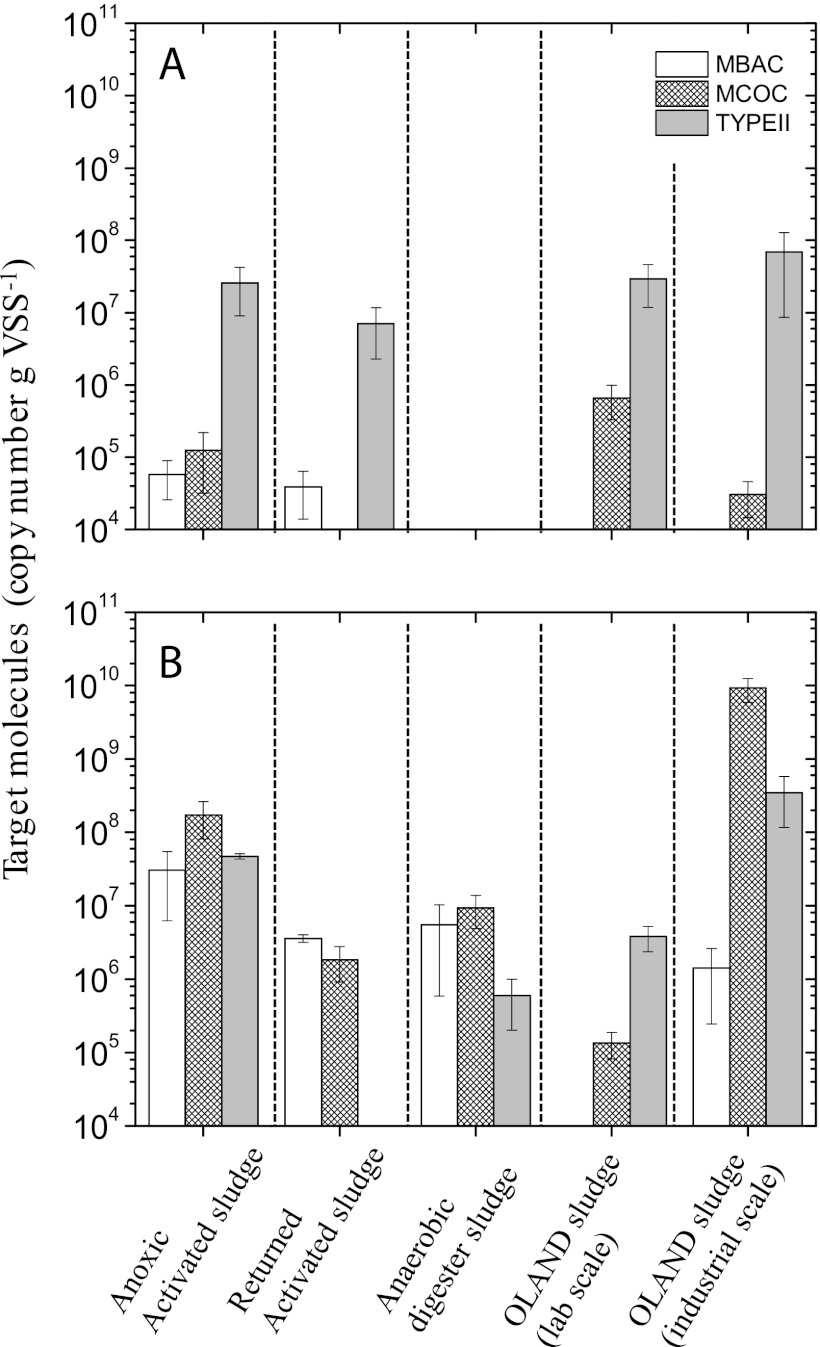
Fig 2.
qPCR analysis of MBAC, MCOC, and TYPEII assays in the starting materials (A) and after 12 days of incubation (B). qPCR was performed in duplicate with each DNA extract (mean ± standard deviation, total n = 6).
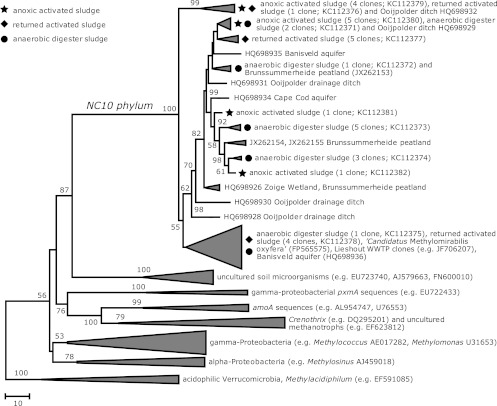
PCR amplification targeting the pmoA gene belonging to NC10 was performed by using a nested approach (12). Consistent with a previous study (34), cloning and sequence analysis revealed the presence of methanotrophs affiliated with “C. Methylomirabilis oxyfera” in activated and anaerobic digester sludges. These sequences clustered within known “C. Methylomirabilis oxyfera”-like pmoA from other environments (Fig. 3). The biomass in the OLAND reactors did not harbor these sequences. The presence of “C. Methylomirabilis oxyfera”-like sequences indicates the potential for anaerobic methane oxidation at the expense of nitrite in the activated and anaerobic digester sludges.
Fig 3.
Phylogenetic tree depicting NC10 bacterial diversity in different compartments of STPs based on pmoA gene sequences. Clustering of nucleotide sequences (351 positions) was inferred by using the neighbor-joining algorithm integrated in MEGA5 (45). Bootstrap support values (1,000 replicates) of greater than 50% are indicated at the nodes. The analysis involved 158 pmoA, amoA, and pxmA gene sequences, but for clarity, they were grouped as shown. The scale bar shows the number of base differences between sequences.
Microarray analysis is able to resolve the methanotrophic composition down to the species level (22) but does not provide the numerical abundances of the methanotrophs detected. Hence, group-specific qPCR assays targeting type Ia (MBAC assay), type Ib (MCOC assay), and type II (TYPEII assay) methanotrophs were performed, with minor modifications as described before (23, 35), to determine the abundance and growth of the active subpopulation in the in vitro incubations (Fig. 2). Before incubation, the number of pmoA gene copies was low, often below or at the detection limit (<104 to 105 copies of target molecule g of volatile suspended solids [VSS]−1). However, the pmoA gene copy number of sequences affiliated with type I methanotrophs markedly increased by 2 to 5 orders of magnitude during aerobic incubation with methane (Fig. 2). The sludge from the OLAND lab scale reactor was the only exception, showing a relatively stable community of type Ib and type II methanotrophs; type Ia was not detected by microarray and qPCR. Type II pmoA was dominant in the starting material, as shown by qPCR and microarray analyses. However, regardless of the different type II community, the respective pmoA copy numbers remained relatively constant or decreased during incubation. With the qPCR assays covering a high percentage of environmental sequences (23), the increase of type Ia/Ib methanotrophs suggests that they are the predominantly active population under the assay conditions used (Fig. 2). Generally, type I methanotrophs were found to be active in other high-methane environments (30, 33, 36–39). The proliferation of methanotrophs after methane addition indicates their potential to mitigate methane emission in wastewater sludges.
While both aerobic and nitrite-driven anaerobic methanotrophs have been detected in wastewater sludge (34, 40), a recent study indicates that aerobic methane oxidation is more important in situ (4). High nitrite-driven anaerobic methane-oxidizing activity has so far been documented only in enrichment cultures of “C. Methylomirabilis oxyfera”-like microorganisms (41–43). Hence, we focused on aerobic methane oxidation in the natural community. Methane uptake was detected in all sludges (Table 1; see Fig. S3 in the supplemental material). Total methane consumption was highest in the anoxic activated sludge, while the other types of sludge exhibited relatively lower methane uptake (Table 1). The anaerobic digester sludge and activated sludge and OLAND reactors receiving methane-rich influent are compartments with large methane loads. The potential for aerobic methane oxidation thus signifies the role of methanotrophs to mitigate methane emission in these compartments if/when electron acceptors are not limiting. Interestingly, the anaerobic digester sludge and OLAND reactor biomass exhibited methane uptake despite the potential inhibitory effects induced by (by)products of ammonia oxidation (31, 32, 44). With a relatively stable methanotrophic population size in the OLAND lab scale reactor (Fig. 2), the higher methane uptake suggests an increase in cell-specific activity. There was, however, a longer lag phase (Table 1) before the onset of activity in the OLAND sludge. Moreover, the decrease in nutrient concentrations after incubation (see Fig. S4 in the supplemental material) coincides with the growth of the methanotrophic population. The relatively fast response in activity (<4 days) and growth in the activated and anaerobic digester sludges indicates the presence of an active aerobic methanotrophic population that acts to attenuate methane emission when conditions turn permissive in STPs.
We investigated STPs to determine the methanotrophic potential in wastewater sludge and provide a first insight into methanotroph ecology in this environment. We demonstrated that the aerobic methanotrophic communities in STPs are diverse and that they potentially attenuate methane emission from this environment. We confirmed the occurrence of “C. Methylomirabilis oxyfera”-like methanotrophs in wastewater sludge. Further, results suggest that the community composition was driven by conditions specific to the processes in STPs.
Nucleotide sequence accession numbers.
Representative pmoA gene sequences were deposited in GenBank under accession numbers KC112371 through KC112382.
ACKNOWLEDGMENTS
We thank Tim Lacoere for excellent technical assistance and Karen De Roy and Pieter van den Abbeele for proofreading the manuscript. We extend our gratitude to Levente Bodrossy (CSIRO, Tasmania, Australia) for introducing us to microarray analysis and to Claudia Lüke (Max Planck Institute, Marburg, Germany) for assistance with microarray analysis. We also thank Willy Verstraete, Brendo Meulman (Desah, Sneek, Netherlands), and Jo De Vrieze for assistance with sampling.
A.H. and N.B. are supported by research grants from the Geconcerteerde Onderzoeksactie of Ghent University (BOF09/GOA/005). S.E.V. is financially supported by the Research Foundation Flanders (Fonds Wetenschappelijk Onderzoek). K.F.E. is supported by a grant from the Darwin Center for Biogeosciences (project 142.16.3071).
Footnotes
Published ahead of print 15 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03426-12.
REFERENCES
- 1. International Panel on Climate Change 2007. Summary for policymakers. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 2. Conrad R. 2009. The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1:285–292 [DOI] [PubMed] [Google Scholar]
- 3. Verstraete W, Vlaeminck SE. 2011. ZeroWasteWater: short-cycling of wastewater resources for sustainable cities of the future. Int. J. Sustain. Dev. World Ecol. 18:253–264 [Google Scholar]
- 4. Daelman MRJ, van Voorthuizen EM, van Dongen U, Volcke EIP, van Loosdrecht MCM. 2012. Methane emission during municipal wastewater treatment. Water Res. 46:3657–3670 [DOI] [PubMed] [Google Scholar]
- 5. Trotsenko YA, Murrell JC. 2008. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63:183–229 [DOI] [PubMed] [Google Scholar]
- 6. Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol. Rev. 34:496–531 [DOI] [PubMed] [Google Scholar]
- 7. Stoecker K, Bendinger B, Schoning B, Nielsen PH, Nielsen JL, Baranyi C, Toenshoff ER, Daims H, Wagner M. 2006. Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 103:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vigliotta G, Nutricati E, Carata E, Tredici SM, De Stefano M, Pontieri P, Massardo DR, Prati MV, De Bellis L, Alifano P. 2007. Clonothrix fusca Roze 1896, a filamentous, sheathed, methanotrophic γ-proteobacterium. Appl. Environ. Microbiol. 73:3556–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548 [DOI] [PubMed] [Google Scholar]
- 10. Strous M. 2011. Beyond denitrification: alternative routes to dinitrogen. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 11. McDonald IR, Bodrossy L, Chen Y, Murrell JC. 2008. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 74:1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luesken FA, Zhu BL, van Alen TA, Butler MK, Diaz MR, Song B, den Camp H, Jetten MSM, Ettwig KF. 2011. pmoA primers for detection of anaerobic methanotrophs. Appl. Environ. Microbiol. 77:3877–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Bares AM, Panikov NS, Tiedje JM. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955–969 [DOI] [PubMed] [Google Scholar]
- 14. Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh SN. 2011. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 61:2456–2463 [DOI] [PubMed] [Google Scholar]
- 15. Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland NK, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1:293–306 [DOI] [PubMed] [Google Scholar]
- 16. Sharp CE, Stott MB, Dunfield PF. Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Front. Microbiol. 3:303 doi:10.3389/fmicb.2012.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Little CD, Palumbo AV, Herbes SE, Lidstrom ME, Tyndall RL, Gilmer PJ. 1988. Trichloroethylene biodegradation by a methane-oxidizing bacterium. Appl. Environ. Microbiol. 54:951–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mussmann M, Brito I, Pitcher A, Damste JSS, Hatzenpichler R, Richter A, Nielsen JL, Nielsen PH, Muller A, Daims H, Wagner M, Head IM. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:16771–16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sauder LA, Peterse F, Schouten S, Neufeld JD. 2012. Low-ammonia niche of ammonia-oxidizing archaea in rotating biological contactors of a municipal wastewater treatment plant. Environ. Microbiol. 14:2589–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vlaeminck SE, Terada A, Smets BF, Van der Linden D, Boon N, Verstraete W, Carballa M. 2009. Nitrogen removal from digested black water by one-stage partial nitritation and anammox. Environ. Sci. Technol. 43:5035–5041 [DOI] [PubMed] [Google Scholar]
- 21. Vlaeminck SE, Terada A, Smets BF, De Clippeleir H, Schaubroeck T, Bolca S, Demeestere L, Mast J, Boon N, Carballa M, Verstraete W. 2010. Aggregate size and architecture determine microbial activity balance for one-stage partial nitritation and anammox. Appl. Environ. Microbiol. 76:900–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bodrossy L, Stralis-Pavese N, Murrell JC, Radajewski S, Weilharter A, Sessitsch A. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566–582 [DOI] [PubMed] [Google Scholar]
- 23. Ho A, Lüke C, Frenzel P. 2011. Recovery of methanotrophs from disturbance: population dynamics, evenness and functioning. ISME J. 5:750–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. R Development Core Team 2012. R: a language and environment for statistical computing, 2.15.1. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 25. Rahman MT, Crombie A, Chen Y, Stralis-Pavese N, Bodrossy L, Meir P, McNamara NP, Murrell JC. 2011. Environmental distribution and abundance of the facultative methanotroph Methylocella. ISME J. 5:1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reim A, Lüke C, Krause S, Pratscher J, Frenzel P. 2012. One millimetre makes the difference: high-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil. ISME J. 6:2128–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baani M, Liesack W. 2008. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc. Natl. Acad. Sci. U. S. A. 105:10203–10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tchawa Yimga M, Dunfield PF, Ricke P, Heyer H, Liesack W. 2003. Wide distribution of a novel pmoA-like gene copy among type II methanotrophs, and its expression in Methylocystis strain SC2. Appl. Environ. Microbiol. 69:5593–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lüke C, Frenzel P. 2011. Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl. Environ. Microbiol. 77:6305–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Noll M, Frenzel P, Conrad R. 2008. Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol. Ecol. 65:125–132 [DOI] [PubMed] [Google Scholar]
- 31. Poret-Peterson AT, Graham JE, Gulledge J, Klotz MG. 2008. Transcription of nitrification genes by the methane-oxidizing bacterium, Methylococcus capsulatus strain Bath. ISME J. 2:1213–1220 [DOI] [PubMed] [Google Scholar]
- 32. Campbell MA, Nyerges G, Kozlowski JA, Poret-Peterson AT, Stein LY, Klotz MG. 2011. Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol. Lett. 322:82–89 [DOI] [PubMed] [Google Scholar]
- 33. Ho A, Kerckhof F-M, Lüke C, Reim A, Krause S, Boon N, Bodelier PLE. 13 August 2012, posting date Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ. Microbiol. Rep. (Epub ahead of print.) doi:10.1111/j.1758–2229.2012.00370.x [DOI] [PubMed] [Google Scholar]
- 34. Luesken FA, van Alen TA, van der Biezen E, Frijters C, Toonen G, Kampman C, Hendrickx TLG, Zeeman G, Temmink H, Strous M, den Camp H, Jetten MSM. 2011. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl. Microbiol. Biotechnol. 92:845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kolb S, Knief C, Stubner S, Conrad R. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu QF, Noll M, Abraham WR, Lu YH, Conrad R. 2008. Applying stable isotope probing of phospholipid fatty acids and rRNA in a Chinese rice field to study activity and composition of the methanotrophic bacterial communities in situ. ISME J. 2:602–614 [DOI] [PubMed] [Google Scholar]
- 37. Kip N, van Winden JF, Pan Y, Bodrossy L, Reichart GJ, Smolders AJP, Jetten MSM, Damste JSS, Op den Camp HJM. 2010. Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat. Geosci. 3:617–621 [Google Scholar]
- 38. Dumont MG, Pommerenke B, Casper P, Conrad R. 2011. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ. Microbiol. 13:1153–1167 [DOI] [PubMed] [Google Scholar]
- 39. Ho A, Lüke C, Cao ZH, Frenzel P. 2011. Ageing well: methane oxidation and methane oxidizing bacteria along a chronosequence of 2000 years. Environ. Microbiol. Rep. 3:738–743 [DOI] [PubMed] [Google Scholar]
- 40. Boon N, de Windt W, Verstraete W, Top EM. 2000. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101–112 [DOI] [PubMed] [Google Scholar]
- 41. Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, Schouten S, Damste JSS, Op den Camp HJM, Jetten MSM, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921 [DOI] [PubMed] [Google Scholar]
- 42. Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MSM, Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75:3656–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu SH, Zeng RJ, Burow LC, Lant P, Keller J, Yuan ZG. 2009. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ. Microbiol. Rep. 1:377–384 [DOI] [PubMed] [Google Scholar]
- 44. Bodelier PLE, Laanbroek HJ. 2004. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol. Ecol. 47:265–277 [DOI] [PubMed] [Google Scholar]
- 45. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]