Abstract
Parasitic chytrid fungi can inflict significant mortality on cyanobacteria but frequently fail to keep cyanobacterial dominance and bloom formation in check. Our study tested whether oligopeptide production, a common feature in many cyanobacteria, can be a defensive mechanism against chytrid parasitism. The study employed the cyanobacterial strain Planktothrix NIVA-CYA126/8 and its mutants with knockout mutations for microcystins, anabaenopeptins, and microviridins, major oligopeptide classes to be found in NIVA-CYA126/8. Four chytrid strains were used as parasite models. They are obligate parasites of Planktothrix and are unable to exploit alternative food sources. All chytrid strains were less virulent to the NIVA-CYA126/8 wild type than to at least one of its oligopeptide knockout mutants. One chytrid strain even failed to infect the wild type, while exhibiting considerable virulence to all mutants. It is therefore evident that producing microcystins, microviridins, and/or anabaenopeptins can reduce the virulence of chytrids to Planktothrix, thereby increasing the host's chance of survival. Microcystins and anabaenopeptins are nonribosomal oligopeptides, while microviridins are produced ribosomally, suggesting that Planktothrix resists chytrids by relying on metabolites that are produced via distinct biosynthetic pathways. Chytrids, on the other hand, can adapt to the oligopeptides produced by Planktothrix in different ways. This setting most likely results in an evolutionary arms race, which would probably lead to Planktothrix and chytrid population structures that closely resemble those actually found in nature. In summary, the findings of the present study suggest oligopeptide production in Planktothrix to be part of a defensive mechanism against chytrid parasitism.
INTRODUCTION
Cyanobacteria are arguably the most successful photoautotrophic organisms in freshwater systems and a major challenge to global water management. They are the dominant phytoplankter in many lakes and large rivers, where they can deteriorate water quality by forming harmful blooms. Some taxa are favored by the anthropogenic eutrophication of aquatic systems and by global warming, allowing these organisms to proliferate in a growing number of water bodies worldwide (1). The success of cyanobacteria is typically attributed to adaptations that ensure a competitive advantage in utilization of growth resources (2, 3) and to a low susceptibility to zooplankton grazing (4). However, freshwater cyanobacteria are also targeted by potent pathogens (5, 6), and it has yet to be explained why these frequently fail to keep cyanobacterial dominance and bloom formation in check.
A major but almost overlooked group of parasites with the potential to inflict significant mortality on most cyanobacteria are the zoosporic true fungi, often called chytrids (phylum Chytridiomycota) (6, 7, 8). Chytrids employ chemotactic zoospores to find a host and intracellular rhizoids to extract nutrients from it (9, 10). In this process, infected host cells are irreversibly damaged. Finally, a new generation of zoospores is formed in sporangia. Chytrids infecting cyanobacteria are obligate parasites with narrow host ranges (7). In nature, chytrid infection of cyanobacteria is considered omnipresent (11) and can burst into epidemics (6).
We recently found a correlation between chytrid virulence to 35 cyanobacterial strains of the genus Planktothrix and their cellular composition of bioactive oligopeptides (12). This led to the working hypothesis that oligopeptide production might be a defensive mechanism against chytrid parasitism. Many freshwater cyanobacteria and in particular the bloom-forming taxa are rich sources of oligopeptides with the ability to inhibit key enzymes (13). Of the several hundreds of compounds described to date, many can be assigned to a few major structural classes, including the anabaenopeptins, the microcystins, and the microviridins. Cyanobacterial strains often contain several oligopeptide classes at the same time, all formed separately by nonribosomal peptide synthetases (14) or through posttranslational modification of ribosomally synthesized precursor peptides (15). Multiple variants of a class may occur if a peptide synthetase exhibits relaxed substrate specificity. Differences in the assortment of oligopeptide synthetase gene clusters and mutations within these clusters cause a considerable diversity among conspecific strains with regard to their qualitative cellular oligopeptide composition (13, 16). Once produced, oligopeptides remain largely intracellular (17), where they may accumulate well beyond the saturation level of the cyanobacterial cytoplasm (18).
The adaptive value of cyanobacterial oligopeptide production is, despite decades of intensive research, still ambiguous. Suggestions for possible functions include a role in interaction between cyanobacterial cells (19, 20) and a function in nutrient acquisition or nutrient storage (21), though most authors tend to see oligopeptides as candidates for allelochemicals (22). The latter idea finds some support in the toxicity of certain oligopeptides to herbivorous grazers (23, 24), although it is not clear whether this toxic effect, which requires ingestion and digestion of the oligopeptide-producing cells, can be of advantage to cyanobacteria. The search for the adaptive value of oligopeptide production is also of interest to water management. Some oligopeptides, namely, those belonging to the class of microcystins, are toxic to warm-blooded animals and have been implicated in numerous poisonings of wildlife, livestock, and humans worldwide (1). Any information on the drivers of oligopeptide production in general and microcystin production in particular is therefore crucial to global water management.
Here we tested whether cyanobacterial oligopeptide production can be a defensive mechanism against chytrid parasitism. We used a simple but unique experimental setup that employed the bloom-forming cyanobacterium NIVA-CYA126/8 as a host model. NIVA-CYA126/8 is the first example of a cyanobacterium with a reasonably complex secondary metabolism for which knockout mutants for all of its major oligopeptide classes exist (14, 15, 25). These mutants differ from the wild type by their inability to produce microcystins, anabaenopeptins, or microviridins only, and so comparing the mutants with their wild type is one of the most direct ways to identify the significance of certain oligopeptide classes to biotic interactions. Anabaenopeptins, microviridins, and microcystins are among the most widespread oligopeptide classes and occur in most cyanobacterial orders and in almost all Planktothrix strains (26, 27). Four chytrid strains, Chy-Kol2008, Chy-Lys2009, Chy-Stein2010, and Chy-Hal2010, were used as parasite models. They are obligate parasites, which are specific to Planktothrix, and are unable to utilize alternative food sources, making them dependent on a host with an apparent mandatory oligopeptide production (26, 27). Hence, the likelihood that chytrids infecting Planktothrix are exposed to oligopeptides in nature in the same way as in our set of infection experiments is high.
MATERIALS AND METHODS
Origin and description of test organisms.
A detailed description of all Planktothrix cultures that were used in this study is given in Fig. 1. Planktothrix NIVA-CYA126/8 produces large amounts of two microcystins (MC-[Asp3]-RR and MC-[Asp3]-LR), two anabaenopeptins (anabaenopeptins 908 and 915), and microviridin K (25). In addition, smaller amounts of cyanopeptolins and aeruginosins can be found (25, 28). The genetic manipulation of NIVA-CYA126/8 and the resulting mutants with knockout mutations for microcystins (NIVA-CYA126/8 Mcy−), anabaenopeptins (NIVA-CYA126/8 Apn−), and microviridin K (NIVA-CYA126/8 Mvd−) were described earlier (14, 15, 25). Briefly, mutants were obtained by electroporation of NIVA-CYA126/8 with a mutant version of one of the peptide synthesis genes, including recombinative replacement of the wild-type copy of this gene. The respective gene was insertionally inactivated using a chloramphenicol acetyltransferase gene. The insertion resulted in a specific and complete inactivation of the respective peptide synthesis, and culture experiments showed that there was no effect on the production of other oligopeptides, either qualitatively or quantitatively. The wild type and mutants of NIVA-CYA126/8 share the same genotype except for the insertion region.
Fig 1.
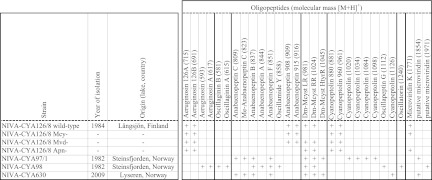
Properties of Planktothrix cultures that were used in this study. The figure shows for each strain its origin, its date of isolation, and its cellular oligopeptide composition. For each oligopeptide, its name and molecular weight (in parentheses) are given. The oligopeptides are sorted by structural class. Mcyst, microcystin. References: NIVA-CYA126/8 and mutants, 14, 15, and 25; all other Planktothrix strains, 27.
Prior to the present study, the inactivity of the mutated peptide synthetases was confirmed by using an oligopeptide screening method described by Rohrlack and coworkers (27). Briefly, oligopeptides were extracted from filters with cultured cyanobacterial material after lyophilization using 50% methanol. For the detection and identification of oligopeptides, liquid chromatography-mass spectroscopy (MS) was used. The instrumental setup included a Waters Acquity ultra-performance liquid chromatography (UPLC) system equipped with a Waters Atlantis C18 column (2.1 by 150 mm, 5-μm particle size) and directly coupled to a Waters Quattro Premier XE tandem quadrupole MS/MS detector (entire system from Waters Norge, Oslo, Norway). The UPLC system was set to deliver a linear gradient from 10% to 45% acetonitrile in water, both containing 0.1% formic acid, within 10 min at a flow rate of 0.25 ml min−1. Screening for oligopeptides was done in total scanning mode for the mass range of 500 to 2,000 Da. Mass signals were identified in fragmentation experiments using the daughter ion scanning mode of the instrument. Fragments were identified by comparison with fragment patterns of available standard material or of compounds identified in earlier studies.
The chytrid strains Chy-Kol2008, Chy-Lys2009, Chy-Hal2010, and Chy-Stein2010 were isolated from Planktothrix-dominated lakes in southern Norway as described earlier (12). The four chytrids have identical morphologies and infection patterns, which are in agreement with those described by Canter and Lund (9) for Rhizophidium megarrhizum. The chytrids are obligate parasites that are specific to Planktothrix and exhibit distinct host strain preferences. It was therefore necessary to employ three Planktothrix strains (NIVA-CYA97/1, NIVA-CYA98, and NIVA-CYA630) as hosts for chytrid cultures and as positive controls in infection experiments.
All organisms of the present study are available from the Norwegian University of Life Sciences. Planktothrix NIVA-CYA126/8 (wild type), NIVA-CYA97/1, NIVA-CYA98, and NIVA-CYA630 were originally supplied by the Norwegian Institute for Water Research Culture Collection of Algae.
Culturing of Planktothrix.
All Planktothrix strains and mutant cell lines were grown in sterile BG11 medium (Sigma-Aldrich Norway, Oslo, Norway) as nonaxenic, semicontinuous cultures. The cultures were maintained under continuous light (13 μmol photons m−2 s−1) supplied by warm-white fluorescent lamps, and they were shaken continuously. The temperature was kept at 22 ± 1°C. The cultures were diluted every other day at the same time to a final optical density of 0.06 (5-cm standard cuvette, 800 nm). Under these conditions, the NIVA-CYA126/8 wild type and mutants with knockout mutations for microcystins and microviridins exhibited the same growth rate of 0.34 day−1. The anabaenopeptin knockout mutant was growing slightly slower at 0.31 day−1. The other Planktothrix strains were exhibiting growth rates of 0.30 day−1 (NIVA-CYA98 and -CYA630) and 0.25 day−1 (NIVA-CYA97/1). To ensure a complete adaptation of all Planktothrix strains and mutant cell lines to the same environmental conditions, they were maintained at a constant growth rate for at least 10 days before they were used in experiments.
Culturing of chytrid strains.
Chy-Hal2010 and Chy-Kol2008 were cultured using Planktothrix NIVA-CYA630 and NIVA-CYA98 (initial optical density of 0.1) as hosts, respectively, in 50-ml culture vessels at 22 ± 1°C. Continuous light at a photon flux density of 5 μmol m−2 s−1 was supplied by warm-white fluorescent tubes. Chy-Lys2009 and Chy-Stein2010 were cultured in the same way but with Planktothrix NIVA-CYA97/1 as the host. Cultures were shaken and microscopically inspected once a day. They were used in infection experiments when the prevalence of infection had exceeded 50%, typically after 6 to 8 days of exposure. As with Planktothrix, all handling of the chytrid cultures was under sterile conditions.
Infection experiments.
Infection experiments aimed to compare the levels of virulence of chytrids to the wild-type NIVA-CYA126/8 and its oligopeptide knockout mutants. The experiments were run in sterile 24-well microtiter plates by mixing in each well 2 ml Planktothrix suspension, at an initial optical density of 0.1, with 0.1 ml chytrid culture. All tests were run in quadruplicates. The microtiter plates were kept at 22 ± 1°C and continuous light at a photon flux density of 5 μmol m−2 s−1. At day 6, the prevalence of infection, defined as the percentage of host filaments infected, was determined for each replicate by light microscopic inspection of 100 Planktothrix filaments at a magnification of ×120. Only filaments carrying at least one sporangium were counted as successfully infected by chytrids. Filaments of NIVA-CYA126/8 and its mutants visually differ from all Planktothrix strains that were used to culture chytrids by their small filament diameter. This made it possible to distinguish NIVA-CYA126/8 filaments from those few that were added with chytrid cultures during startup of the infection experiments. Since infected hosts eventually die, the prevalence of infection after 6 days of exposure is used as a proxy for the virulence of a given chytrid strain to a given host. The Planktothrix strain NIVA-CYA97/1 served as a positive control in experiments with chytrid strains Chy-Lys2009 and Chy-Stein2010, while Planktothrix strains NIVA-CYA98 and NIVA-CYA630 were used as positive controls in experiments with Chy-Kol2008 and Chy-Hal2010, respectively. The positive controls were used to prove viability of chytrid strains, not to estimate the maximal virulence of a chytrid strain.
Statistics.
Mean values of the prevalence of infection (dependent variable) for host-parasite combinations (independent variable) were compared by the one-sided Mann-Whitney U test at a significance level of 0.05. This test was used because the data originated from independent observations and did not show normal distribution.
RESULTS AND DISCUSSION
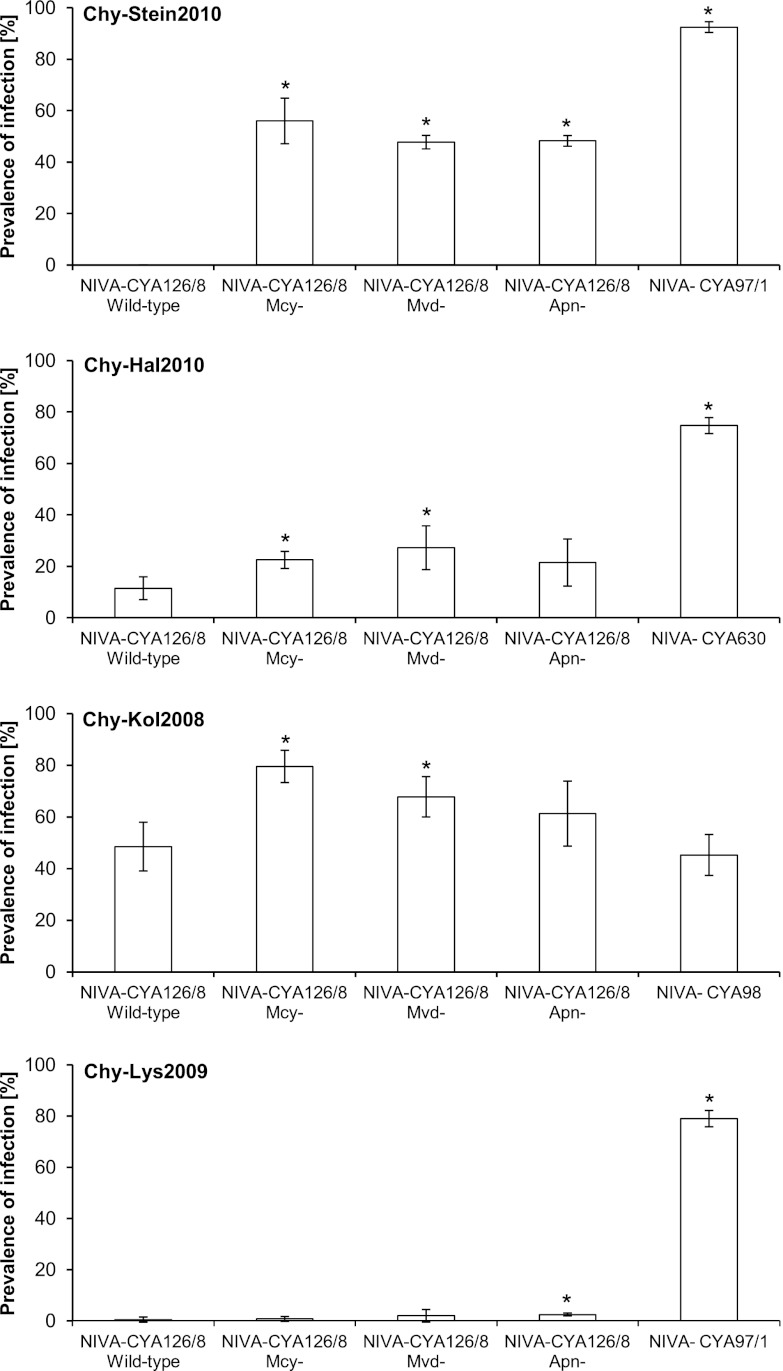
The comparison of the wild-type NIVA-CYA126/8 with its oligopeptide knockout mutants in infection experiments returned unambiguous results (Fig. 2). Chytrid strain Chy-Stein2010, for example, was unable to infect the wild-type NIVA-CYA126/8, while it showed considerable virulence to all NIVA-CYA126/8 mutants. Chytrids Chy-Hal2010 and Chy-Kol2008 were significantly less virulent to the wild-type NIVA-CYA126/8 than to the mutants with knockout mutations for the production of microcystins and microviridins. Since wild-type NIVA-CYA126/8 and the mutants share the same genotype except for mutations in oligopeptide synthetase genes, the higher chytrid virulence to NIVA-CYA126/8 mutants can be explained only by their inability to synthesize certain oligopeptides. It is therefore evident that producing microcystins, microviridins, and/or anabaenopeptins can reduce the virulence of chytrids, thereby increasing the host's chance of survival. It is also obvious that a given Planktothrix strain can produce multiple oligopeptide classes and structural variants with antichytrid activity at a time. Planktothrix may therefore resist chytrids by relying on the combined action of multiple factors. Here it is important to keep in mind that while microcystins and anabaenopeptins are nonribosomal peptides (14), microviridins are produced through posttranslational modification of ribosomally synthesized precursor peptides (15). Planktothrix may therefore control chytrid infection by employing oligopeptides that are produced via very different biosynthetic pathways. Moreover, there is reason to believe that the antichytrid defense of Planktothrix utilizes an even broader range of factors, since the resistance of the wild-type NIVA-CYA126/8 to chytrid strain Chy-Lys2009 could not be explained by microcystins, anabaenopeptins, and microviridins alone (Fig. 2).
Fig 2.
Results of infection experiments. Each diagram shows the prevalence of infection (defined as the percentage of host filaments infected) for the Planktothrix wild-type NIVA-CYA126/8 and its knockout mutants after 6 days of exposure to one of the four chytrid strains (Chy-Stein2010, Chy-Hal2010, Chy-Kol2008, and Chy-Lys2009). The mutant cultures are labeled Mcy− (microcystin knockout), Mvd− (microviridin K knockout), and Apn− (anabaenopeptin knockout). The Planktothrix strains NIVA-CYA97/1, NIVA-CYA98, and NIVA-CYA630 served as positive controls to demonstrate chytrid viability. For each host-parasite combination (independent variable), the prevalence of infection (dependent variable) is shown as the mean value with standard deviation for 4 replicates. Statistically significant differences from results from the wild-type NIVA-CYA126/8 setups are indicated by asterisks. Typically, the mutant cell lines exposed to a given chytrid showed the same prevalence of infection, which differed significantly from that of the respective positive-control strain. However, in experiments with the chytrids Chy-Kol2008 and Chy-Lys2009, the anabaenopeptin and microcystin knockout mutants differed significantly in their prevalence of infection.
Most microcystins are inhibitors of eukaryotic protein phosphatases 1 and 2A (13). At least the latter enzyme occurs in chytrids (UniprotKB accession number F4P5U1), suggesting that the antichytrid activity of microcystins may be due to an inhibition of chytrid protein phosphatases. Many other oligopeptide classes, including anabaenopeptins and microviridins, comprise structural variants targeting a diverse range of proteolytic and other enzymes (29, 30). Parasitic chytrids have absorptive nutrition. Consequently, they probably release proteolytic enzymes from their rhizoids into the host's cytoplasm to extract nutrients. In fact, several studies showed that parasitic fungi in general (31) and chytrids in particular (32, 33) utilize proteases to digest their hosts. Any chytrid proteases that are released into the cytoplasm of Planktothrix will come in direct contact with anabaenopeptins and microviridins, which, like most oligopeptides, largely remain intracellular (17). It is therefore reasonable to assume that anabaenopeptins and microviridins restrain chytrids by inhibiting chytrid proteases that act as virulence factors. If true, this would link many of the known oligopeptides to the proposed antichytrid defense of Planktothrix, since the ability to inhibit proteolytic enzymes is widespread among oligopeptides outside the class of microcystins (30).
Parasitic chytrid fungi can rapidly adapt to suboptimal hosts, and the host's main option to counter this high level of adaptability is diversification (34). The parallel production of multiple factors with antichytrid activity may be one way for Planktothrix to diversify its defense and so to slow parasite adaptation. Moreover, chytrids typically coevolve with their hosts (35). Such a coevolution normally leads to diversification in host populations, which, in turn, may trigger diversification in chytrid populations (34, 35). Hence, if Planktothrix possesses an antichytrid defense involving oligopeptides, a host-parasite coevolution should result in a subdivision of Planktothrix populations into chemotypes with different sets of oligopeptides and the cooccurrence of these chemotypes with chytrid lineages exhibiting distinct chemotype preferences. These predictions are in agreement with what has been found to occur in Norwegian waters (12). The occurrence of chytrid lineages with distinct Planktothrix chemotype preferences suggests that chytrid strains infecting Planktothrix can adapt to oligopeptides produced by Planktothrix in different ways. This is supported by the observation that our four chytrid strains differ considerably in their virulence to NIVA-CYA126/8 and its mutants.
The above findings and considerations allow drawing a number of conclusions on the antichytrid defensive system of Planktothrix. Due to differences in bioactivity between and within oligopeptides classes (29, 30), the antichytrid capability of a given Planktothrix strain probably depends on both the oligopeptide classes and the structural variants it produces. Differences in bioactivity also suggest that oligopeptide classes and some structural variants affect chytrids in an independent fashion. We therefore propose that Planktothrix possesses an antichytrid defensive system which is based on the combined effects of independently acting oligopeptides and at least one yet-unknown nonoligopeptide factor. If so, the total defensive power of Planktothrix toward a given chytrid strain is determined by either the sum or the synergy of the effects caused by individual elements of the defensive system.
The situation is complicated by the finding that chytrid strains can differ in their adaptation to oligopeptides. The outcome of a host-parasite encounter therefore depends on both the defensive system of the respective Planktothrix strain and the adaptations of the particular parasite to the host's defensive system or to certain elements of it. The Planktothrix control strains NIVA-CYA97/1, NIVA-CYA98, and NIVA-CYA630, for example, are rich in oligopeptides but nevertheless proved highly susceptible to our chytrid strains. We needed positive controls to demonstrate parasite viability, and so we might have selected Planktothrix strains with a high number of oligopeptides to which our chytrids are well adapted. Planktothrix NIVA-CYA126/8 is equally rich in oligopeptides as the control strains but nevertheless proved much less susceptible to most of our chytrid strains. Several of its oligopeptides are effective against our chytrids, as suggested by the results of infection experiments with the wild-type NIVA-CYA126/8 and mutants. Of these effective oligopeptides, only two (MC-[Asp3]-RR and MC-[Asp3]-LR) also occur in the positive-control strains, explaining why these control strains are more susceptible to most of our chytrids than all variants of NIVA-CYA126/8. Here it must be kept in mind that the probably occurring reciprocal adaptation of host and parasite, the vast number of possible oligopeptide combinations, and a yet-unknown number of other Planktothrix factors with antichytrid activity allow for a tremendous diversity in the outcome of Planktothrix-chytrid interactions.
A defensive system may be constitutive or inducible. Both types may involve secondary metabolites (36). When chytrids infect Planktothrix, they penetrate cell walls and lyse cells (9). This must result in leakage of oligopeptides into the surrounding medium. Schatz and coworkers (20) found that exposure of cyanobacterial cells to dissolved oligopeptides causes an upregulation of oligopeptide production. This upregulation, which most likely follows a chytrid-mediated leakage of oligopeptides into the medium, may be seen as part of an inducible defense. However, whether chytrid enzymes are inhibited by Planktothrix oligopeptides depends on how much of the effective oligopeptides is dissolved in the host's cytoplasm. In many cases, the cytoplasm of cyanobacteria is saturated or oversaturated with oligopeptides already in pure laboratory cultures (18), even in the absence of contact with any pathogen. It is therefore unlikely that an upregulation of oligopeptide production would significantly improve the antichytrid defense. This suggests that the putative antichytrid defense of Planktothrix may be constitutive.
Studies on chytrids infecting diatoms showed that the host may limit parasite growth by hypersensitivity, i.e., by programmed death of cells that are exposed to the parasites (35). We were unable to observe a similar effect in experiments with Planktothrix. As mentioned, chytrids can rapidly adapt to suboptimal hosts (34). On the other hand, an adaptation of a clonal chytrid culture from one generation to another has, to our knowledge, never been described. This may mean that chytrid adaptation is driven mainly by changes in the genetic code which take time.
In summary, the results of infection experiments and the above considerations on Planktothrix-chytrid coevolution suggest that oligopeptide production by Planktothrix is a part of a defensive mechanism against chytrid parasitism. This suggestion is supported by the previously reported correlation between chytrid virulence to 35 Planktothrix strains and their oligopeptide composition (12). Our work does not exclude the possibility that oligopeptides may serve additional metabolic or allelopathic functions. In fact, the finding that the antichytrid defense of Planktothrix may include both ribosomal and nonribosomal oligopeptides, i.e., metabolites that probably evolved independently, makes it unlikely that oligopeptides originally evolved as antichytrid agents. Instead, already-existing oligopeptides may have been “recruited” for an antichytrid defensive system when the occurrence of parasitic chytrids in the Devonian age (37) or later confronted Planktothrix with a new group of very potent pathogens. The suggested function as antichytrid agents is therefore not in contradiction to the common belief that oligopeptide evolution predated that of eukaryotes (38). Strictly speaking, the findings of the present study cannot be generalized beyond the host-parasite relationship of Planktothrix and chytrids. Nevertheless, due to the widespread occurrence of chytrid parasitism and oligopeptide production in cyanobacteria, working toward such a generalization seems a promising task for future research.
ACKNOWLEDGMENTS
We express our gratitude to Randi Skulberg, Norwegian Institute for Water Research, for supplying chytrid strains and to Marcia Kyle, Norwegian University of Life Sciences, for many constructive comments on this paper.
This work was supported by Norwegian Research Council grant 183360/S30 and by the Austrian Science Fund (FWF-P24070).
Footnotes
Published ahead of print 8 February 2013
REFERENCES
- 1. Codd GA. 2005. CYANONET—a global network for cyanobacterial bloom and toxin risk management. Initial situation assessment and recommendations. Technical documents in hydrology, no. 76. UNESCO, Paris, France [Google Scholar]
- 2. Shapiro J. 1990. Current beliefs regarding dominance of blue-greens: the case for the importance of CO2 and pH. Verh. Int. Ver. Limnol. 24:38–54 [Google Scholar]
- 3. Dokulil MT, Teubner K. 2000. Cyanobacterial dominance in lakes. Hydrobiologia 438:1–12 [Google Scholar]
- 4. Lampert W. 1987. Daphnia. Mem. Ist. Ital. Idrobiol. 45:143–192 [Google Scholar]
- 5. Deng L, Hayes PK. 2008. Evidence for cyanophages active against bloom-forming freshwater cyanobacteria. Freshw. Biol. 53:1240–1252 [Google Scholar]
- 6. Rasconi S, Niquil N, Sime-Ngando T. 2012. Phytoplankton chytridiomycosis: community structure and infectivity of fungal parasites in aquatic ecosystems. Environ. Microbiol. 14:2151–2170 [DOI] [PubMed] [Google Scholar]
- 7. Sparrow FK. 1960. Aquatic phycomycetes. The University of Michigan Press, Ann Arbor, MI [Google Scholar]
- 8. Sigee DC, Selwyn A, Gallois P, Dean AP. 2007. Patterns of cell death in freshwater colonial cyanobacteria during the late summer bloom. Phycologia 46:284–292 [Google Scholar]
- 9. Canter HM, Lund JWG. 1951. Studies on plankton parasites. III. Examples of the interaction between parasitism and other factors determining the growth of diatoms. Ann. Bot. 15:359–371 [Google Scholar]
- 10. Gleason FH, Lilje O. 2009. Structure and function of fungal zoospores: ecological implications. Fungal Ecol. 2:53–59 [Google Scholar]
- 11. Rasconi S, Jobard M, Jouve L, Sime-Ngando T. 2009. Use of Calcofluor White for detection, identification, and quantification of phytoplanktonic fungal parasites. Appl. Environ. Microbiol. 75:2545–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sønstebø JH, Rohrlack T. 2011. Possible implications of chytrid parasitism for population subdivision in freshwater cyanobacteria of the genus Planktothrix. Appl. Environ. Microbiol. 77:1344–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Welker M, von Döhren H. 2006. Cyanobacterial peptides—nature's own combinatorial biosynthesis. FEMS Microbiol. Rev. 30:530–563 [DOI] [PubMed] [Google Scholar]
- 14. Kurmayer R, Christiansen G. 2009. The genetic basis of toxin production in cyanobacteria. Freshw. Rev. 2:31–50 [Google Scholar]
- 15. Philmus B, Christiansen G, Yoshida WY, Hemscheidt TK. 2008. Post-translational modification in microviridin biosynthesis. Chembiochem 9:3066–3073 [DOI] [PubMed] [Google Scholar]
- 16. Christiansen G, Kurmayer R, Liu Q, Börner T. 2006. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl. Environ. Microbiol. 72:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Young FM, Thomson C, Metcalf JS, Lucocq JM, Codd GA. 2005. Immunogold localisation of microcystins in cryosectioned cells of Microcystis. J. Struct. Biol. 151:208–214 [DOI] [PubMed] [Google Scholar]
- 18. Jüttner F, Lüthi H. 2008. Topology and enhanced toxicity of bound microcystins in Microcystis PCC 7806. Toxicon 51:388–397 [DOI] [PubMed] [Google Scholar]
- 19. Kehr JC, Zilliges Y, Springer A, Disney MD, Ratner DD, Bouchier C, Seeberger PH, Tandeau de Marsac N, Dittmann E. 2006. A mannan binding lectin is involved in cell-cell attachment in a toxic strain of Microcystis aeruginosa. Mol. Microbiol. 59:893–906 [DOI] [PubMed] [Google Scholar]
- 20. Schatz D, Keren Y, Vardi A, Sukenik A, Carmeli S, Börner T, Dittmann E, Kaplan A. 2007. Towards clarification of the biological role of microcystins, a family of cyanobacterial toxins. Environ. Microbiol. 9:965–970 [DOI] [PubMed] [Google Scholar]
- 21. Kotak BG, Lam AKY, Prepas EE, Hrudey SE. 2000. Role of chemical and physical variables in regulating microcystin-LR concentration in phytoplankton of eutrophic lakes. Can. J. Fish. Aquat. Sci. 57:1584–1593 [Google Scholar]
- 22. Christoffersen K. 1996. Ecological implications of cyanobacterial toxins in aquatic food webs. Phycologia 35:42–50 [Google Scholar]
- 23. Blom JF, Bister B, Bischoff D, Nicholson G, Jung G, Süssmuth RD, Jüttner F. 2003. Oscillapeptin J, a new grazer toxin of the freshwater cyanobacterium Planktothrix rubescens. J. Nat. Prod. 66:431–434 [DOI] [PubMed] [Google Scholar]
- 24. Rohrlack T, Christoffersen K, Dittmann E, Nogueira I, Vasconselos V, Börner T. 2005. Ingestion of microcystins by Daphnia: intestinal uptake and toxic effects. Limnol. Oceanogr. 50:440–448 [Google Scholar]
- 25. Christiansen G, Philmus B, Hemscheidt T, Kurmayer R. 2011. Genetic variation of adenylation domains of the anabaenopeptin synthesis operon and evolution of substrate promiscuity. J. Bacteriol. 193:3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Welker M, Christiansen G, von Döhren H. 2004. Diversity of coexisting Planktothrix (Cyanobacteria) chemotypes deduced by mass spectral analysis of microystins and other oligopeptides. Arch. Microbiol. 182:288–298 [DOI] [PubMed] [Google Scholar]
- 27. Rohrlack T, Skulberg R, Skulberg OM. 2009. Distribution of oligopeptide chemotypes of the cyanobacterium Planktothrix and their persistence in selected lakes in Fennoscandia. J. Phycol. 45:1259–1265 [DOI] [PubMed] [Google Scholar]
- 28. Okumura HS, Philmus B, Portmann C, Hemscheidt TK. 2009. Homotyrosine-containing cyanopeptolins 880 and 960 and anabaenopeptins 908 and 915 from Planktothrix agardhii CYA 126/8. J. Nat. Prod. 72:172–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rohrlack T, Christoffersen K, Hansen PE, Zhang W, Czarnecki O, Henning M, Fastner J, Erhard M, Neilan BA, Kaebernick M. 2003. Isolation, characterization, and quantitative analysis of microviridin J, a new Microcystis metabolite toxic to Daphnia. J. Chem. Ecol. 29:1757–1770 [DOI] [PubMed] [Google Scholar]
- 30. Rouhiainen L. 2004. Ph.D. thesis University of Helsinki, Helsinki, Finland [Google Scholar]
- 31. St. Leger RJ, Joshi L, Roberts DW. 1997. Adaptation of proteases and carbohydrases of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology 143:1983–1992 [DOI] [PubMed] [Google Scholar]
- 32. Krarup T, Olson LW, Heldt-Hansen HP. 1994. Some characteristics of extracellular proteases produced by members of the Chytridiales and the Spizellomycetales (Chytridiomycetes). Can. J. Microbiol. 40:106–112 [Google Scholar]
- 33. Piotrowski JS, Annis SL, Longcore JE. 2004. Physiology of Batrachochytrium dendrobatidis, a chytrid pathogen of amphibians. Mycologia 96:9–15 [PubMed] [Google Scholar]
- 34. De Bruin A, Ibelings BW, Kagami M, Mooij WM, Van Donk E. 2008. Adaptation of the fungal parasite Zygorhizidium planktonicum during 200 generations of growth on homogeneous and heterogeneous populations of its host, the diatom Asterionella formosa. J. Eukaryot. Microbiol. 55:69–74 [DOI] [PubMed] [Google Scholar]
- 35. Gsell SG. 2013. Ph.D. thesis University of Utrecht, Utrecht, The Netherlands [Google Scholar]
- 36. Kaplan I, Halitschke R, Kessler A, Sardanelli S, Denno RF. 2008. Constitutive and induced defenses to herbivory in above and below plan tissues. Ecology 89:392–406 [DOI] [PubMed] [Google Scholar]
- 37. Taylor TN, Remy W, Hass H. 1994. Allomyces in the Devonian. Nature 367:601 [Google Scholar]
- 38. Rantala A, Fewer DP, Hisbergues M, Rouhiainen L, Vaitomaa J, Börner T, Sivonen K. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. U. S. A. 101:568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]