Abstract
Defensins are small antimicrobial peptides (AMPs) that play an important role in the innate immune system of mammals. Since the effect of mycotoxin contamination of food and feed on the secretion of intestinal AMPs is poorly understood, the aim of this study was to elucidate the individual and combined effects of four common Fusarium toxins, deoxynivalenol (DON), nivalenol (NIV), zearalenone (ZEA), and fumonisin B1 (FB1), on the mRNA expression, protein secretion, and corresponding antimicrobial effects of porcine β-defensins 1 and 2 (pBD-1 and pBD-2) using a porcine jejunal epithelial cell line, IPEC-J2. In general, upregulation of pBD-1 and pBD-2 mRNA expression occurred following exposure to Fusarium toxins, individually and in mixtures (P < 0.05). However, no significant increase in secreted pBD-1 and pBD-2 protein levels was observed, as measured by enzyme-linked immunosorbent assay (ELISA). Supernatants from IPEC-J2 cells exposed to toxins, singly or in combination, however, possessed significantly less antimicrobial activity against Escherichia coli than untreated supernatants. When single toxins and two-toxin combinations were assessed, toxicity effects were shown to be nonadditive (including synergism, potentiation, and antagonism), suggesting interactive toxin effects when cells are exposed to mycotoxin combinations. The results show that Fusarium toxins, individually and in mixtures, activate distinct antimicrobial defense mechanisms possessing the potential to alter the intestinal microbiota through diminished antimicrobial effects. Moreover, by evaluating toxin mixtures, this improved understanding of toxin effects will enable more effective risk assessments for common mycotoxin combinations observed in contaminated food and feed.
INTRODUCTION
Antimicrobial peptides (AMPs) are effector molecules of innate immunity with direct antimicrobial activity (1). AMPs can also be mediators of inflammation, influencing a variety of processes, such as proliferation, adaptive immune system regulation, wound repair, cytokine and histamine release, chemotaxis, and protease and antiprotease balance (1–8). To date, hundreds of AMPs identified from vertebrates, invertebrates, plants, and fungi have been characterized, with sequence data deposited in the Antimicrobial Sequences Database (AMSDb), which is publically available (http://www.bbcm.univ.trieste.it/~tossi/amsdb.html) (9).
One of the major AMP subclasses includes the β-defensin family. The members are mainly expressed in epithelial cells of organs exposed to the external environment, such as the skin, gastrointestinal tract, and respiratory tract (10). Dynamic regulation of β-defensins has been shown in various models of gastrointestinal illnesses and inflammation. For example, Campylobacter jejuni was shown to induce bactericidal human β-defensins 2 and 3 (hBD-2 and hBD-3) in intestinal epithelial cells (IECs) (11). Reports have also demonstrated that Salmonella infection differentially affected the expression of porcine β-defensin 1 (pBD-1) and pBD-2 mRNA transcript levels in porcine ileum and jejunum epithelial cells (10, 12).
Fusarium spp. are commonly recovered from cereals grown in temperate areas of America, Europe, and Asia (13). Fusarium toxins elicit a wide spectrum of toxic effects, including the capacity to modify normal immune functions in both humans and animals (14). The most important Fusarium toxins potentially affecting mammalian health are zearalenone (ZEA), fumonisin B1 (FB1), and trichothecenes, such as deoxynivalenol (DON) and nivalenol (NIV). In crops and associated food products, the presence of multiple mycotoxin types is commonly observed around the world, making Fusarium toxins a serious public health concern. However, investigations on the combined effects of mycotoxins observed in the food supply are scarce. The few studies performed have been based on endpoints that include inhibition of protein and DNA synthesis, DNA methylation and fragmentation, cell viability, and proliferation (15–20). However, the combined effects of mycotoxins on intestinal immunity are currently poorly understood. Previous experiments have shown that exposure to several mycotoxins may increase susceptibility to experimental or natural mucosal infections by inducing bacterial translocation and colonization across the intestinal epithelium (21, 22), but no data are available examining the role mycotoxin combinations may play in intestinal infections. Reports have been done demonstrating that the antimicrobial activity of AMPs, such as β-defensins, may be associated with bacterial populations in the intestine, which naturally present a barrier limiting undesirable gut mucosal infections (23–25). Accordingly, we hypothesized that a mixture of naturally cooccurring Fusarium toxins that can be observed in food exert significant effects on the synthesis and secretion of AMPs compared to their effects as individual toxins. Such effects could have a significant influence on microbial survival in the gut. This study used a porcine intestinal epithelial cell line, IPEC-J2 (25), to investigate the individual and interactive effects of DON, NIV, ZEA, and FB1 on the mRNA expression and secretion of pBD-1 and pBD-2 and examined antimicrobial effects observed in toxin-exposed cell supernatants.
MATERIALS AND METHODS
Chemicals and reagents.
All mycotoxins (DON, NIV, ZEA, and FB1), dimethyl sulfoxide (DMSO), phosphate-buffered saline (PBS), Luria-Bertani (LB) broth, and agar were obtained from Sigma Chemical Company (St. Louis, MO). Dulbecco's modified Eagle medium (DMEM)–Ham's F-12 (1:1) and fetal bovine serum (FBS) were provided by Gibco-Life Technology (Eggenstein, Germany). RNAiso Plus was purchased from TaKaRa (Dalin, China). SuperScript III First-Strand Synthesis SuperMix was supplied by Invitrogen Life Technologies (Carlsbad, CA). Fast SYBR green master mix was obtained from Applied Biosystems (Foster City, CA). Commercial enzyme-linked immunosorbent assay (ELISA) kits for pBD-1 and pBD-2 were purchased from Uscn Life Science Inc. (Wuhan, China).
Cell line and culture conditions.
IPEC-J2 is a nontransformed intestinal cell line originally derived from the jejunums of neonatal, unsuckled piglets (26) and was a kind gift of Per Torp Sangild (Department of Human Nutrition/Clinical Nutrition, Faculty of Life Science, University of Copenhagen, Copenhagen, Denmark). Cells (passages 80 to 92) were maintained in DMEM–Ham's F-12 (1:1) containing high glucose (4.5 g/liter) supplemented with 10% FBS and incubated at 37°C in an atmosphere of 5% CO2-95% air mixture. All cells were screened for mycoplasma contamination prior to use (Lonza, Basel, Switzerland).
Preliminary mycotoxin concentration response experiment.
Fusarium toxin (DON, NIV, ZEA, and FB1) concentrations were optimized previously by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (L. Y. M. Wan, P. C. Turner, and E. El-Nezami, submitted for publication). Identified concentrations for toxins, individually and in combination, were used to examine the impact on mRNA expression, secreted-protein levels, and associated antimicrobial effects, as described below.
Identification of mycotoxin combination treatments.
To minimize the number of possible toxin combinations (i.e., all possible combinations for every concentration of the respective toxin), an inscribed central composite design was used (27). It incorporated a fractional factorial approach with four factors (i.e., DON, NIV, ZEA, and FB1) and has been used similarly previously (Wan et al. submitted). Through this approach, the number of toxin combination treatments was reduced from 44 to 16. The design matrix is presented in Table 1.
Table 1.
Design matrix for four Fusarium toxins
| Treatment combination no. | DON (0.5 or 2 μM) | NIV (0.5 or 2 μM) | ZEA (10 or 40 μM) | FB1 (20 or 40 μM) |
|---|---|---|---|---|
| 1 | 0a | 0 | 0 | 0 |
| 2 | 1b | 0 | 0 | 0 |
| 3 | 0 | 1 | 0 | 0 |
| 4 | 0 | 0 | 1 | 0 |
| 5 | 0 | 0 | 0 | 1 |
| 6 | 1 | 1 | 0 | 0 |
| 7 | 1 | 0 | 1 | 0 |
| 8 | 1 | 0 | 0 | 1 |
| 9 | 0 | 1 | 1 | 0 |
| 10 | 0 | 1 | 0 | 1 |
| 11 | 0 | 0 | 1 | 1 |
| 12 | 1 | 1 | 1 | 0 |
| 13 | 1 | 1 | 0 | 1 |
| 14 | 1 | 0 | 1 | 1 |
| 15 | 0 | 1 | 1 | 1 |
| 16 | 1 | 1 | 1 | 1 |
1, mycotoxin treatment.
0, no mycotoxin treatment.
qPCR.
IPEC-J2 cells were seeded at a density of 5 × 105 CFU/well in 6-well culture plates (Costar, Corning, NY) and allowed to adhere for 24 h. Typically, confluence was achieved after 3 to 4 days, with media being replaced three times weekly; for all experiments, cells were used within 14 days from seeding (26). The cells were washed with PBS and treated with Fusarium toxins in serum-free medium for 48 h. Total RNA was extracted using RNAiso Plus according to the manufacturer's instructions. RNA was resuspended in 30 μl of nuclease-free water and stored at −80°C. RNA concentrations were measured using a NanoDrop ND-1000 Spectrophotometer (Nano-Drop Technologies, Wilmington, DE). Prior to use in quantitative PCR (qPCR), RNA quality was determined by ensuring a value of >1.8 for the A260/A280 ratio. cDNA was prepared from 1 μg of total RNA using SuperScript III First-Strand Synthesis SuperMix according to the manufacturer's instructions. qPCR was performed to quantify mRNA transcript levels for pBD-1 and pBD-2 relative to the expression of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) endogenous housekeeping control gene. All samples were run on a StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA) using 1 μl of cDNA and Fast SYBR green master mix, with final primer concentrations of 0.5 μM per primer in a final volume of 20 μl. Porcine specific cytokine primers (Table 2) were generated from published GenBank sequences using Primer Express software (Applied Biosystems). Samples were centrifuged briefly and thermocycled using the default fast program (40 cycles of 95°C for 3 s, 60°C for 30 s). Relative changes in gene expression levels of pBD-1 and pBD-2 in cultured jejunal enterocytes resulting from mycotoxin treatments were normalized against GAPDH using the 2−ΔΔCT method as described previously (28). Experiments were repeated four times independently, with each treatment performed in triplicate.
Table 2.
Primer sequences for quantification of pBD-1, pBD-2, and GAPDH by qPCR
| Primer set | Product length (bpa) | Primer sequence (5′–3′) |
Accession no. | |
|---|---|---|---|---|
| Forward | Reverse | |||
| pBD-1 | 141 | CTCCTCCTTGTATTCCTCCT | GGTGCCGATCTGTTTCAT | NM_213838 |
| pBD-2 | 148 | GACTGTCTGCCTCCTCTC | GGTCCCTTCAATCTGTTG | NM_214442 |
| GAPDH | 120 | ATGGTGAAGGTC GGAGTG | GTAGTGGAGGTCAATGAAGG | NM_001206359 |
bp, base pairs.
ELISA analysis of pBD-1 and pBD-2.
Protein levels of pBD-1 and pBD-2 in cell supernatants were analyzed using a commercial ELISA kit for pBD-1 and pBD-2 according to the manufacturer's instructions. All samples were run in duplicate, with experiments repeated three times independently for each treatment.
Analysis of antibacterial activity.
To examine the antibacterial effects of toxin treatments on exposed IPEC-J2 cells, a clinical Escherichia coli strain, ATCC 25922, kindly provided by W. C. Yam, Department of Microbiology, University of Hong Kong, was employed. In short, all tests were performed with E. coli grown at 37°C in LB broth to an optical density at 600 nm of 0.4. Cells were centrifuged at 2,060 × g (Beckman Coulter, Fullerton, CA; GS-6R centrifuge) for 15 min and resuspended in sterile phosphate-buffered saline (PBS) at a final concentration of 1.0 × 107 CFU/ml. A 0.5-ml aliquot was mixed with 0.5 ml of supernatant obtained from IPEC-J2 cells exposed or unexposed to the respective toxin treatments. Following incubation for 2 h at 37°C with shaking (200 rpm), serial dilutions were plated on LB agar, with CFU counted after 18 h at 37°C. Relative changes in CFU following treatment were calculated as (CFU after mycotoxin-treated cell supernatant incubation)/(CFU after control cell supernatant incubation).
Statistical analyses.
Results of qPCR and antimicrobial effects were expressed as the mean ± standard error of the mean (SEM) of four individual experiments. All data analyses were performed using the SPSS statistical package (SPSS version 20.0 for Windows; SPSS Inc., Chicago, IL). Data were first evaluated for normality with the Shapiro-Wilk and Levene's variance homogeneity test. The data were not normally distributed for pBD-1 and pBD-2 mRNA and protein levels but were normally distributed for CFU counts. One-way analysis of variance (ANOVA) with the Kruskal-Wallis test, followed by the Mann-Whitney U test, was used to identify significant differences for nonparametric data. One-way ANOVA with Dunnett's multiple-comparison test was used for analyzing parametric data. Differences were considered to be statistically significant when P values were less than 0.05. Differences in pBD-1 and pBD-2 protein levels for three independent experiments were analyzed using a scatter plot.
Univariate analyses of variance were performed to determine if there were any associations between different toxin treatments and bacterial survival. Effects with P values of less than 0.05 at the 95% confidence interval were regarded as significant, suggesting potential interactive effects (either synergistic or less than additive) of different Fusarium toxins detected in the bacterial-survival assay, whereas effects with P values of >0.05 at the 95% confidence interval were considered nonsignificant. A lack of interaction indicates the effects are additive (i.e., combined effects would be the sum of their individual effects) (29). Correlations between pBD-1 and pBD-2 gene expression, supernatant protein levels, and antibacterial effects of supernatants of IPEC-J2 cells treated with cytotoxic and noncytotoxic concentrations of DON, NIV, ZEA, and FB1, individually and in mixtures, were assessed by Pearson's (parametric) and Spearman's (nonparametric) correlations.
RESULTS
Individual and combined effects of mycotoxins on pBD-1 and pBD-2 mRNA expression in IPEC-J2 epithelial cells.
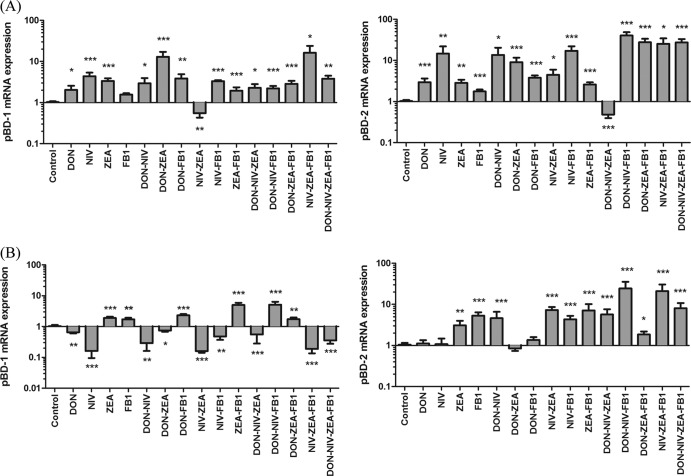
Levels of mRNA expression of pBD-1 and pBD-2 in IPEC-J2 cells following exposure to DON, NIV, ZEA, or FB1 are presented in Fig. 1. Relative levels of pBD-1 were upregulated in IPEC-J2 cells upon treatment with cytotoxic concentrations (expressed as toxin and concentration in μM, e.g., DON 2 represents DON at 2 μM) of Fusarium toxins alone (DON 2, NIV 2, or ZEA 40) or in any combination of DON 2, NIV 2, ZEA 40, and FB1 40, except NIV 2-ZEA 40. When cells were treated with noncytotoxic concentrations of DON 0.5, NIV 0.5, ZEA 10, and FB1 20, relative levels of pBD-1 mRNA were significantly (P < 0.05) downregulated in DON 0.5 and NIV 0.5 alone and in mixtures of DON 0.5-NIV 0.5, DON 0.5-ZEA 10, NIV 0.5-ZEA 10, NIV 0.5-FB1 20, DON 0.5-NIV 0.5-ZEA 10, NIV 0.5-ZEA 10-FB1 20, and DON 0.5-NIV 0.5-ZEA 10-FB1 20 but were upregulated in ZEA 10 and FB1 20 alone and mixtures of DON 0.5-FB1 20, ZEA 10-FB1 20, DON 0.5-NIV 0.5-FB1 20, and DON 0.5-ZEA 10-FB1 20.
Fig 1.
Relative abundances of pBD-1 and pBD-2 mRNAs from porcine IPEC-J2 cells isolated 48 h following treatment, individually or in combination, with cytotoxic (A) and noncytotoxic (B) concentrations of DON, NIV, ZEA, and FB1. The results are expressed as means (±standard errors of the mean [SEM]; n = 4) relative to the control; *, **, and ***, P < 0.05, 0.01, and 0.001, respectively, compared to the control.
Similarly, pBD-2 mRNA expression was significantly (P < 0.05) upregulated in most of the cytotoxic treatment groups, except for DON 2-NIV 2-ZEA 40 treatment. However, when cells were treated with noncytotoxic concentrations of DON 0.5, NIV 0.5, ZEA 10, and FB1 20, significant upregulation of pBD-2 mRNA expression was observed only in ZEA 10 and FB1 20 alone and in mixtures of DON 0.5-NIV 0.5, NIV 0.5-ZEA 10, NIV 0.5-FB1 20, ZEA 10-FB1 20, DON 0.5-NIV 0.5-ZEA 10, DON 0.5-NIV 0.5-FB1 20, DON 0.5-ZEA 10-FB1 20, NIV 0.5-ZEA 10-FB1 20, and DON 0.5-NIV 0.5-ZEA 10-FB1 20.
ELISA analysis of pBD-1 and pBD-2 protein levels in IPEC-J2 cell culture supernatants.
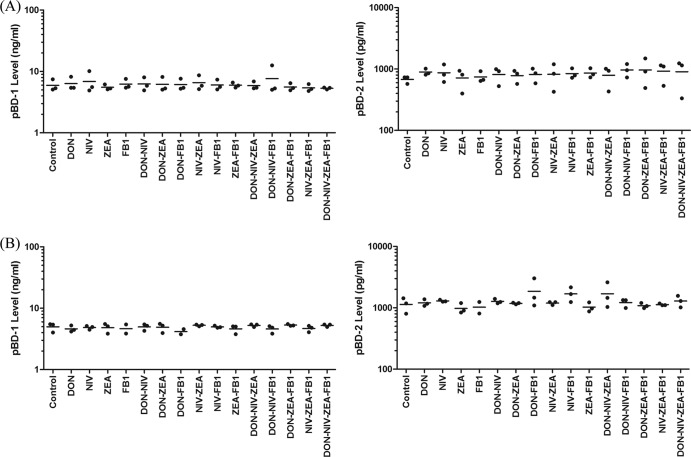
Since the qPCR data revealed changes in mRNA expression of pBD-1 and pBD-2 following exposure to cytotoxic and noncytotoxic concentrations of DON, NIV, ZEA, and FB1, individually and in mixtures, cell supernatants were also measured for their respective protein levels by ELISA. Scatter plots showing the effects of mycotoxins, individually and in mixtures, on pBD-1 and pBD-2 secretion levels are presented in Fig. 2. Differences were not statistically significant for any treatment.
Fig 2.
Scatter plots illustrating the cytotoxic (A) and noncytotoxic (B) effects of DON, NIV, ZEA, and FB1, individually and in mixtures, on pBD-1 and pBD-2 levels in cell supernatants as analyzed by ELISA. Means are indicated by the horizontal lines (n = 3).
Antibacterial activities of cell culture supernatants.
The antibacterial activities of pBD-1 and pBD-2 in cell culture supernatants were evaluated by plate counting. When IPEC-J2 cells were treated with cytotoxic concentrations of DON 2, NIV 2, ZEA 40, and FB1 40, concentrations of E. coli were significantly increased (P < 0.05) in DON 2, NIV 2, and FB1 40 alone and in mixtures of DON 2-NIV 2, NIV 2-FB1 40, ZEA 40-FB1 40, DON 2-NIV 2-ZEA 40, DON 2-NIV 2-FB1 40, DON 2-ZEA 40-FB1 40, NIV 2-ZEA 40-FB1 40, and DON 2-NIV 2-ZEA 40-FB1 40 (Fig. 3A). However, when cells were treated with noncytotoxic concentrations of DON 0.5, NIV 0.5, ZEA 10, and FB1 20, significant increases in the concentrations of E. coli were observed only for NIV alone, NIV 0.5-ZEA 10, DON 0.5-NIV 0.5-FB1 20, NIV 0.5-ZEA 10-FB1 20, and DON 0.5-NIV 0.5-ZEA 10-FB1 20 (Fig. 3B). Such increases in E. coli concentrations demonstrated that treated cell supernatants possessed reduced antibacterial activity.
Fig 3.
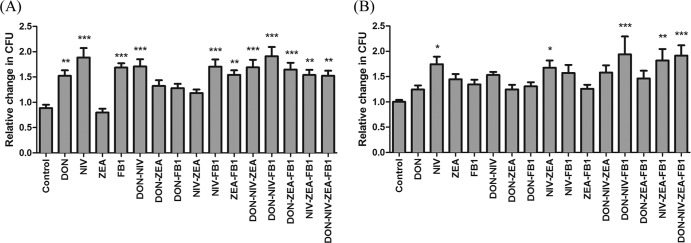
Antibacterial activities of IPEC-J2 cell culture supernatants as determined by numbers of CFU following exposure to cytotoxic (A) and noncytotoxic (B) concentrations of DON, NIV, ZEA, and FB1, individually and in mixtures. The number of E. coli CFU in the control was set at 1. The results are expressed as the relative change in the CFU count (±SEM) compared to the control (n = 4). *, **, and ***, P < 0.05, 0.01, and 0.001, respectively, compared to the control.
In order to determine if there were any interactions among DON, NIV, ZEA, and FB1 affecting E. coli survival, univariate ANOVA was conducted (Table 3). ANOVA provides a powerful statistical tool for tests involving multiple factors and their interactions (30). The results reveal nonadditive interactions in mixtures of NIV 2-FB1 40 (F1,151 = 31.383; P < 0.001), DON 2-NIV 2-FB1 40 (F1,151 = 17.788; P < 0.001), and DON 2-NIV 2-ZEA 40-FB1 40 (F1,151 = 12.432; P = 0.001) (Table 3). No interactions were observed for noncytotoxic concentrations of DON, NIV, ZEA, and FB1 (data not shown).
Table 3.
Results of univariate analyses of multiple-Fusarium-toxin exposure (cytotoxic) on IPEC-J2 cells as analyzed by surviving CFU
| Toxin combination | CFU |
|
|---|---|---|
| F value | P valuea | |
| DON-NIV | 0.506 | 0.478 |
| DON-ZEA | 0.939 | 0.334 |
| DON-FB1 | 0.977 | 0.325 |
| NIV-ZEA | 0.029 | 0.865 |
| NIV-FB1 | 31.383 | <0.001 |
| ZEA-FB1 | 1.874 | 0.173 |
| DON-NIV-ZEA | 3.415 | 0.067 |
| DON-NIV-FB1 | 17.788 | <0.001 |
| DON-ZEA-FB1 | 0.35 | 0.555 |
| NIV-ZEA-FB1 | 1.235 | 0.268 |
| DON-NIV-ZEA-FB1 | 12.432 | 0.001 |
Boldface indicates P < 0.05.
Correlations among pBD-1 and pBD-2 mRNA expression, protein levels, and antibacterial activity.
Upon exposure to cytotoxic concentrations of DON, NIV, ZEA, and FB1, significant positive correlations were observed between pBD-1 and pBD-2 mRNA expression levels (P < 0.001) as determined by qPCR. Significant negative correlations were revealed between pBD-1 and pBD-2 protein levels (P < 0.001). The antibacterial activities of the cell culture supernatants correlated significantly (P = 0.031) with pBD-2 mRNA expression. However, when cells were treated with noncytotoxic concentrations, the only significant correlation was between pBD-1 mRNA expression and pBD-2 levels (P = 0.038), though the reasons for this are not clear (Table 4).
Table 4.
Correlations among pBD-1 and pBD-2 mRNA expression, protein levels, and antibacterial activity for cytotoxic and noncytotoxic concentrations of Fusarium toxins
| Parameter at Fusarium toxin concn: | Valuea |
|||
|---|---|---|---|---|
| pBD-1 |
pBD-2 |
|||
| mRNA | Secretion | mRNA | Secretion | |
| Cytotoxic | ||||
| pBD-1 secretion | −0.041 (0.787) | |||
| pBD-2 mRNA | 0.344 (<0.001) | 0.130 (0.465) | ||
| pBD-2 secretion | 0.003 (0.986) | −0.666 (<0.001) | −0.104 (0. 557) | |
| CFU | 0.015 (0.859) | 0.138 (0.395) | 0.186 (0.031) | 0.086 (0.597) |
| Noncytotoxic | ||||
| pBD-1 secretion | −0.191 (0.204) | |||
| pBD-2 mRNA | 0.003 (0.969) | −0.006 (0. 972) | ||
| pBD-2 secretion | −0.304 (0.038) | −0.289 (0.051) | −0.198 (0.202) | |
| CFU | 0.088 (0.259) | 0.054 (0.720) | −0.054 (0.489) | 0.052 (0.728) |
Values are expressed as correlation coefficients, with P values in parentheses (boldface indicates P < 0.05).
DISCUSSION
This is the first study to demonstrate the individual and combined effects of four commonly occurring Fusarium toxins, DON, NIV, ZEA, and FB1, on the mRNA expression and protein secretion of AMPs pBD-1 and pBD-2. Following treatment with most toxins, mRNA expression was significantly affected for both genes, though cell supernatant levels of pBD-1 and pBD-2 were not significantly changed. In this study, the expression of pBD-1 and pBD-2 was shown to be low and is similar to what has been reported for other cells (10, 24, 31). It has previously been suggested that this basal level of expression may reflect a potential for upregulation of these defensins upon bacterial infection, and upregulation of these defensins has been reported following Salmonella infection (10, 24).
pBD-1 is a well-known porcine homologue of hBD-2 (32). Zhang et al. (8) and Elahi et al. (33) previously showed pBD-1 to be constitutively expressed in different tissues (8). However, induction of pBD-1 mRNA expression may also be stimulated upon exposure to food contaminants or enteric pathogens (10, 33, 34) and is in line with our data, which show pBD-1 mRNA expression was upregulated following exposure to DON, NIV, ZEA, and FB1, individually and in mixtures. Elevated expression of pBD-1 in, for example, IECs may play a potential role in surveillance and maintenance of a homeostatic state of microflora on the mucosal epithelium and may be significant in preventing the development or progression of diseases (33, 35, 36).
pBD-2, on the other hand, is a recently discovered and described porcine defensin found in the intestines of pig species and is speculated to be the porcine orthologue of hBD-1 (24, 37). Results show that mRNA expression of pBD-2 was upregulated in IPEC-J2 cells following exposure to DON, NIV, ZEA, and FB1, individually and in mixtures. It has been reported that upregulation of defensins contributes to the early response to bacterial infections, tissue injury, and inflammation (23). Any disturbance of defensin production may lead to the disruption of microbial homeostasis, potentially contributing to chronic enteric diseases, such as Crohn's disease (11, 38) and inflammatory bowel disease (39).
To date, regulation of pBD-1 and pBD-2 levels has been described at the transcriptional level (10, 12, 24, 31), with little data on posttranscriptional regulation available (40). Thus, we investigated the protein levels of pBD-1 and pBD-2 present in cell supernatants and compared them with our transcriptional data obtained by qPCR. For both proteins, no significant increases in protein levels were observed under our experimental conditions, which were inconsistent with the significant changes observed with mRNA levels. Discrepancies between mRNA expression, protein abundance, and biological effects have been reported in previous studies (41–43). These differences may be explained, at least partly, by posttranscriptional or posttranslational regulatory mechanisms linked to defensin molecule secretion and are likely influenced by protein degradation pathways (44), though this remains to be shown. Moreover, differences between defensin mRNA and protein secretion may also reveal issues associated with detection thresholds for either mRNA or protein, or secretion of the proteins. Despite advances in the technologies for measuring protein abundance in recent decades, the experimental techniques for protein identification and quantification still lag considerably behind the highly sensitive methods available for quantifying mRNA transcript levels (45). However, while mRNA expression values are useful in various applications, such as classification, identification, and prediction of drug-induced toxicities or cancers (46, 47), the results are correlative rather than causative. It is generally recognized that changes in protein levels, even when subtle (i.e., less than what is deemed statistically significant), may have significant biological effects, and this is currently an area in which analytical techniques with increased sensitivity are required (45).
The results of the antibacterial assay indicate the relevance of defensin expression in gastrointestinal immunity (1). Interestingly, exposure of cells to mycotoxins, individually and/or in combination, resulted in significant reductions in antibacterial activity against E. coli, though significant changes in protein levels of pBD-1 and pBD-2 were not observed. It is possible that mycotoxin exposure leads to the production of other antimicrobial metabolites or proteins in cell supernatants that may indirectly contribute to the observed results. Nevertheless, although no significant increases in protein levels for either pBD-1 or pBD-2 were observed, subtle changes may still contribute to and/or reflect dysregulated immunity in the IECs (48), which is supported by the apparent and significant reductions in antibacterial effects associated with toxin-treated cells reported in the current study.
Additionally, in order to determine whether changes in mRNA expression, protein secretion, and/or antibacterial activity may be attributed to cytotoxic effects, noncytotoxic concentrations of DON (0.5 μM), NIV (0.5 μM), ZEA (10 μM), and FB1 (20 μM) were investigated (Wan et al., submitted). The results showed that noncytotoxic concentrations of DON, NIV, ZEA, and FB1 alone or in combination also affected mRNA expression and protein secretion of pBD-1 and pBD-2, as well as antibacterial activity. However, significant upregulation of pBD-1 and pBD-2 mRNA expression and reductions in antibacterial effects were primarily observed in toxin mixtures found to cause cytotoxicity in the MTT assay (Wan et al., submitted). Therefore, it is possible that cytotoxic effects may contribute to the antibacterial activity of IPEC-J2 cells. However, it is currently not clear whether this may have occurred, and if so, what the mechanism(s) may be. Further studies are required to characterize the effects of individual and combined Fusarium toxins on the regulation of pBD-1 and pBD-2 protein levels and the associated antibacterial activities.
There is no information in the literature regarding the combined effect of Fusarium toxins on antibacterial activity in vitro. In this study, significant interactions between treatments in the present analysis would indicate that the addition of more than one mycotoxin has a nonadditive effect, whereas an observed lack of interaction would indicate that effects were additive (29). The results showed that additive effects were noted in most of the toxin mixtures, leading to increases in bacterial survival compared with single treatments of Fusarium toxin. A similar phenomenon was reported by Groten et al. (49) and Tajima et al. (51), who observed additive effects on the inhibition of DNA synthesis when L929 fibroblasts were exposed to multiple Fusarium toxins (DON, NIV, T-2 toxin, ZEA, and FB1), though some synergistic interactions were also reported (49, 51). Additive effects may occur when there is cooccurrence of more than one mycotoxin causing toxicity through the same mechanism of action (52). In our study, interactive effects, which have not been reported previously, were observed in mixtures containing NIV and FB1, though the reason for such interaction remains unknown. This may indicate that these commonly occurring Fusarium toxins, when combined, may interact with each other so that the magnitude of the resulting toxic effects generated may be potentiated or reduced by actions of other toxins (51). Synergistic actions may occur when mycotoxin mixtures act at different stages of the same toxicity pathway or when the presence of one mycotoxin increases the absorption or decreases the metabolic degradation of another (52). Antagonism, on the other hand, may occur when mycotoxins compete with one another for the same target/receptor site (53). However, the present results with multiple Fusarium toxins suggest that Fusarium toxins alone may not predict their effects in natural environments, where combinations of toxins are frequently observed, meaning that investigations of commonly observed toxin mixtures are necessary and would enable more accurate and effective risk assessments (54). To understand the nature of these interactive effects, a molecular-level understanding of mycotoxin-mycotoxin interaction is required in the future in order to develop more effective detoxification and remediation strategies aimed at understanding mycotoxin impacts on animal and human health (55).
In summary, gene expression, protein secretion for pBD-1 and pBD-2, and the consequent antimicrobial impact of cell supernatants of IPEC-J2 cells following exposure to DON, NIV, ZEA, and FB1, individually and in mixtures, were determined. A consequent induction of mRNA for both defensins was observed, though this significant transcriptional induction did not correlate with significant changes in the respective secreted protein levels. Regardless, toxin exposure resulted in reduced antibacterial effects observed in cell supernatants, suggesting exposure to mycotoxins may contribute to microbiome changes in the gastrointestinal environment. Based on our data, Fusarium toxins, either alone or in combination, potentially activate distinct antimicrobial defense mechanisms, which may alter the intestinal microbiota, potentially leading to imbalances impacting the overall health and well-being of animals and humans (56). However, the precise mechanisms by which mycotoxins elicit these effects remain unclear. Further studies using molecular approaches, such as high-throughput mRNA sequencing and proteomics, will be useful to elucidate the mechanistic pathways involved in understanding the dynamic interplay that occurs between Fusarium toxins and IECs (57).
ACKNOWLEDGMENTS
The work was supported by a Hong Kong Research Grant Council grant (number 765810).
We thank P. T. Sanglid from the Department of Human Nutrition/Clinical Nutrition, Faculty of Life Science, University of Copenhagen, Copenhagen, Denmark, for kindly providing the IPEC-J2 cell line. We also thank W. C. Yam from the Department of Microbiology, University of Hong Kong, for providing the E. coli (ATCC 25922) bacterial strain and John Bacon-Shone of the Social Sciences Research Centre of the University of Hong Kong for his statistical advice.
We have no conflicts of interest.
Footnotes
Published ahead of print 25 January 2013
REFERENCES
- 1. Bevins CL, Martin-Porter E, Ganz T. 1999. Defensins and innate host defence of the gastrointestinal tract. Gut 45: 911–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bals R. 2000. Epithelial antimicrobial peptides in host defense against infection. Respir. Res. 1: 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cunliffe RN, Mahida YR. 2004. Expression and regulation of antimicrobial peptides in the gastrointestinal tract. J. Leukoc. Biol. 75: 49–58 [DOI] [PubMed] [Google Scholar]
- 4. Yang D, Liu ZH, Tewary P, Chen Q, de la Rosa G, Oppenheim JJ. 2007. Defensin participation in innate and adaptive immunity. Curr. Pharm. Des. 13: 3131. [DOI] [PubMed] [Google Scholar]
- 5. Diamond G, Beckloff N, Weinberg A, Kisich KO. 2009. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 15: 2377–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andreu D, Rivas L. 1998. Animal antimicrobial peptides: an overview. Biopolymers 47: 415–433 [DOI] [PubMed] [Google Scholar]
- 7. Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3: 710–720 [DOI] [PubMed] [Google Scholar]
- 8. Zhang G, Ross CR, Blecha F. 2000. Porcine antimicrobial peptides: new prospects for ancient molecules of host defense. Vet. Res. 31: 277–296 [DOI] [PubMed] [Google Scholar]
- 9. Tossi A, Sandri L. 2002. Molecular diversity in gene-encoded, cationic antimicrobial polypeptides. Curr. Pharm. Des. 8: 743–761 [DOI] [PubMed] [Google Scholar]
- 10. Veldhuizen EJA, Hendriks HG, Hogenkamp A, van Dijk A, Gaastra W, Tooten PCJ, Haagsman HP. 2006. Differential regulation of porcine β-defensins 1 and 2 upon Salmonella infection in the intestinal epithelial cell line IPI-2I. Vet. Immunol. Immunopathol. 114: 94–102 [DOI] [PubMed] [Google Scholar]
- 11. Zilbauer M, Dorrell N, Boughan PK, Harris A, Wren BW, Klein NJ, Bajaj-Elliott M. 2005. Intestinal innate immunity to Campylobacter jejuni results in induction of bactericidal human beta-defensins 2 and 3. Infect. Immun. 73: 7281–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Veldhuizen EJA, Koomen I, Ultee T, van Dijk A, Haagsman HP. 2009. Salmonella serovar specific upregulation of porcine defensins 1 and 2 in a jejunal epithelial cell line. Vet. Microbiol. 136: 69–75 [DOI] [PubMed] [Google Scholar]
- 13. Creppy EE. 2002. Update of survey, regulation and toxic effects of mycotoxins in Europe. Toxicol. Lett. 127: 19–28 [DOI] [PubMed] [Google Scholar]
- 14. Pestka JJ, Bondy GS. 1990. Alteration of immune function following dietary mycotoxin exposure. Can. J. Physiol. Pharmacol. 68: 1009–1016 [DOI] [PubMed] [Google Scholar]
- 15. Luongo D, De Luna R, Russo R, Severino L. 2008. Effects of four Fusarium toxins (fumonisin B1, α-zearalenol, nivalenol and deoxynivalenol) on porcine whole-blood cellular proliferation. Toxicon 52: 156–162 [DOI] [PubMed] [Google Scholar]
- 16. Luongo D, Severino L, Bergamo P, De Luna R, Lucisano A, Rossi M. 2006. Interactive effects of fumonisin B1 and α-zearalenol on proliferation and cytokine expression in Jurkat T cells. Toxicol. In Vitro 20: 1403–1410 [DOI] [PubMed] [Google Scholar]
- 17. Kouadio JH, Dano SD, Moukha S, Mobio TA, Creppy EE. 2007. Effects of combinations of Fusarium mycotoxins on the inhibition of macromolecular synthesis, malondialdehyde levels, DNA methylation and fragmentation, and viability in Caco-2 cells. Toxicon 49: 306–317 [DOI] [PubMed] [Google Scholar]
- 18. Kouadio JH, Mobio TA, Baudrimont I, Moukha S, Dano SD, Creppy EE. 2005. Comparative study of cytotoxicity and oxidative stress induced by deoxynivalenol, zearalenone or fumonisin B1 in human intestinal cell line Caco-2. Toxicology 213: 56–65 [DOI] [PubMed] [Google Scholar]
- 19. Kubena LF, Edrington TS, Harvey RB, Buckley SA, Phillips TD, Rottinghaus GE, Casper HH. 1997. Individual and combined effects of fumonisin B1 present in Fusarium moniliforme culture material and T-2 toxin or deoxynivalenol in broiler chicks. Poult. Sci. 76: 1239–1247 [DOI] [PubMed] [Google Scholar]
- 20. Boeira LS, Bryce JH, Stewart GG, Flannigan B. 2000. The effect of combinations of Fusarium mycotoxins (deoxynivalenol, zearalenone and fumonisin B1) on growth of brewing yeasts. J. Appl. Microbiol. 88: 388–403 [DOI] [PubMed] [Google Scholar]
- 21. Maresca M, Yahi N, Younès-Sakr L, Boyron M, Caporiccio B, Fantini J. 2008. Both direct and indirect effects account for the pro-inflammatory activity of enteropathogenic mycotoxins on the human intestinal epithelium: stimulation of interleukin-8 secretion, potentiation of interleukin-1α effect and increase in the transepithelial passage of commensal bacteria. Toxicol. Appl. Pharm. 228: 84–92 [DOI] [PubMed] [Google Scholar]
- 22. Oswald IP, Desautels C, Laffitte J, Fournout S, Peres SY, Odin M, Le Bars P, Le Bars J, Fairbrother JM. 2003. Mycotoxin fumonisin B1 increases intestinal colonization by pathogenic Escherichia coli in pigs. Appl. Environ. Microbiol. 69: 5870–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Veldhuizen EJA, Rijnders M, Claassen EA, van Dijk A, Haagsman HP. 2008. Porcine α-defensin 2 displays broad antimicrobial activity against pathogenic intestinal bacteria. Mol. Immunol. 45: 386–394 [DOI] [PubMed] [Google Scholar]
- 24. Veldhuizen EJA, van Dijk A, Tersteeg MHG, Kalkhove SIC, van der Meulen J, Niewold TA, Haagsman HP. 2007. Expression of α-defensins pBD-1 and pBD-2 along the small intestinal tract of the pig: lack of upregulation in vivo upon Salmonella typhimurium infection. Mol. Immunol. 44: 276–283 [DOI] [PubMed] [Google Scholar]
- 25. Witthöft T, Pilz CS, Fellermann K, Nitschke M, Stange EF, Ludwig D. 2005. Enhanced human α-defensin-2 (hBD-2) expression by corticosteroids is independent of NF-κB in colonic epithelial cells (Caco 2). Dig. Dis. Sci. 50: 1252–1259 [DOI] [PubMed] [Google Scholar]
- 26. Schierack P, Nordhoff M, Pollmann M, Weyrauch K, Amasheh S, Lodemann U, Jores J, Tachu B, Kleta S, Blikslager A, Tedin K, Wieler L. 2006. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 125: 293–305 [DOI] [PubMed] [Google Scholar]
- 27. Heussner AH, Dietrich DR, O'Brien E. 2006. In vitro investigation of individual and combined cytotoxic effects of ochratoxin A and other selected mycotoxins on renal cells. Toxicol. In Vitro 20: 332–341 [DOI] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 29. Boone MD. 2008. Examining the single and interactive effects of three insecticides on amphibian metamorphosis. Environ. Toxicol. Chem. 27: 1561–1568 [DOI] [PubMed] [Google Scholar]
- 30. Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26: 32–46 [Google Scholar]
- 31. Mariani V, Palermo S, Fiorentini S, Lanubile A, Giuffra E. 2009. Gene expression study of two widely used pig intestinal epithelial cell lines: IPEC-J2 and IPI-2I. Vet. Immunol. Immunopathol. 131: 278–284 [DOI] [PubMed] [Google Scholar]
- 32. Dybvig T, Facci M, Gerdts V, Wilson H. 2011. Biological roles of host defense peptides: lessons from transgenic animals and bioengineered tissues. Cell Tissue Res. 343: 213–225 [DOI] [PubMed] [Google Scholar]
- 33. Elahi S, Buchanan RM, Attah-Poku S, Townsend HGG, Babiuk LA, Gerdts V. 2006. The host defense peptide beta-defensin 1 confers protection against Bordetella pertussis in newborn piglets. Infect. Immun. 74: 2338–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang G, Hiraiwa H, Yasue H, Wu H, Ross CR, Troyer D, Blecha F. 1999. Cloning and characterization of the gene for a new epithelial α-defensin. J. Biol. Chem. 274: 24031–24037 [DOI] [PubMed] [Google Scholar]
- 35. Hancock REW. 1997. Peptide antibiotics. Lancet 349: 418–422 [DOI] [PubMed] [Google Scholar]
- 36. Moser C, Weiner DJ, Lysenko E, Bals R, Weiser JN, Wilson JM. 2002. α-Defensin 1 contributes to pulmonary innate immunity in mice. Infect. Immun. 70: 3068–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sang Y, Patil A, Zhang G, Ross C, Blecha F. 2006. Bioinformatic and expression analysis of novel porcine α-defensins. Mamm. Genome 17: 332–339 [DOI] [PubMed] [Google Scholar]
- 38. Wehkamp J, Fellermann K, Herrlinger KR, Bevins CL, Stange EF. 2005. Mechanisms of disease: defensins in gastrointestinal diseases. Nat. Clin. Pract. Gastroenterol. Hepatol. 2: 406–415 [DOI] [PubMed] [Google Scholar]
- 39. Aldhous M, Noble C, Satsangi J. 2009. Dysregulation of human α-defensin-2 protein in inflammatory bowel disease. PLoS One 4: e6285 doi:10.1371/journal.pone.0006285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J, Deng J, Li Y, Yang Q. 2011. The effect of Lactobacillus on the expression of porcine α-defensin-2 in the digestive tract of piglets. Livest. Sci. 138: 259–265 [Google Scholar]
- 41. Hertzel AV, Smith LA, Berg AH, Cline GW, Shulman GI, Scherer PE, Bernlohr DA. 2006. Lipid metabolism and adipokine levels in fatty acid-binding protein null and transgenic mice. Am. J. Physiol. Endocrinol. Metab. 290: E814–E823 [DOI] [PubMed] [Google Scholar]
- 42. Hodin CM, Verdam FJ, Grootjans J, Rensen SS, Verheyen FK, Dejong CHC, Buurman WA, Greve JW, Lenaerts K. 2011. Reduced Paneth cell antimicrobial protein levels correlate with activation of the unfolded protein response in the gut of obese individuals. J. Pathol. 225: 276–284 [DOI] [PubMed] [Google Scholar]
- 43. Okada H, Nakajima T, Sanezumi M, Ikuta A, Yasuda K, Kanzaki H. 2000. Progesterone enhances interleukin-15 production in human endometrial stromal cells in vitro. J. Clin. Endocrinol. Metab. 85: 4765–4770 [DOI] [PubMed] [Google Scholar]
- 44. Ganz T. 2007. Biosynthesis of defensins and other antimicrobial peptides, p 62–76 Ciba Foundation Symposium 186: antimicrobial peptides. John Wiley & Sons, Ltd., Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 45. Greenbaum D, Colangelo C, Williams K, Gerstein M. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiang Y, Gerhold D, Holder D, Figueroa D, Bailey W, Guan P, Skopek T, Sistare F, Sina J. 2007. Diagnosis of drug-induced renal tubular toxicity using global gene expression profiles. J. Transl. Med. 5: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286: 531–537 [DOI] [PubMed] [Google Scholar]
- 48. Fukata M, Michelsen KS, Eri R, Thomas LS, Hu B, Lukasek K, Nast CC, Lechago J, Xu R, Naiki Y, Soliman A, Arditi M, Abreu MT. 2005. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G1055–G1065 [DOI] [PubMed] [Google Scholar]
- 49. Groten JP, Tajima O, Feron VJ, Schoen ED. 1998. Statistically designed experiments to screen chemical mixtures for possible interactions. Environ. Health Perspect. 106: 1361–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51. Tajima O, Schoen ED, Feron VJ, Groten JP. 2002. Statistically designed experiments in a tiered approach to screen mixtures of Fusarium mycotoxins for possible interactions. Food Chem. Toxicol. 40: 685–695 [DOI] [PubMed] [Google Scholar]
- 52. Cavaliere C, Foglia P, Pastorini E, Samperi R, Laganà A. 2005. Development of a multiresidue method for analysis of major Fusarium mycotoxins in corn meal using liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 19: 2085–2093 [DOI] [PubMed] [Google Scholar]
- 53. Ruiz MJ, Franzova P, Juan-García A, Font G. 2011. Toxicological interactions between the mycotoxins beauvericin, deoxynivalenol and T-2 toxin in CHO-K1 cells in vitro. Toxicon 58: 315–326 [DOI] [PubMed] [Google Scholar]
- 54. Speijers GJA, Speijers MHM. 2004. Combined toxic effects of mycotoxins. Toxicol. Lett. 153: 91–98 [DOI] [PubMed] [Google Scholar]
- 55. Fink-Gremmels J. 1999. Mycotoxins: their implications for human and animal health. Vet. Q. 21: 115–120 [DOI] [PubMed] [Google Scholar]
- 56. Aziz Q, Doré J, Emmanuel A, Guarner F, Quigley EMM. 2013. Gut microbiota and gastrointestinal health: current concepts and future directions. Neurogastroenterol. Motil. 25: 4–15 [DOI] [PubMed] [Google Scholar]
- 57. Giacometti J, Tomljanovic AB, Josic D. 18 October 2012. Application of proteomics and metabolomics for investigation of food toxins. Food Res. Int. doi:10.1016/j.foodres.2012.10.019 [Google Scholar]