Abstract
Small-subunit (SSU) rRNA gene sequences associated with the phylum Armatimonadetes were analyzed using multiple phylogenetic methods, clarifying both the phylum boundary and the affiliation of previously ambiguous groupings. Here we define the Armatimonadetes as 10 class-level groups and reclassify two previously associated groups as candidate divisions WS1 and FBP.
TEXT
Candidate division OP10, first identified in 1998 (1), was renamed as the phylum Armatimonadetes with the characterization of the type strain Armatimonas rosea YO-36T (2). Hundreds of phylotypes from a broad range of environmental niches have been added to the group since its inception (1, 3); as of April 2012, 568 sequences in the SILVA database alone were classified as Armatimonadetes or OP10, and 364, 568, and 653 small-subunit (SSU) rRNA gene sequences were classified as either Armatimonadetes or candidate division OP10 in the EMBL, SILVA, and RDP databases, respectively. Using the SILVA database numbers, the sequences have a maximum SSU rRNA gene sequence dissimilarity of ∼29%, a significantly diverse phylum compared to the average phylum diversity of 19.3% (4). However, the phylogeny of Armatimonadetes is still poorly defined, as recent publications (2, 5–9) have not been able to agree upon a consistent and well-supported consensus tree. These publications have used a variety of methods and outgroup/ingroup sequence selections, which has resulted in the description of between 4 and 12 (in some cases poorly supported) subgroupings. Currently, it is difficult to compare published tree topologies and group nomenclatures due to the inconsistent application of SSU rRNA gene sequences and the application of different methods to generate phylogenetic frameworks for the former candidate division OP10/Armatimonadetes. Thus, the phylogenetic diversity and subdivisional architecture of Armatimonadetes have remained uncertain.
In order to clarify the phylogenetic relationships within the Armatimonadetes and establish well-supported taxon boundaries, we used multiple phylogenetic methods to (i) confirm the division boundaries and (ii) define the subphylum level group structure(s), including the previously identified classes Armatimonadia (2) and Chthonomonadetes (8) and the recently validated class Fimbriimonadia (7).
For phylogenetic inference, near-full-length SSU rRNA genes putatively identified as Armatimonadetes or OP10 were selected from the SILVA SSU NR (nonredundant) Release 108 database (10) for initial sequence alignment. This ingroup data set consisted of 492 sequences. The outgroup consisted of 46 sequences, which were a combination of the three outgroup sets described by Dalevi et al. (5), named OP10A, OP10B, and OP10C. The combined outgroup was replicated with only minor adjustments: if sequences were not included in the SILVA NR database, the most closely related sequences in the SILVA SSU NR database were used. All sequence accession numbers here are listed in Data Set S1 in the supplemental material. The subdivisional group numbering (groups 1 to 12) recently defined by Dunfield et al. (6) was used as a default, nonpresumptuous naming scheme for hypothesis testing. Monophyletic groupings, once established, were subsequently given unique identifiers based on the name of the first validly published phylotype within said group, as described in the Greengenes database (11).
Two applications of neighbor-joining (NJ) and of maximum-likelihood (ML) methods and one application of Bayesian inference (BI) were utilized to define the nodal support for class level groupings, the details of which are described in File S1 in the supplemental material. Of 492 OP10/Armatimonadetes sequences extracted from the SILVA database, 15 flagged as chimeric using Pintail (12) were identified (see Data Set S1 in the supplemental material) and excluded from further calculations.
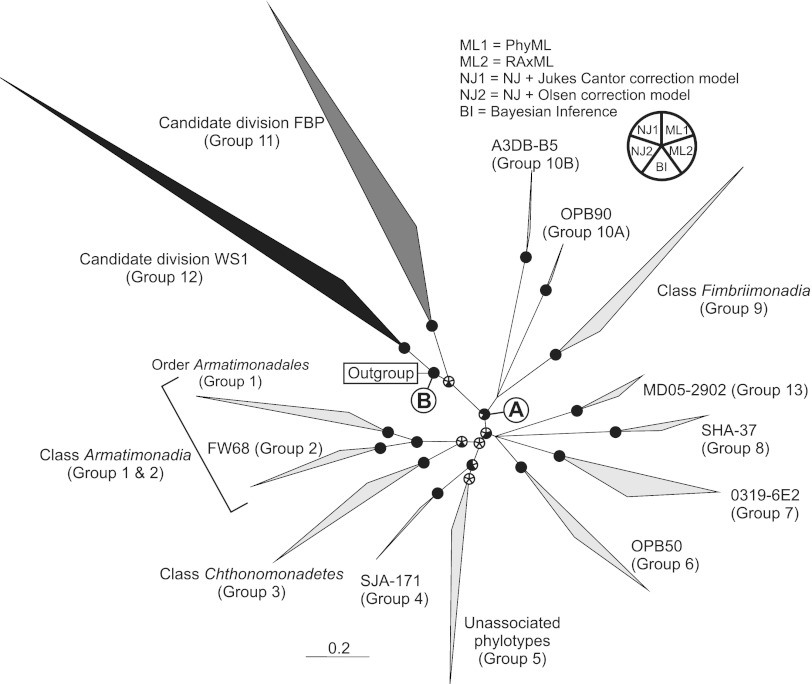
A radial consensus phylogenetic tree summarizing the final grouping nomenclature and relationships is presented in Fig. 1 along with the support of the nodes by the five methods used. Support values of the groupings as well as tested hypothetical combinations of these groupings are presented in Data Set S2 in the supplemental material. A total of 12 monophyletic groupings were defined from the Armatimonadetes/OP10 ingroup data set, which included two proposed candidate divisions and 10 class-level divisions (Fig. 1). Bipartition support values of groups within Armatimonadetes confirm the monophyly of the previously described classes Armatimonadia (group 1 to 2), Chthonomonadetes (group 3), and Fimbriimonadia (group 9), as defined by the respective authors (2, 7, 8). The boundary of the class Armatimonadia (group 1) should also include group 2 due to the strong support for monophyly of the combined “supergroup” (groups 1 and 2); this amalgamation is consistent with the variety of ecological niches reported previously, including temperate soils, fresh water, human skin, and microbial biofilms (6). Furthermore, we suggest that that group 1 of the expanded class Armatimonadia (2, 6–8) now represent the order Armatimonadales (Fig. 1).
Fig 1.
Unrooted consensus tree showing the phylum Armatimonadetes and affiliated groups. The groups were based those described by Dunfield et al. (6); the phylotypes (accession numbers) within each group are listed in Data Set S1 in the supplemental material. The analyses of support for the groups are detailed in Data Set S2. The nodal support here displays supports by five phylogenetic methods (two NJ, two ML, and one BI analysis), which are represented by pie charts on the individual bifurcation nodes. Support is defined as a ≥70% bootstrap proportion value (NJ and ML) and a ≥95% posterior probability value (BI). A node supported by the specific method has the corresponding portion of the pie chart shaded black. Multifurcations were manually introduced into nodes without support from any of the methods (15). Taxon designations are shown based on the position of type strains and the supported monophyly of nodes. On the consensus tree, node A represents the phylum Armatimonadetes and node B represents the superphylum resulting from the inclusion of Armatimonadetes with the candidate divisions FBP (group 11) and WS1 (group 12). The scale bar represents 0.2 nucleotide substitution per site.
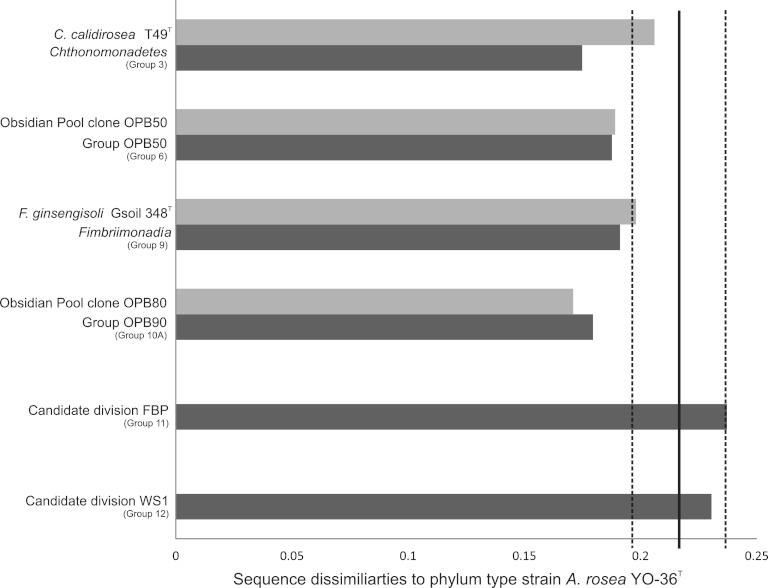
Figure 1 shows two deeply branching nodes, A and B, with strong monophyly support. Node B included all the ingroup sequences and was supported by all five of the phylogenetic methods. Node A, which was supported by four of five methods, included the original OP10 phylotypes and all three described Armatimonadetes classes but consistently excluded groups 11 and 12. The phylogenetic distance of these two groups from the phylum type strain, strong and reproducibly monophyletic support (3), and distinct environmental distributions (6) indicate that groups 11 and 12 should be considered candidate divisions distinct from Armatimonadetes. Sequence dissimilarity analysis (Fig. 2) corroborates this assertion, as maximum and mean similarities equal (group 12) or exceed (group 11) the upper limit (95% confidence interval) of the previously published average maximum bacterial phylum boundary (21.6% ± 2.0%) (4). Although group 12 was recently included within the Armatimonadetes in the SILVA database, it was previously identified as candidate division WS1 (13). Here we term group 11 as candidate division FBP based on the earliest published, near-full-length (>1.4-kb) phylotype, clone FBP249 (AY250868) (14). The two candidate divisions also share a well-supported relationship with Armatimonadetes (node B), a relationship which would therefore likely represent the basal node for a putative highly divergent superphylum.
Fig 2.
Bar graph displaying sequence dissimilarities between Armatimonadetes type strain A. rosea YO-36T and key phylotypes (Armatimonadetes isolates and the original OP10 clones). The graph also displays mean sequence dissimilarities between A. rosea YO-36T and groups containing the key phylotypes Chthonomonadetes (group 3), OPB50 (group 6), Fimbriimonadia (group 9), and OPB90 (group 10A), candidate division FBP (group 11), and candidate division WS1 (group 12). The phylotypes (and accession numbers) within each group are listed in Data Set S1 in the supplemental material. The solid vertical line marking the value of 0.216 indicates the average maximum phylum sequence dissimilarity calculated by Yarza et al. (4). The flanking dotted line indicates the 95% confidence interval (±0.02).
In this study, applying multiple methods to discern phylogenetic relationships within Armatimonadetes has highlighted problematic sequences as well as nodes with inconsistent support values. Despite extensive analyses, the phylogenetic resolution of Armatimonadetes should be considered ongoing and is still limited by factors such as the limits of phylogenetic signals within the current SSU rRNA gene data sets. Evidence suggests the independent monophyly of the groups OPB90 (group 10A) and A3DB-B5 (group 10B), although the two groups were previously associated and the clones in both groups were found only in geothermal environments (6); whether this is a result of convergent evolution or limitations in the data set available awaits future investigations. Group 5 was also unsupported as a monophyletic group, but unlike with group 10, we were unable to identify any clearly distinguishable monophyletic subgroups. Therefore, its incumbents should be considered a collection of unassociated phylotypes.
In summary, the data presented in this study clarify the phylogenetic architecture of the phylum Armatimonadetes. The research confirms the monophyly of the three described Armatimonadetes classes along with seven additional class-level groupings and defines the basal node of the phylum. In addition, two clades (WS1 and FBP) were demonstrated to be consistently monophyletic and hence were confirmed as candidate divisions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Sarah Beanland Memorial Scholarship and Geothermal Resources of New Zealand (GRN) funding at GNS Science.
Footnotes
Published ahead of print 1 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03333-12.
REFERENCES
- 1.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamaki H, Tanaka Y, Matsuzawa H, Muramatsu M, Meng X-Y, Hanada S, Mori K, Kamagata Y. 2011. Armatimonas rosea gen. nov., sp. nov., of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10. Int. J. Syst. Evol. Microbiol. 61:1442–1447 [DOI] [PubMed] [Google Scholar]
- 3.Hugenholtz P, Goebel BM, Pace NR. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarza P, Ludwig W, Euzeby J, Amann R, Schleifer KH, Glockner FO, Rossello-Mora R. 2010. Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses. Syst. Appl. Microbiol. 33:291–299 [DOI] [PubMed] [Google Scholar]
- 5.Dalevi D, Hugenholtz P, Blackall LL. 2001. A multiple-outgroup approach to resolving division-level phylogenetic relationships using 16S rDNA data. Int. J. Syst. Evol. Microbiol. 51:385–391 [DOI] [PubMed] [Google Scholar]
- 6.Dunfield PF, Tamas I, Lee KC, Morgan XC, McDonald IR, Stott MB. 2012. Electing a candidate: a speculative history of the bacterial phylum OP10. Environ. Microbiol. 14:3069–3080 [DOI] [PubMed] [Google Scholar]
- 7.Im W-T, Hu Z-Y, Kim K-H, Rhee S-K, Meng H, Lee S-T, Quan Z-X. 2012. Description of Fimbriimonas ginsengisoli gen. nov., sp. nov. within the Fimbriimonadia class nov., of the phylum Armatimonadetes. Antonie van Leeuwenhoek 102:307–317 [DOI] [PubMed] [Google Scholar]
- 8.Lee KC, Dunfield PF, Morgan XC, Crowe MA, Houghton KM, Vyssotski M, Ryan JL, Lagutin K, McDonald IR, Stott MB. 2011. Chthonomonas calidirosea gen. nov., sp. nov., an aerobic, pigmented, thermophilic micro-organism of a novel bacterial class, Chthonomonadetes classis nov., of the newly described phylum Armatimonadetes originally designated candidate division OP10. Int. J. Syst. Evol. Microbiol. 61:2482–2490 [DOI] [PubMed] [Google Scholar]
- 9.Portillo MC, Gonzalez JM. 2008. Members of the candidate division OP10 are spread in a variety of environments. World J. Microbiol. Biotechnol. 25:347–353 [Google Scholar]
- 10.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dojka MA, Hugenholtz P, Haack SK, Pace NR. 1998. Microbial diversity in a hydrocarbon- and chlorinated solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Torre JR, Goebel BM, Friedmann EI, Pace NR. 2003. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 69:3858–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peplies J, Kottmann R, Ludwig W, Glockner FO. 2008. A standard operating procedure for phylogenetic inference (SOPPI) using (rRNA) marker genes. Syst. Appl. Microbiol. 31:251–257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.