Abstract
To determine if hospital effluent input has an ecological impact on downstream aquatic environment, antibiotic resistance in Enterococcus spp. along a medical center-retirement home-wastewater treatment plant-river continuum in France was determined using a culture-based method. Data on antibiotic consumption among hospitalized and general populations and levels of water contamination by antibiotics were collected. All isolated enterococci were genotypically identified to the species level, tested for in vitro antibiotic susceptibility, and typed by multilocus sequence typing. The erm(B) and mef(A) (macrolide resistance) and tet(M) (tetracycline resistance) genes were detected by PCR. Along the continuum, from 89 to 98% of enterococci, according to the sampled site, were identified as Enterococcus faecium. All E. faecium isolates from hospital and retirement home effluents were multiply resistant to antibiotics, contained erm(B) and mef(A) genes, and belonged to hospital-adapted clonal complex 17 (CC17). Even though this species remained dominant in the downstream continuum, the relative proportion of CC17 isolates progressively decreased in favor of other subpopulations of E. faecium that were more diverse, less resistant to antibiotics, and devoid of the classical macrolide resistance genes and that belonged to various sequence types. Antibiotic concentrations in waters were far below the MICs for susceptible isolates. CC17 E. faecium was probably selected in the gastrointestinal tract of patients under the pressure of administered antibiotics and then excreted together with the resistance genes in waters to progressively decrease along the continuum.
INTRODUCTION
Enterococci are Gram-positive, opportunistic bacteria that inhabit the gastrointestinal tracts of humans and many animals. They are also widely present as fecal contaminants of food and as starter cultures for the production of cheese and fermented sausages. In humans, Enterococcus faecalis and Enterococcus faecium are among the major causes of nosocomial infections worldwide. They are increasingly isolated from the bloodstream, urinary tract, and surgical sites. Although E. faecalis remains the predominant species, the proportion of E. faecium in clinical isolates has markedly increased (1). A reason for the increasing frequency of E. faecium as a cause of various infections could be its propensity for acquisition of antibiotic resistance genes. In particular, the prevalence of vancomycin-resistant enterococci (VRE) has been on the rise in the last decades. The antimicrobial pressure in a hospital environment would be the driving force for its selection. It is now considered that a distinct subpopulation consisting of hospital-adapted, ampicillin-resistant E. faecium strains has been a key factor for the successful spread of multiply antibiotic-resistant E. faecium strains (resistant or not resistant to vancomycin) in hospitals. Molecular epidemiological surveys using DNA sequence typing (multilocus sequence typing [MLST]) and phylogenetic analysis have shown that this subpopulation belongs to a limited number of sequence types (ST) that could be grouped in a clonal complex designated CC17 (2, 3). Clones belonging to the CC17 lineage are mostly characterized by ampicillin and fluoroquinolone resistance (4) and possess a pathogenicity island harboring the putative virulence genes esp and hylEfm coding for an enterococcal surface protein and a hyaluronidase, respectively (5).
Massive antibiotic use in both human and veterinary medicines is considered the cause of the emergence of the bacterial resistance that is regarded as a major problem for public health on a worldwide scale. In Europe, about 10,000 tons of antibiotics are consumed per year (6), equally in veterinary usage (growth promotion to 2006 [prophylaxis and treatment]) and in human medicine. After antibiotic consumption, large proportions of the administered antibiotics are excreted into the environment via urine and feces in metabolized and nonmetabolized forms or coupled with polar molecules (7). Thus, those antibiotic residues that contaminate aquatic environments (8) may play a role in selection of antibiotic-resistant bacteria. However, this is a controversial issue, in part because the ecology of bacteria and their resistance genes in the agricultural and urban environment is poorly documented.
In addition to putative selection pressure related to the input of antibiotics and their degradation products in waters, antibiotic-resistant fecal bacteria selected in humans and animals and released in effluents may also contaminate aquatic environments (9). In developed countries, the most important sources of antibiotic-resistant fecal bacteria are effluents of wastewater treatment plants (WWTP) (10). The second well-known contribution of antibiotic-resistant fecal bacteria is due to both soil leaching and runoffs, for which the quantities and the frequencies depend on the hydrology and the use of the watershed (11). Once they are released in environmental water, survival of enterococci depends on biotic and abiotic factors, mainly, their ability to overcome environmental stresses (oligotrophy, sunlight, salinity), the viral lysis, and protozoan predation. Moreover, persistence of enterococci in water is greatly influenced by their association with organic matter, such as biofilms on copepods or plankton (12, 13). However, little is currently known about the evolution of the contamination by both antibiotic compounds and antibiotic-resistant bacteria in aquatic environments from the source of contamination to the river.
Recently, a multidisciplinary study associating chemists, hydrologists, and microbiologists was launched to investigate the relationship between the presence of antibiotics and that of antibiotic-resistant Escherichia coli in waters along a medical center-WWTP-river continuum (4 km) during a period of influenza outbreak leading to increased antibiotic prescription by general practitioners (14). In this study, we used the same sampling data sets to obtain a deeper understanding of the changes of enterococcal populations and of their resistance to antibiotics along this continuum.
MATERIALS AND METHODS
Study sites and sampling strategy.
Contamination by antibiotics and by the antibiotic-resistant enterococci was investigated along a continuum formed by a medical center, a retirement home, a WWTP, and a river in the northwest of France (Fig. 1). During a period of high epidemicity corresponding to maximal antibiotic use (influenza outbreak, December 2009), samples were collected along the pathway from medical center effluents to a river (Table 1) using autosamplers (ISCO 6700s; Roucaire, Courtaboeuf, France). One sample was collected from each site. Effluent samples from the hospital (sampling site no. 1) and the retirement home (sampling site no. 2) were collected at two discharge points before the main sewer. Another site, located 4 km from the medical center, was the town's wastewater treatment plant (WWTP), which collects the wastewaters of the medical center and of 9,058 inhabitants. The WWTP treatment consisted of a primary treatment with a screen, an aerated grit-removal tank, and a primary clarifier. The secondary treatment consisted of an activated sludge system and a second clarifier. Mean daily wastewater samples were collected at the entrance (sampling site no. 3) and the outlet (sampling site no. 4) of the WWTP. Sampling site no. 5, located 4.1 km from the medical center, was the WWTP discharge in the river (located at 4 m from the WWTP outlet). Finally, the upstream of the river (sampling site no. 6) before the discharge of the WWTP was sampled independently. All water samples were collected during a time period which extended from 16 h to 24 h depending on the site. For each site, 1 liter was collected every hour and 250 ml of the pooled sample was used for analysis. Samples were collected in polyethylene flasks for microbiological analyses and in 4 glass and 2 amber glass flasks for chemical analyses. Samples were stored at 4 to 6°C, and microbiological analysis was carried out within 8 h. For chemical analysis, samples were first filtered and then frozen at −20°C until analysis was performed.
Fig 1.
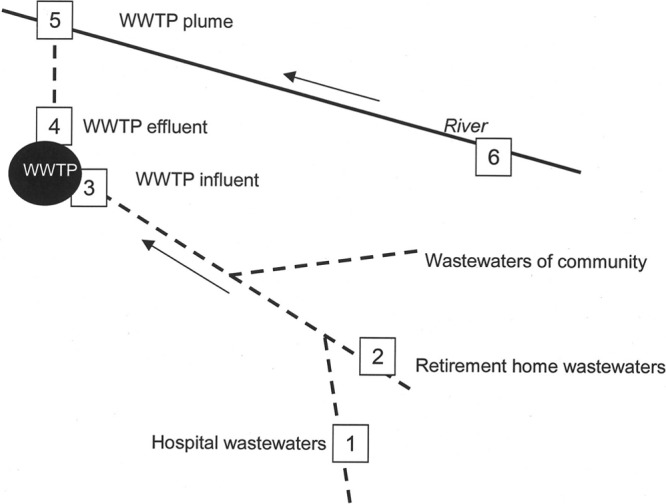
Sampled sites. The sampled sites are indicated by numbers. The water flow is shown by arrows. Dashed lines indicate pipes. (Adapted from reference 14 with permission [copyright 2012 American Chemical Society].)
Table 1.
Contamination of water by enterococci along the medical center-WWTP-river continuum
| Parameter | Value for indicated sampled site |
||||
|---|---|---|---|---|---|
| Retirement homea | Hospitalb | WWTP |
River (WWTP plume) | ||
| Influent | Effluent | ||||
| Anthropic pressure | 180 beds | 87 beds | 9,058 inhabitants | 9,058 inhabitants | nrc |
| Density of enterococci (CFU · 100 ml) | (2.6 ± 1.6) × 105 | (6.5 ± 0.1) × 106 | (7.0 ± 0.4) × 105 | (1.4 ± 0.2) × 104 | (3.7 ± 1.0) × 102 |
| No. of tested enterococci | 49 | 48 | 47 | 46 | 45 |
| Proportion of E. faecium (%) | 98 | 98 | 96 | 91 | 89 |
| Proportion of E. faecalis (%) | 0 | 2 | 0 | 0 | 0 |
| Proportion of E. durans (%) | 0 | 0 | 2 | 0 | 7 |
| Proportion of E. hirae (%) | 2 | 0 | 2 | 9 | 4 |
Mean residence time, 10 years.
Mean residence time, 4 to 28 days.
nr, not relevant.
Detection and quantification of antibiotics in water samples.
The chemical analysis, using solid-phase extraction (SPE) coupled with liquid chromatography-tandem mass spectrometry (LC-MS-MS) as previously described (14), was performed to determine levels of contamination by 34 antibiotics and the relationship with antibiotic use. The chosen antibiotics were 7 penicillins, 5 cephalosporins, 6 fluoroquinolones, 2 quinolones, 4 tetracyclines, 10 sulfonamides, a dihydrofolate reductase inhibitor (trimethoprim), and 5 macrolides (14). The antibiotic consumption data were obtained from pharmacists both in the hospital and in the community (6 pharmacies) (14).
Isolation of enterococci.
Enterococci were counted using membrane filtration methods (HA047 filter; Millipore, Bedford, MA) (0.45-μm pore size). Filters were placed on a selective chromogenic agar specific for enterococci (RAPID'Enterococcus; Bio-Rad, Marnes-la-Coquette, France) and incubated for 48 h at 44°C. Blue colonies were then isolated on a Slanetz medium incubated for 48 h at 44°C. The threshold value for counting in water was 10 CFU per 100 ml. A maximum of 50 randomly selected colonies were studied for each sample site. Enterococcal isolates were stored at −80°C for further studies. A total of 240 samples of enterococci isolated along the hospital continuum were thus collected.
Identification of enterococci and antibiotic susceptibility testing.
Identification of enterococci was performed as follows. Enterococci were identified by using API Strept galleries (bioMérieux, La-Balme-les-Grottes, France). Identification of E. faecalis, E. faecium, E. gallinarum, and E. casseliflavus was confirmed by a multiplex PCR using primers specific for the ddl genes (encoding a bacterial ligase) of these species, as previously described (15). For the other enterococcal species that yielded negative PCR results, in particular, Enterococcus hirae, Enterococcus durans, and Enterococcus avium, a DNA fragment internal to the sodA gene was amplified and sequenced, as previously described (16).
Susceptibility of enterococci to antibiotics was tested by the agar diffusion method according to the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM) (http://www.sfm-microbiologie.org). The antibiotic disks and Mueller-Hinton medium were from Bio-Rad, and the tested antibiotics were ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, kanamycin, lincomycin, linezolid, rifampin, streptomycin, tetracycline, and vancomycin. Bacteria were classified as susceptible, intermediate, or resistant according to the CA-SFM recommendations. Isolates were considered highly resistant to an antibiotic when they grew at the contact of the corresponding antibiotic disk. E. faecalis CIP103214 (ATCC 29212) and Staphylococcus aureus ATCC 25923 were used as quality control strains.
PCR of resistance genes.
Isolates resistant to antimicrobials were tested for the presence of resistance genes by PCR. Specific oligonucleotide primers previously described were used to amplify the major resistance genes of resistance to macrolides and tetracyclines usually detected in clinical isolates of enterococci (17, 18). The erm(B) gene (encoding a ribosomal methylase) and the mef(A) gene (encoding an efflux pump belonging to the major facilitator superfamily [MFS]), conferring resistance to macrolides, and the tet(M) gene, conferring resistance to tetracyclines, were amplified. Strains of Streptococcus pneumoniae HM30 [erm(B), tet(M)] and S. pneumoniae [mef(A)] were used as PCR positive controls. PCR conditions were as previously described (17, 19).
Multilocus sequence typing.
MLST assays based on polymorphism of seven housekeeping genes (atpA, ddl, gdh, purK, gyd, pstS, and adk) were performed as previously described (2). Different sequences were assigned allele numbers and different allelic profiles were assigned sequence types (STs) based on the E. faecium MLST database (http://efaecium.mlst.net/). The e-burst software was used to define clonal complexes (20).
Statistical analysis.
The percentages of enterococci resistant to ampicillin and of those carrying the erm(B) or mef(A) genes in the water samples were compared using Fisher's exact test. Tests were carried out using GraphPad software (GraphPad, San Diego, CA). A P value of ≤0.05 was considered statistically significant.
RESULTS
Water contamination by enterococci.
Water contamination by enterococci was investigated along the hospital-retirement home-WWTP-river continuum (Fig. 1). Abundance of Enterococcus along the continuum decreased from 6.5 × 106 CFU 100 ml−1 in the hospital effluent to 3.7 × 102 CFU 100 ml−1 in the WWTP plume (river), with flows of 1 m3 h−1 (hospital effluents) to 23.6 m3 s−1 (river) (Table 1). The species diversity was low, and E. faecium was largely predominant throughout the continuum (Table 1), with a few isolates of E. faecalis, Enterococcus durans, and Enterococcus hirae.
Resistance to antibiotics in enterococci.
Along the continuum, no isolate was resistant to vancomycin or to linezolid. One hundred percent and 87.5% of isolates (all enterococcal species combined) were resistant to penicillins in hospital and retirement home effluents, respectively (Table 2). In contrast, only 19.1% and 19% of isolates from WWTP inflow and effluent were resistant to ampicillin, respectively (P < 0.0001). Finally, in the receiving river (sampled site no. 5), the frequency of resistance to ampicillin was only 4% (P < 0.0001 compared to the hospital and the retirement home; P = 0.012 compared to the WWTP). Similarly, high-level resistance to fluoroquinolones, defined by growth at the contact of the ciprofloxacin disk as determined by the disk-diffusion method, was detected in 100% and 75% of enterococcal isolates from the retirement home and hospital effluents, respectively, and 15.6%, 17%, and 5% of the isolates from the WWTP inflow, WWTP effluents, and river, respectively. No isolate from the hospital effluents was resistant to tetracyclines, whereas small proportions of resistant isolates were detected in the effluents of the retirement home (21%) and the influents and effluents of the WWTP (influent, 28%, effluent, 26%) and in the river (24%). The proportion of isolates not susceptible to erythromycin exceeded 70% throughout the continuum (Table 2). However, differences in the levels of resistance to erythromycin were observed, depending on the sampled site. All nonsusceptible isolates from the hospital and retirement home effluents displayed high-level resistance to erythromycin (defined by growth at the contact of the erythromycin disk), whereas most from WWTP effluents and river were intermediately resistant to erythromycin. Ampicillin-resistant isolates from hospital and retirement home effluents were nearly all coresistant to erythromycin and ciprofloxacin. The changes in multiple resistances to antibiotics (resistance to high levels of ampicillin, fluoroquinolones, and erythromycin) were mostly observed in the E. faecium isolates that were predominant throughout the continuum, suggesting corresponding variations in subpopulations of E. faecium.
Table 2.
Contamination of water by antibiotics and percentages of resistance to antibiotics in enterococci
| Antibiotic class | Value for sample from indicated site |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. 1; hospital |
No. 2; retirement home |
No. 3; WWTP inflow |
No. 4; WWTP effluent |
No. 5; WWTP plume |
||||||
| Concn (ng/ml) | % of resistant enterococci | Concn (ng/ml) | % of resistant enterococci | Concn (ng/ml) | % of resistant enterococci | Concn (ng/ml) | % of resistant enterococci | Concn (ng/ml) | % of resistant enterococci | |
| Penicillins | 0 | 100.0 | 0.7 | 87.5 | 0.0 | 19.1 | 0.0 | 19.0 | 0.0 | 4.0a,b |
| Cephalosporins | 161 | —c | 0.0 | — | 0.0 | — | 0.0 | — | 0.0 | — |
| Fluoroquinolones | 141 | 75.0 | 59 | 100.0 | 0.4 | 15.6 | 0.14 | 17.0 | 0.001 | 5.0 |
| Tetracyclines | 1 | 0.0 | 0 | 21.0 | 0.15 | 28.0 | 0.01 | 26.0 | 0.0 | 24.0 |
| Macrolides | 7.9 | 72.0 | 21.9 | 87.0 | 3.5 | 77.0 | 2.1 | 80.0 | 0.007 | 72.0 |
P < 0.0001 versus hospital and retirement home.
P = 0.012 versus WWTP.
—, antibiotic resistance not tested.
Macrolide and tetracycline resistance genes in enterococci.
Since E. faecium was the majority species, all subsequent studies were done on isolates belonging to this species. The mechanisms explaining the resistance displayed by the great majority of E. faecium differed according to the sampled site (Table 3). Detection by PCR of the antibiotic resistance gene content of isolates showed that the erm(B) gene was present in more than 75% of isolates in hospital and retirement home effluents. The prevalence of this gene decreased along the continuum to reach only 6.7% of isolates from the river. The efflux gene mef(A), alone or in combination with another relevant gene(s), was more prevalent in the WWTP influent and effluent (43.8% in influent and 57.9% in effluent) than in the hospital and retirement home effluents (25% and 4.8%, respectively). Obviously, mechanisms of resistance other than ribosomal methylation by an Erm(B) methylase or efflux by a Mef(A) pump were responsible for resistance to macrolides in the majority of isolates from WWTP effluents and river.
Table 3.
Resistance to erythromycin and distribution of macrolide resistance genes according to site
| Site or source | % isolates not susceptible to erythromycina | % isolates with indicated antibiotic resistance gene(s) |
|||
|---|---|---|---|---|---|
| erm(B) | mef(A) | erm(B) + mef(A) | Other | ||
| No. 1; hospital | 72 | 75 | 0 | 25 | 0 |
| No. 2; retirement home | 87 | 85.7 | 4.8 | 0 | 9.5 |
| No. 3; WWTP influent | 77 | 12.5 | 18.8 | 25 | 43.7b |
| No. 4; WWTP effluent | 80 | 15.8 | 47.4 | 10.5 | 26.3c |
| No. 5; WWTP plume | 72 | 6.7 | 6.7 | 0 | 86.7d |
Intermediate and resistant isolates.
P < 0.0001 versus hospital and retirement home.
P < 0.0001 versus hospital; P = 0.028 versus retirement home; P = 0.053 versus WWTP influent.
P < 0.0001 versus hospital, retirement home, WWTP influent, and WWTP effluent.
Resistance to tetracyclines was mostly explained by the presence of the tet(M) gene present in 100% of tetracycline-resistant isolates of retirement home effluents, 92.3% and 83% of WWTP influent and effluent isolates, respectively, and 86.7% of river isolates. Few E. faecium isolates were resistant to high levels of gentamicin (hospital effluents, n = 5; WWTP influent, n = 3; WWTP effluent, n = 2; river, n = 1), except in the retirement home (n = 10).
Sequence types.
Twelve to 15 E. faecium isolates per site were studied by MLST (Table 4). All isolates from the hospital and retirement home effluents belonged to the ST78 that is part of clonal complex CC17, typically including hospital-adapted and outbreak-associated isolates. The presence of ST78 or of other STs belonging to the CC17 complex was also detected in the downstream continuum, but the isolates were in a minority. In contrast, E. faecium isolates from the downstream continuum belonged to various STs, including some that have never been reported before. The greater diversity was observed in the river. Some STs of isolates from the WWTP and river were grouped in sequence types (ST266, ST29, and ST178) that were previously characterized by phylogenetic analysis as associated with poultry and pigs (3).
Table 4.
Sequence types of E. faecium isolates determined by MLSTa
| Site or source (no. of studied isolates) | Sequence type(s) (no. of isolates) |
|||
|---|---|---|---|---|
| Hospital adapted (CC17 types) | Poultry | Human commensal, pig | Other | |
| No. 1; hospital (13) | ST78 (13) | |||
| No. 2; retirement home (15) | ST78 (15) | |||
| No. 3; WWTP influent (14) | ST18, ST78 | ST291, ST264, ST330 | ||
| No. 4; WWTP effluent (14) | ST78 (3), ST18 | ST178 | ST22, ST32 (3), STnew1, STnew2 (2), STnew3 | |
| No. 5; WWTP plume (12) | ST78, ST402, ST165 | ST266, ST29 (2) | ST22, ST32 (2), ST12, ST39, ST78, ST165, ST253, ST402, STnew4 (2) | |
Assignment to host reservoir was according to Willems et al. (3).
Antibiotic consumption and contamination of water by antibiotics.
Antimicrobial consumption data have been presented in detail elsewhere (14), except those for macrolides, presented herein for the hospital and the retirement home, and can be summarized as follows (Table 5). During the study period, three antibiotic families were mostly prescribed at the hospital: β-lactams (penicillins and cephalosporins), quinolones, and macrolides. In addition, a few sulfonamides and tetracyclines were also prescribed. At the retirement home, β-lactams, quinolones, and, to a lesser extent, macrolides were prescribed. The most prescribed antibiotic family (penicillins) was detected in effluents at low concentrations, which can be explained by the important degradation of penicillins in water, essentially by hydrolysis (21). In contrast, quinolones, cephalosporins, and macrolides, less prescribed but more stable, were detected in hospital effluents at relatively high concentrations of, respectively, 141 ng/ml, 161 ng/ml, and 7.5 ng/ml (Table 3). Of note, although not or minimally prescribed, tetracyclines and sulfonamides were detected in the hospital effluents and in the retirement home effluents, respectively. The presence of these molecules could be explained by uncontrolled inputs such as those from staff members (100 persons per day) and/or visitors.
Table 5.
Antibiotic consumption
| Antibiotic(s) | Antibiotic consumption (g) |
||
|---|---|---|---|
| Hospitala | Retirement homea | Communityb | |
| Glycopeptide (vancomycin) | 0 | 0 | NAc |
| β-Lactams | |||
| Ampicillin/amoxicillin | 1,930 | 544 | 258,430 |
| Cloxacillin | 0 | 16 | 736 |
| Cephalosporins | 453 | 550 | 3,444 |
| Fluoroquinolones | 115 | 0 | 980 |
| Tetracyclines | 1.1 | 0 | 1,562 |
| Macrolides | 75 | 20 | NA |
Antibiotic consumption 1 month before sampling.
Antibiotic consumption from October 2009 to March 2010.
NA, not available.
DISCUSSION
Enterococci and E. coli are used as fecal indicator bacteria, reflecting the human health risk in drinking and recreational waters (22). In this study, the fate of enterococci resistant to antibiotics in relation to the presence of antibiotics in water was monitored from a medical center to a river.
E. faecium was largely found throughout the continuum as the dominant species. The underrepresentation of the other species, in particular, E. faecalis, was unexpected. However, sampling of the river (site no. 6; Fig. 1) upstream from the human continuum using the same methodology showed, as expected, a greater biodiversity and a predominance of E. faecalis, with a 50% proportion of E. faecalis, 20% of E. faecium, 14% of E. casseliflavus, 7% of E. durans, and 2% of E. avium (data not shown). Therefore, the predominance of E. faecium appeared to be particular to this continuum. However, since sampling was performed only in December, we cannot rule out the influence of seasonal variations not only on the amounts of enterococci in water but also on the relative proportions of the various enterococcal species (23). In a recent study, persistence of enterococci in winter months in water was dependent upon the bacterial species (24). In particular, E. hirae was able to replicate in the environment at a high rate even in winter. However, this species was not prevalent in our study. The predominance of E. faecium was related to human activity in certain studies. In Canadian surveys, it has been shown that E. faecalis was dominant in waters of agricultural areas and in domesticated mammals and birds and wildlife feces, whereas E. faecium was dominant among wastewater isolates (25, 26). In a municipal WWTP from Poland, E. faecium was also predominant (60.8% of enterococci) (27). Finally, in a recent study, E. faecium represented the vast majority of enterococci that were detected in the sediments of the lake of Geneva (Switzerland), which is subjected to extensive microbial contamination by WWTP outlet discharges and rivers (28).
In our study, E. faecium isolates from the effluents of hospital and retirement home were highly resistant to penicillins, cephalosporins (intrinsic resistance), and fluoroquinolones. Factors that might lead to selection of particular groups of antibiotic-resistant enterococci in the environment are (i) antibiotic consumption by humans and animals that results in excretion of resistant bacteria, mostly in the form of feces, and (ii) the presence of antibiotics in water that can create or maintain the selective pressure. The concentration of fluoroquinolones (0.14 ng/ml) and cephalosporins (0.16 ng/ml) in hospital effluents was far below the MICs for susceptible enterococci (MIC greater than 16 μg/ml for cephalosporins and equal to 1 or 2 μg/ml for ciprofloxacin, the most active fluoroquinolone [29]) and was much farther below those for resistant enterococci. However, it has been postulated that concentrations below MICs may have selective effects leading to enrichment of the environment by antibiotic-resistant bacteria (30, 31). In a recent publication, minimum selective concentrations for resistant E. coli and Salmonella spp. were 100 pg/ml and 15 ng/ml, for ciprofloxacin and tetracycline, respectively (32). These concentration levels were detected in the hospital and retirement home effluents and may have contributed to select resistant strains. In contrast, in the downstream effluents, antibiotic concentrations were very low and a contribution of the free antibiotic concentrations in water to maintaining a selective pressure against the multiply resistant enterococci was unlikely. However, we cannot exclude the possibility of the presence of higher concentrations of antibiotics associated with sediments or particles in waters as previously reported for quinolones in marine sediments (33). Most likely, antibiotic-resistant bacteria have been selected in the gut of patients by these antibiotics highly prescribed in these health care settings. The resistant bacteria would then have been excreted in effluents. However, no screening of hospitalized patients' stools has been carried out to confirm this hypothesis and no E. faecium outbreak has been reported by the hospital. The typing of all E. faecium isolates from the hospital and the retirement home as ST78, a typical sequence type of complex CC17 associated with hospital outbreaks, supports this hypothesis. The reason for the high frequency of high-level resistance to gentamicin in enterococci from retirement home compared to the low frequency in isolates from the other sampled sites, including the hospital, remained unknown. Although aminoglycosides were rarely prescribed in the retirement home (data not shown), resistance of isolates to high levels of gentamicin might have been coselected by β-lactams or fluoroquinolones and then spread.
Although E. faecium isolates were multiply resistant to antibiotics, none was resistant to vancomycin. This might be related to a low prevalence of resistance to vancomycin in France (34) (EARSS web site; http://www.ecdc.europa.eu/en/activities/surveillance/EARS-Net/database/Pages/database.aspx), in contrast to other European countries and the United States (35). Recently, in Ireland, where the prevalence of resistance to vancomycin in clinical isolates of E. faecium reaches 44%, VRE were detected in the outflow of a wastewater treatment plant and in a single rural drinking water supply (36). A similar observation was reported for wastewaters from the south coast of England (37) and in Portugal (38), two countries with a high prevalence of VRE.
Our study showed that there were no significant differences in the proportions of E. faecium between the inflow and the effluent from the WWTP and therefore that wastewater treatment did not result in a specific removal of E. faecium. The same observation has been made previously for VRE (38, 39). However, it should be noted that the WWTP inflow contained a decreased proportion of CC17 E. faecium compared to the medical center effluents, in relation either to a poor survival of CC17 isolates in water or more likely to dilution by other types of E. faecium. A greater diversity of E. faecium STs was observed, and only a few isolates were resistant to ampicillin and fluoroquinolones. Except for CC17 isolates, no link to a particular host based on clonal groups could be found.
Downstream, in the river, a greater diversity of enterococcal species and of E. faecium STs was noted. Some E. faecium isolates displayed STs belonging to clonal complexes associated with pig isolates (3). The percentages of resistance to ampicillin and to high levels of ciprofloxacin continued to decrease, although not significantly compared to those seen with isolates from the WWTP. However, CC17 E. faecium isolates still persisted in the WWTP plume, but at very low counts. Again, a dilution effect likely contributed to the relative disappearance of the multiply resistant isolates. It should be noted that although the percentage of resistance to macrolides remained high throughout the continuum (Table 3), the level of resistance progressively decreased from high to intermediate.
The persistence in waters of bacteria with mobile resistance genes presents a hazard of transfer of resistance genes to pathogenic bacteria and further dissemination. Resistance to ampicillin and to fluoroquinolones in enterococci is considered nontransferable, since it is commonly related to mutations in chromosomal genes, namely, the pbp5 gene and the gyrA and parC genes, respectively. In contrast, resistance to macrolides and tetracyclines in enterococci from humans and animals is often borne by mobile genetic elements bearing the erm(B) and tet(M) genes, respectively (18, 39), and may be acquired by other Gram-positive pathogens. Beyond the snapshot provided by the study of antibiotic resistance genes at a given moment in specific reservoirs, the occurrence and the fate of the antibiotic resistance gene flux in nature should be monitored, which is currently difficult to perform. In the hospital and retirement home effluents, high-level resistance to erythromycin is mostly explained by the presence of the classical erm(B) gene alone or combined with the efflux mef(A) gene. These genes are shared by other Gram-positive human pathogens, such as Streptococcus pneumoniae and Streptococcus pyogenes (40). In the rest of the continuum, the prevalence of these genes decreased together with the prevalence of ampicillin-resistant, ciprofloxacin-resistant isolates. In the WWTP plume, these genes rarely accounted for resistance to erythromycin, which was still expressed by a majority of isolates. The other mechanisms responsible for resistance to erythromycin were not investigated in this study.
Our data are consistent with those obtained previously for E. coli in the same continuum at the same period (14). Antibiotic-resistant E. coli strains were detected in the medical center effluents that originated from patients. Along the continuum, the occurrence of E. coli resistant to antibiotics and those carrying class 1 integrons significantly decreased in water samples.
In conclusion, we have observed in hospital and retirement home effluents the presence of ampicillin-resistant, ciprofloxacin-resistant enterococci bearing acquired macrolide resistance genes, with an overwhelming representation of E. faecium belonging to the CC17 complex. These bacteria would have been selected in the gastrointestinal tract of patients by the selective pressure of administered antibiotics and then excreted together with the resistance genes into waters where they may be initially maintained by low antibiotic levels. Although the species E. faecium remained dominant in the downstream continuum, the proportion of CC17 spontaneously started to decrease before passage through the WWTP in favor of other subpopulations of E. faecium that were more diverse, less resistant to antibiotics, and devoid of the classical macrolide resistance genes and that belonged to various STs. Their original host could not be identified in most cases, although some isolates probably came from animal reservoirs of pigs and poultry. Although free antibiotics were detected in downstream waters, their barely detectable levels would be unlikely to allow selection of resistant enterococci.
ACKNOWLEDGMENTS
This work was supported by the scientific program FLASH-Seine Aval (http://seine-aval.crihan.fr/web/), by EC2CO CNRS, and by a grant from the Ministry of Research to EA4655. K.O. held a research grant (SFR SCALE) from Haute Normandie regional council (France).
We thank Michel Simon, Caroline Bance, and Michel Auzou for excellent technical assistance. We thank also Michel Leroux, Aurélie Lamy, Yvon Goarvot, Sophie Coté, and downtown pharmacists for antibiotic consumption data and the people who gave us access to the medical center and to the WWTP.
Footnotes
Published ahead of print 1 February 2013
REFERENCES
- 1.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10:266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, Van Embden JD, Willems RJ. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Top J, Willems R, Bonten M. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297–308 [DOI] [PubMed] [Google Scholar]
- 5.van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, Schapendonk CM, Hendrickx AP, Nijman IJ, Bonten MJ, Tettelin H, Willems RJ. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239 doi:10.1186/1471-2164-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kümmerer K. 2003. Significance of antibiotics in the environment. J. Antimicrob. Chemother. 52:5–7 [DOI] [PubMed] [Google Scholar]
- 7.Chambers HF. 2001. Antimicrobial agents: protein synthesis inhibitors and miscellaneous antibacterial agents, p 1239–1271. In Brunton LL, Chabner BA, Knollmann BC. (ed), Goodman and Gilman's the pharmacological basis of therapeutics. McGraw-Hill, New York, NY [Google Scholar]
- 8.Zuccato E, Castiglioni S, Bagnati R, Melis M, Fanelli R. 2010. Source, occurrence and fate of antibiotics in the Italian aquatic environment. J. Hazard. Mater. 179:1042–1048 [DOI] [PubMed] [Google Scholar]
- 9.Salyers AA, Gupta A, Wang Y. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes.Trends Microbiol. 12:412–416 [DOI] [PubMed] [Google Scholar]
- 10.Reinthaler FF, Posch J, Feierl G, Wüst G, Haas D, Ruckenbauer G, Mascher F, Marth E. 2003. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 37:1685–1690 [DOI] [PubMed] [Google Scholar]
- 11.Cristian T, Schneider RJ, Fäber H, Skutlarek D, Meyer MT, Goldbach HE. 2003. Determination of antibiotic residues in manure, soil, and surface waters. Acta Hydrochim. Hydrobiol. 31:36–44 [Google Scholar]
- 12.Mote BL, Turner JW, Lipp EK. 2012. Persistence and growth of the fecal indicator bacteria enterococci in detritus and natural estuarine plankton communities. Appl. Environ. Microbiol. 78:2569–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Signoretto C, Burlacchini G, Pruzzo C, Canepari P. 2005. Persistence of Enterococcus faecalis in aquatic environments via surface interactions with copepods. Appl. Environ. Microbiol. 71:2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberlé K, Capdeville MJ, Berthe T, Budzinski H, Petit F. 2012. Evidence for a complex relationship between antibiotics and antibiotic-resistant Escherichia coli: from medical center patients to a receiving environment. Environ. Sci. Technol. 46:1859–1868 [DOI] [PubMed] [Google Scholar]
- 15.Dutka-Malen S, Evers S, Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poyart C, Quesnes G, Trieu-Cuot P. 2000. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J. Clin. Microbiol. 38:415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angot P, Vergnaud M, Auzou M, Leclercq R. 2000. Macrolide resistance phenotypes and genotypes in French clinical isolates of Streptococcus pneumoniae. Observatoire de Normandie du Pneumocoque. Eur. J. Clin. Microbiol. Infect. Dis. 19:755–758 [DOI] [PubMed] [Google Scholar]
- 18.Petsaris O, Miszczak F, Gicquel-Bruneau M, Perrin-Guyomard A, Humbert F, Sanders P, Leclercq R. 2005. Combined antimicrobial resistance in Enterococcus faecium isolated from chickens. Appl. Environ. Microbiol. 71:2796–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aminov RI, Garrigues-Jeanjean N, Mackie RI. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Längin A, Alexy R, König A, Kümmerer K. 2009. Deactivation and transformation products in biodegradability testing of beta-lactams amoxicillin and piperacillin. Chemosphere 75:347–354 [DOI] [PubMed] [Google Scholar]
- 22.WHO 2004. WHO guidelines for drinking-water quality, p 515. Recommendations, 3rd ed, vol 1 World Health Organization, Geneva, Switzerland [Google Scholar]
- 23.Pan X, Jones KD. 2012. Seasonal variation of fecal indicator bacteria in storm events within the US stormwater database. Water Sci. Technol. 65:1076–1080 [DOI] [PubMed] [Google Scholar]
- 24.Bonjoch X, García-Aljaro C, Blanch AR. 2011. Persistence and diversity of faecal coliform and enterococci populations in faecally polluted waters. J. Appl. Microbiol. 111:209–215 [DOI] [PubMed] [Google Scholar]
- 25.Lanthier M, Scott A, Lapen DR, Zhang Y, Topp E. 2010. Frequency of virulence genes and antibiotic resistances in Enterococcus spp. isolates from wastewater and feces of domesticated mammals and birds, and wildlife. Can. J. Microbiol. 56:715–729 [DOI] [PubMed] [Google Scholar]
- 26.Lanthier M, Scott A, Zhang Y, Cloutier M, Durie D, Henderson VC, Wilkes G, Lapen DR, Topp E. 2011. Distribution of selected virulence genes and antibiotic resistance in Enterococcus species isolated from the South Nation River drainage basin, Ontario, Canada. J. Appl. Microbiol. 110:407–421 [DOI] [PubMed] [Google Scholar]
- 27.Łuczkiewicz A, Jankowska K, Fudala-Książek S, Olańczuk-Neyman K. 2010. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 44:5089–5097 [DOI] [PubMed] [Google Scholar]
- 28.Thevenon F, Regier N, Benagli C, Tonolla M, Adatte T, Wildi W, Poté J. 2012. Characterization of fecal indicator bacteria in sediments cores from the largest freshwater lake of Western Europe (Lake Geneva, Switzerland). Ecotoxicol. Environ. Saf. 78:50–56 [DOI] [PubMed] [Google Scholar]
- 29.Varon E. 2009. Quinolones and Gram-positive bacteria, p 243–259. In Courvalin P, Leclercq R, Rice L. (ed), Antibiogram. ASM Press, Washington, DC [Google Scholar]
- 30.Liu A, Fong A, Becket E, Yuan J, Tamae C, Medrano L, Maiz M, Wahba C, Lee C, Lee K, Tran KP, Yang H, Hoffman RM, Salih A, Miller JH. 2011. Selective advantage of resistant strains at trace levels of antibiotics: a simple and ultrasensitive color test for detection of antibiotics and genotoxic agents. Antimicrob. Agents Chemother. 55:1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baquero F, Negri MC, Morosini MI, Blazquez J. 1998. Antibiotic-selective environments. Clin. Infect. Dis. 27:S5–S11 [DOI] [PubMed] [Google Scholar]
- 32.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158 doi:10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu WH, Zhang G, Wai OWH, Zou SC, Li XD. 2009. Transport and adsorption of antibiotics by marine sediments in a dynamic environment. J. Soils Sediments 9:364–373 [Google Scholar]
- 34.Bourdon N, Fines-Guyon M, Thiolet JM, Maugat S, Coignard B, Leclercq R, Cattoir V. 2011. Changing trends in vancomycin-resistant enterococci in French hospitals, 2001–08. J. Antimicrob. Chemother. 66:713–721 [DOI] [PubMed] [Google Scholar]
- 35.Ramsey AM, Zilberberg MD. 2009. Secular trends of hospitalization with vancomycin-resistant enterococcus infection in the United States, 2000–2006. Infect. Control Hosp. Epidemiol. 30:184–186 [DOI] [PubMed] [Google Scholar]
- 36.Morris D, Galvin S, Boyle F, Hickey P, Mulligan M, Cormican M. 2012. Enterococcus faecium of the vanA genotype in rural drinking water, effluent, and the aqueous environment. Appl. Environ. Microbiol. 78:596–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caplin JL, Hanlon GW, Taylor HD. 2008. Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ. Microbiol. 10:885–892 [DOI] [PubMed] [Google Scholar]
- 38.Araújo C, Torres C, Silva N, Carneiro C, Gonçalves A, Radhouani H, Correia S, da Costa PM, Paccheco R, Zarazaga M, Ruiz-Larrea F, Poeta P, Igrejas G. 2010. Vancomycin-resistant enterococci from Portuguese wastewater treatment plants. J. Basic Microbiol. 50:605–609 [DOI] [PubMed] [Google Scholar]
- 39.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 16:541–554 [DOI] [PubMed] [Google Scholar]
- 40.Leclercq R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482–492 [DOI] [PubMed] [Google Scholar]


