Abstract
We grew Pseudomonas aeruginosa in LB and artificial sputum medium (ASM) (filtered and unfiltered) and quantified metabolite utilization and excretion by nuclear magnetic resonance (NMR) spectroscopy (metabolic footprinting or extracellular metabolomics). Utilization rates were similar between media, but there were differences in excretion—e.g., acetate was produced only in unfiltered ASM.
TEXT
The opportunistic pathogen Pseudomonas aeruginosa is the major cause of morbidity and mortality for cystic fibrosis (CF) patients (1). In order to better understand P. aeruginosa infection, it is important to create model conditions that aim to mimic those in the CF lung, such as media designed to mimic the nutritional environment of sputum from CF patients. These include artificial sputum medium (ASM) (2), a complex medium made up of porcine mucin, DNA, and amino acids, and synthetic cystic fibrosis medium (SCFM) (3), a defined medium containing mainly amino acids and lactate. In addition, many studies use a more-conventional rich medium, like lysogeny broth (LB) (4), which is based on tryptone and yeast extract. All of these media support growth of P. aeruginosa to high cell numbers, but the question remains how the differences in composition alter the physiology and metabolism of the inoculated bacteria and, hence, the relevance to CF infections. Generally, approaches to validating this have depended on indirect means to assess metabolism—for instance, profiling gene transcription (3). However, transcriptional data are often insufficient to capture the actual metabolic changes that occur as a result of perturbing a bacterial cell. Because of this, we wanted to examine the actual changes in metabolite utilization by P. aeruginosa in a comparison of three different complex media, LB, ASM (oASM, prepared according to its original recipe [2]), and a filtered version of ASM (fASM, prepared according to the method of Kirchner et al. [5]). We have previously investigated metabolite utilization in SCFM in depth (6), so we did not include this medium again here.
We grew wild-type P. aeruginosa PA14 under aerobic conditions in batch culture for 24 h (growth conditions described in detail in reference 6) and sampled 0.6 ml of culture at nine time points (before inoculation, hourly from 2 to 8 h, and then at 24 h). Of these samples, 0.1 ml was used for measuring the optical density (OD) for LB and fASM samples (oASM is turbid and the OD cannot be used to monitor cell growth). The supernatants were then analyzed by nuclear magnetic resonance (NMR) spectroscopy as detailed in Behrends et al. (7). The resulting spectra were assigned and integrated for each medium. In total, we were able to assign 19 compounds detectable across all media, of which 18 were able to be quantified (Table 1) (histidine was detected but not integrated here because of pH-induced resonance frequency shifts between spectra) and normalized to compound levels in the uninoculated media. Four further metabolites were able to be identified in at least two media. Finally, the metabolite concentrations were fitted using nonlinear (sigmoid) models, a process which allows between-media comparisons of uptake and excretion time for individual compounds (6).
Table 1.
Fitted compound half-life values derived from sigmoid fits of concentration data for metabolites found across all growth media
| Metabolite |
t1/2 (h) on each mediuma |
|||||||
|---|---|---|---|---|---|---|---|---|
| oASM |
fASM |
LB |
oASM (PAO1) |
|||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Asparagine | 3.55 | 0.28 | 1.98 | 0.19 | 1.03 | 0.03 | 2.91 | 0.25 |
| Aspartate | 4.15 | 0.20 | 4.17 | 0.07 | 3.02 | 0.16 | 4.19 | 0.31 |
| Alanine | 4.96 | 0.56 | 3.41 | 0.06 | 2.74 | 0.05 | 4.24 | 0.11 |
| Glutamate | 5.11 | 0.33 | 4.24 | 0.03 | 3.03 | 0.10 | 4.42 | 0.11 |
| Isoleucine | 8.56 | 0.18 | 8.62 | 0.75 | >12 | ND | 5.99 | 1.17 |
| Leucine | 9.39 | 0.78 | 7.25 | 0.06 | >12 | ND | 5.99 | 1.17 |
| Lysine | 10.12 | 1.85 | 8.12 | 0.54 | >12 | ND | 7.36 | 0.58 |
| Glycine | 10.16 | 0.73 | 8.99 | 0.57 | >12 | ND | 8.90 | 0.51 |
| Threonine | 10.48 | 0.71 | 8.13 | 0.12 | 9.07 | 0.28 | 8.34 | 0.71 |
| Phenylalanine | 10.66 | 0.20 | 7.91 | 0.05 | 11.14 | 0.00 | 8.23 | 0.40 |
| Tryptophan | 11.55 | 1.65 | 8.09 | 0.12 | 9.19 | 0.27 | >12 | ND |
| Tyrosine | >12 | ND | 10.92 | 1.52 | >12 | ND | 11.83 | 1.01 |
| Methionine | >12 | ND | >12 | ND | >12 | ND | >12 | ND |
| Valine | >12 | ND | >12 | ND | >12 | ND | 9.51 | 0.33 |
ND, not determined.
Metabolite uptake is tightly controlled and broadly comparable in rich media.
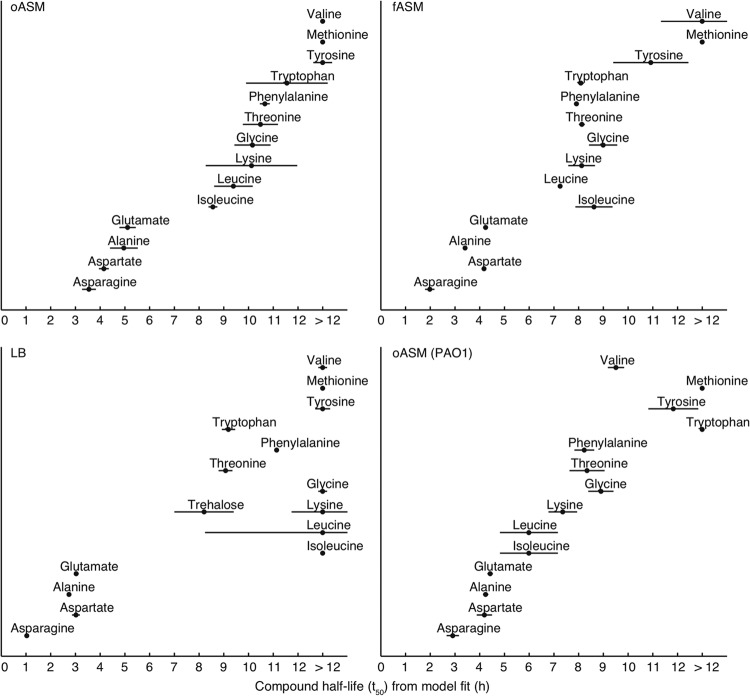
There are several possible ways in which medium composition can alter bacterial physiology and, therefore, the bacterial interaction with the medium. Changes can be qualitative, i.e., the same strain utilizes/excretes a given compound from/into one medium but not another, or quantitative, i.e., the medium composition affects the dynamics of utilization/excretion. By nonlinear fitting, the time dimension of a data set is compressed to produce biologically meaningful sigmoid parameters that can detect both qualitative and quantitative changes—e.g., the t1/2 value corresponds to the time at which half of the compound has been taken up (a compound's half-life). Some metabolites, though, e.g., S-oxo-methionine, had complex utilization profiles, and so standard sigmoid models were not able to be fitted (Fig. 1). In total, 14 metabolites were successfully fitted (Table 1). For these compounds, there were no differences between the media after 24 h of growth, as all detectable amino acids (with the exception of methionine, which was not taken up from any medium) were completely depleted after 24 h. In addition, the dynamics of uptake were similar across all media at first glance, as the orders in which metabolites were taken up were broadly comparable; the most “outlying” medium, unsurprisingly, was LB: both of the ASM were more similar to each other than to LB (Fig. 2). The two ASM had essentially identical orders of metabolite uptake, but metabolites were taken up slightly more quickly from fASM than from oASM, possibly because oASM also contains alternative carbon sources (macromolecules, lipid droplets). In addition, we profiled P. aeruginosa PAO1 growth on oASM in order to permit comparison with earlier studies. While there certainly were some differences from PA14, e.g., valine was used earlier by PAO1, the overall patterns of metabolite usage of the two backgrounds were similar (Fig. 2).
Fig 1.
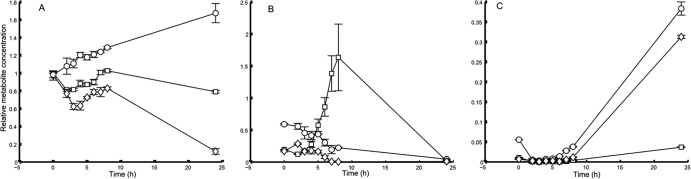
Excreted metabolites indicate differences in Pseudomonas aeruginosa PA14 fermentative metabolism when grown on filtered or unfiltered artificial sputum medium. (A) S-oxo-methionine; (B) acetate; (C) formate. Squares indicate unfiltered ASM, diamonds indicate filtered ASM, and circles indicate LB. Concentrations represent integral values of characteristic resonances relative to the internal standard sodium 3-trimethylsilyl-1-propanesulfonate-d6. Error bars indicate the standard deviation (SD) of the mean.
Fig 2.
The orders of uptake of metabolites are similar but not identical between rich media. The t1/2 (i.e., the time taken for half of the compound to be consumed) is represented by dots, and the period during which the compound is taken up at the maximum rate is represented by horizontal lines; both of these are based on fitting sigmoid models to metabolite concentration data. (A) Unfiltered ASM; (B) filtered ASM; (C) LB.
In agreement with previous studies (6, 8), there was a set of compounds—asparagine, aspartate, alanine, and glutamate (and, in the case of oASM and fASM, also glutamine)—that were taken up early on in growth (Table 1). For these “early-uptake” compounds, the t1/2 values increased from LB to fASM to oASM, i.e., quickest uptake from LB and slowest from oASM. Surprisingly, this order of uptake was reversed for the late-uptake metabolites. Threonine, phenylalanine, and tryptophan had t1/2 values of less than 12 h for all media. In contrast, glycine, lysine, isoleucine, and valine were taken up only after 12 h from LB but at between 7 and 11 h from the ASM (Table 1; Fig. 2). Compound uptake in Pseudomonas is tightly controlled by catabolite repression, which affects amino acids and other organic acids, as well as carbohydrates (8–10). The most likely explanation for the surprising delay in the utilization of late-uptake metabolites from LB is catabolite repression by a metabolite found in LB but not in the two ASM. Trehalose is a good candidate for this metabolite: it is present in LB but not ASM and was taken up after the early-uptake but before the late-uptake metabolites (Fig. 2). There is little information on catabolite repression by trehalose for Pseudomonas species (10). In order to test this, we repeated the experiment but with trehalose spiked into oASM at a 2 mM concentration. This demonstrated that trehalose is not, in fact, causing the catabolite repression: the orders of metabolite uptake were essentially identical for the two ASM, regardless of the presence of trehalose (data not shown). The putative repressing compound in LB remains to be elucidated.
Metabolite excretion differs between media.
In contrast to uptake, compound excretion was noticeably different between the media. The biggest difference was found when comparing oASM and fASM: acetate was found in significant amounts only in the oASM samples (Fig. 1). As the media differ only in a final filtration step, the filtration must remove one or several acetogenic substrates. The turbidity of oASM medium is (at least partly) because of suspended lipid droplets. The lipid fatty acids are metabolized via acetyl coenzyme A (acetyl-CoA), and so the acetate seen in oASM is potentially the product of overflow metabolism to protect the CoA pool of the cell (11). To test this, we repeated the experiment with oASM but omitted the egg yolk (a major source of lipids). In non-egg-yolk-containing ASM, the acetate production was significantly reduced compared to that in standard oASM (at maximum, after 8 h, levels were at 40% of levels in oASM; P < 0.02; t test) but was still significantly higher than that in fASM (after 8 h, levels were 15× higher; P < 0.01; t test). Therefore, we conclude that the egg yolk is an acetogenic substrate in oASM but is not the only one. These are potentially important differences between the two versions of ASM, given that acetate excretion by P. aeruginosa CF clinical isolates has been linked to the length of infection (and, hence, metabolic adaptation to lung conditions) (12). Formate excretion also differed between the media, with levels in fASM and LB higher than those in oASM (Fig. 1).
In addition to fermentation products, several low-concentration metabolites were detectable above the baseline after 24 h of incubation, and some of these were clearly different between fASM and oASM (data not shown). In-depth characterization of these compounds is beyond the scope of our current study, but results indicate that there may be other specific medium-dependent metabolic differences which may well be related to cellular signaling, for instance. In summary, direct analysis of changing exometabolomic profiles can highlight bacterial responses to different media; Pseudomonas aeruginosa metabolite uptake is broadly comparable across different rich media (and very similar between the filtered and unfiltered versions of ASM), but changes in metabolite excretion indicate that there are also differences in cellular metabolism between media.
ACKNOWLEDGMENTS
We thank the anonymous reviewers of our manuscript, who made helpful suggestions for improving the paper.
Footnotes
Published ahead of print 25 January 2013
REFERENCES
- 1. Davies JC, Alton EWFW, Bush A. 2007. Cystic fibrosis. BMJ 335: 1255–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sriramulu DD, Lünsdorf H, Lam JS, Römling U. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54: 667–676 [DOI] [PubMed] [Google Scholar]
- 3. Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189: 8079–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kirchner S, Fothergill JL, Wright EA, James CE, Mowat E, Winstanley C. 2012. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J. Vis. Exp. 2012: e3857 doi:10.3791/3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Behrends V, Ebbels TMD, Williams HD, Bundy JG. 2009. Time-resolved metabolic footprinting for nonlinear modeling of bacterial substrate utilization. Appl. Environ. Microbiol. 75: 2453–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Behrends V, Williams KJ, Jenkins VA, Robertson BD, Bundy JG. 2012. Free glucosylglycerate is a novel marker of nitrogen stress in Mycobacterium smegmatis. J. Proteome Res. 11: 3888–3896 [DOI] [PubMed] [Google Scholar]
- 8. Moreno R, Martínez-Gomariz M, Yuste L, Gil C, Rojo F. 2009. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9: 2910–2928 [DOI] [PubMed] [Google Scholar]
- 9. Browne P, Barret M, O'Gara F, Morrissey JP. 2010. Computational prediction of the Crc regulon identifies genus-wide and species-specific targets of catabolite repression control in Pseudomonas bacteria. BMC Microbiol. 10: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 34: 658–684 [DOI] [PubMed] [Google Scholar]
- 11. Wolfe AJ. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69: 12–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Behrends V, Ryall B, Zlosnik JEA, Speert DP, Bundy JG, Williams HD. 2013. Metabolic adaptations of Pseudomonas aeruginosa during cystic fibrosis chronic lung infections. Environ. Microbiol. 15: 398–408 [DOI] [PubMed] [Google Scholar]