Abstract
In the United States, the blaCMY-2 gene contained within incompatibility type A/C (IncA/C) plasmids is frequently identified in extended-spectrum-cephalosporin-resistant (ESCr) Escherichia coli strains from both human and cattle sources. Concerns have been raised that therapeutic use of ceftiofur in cattle may increase the prevalence of ESCr E. coli. We report that herd ESCr E. coli fecal and hide prevalences throughout the residency of cattle at a feedlot, including during the period of greatest ceftiofur use at the feedlot, were either not significantly different (P ≥ 0.05) or significantly less (P < 0.05) than the respective prevalences at arrival. Longitudinal sampling of cattle treated with ceftiofur demonstrated that once the transient increase of ESCr E. coli shedding that follows ceftiofur injection abated, ceftiofur-injected cattle were no more likely than untreated members of the same herd to shed ESCr E. coli. Pulsed-field gel electrophoresis (PFGE) genotyping, antibiotic resistance phenotyping, screening for presence of the blaCMY-2 gene, and plasmid replicon typing were performed on 312 ESCr E. coli isolates obtained during six sampling periods spanning the 10-month residence of cattle at the feedlot. The identification of only 26 unique PFGE genotypes, 12 of which were isolated during multiple sampling periods, suggests that clonal expansion of feedlot-adapted blaCMY-2 E. coli strains contributed more to the persistence of blaCMY-2 than horizontal transfer of IncA/C plasmids between E. coli strains at this feedlot. We conclude that therapeutic use of ceftiofur at this cattle feedlot did not significantly increase the herd prevalence of ESCr E. coli.
INTRODUCTION
Extended-spectrum cephalosporins (ESC) are critically important to human medicine and are frequently prescribed for the treatment of invasive Escherichia coli and Salmonella enterica infections (1–3). The blaCMY-2 gene, encoding the AmpC-like β-lactamase CMY-2, is frequently harbored by large (>120-kbp) incompatibility type A/C (IncA/C) plasmids in ESC-resistant (ESCr) E. coli and ESCr S. enterica strains isolated from human and animal sources in the United States (4–13). IncA/C plasmids are considered broad-host-range plasmids since they have been identified in many bacterial species, including Aeromonas, Escherichia, Klebsiella, Photobacterium, Salmonella, Vibrio, and Yersinia (14, 15). IncA/C plasmids possess conserved backbone sequences, but the sequences of genetic element insertions carrying antibiotic resistance genes are often divergent (14, 16–18). However, the insertions carrying genes conferring resistance to tetracyclines (tetA), phenicols (floR), and streptomycin (aadA2) are generally conserved between blaCMY-2 IncA/C plasmids harbored in E. coli and S. enterica hosts (17, 18). Thus, it has been hypothesized that commensal E. coli populations in the lower gastrointestinal systems of cattle may serve as a reservoir of blaCMY-2 IncA/C plasmids, which could then be transferred to more virulent food-borne pathogens, including S. enterica (12, 18).
Ceftiofur (TIO) is an ESC approved for use in cattle to treat several illnesses, including bovine respiratory disease complex. The critical importance of ESC to human medicine along with concerns that agricultural use of TIO may contribute to the occurrence of human ESCr infections factored into the European Food Safety Authority recommendations to severely restrict or eliminate TIO use in animal agriculture (19). Injection with TIO has been demonstrated to transiently increase the fecal concentrations of ESCr E. coli and blaCMY-2 in individual treated cattle (20–22). The long-term impact of therapeutic TIO injection of cattle on herd prevalence of ESCr E. coli is unclear since only two studies have correlated TIO use (in dairy cattle herds sampled once or twice) to ESC susceptibilities of commensal E. coli. One study found an association between TIO use and isolation of E. coli with reduced susceptibility to the ESC ceftriaxone (23), while the other study found no association between extent of TIO use and ESCr E. coli prevalence (24).
The factors contributing to long-term maintenance of ESCr E. coli and the blaCMY-2 gene in the absence of TIO use are unclear. In vitro experiments have demonstrated that carriage of blaCMY-2 IncA/C plasmids imposes a fitness cost on the host bacteria, leading to the conclusion that long-term maintenance of IncA/C plasmids requires selective pressure (25). A mathematical model of ESCr E. coli populations in cattle suggests that E. coli strains harboring blaCMY-2 IncA/C plasmids could persist during periods of low selective pressure even if they grow slower than other commensal E. coli strains if frequent horizontal transfer of blaCMY-2 IncA/C plasmids occurs or a sufficient fraction of E. coli bacteria ingested by cattle contain blaCMY-2 IncA/C plasmids (26). Both E. coli and S. enterica harboring blaCMY-2 IncA/C plasmids have been isolated from diverse animal hosts in disparate geographic locations with varied exposure to antibiotics; thus, an undefined combination of selective pressures, biological mechanisms, and population dynamics must ameliorate the fitness cost of blaCMY-2 IncA/C plasmid carriage (18). Isolation and characterization of ESCr E. coli strains that persist in cattle production environments, in the absence of TIO use, is required to further our understanding of the factors that may contribute to the persistence of antibiotic resistance.
The population of 763 cattle used in this study was raised on pasture at the Roman L. Hruska U.S. Meat Animal Research Center (USMARC) from birth until weaning, when they were transferred to the on-site feedlot. The USMARC maintains detailed individual cattle health records, including all antibiotic treatments. This population of cattle was not treated with subtherapeutic levels of antibiotics. TIO was the preferred antibiotic for the treatment of bovine respiratory disease complex (shipping fever), infectious pododermatitis (foot rot), and infectious keratoconjunctivitis (pink eye). Typically, cattle at the USMARC feedlot are most susceptible to disease during the 4 to 6 weeks following weaning and introduction to the feedlot. The most concentrated use of therapeutic antibiotics occurs during this time, termed the “period of increased disease susceptibility.” Following this period, occurrences of illness and antibiotic use typically decline (Shuna A. Jones [USMARC Veterinary Medical Officer], personal communication).
This population of cattle presented an opportunity for a longitudinal study on the effects of TIO use on the prevalence and persistence of ESCr E. coli. This cattle population also presented an opportunity for the isolation and characterization of ESCr E. coli prevalent in this cattle population during periods of limited TIO use. Thus, the goals of this study were to (i) determine the fecal and hide prevalences of ESCr E. coli for the cattle population from feedlot arrival until shortly before harvest, (ii) determine the fecal and hide prevalences of ESCr E. coli for the cattle injected with TIO during the period of increased disease susceptibility from the day of treatment to shortly before harvest, and (iii) characterize ESCr E. coli cattle isolates obtained throughout the study with pulsed-field gel electrophoresis (PFGE) genotyping, antibiotic resistance phenotyping, plasmid size analysis, plasmid replicon typing, and screening for presence of the blaCMY-2 gene.
MATERIALS AND METHODS
Study population and cattle sampling.
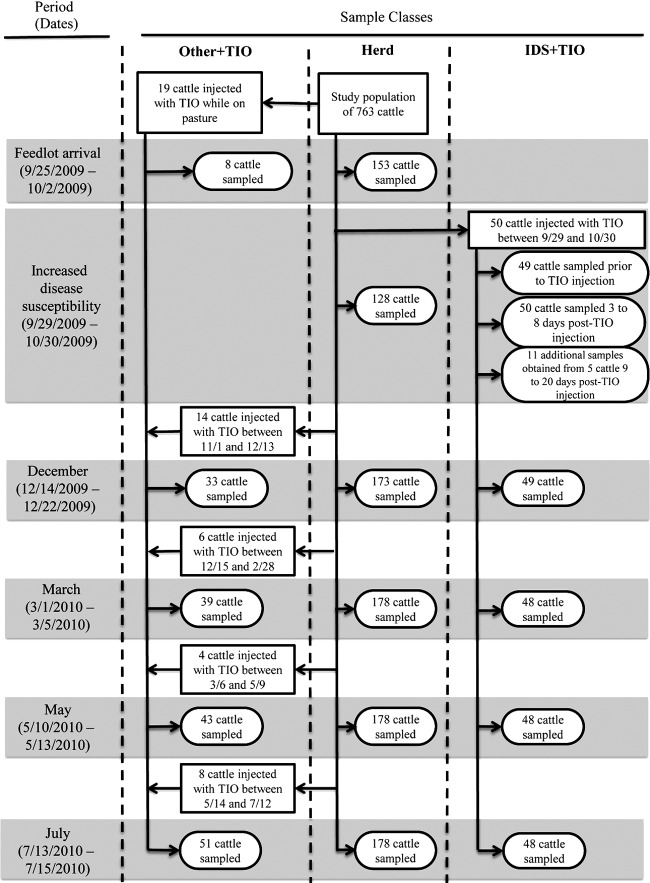
The study population consisted of 763 cattle (403 steers and 360 heifers) born between 22 March 2009 and 16 June 2009 at USMARC and raised on pasture at USMARC until weaned, when they were transferred to the USMARC feedlot between the dates of 25 September and 2 October 2009. The cattle then resided at the feedlot until July 2010, when they were transported to harvest. Detailed records of all antibiotics administered to study animals throughout their life span were maintained, and no antibiotics were included in feed. Samples were obtained during six periods, defined as follows: feedlot arrival (25 September 2009 to 2 October 2009), increased disease susceptibility (29 September 2009 to 30 October 2009), December 2009 (14 December 2009 to 22 December 2009), March 2010 (1 March 2010 to 5 March 2010), May 2010 (10 May 2010 to 13 May 2010), and July 2010 (13 July 2010 to 15 July 2010). Dates of the feedlot arrival and increased disease susceptibility periods overlapped since cattle were introduced to the feedlot (and samples taken) over 5 days (on 25, 29, and 30 September 2009 and 1 and 2 October 2009), while therapeutic TIO injections at the feedlot began on 29 September 2009, before the entire studied population arrived at the feedlot. Three classes of samples were obtained during this study and are described below, and the sampling scheme is illustrated in Fig. 1.
Fig1.
Flow diagram of sampling scheme. Gray boxes indicate sampling periods. Vertical dashed line lines delineate sample classes. Other+TIO, cattle injected with ceftiofur at times other than during the increased disease susceptibility period. IDS+TIO, cattle injected with ceftiofur during the increased disease susceptibility period.
(i) Herd samples.
At least 20% of the population of each pen was sampled during the feedlot arrival (n = 153), December 2009 (n = 173), March 2010 (n = 178), May 2010 (n = 178), and July 2010 (n = 178) periods. Cattle were selected for sampling using a random-number generator. During the period of increased disease susceptibility, 128 samples were obtained over 4 days (9, 16, 23, and 30 October 2009); on each of these days 32 cattle were sampled, two from each of the 16 pens containing the study population. Cattle previously injected with TIO were excluded from herd sampling during all six periods.
(ii) Cattle injected with TIO during the period of increased disease susceptibility.
Fifty cattle were injected with TIO during the increased disease susceptibility period and were designated “IDS+TIO” cattle. Pre-TIO injection samples were obtained from 49 of these cattle. All cattle injected with TIO during the increased disease susceptibility period were held in a hospital pen until a follow-up health examination that occurred 3 to 8 days following injection. Post-TIO injection samples were obtained from all 50 IDS+TIO cattle during these follow-up health examinations. Following completion of the follow-up health examination, all cattle were returned to the pens they originally resided in, except for five cattle that remained in the hospital pen since they required additional observation. A total of 11 additional post-TIO injection samples were obtained from these five cattle during additional health examinations that occurred between 9 and 20 days after the first TIO injection. During the December 2009 period, samples were obtained from 49 of the 50 IDS+TIO cattle since one of the injected cattle died prior to the December 2009 sampling. During the March 2010, May 2010, and June 2010 periods, samples were obtained from 48 of the 50 IDS+TIO cattle since one of the injected cattle died prior to the March 2010 sampling.
(iii) Cattle injected with TIO at times other than during the period of increased disease susceptibility.
Fifty-one cattle were injected with TIO at times other than during the period of increased disease susceptibility and were termed “Other+TIO” cattle. Nineteen cattle were injected with TIO prior to arrival at the feedlot, and samples were recovered from eight of these cattle during the feedlot arrival period. Fourteen additional cattle were injected with TIO between 1 November 2009 and 13 December 2009, for a total of 33 Other+TIO cattle, and all 33 were sampled during the December 2009 period. Six additional cattle were injected with TIO between 23 December 2009 and 28 February 2010, for a total of 39 Other+TIO cattle, and all 39 were sampled during the March 2010 period. Four additional cattle were injected with TIO between 6 March 2010 and 9 May 2010, for a total of 43 Other+TIO cattle, and all 43 were sampled during the May 2010 period. Eight additional cattle were injected with TIO between 14 May 2010 and 14 July 2010, for a total of 51 Other+TIO cattle, and all 51 were sampled during the July 2010 period.
Therapeutic antibiotic administration.
During the life spans of the study cattle (March 2009 to July 2010), there were 157 occasions of therapeutic antibiotic administration (Table 1). One hundred thirty-seven cattle were injected with antibiotics; 16 cattle were injected with antibiotics on more than one occasion. During the life spans of the study cattle, there were 110 therapeutic TIO injections (Table 1). One hundred one cattle were injected with TIO; eight cattle received multiple TIO injections. The month with the highest number of TIO injections, 52, was October 2009. During November 2009, there were 14 TIO injections administered. From December 2009 through July 2010, 24 injections of TIO were administered. There were 47 administrations of other therapeutic antibiotics; the highest frequencies occurred in July 2009, with 16, and September 2009, with 13 (Table 1).
Table 1.
Individual therapeutic antibiotic treatments administered to members of the studied herd of 763 cattle
| Mo and yr | No. of injections of indicated antibiotic(s)a |
Total no. of antibiotic injections | No. of animals injected with antibiotics | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ceftiofur | Cloxacillin and oxytetracycline | Enrofloxacin | Florfenicol | Florfenicol and sulfadimethoxine | Oxytetracycline | Penicillin G | Tulathromycin | |||
| March 2009 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| April 2009 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 2 |
| May 2009 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| June 2009 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 2 |
| July 2009 | 3 | 10 | 0 | 3 | 1 | 2 | 0 | 0 | 19 | 18b |
| August 2009 | 6 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 10 |
| September 2009 | 10 | 8 | 0 | 0 | 0 | 3 | 2 | 0 | 23 | 23 |
| October 2009 | 52 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 53 | 51c |
| November 2009 | 14 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 16 | 15d |
| December 2009 | 4 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 6 | 6 |
| January 2010 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 5 | 5 |
| February 2010 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 6 |
| March 2010 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 |
| April 2010 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| May 2010 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 4 |
| June 2010 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 |
| July 2010 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 |
| Total over life spans of herd cattle (March 2009 to July 2010) | 110 | 23 | 1 | 5 | 2 | 12 | 2 | 2 | 157 | 137e |
Two antibiotics listed in the same column indicate injection of the same animal with both antibiotics simultaneously.
In July 2009 an animal was injected with ceftiofur and then 3 days latter injected with oxytetracycline.
Two animals each received two ceftiofur injections in October 2009.
One animal received two ceftiofur injections in November 2009.
One hundred thirty-seven cattle were injected with antibiotics, 16 cattle were injected with antibiotics on more than one occasion.
Prevalence and enumeration of ESCr E. coli.
Sample collection occurred while cattle were restrained in a squeeze chute during vaccination, health examination, therapeutic treatment, or routine weighing. Fecal samples were collected by inserting a foam-tipped swab (catalog no. 10812-022; VWR International, Buffalo Grove, IL) 3 to 5 cm into the anus of each animal. Immediately following fecal sample collection, the swab was placed into 4 ml of tryptic soy broth (TSB) (Becton Dickinson, Sparks, MD). Hide samples (1,000 cm2) were collected from each animal behind the right shoulder with a sterile sponge (Nasco, Fort Atkinson, WI) prewetted with 20 ml of buffered peptone water (BPW) (Becton Dickinson). Immediately following hide sampling, the sponge was placed into a sterile bag. Fecal and hide samples were collected by different people, and both changed gloves following each sample. Samples were processed within 4 h of sampling. Fecal samples were suspended by vortexing at maximum speed for 30 s. A 1-ml aliquot of suspended fecal matter was removed and serially diluted in TSB. Hide sponge samples were hand massaged for 15 s, and a 1-ml aliquot of suspension was removed and serially diluted in BPW. Selected dilutions were spiral plated onto MacConkey agar (Becton Dickinson, Sparks, MD) containing no antibiotics (MAC) and onto MacConkey agar supplemented with 4 mg liter−1 of cefotaxime (MAC+CTX). Cefotaxime was obtained from Sigma Co. (St. Louis, MO). Plates were incubated overnight at 37°C. Pink to red colonies on MAC plates were enumerated as lactose-fermenting coliforms. Pink to red colonies on MAC+CTX were enumerated as presumptive ESCr E. coli.
Confirmation of presumptive ESCr E. coli isolates.
Up to six presumptive ESCr E. coli isolates per sample were streaked onto MAC+CTX plates and incubated at 37°C overnight. From each MAC+CTX streak plate, a single isolated pink to red colony was selected and streaked onto a Trypticase soy agar (TSA) plate (Becton Dickinson). One isolated colony from each TSA streak was inoculated into a 0.7-ml tryptic soy broth (TSB) (Becton Dickinson) culture contained in a 96-well block. Inoculated blocks were incubated overnight at 37°C, followed by the addition of glycerol to a final concentration of 15% to allow preservation at −80°C. Prior to freezing, each culture was stamped onto five 150-mm TSA plates with a 96-pin Boekel microplate replicator (Boekel Scientific, Feasterville, PA) to screen for antibiotic resistance. The five plates were supplemented with antibiotics as follows: no additional antibiotics, 32 mg liter−1 ampicillin (AMP), 4 mg liter−1 CTX, 64 mg liter−1 kanamycin (KAN), 32 mg liter−1 nalidixic acid (NAL), or 32 mg liter−1 tetracycline (TET). All antibiotics were obtained from Sigma Co. Plates were incubated overnight at 37°C, and isolates were grouped into categories based on their growth on the screened antibiotics. From each sample, at least one isolate from each category was selected for biochemical confirmation of E. coli and PFGE. Biochemical confirmation of E. coli was performed using the Sensititre broth microdilution system and Gram-negative identification plates (TREK Diagnostic Systems, Cleveland, Ohio) according to the manufacturers' instructions. Samples with one or more presumptive ESCr E. coli isolates confirmed as E. coli were designated ESCr E. coli prevalent.
PFGE.
Pulsed-field gel electrophoresis (PFGE) analysis was performed according to the protocol developed by the Centers for Disease Control and Prevention (27). Agarose-embedded DNA was digested with XbaI (New England BioLabs, Beverly, MA). Banding patterns were either classified as unique or grouped into clusters based on ≥90% homology using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium), employing the Dice similarity coefficient with a 1.5% band position tolerance in conjunction with the unweighted-pair group method using arithmetic averages for clustering.
Antibiotic susceptibility determinations.
Antibiotic susceptibility testing was performed using the Sensititre broth microdilution system and CMV1AGNF plates (TREK Diagnostic Systems) to determine the MIC for each of 15 antibiotic agents. The antimicrobials and breakpoints for resistance in this panel were as follows: amikacin (AMI), ≥64 μg ml−1; amoxicillin-clavulanic acid (AMC) ≥32/16 μg ml−1; AMP, ≥32 μg ml−1; cefoxitin (FOX), ≥32 μg ml−1; TIO, ≥8 μg ml−1; ceftriaxone (AXO), ≥4 μg ml−1; chloramphenicol (CHL), ≥32 μg ml−1; ciprofloxacin (CIP), ≥4 μg ml−1; gentamicin (GEN), ≥16 μg ml−1; KAN, ≥64 μg ml−1; NAL, ≥32 μg ml−1; streptomycin (STR), ≥64 μg ml−1; sulfisoxazole (FIS), ≥512 μg ml−1; TET, ≥16 μg ml−1; and trimethoprim-sulfamethoxazole (COT), ≥4/76 μg ml−1. Isolates resistant to three or more classes of antibiotics were considered to be multidrug resistant (MDR). The antibiotic classes were as follows: aminoglycoside (AMI, GEN, KAN, and STR), β-lactam/β-lactamase inhibitor combination (AMC), cephem (FOX, TIO, and AXO), folate pathway inhibitor (FIS and COT), penicillin (AMP), phenicol (CHL), quinolone (CIP and NAL), and tetracycline (TET). The following organisms were used as quality control strains in the antimicrobial sensitivity assays: Pseudomonas aeruginosa ATCC 27853, E. coli ATCC 25922, and Staphylococcus aureus ATCC 25923.
Detection of the blaCMY-2 gene and 18 plasmid incompatibility group replicons.
Template DNA for PCR detection of the blaCMY-2 gene and 18 plasmid incompatibility group (Inc) replicons was prepared by combining 10 μl of overnight culture of an isolated colony with 100 μl of BAX lysis buffer (DuPont Qualicon Inc., Wilmington, DE). Mixtures were incubated at 37°C for 20 min, followed by incubation at 95°C for 10 min. Lysates were then cooled to room temperature on ice. Lysates were subjected to PCR using the primers and conditions described by Kozak et al. (28) to determine the presence or absence of the blaCMY-2 gene. Lysates were subjected to multiplex PCRs using the primers and conditions described by Johnson et al. (29) to determine the presence or absence of 18 plasmid Inc replicons (IncA/C, IncB/O, IncFIA, IncFIIA, IncFIB, IncFIC, IncFrep, IncHI1, IncHI2, IncI1, IncK/B, IncL/M, IncN, IncP, IncT, IncW, IncX, and IncY).
Plasmid size analysis.
Plasmids were isolated using the method described by Kado and Liu (30). Isolated plasmids were subjected to agarose gel electrophoresis on 1% Tris-borate-EDTA agarose gels run for 4.5 h at 10 V cm−1. Plasmid sizes were estimated relative to the BAC-Tracker supercoiled DNA ladder (catalog no. BT010950; Epicentre Technologies Corp., Madison, WI) and the supercoiled DNA ladder, 2 to 10 kb (catalog no. G6231; Promega Corp., Madison, WI).
Statistics.
Period ESCr E. coli prevalence values within a sample class (herd, IDS+TIO, or Other+TIO) and sample type (fecal or hide) were compared using Pearson's χ2 with Bonferroni's correction for multiple comparisons, with P values of <0.05 considered significant. ESCr E. coli prevalence values from the same period were compared between sample classes (herd versus IDS+TIO and herd versus Other+TIO) using a two-tailed Fisher exact test, with P values of <0.05 considered significant. Comparisons of ESCr E. coli prevalence values from the same period between sample types (fecal versus hide samples) were performed using a two-tailed Fisher exact test, with P values of <0.05 considered significant. All comparisons were performed using the Compare2 program of the WinPepi (ver. 11.7) package (31).
RESULTS
Herd prevalences and concentrations of ESCr E. coli.
Only cattle that had not received TIO injections were sampled to determine the herd prevalences and concentrations of ESCr E. coli. When cattle arrived at the feedlot, the herd ESCr E. coli prevalencesfecal prevalence was 3.9% (Table 2). Subsequent herd ESCr E. coli fecal prevalences ranged from 1.7 to 11.2%, but none of these prevalences differed significantly from the prevalence at arrival. The highest herd fecal ESCr E. coli prevalence (11.2%) occurred in July 2010 and was significantly higher (P < 0.05) than the December 2009, March 2010, and May 2010 (Table 2). Overall, 988 herd fecal samples were obtained during this study, and concentrations of ESCr E. coli that were ≥2.00 log CFU/swab were obtained from 22 samples (2.2%) (Table 2). These ESCr E. coli bacteria constituted only a small fraction of the total fecal lactose-fermenting coliforms shed; lactose-fermenting coliforms were enumerated from 984 of the 988 (99.6%) of the herd fecal samples, and mean lactose-fermenting coliform concentrations by sample period ranged from 4.84 to 6.00 log CFU/swab (data not shown).
Table 2.
Qualitative and quantitative evaluation of ESCr E. coli for a cattle herd residing at a feedlot
| Period | No. sampled | Fecal samples |
Hide samples |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % ESCr E. coli prevalence (frequency)a | No. of samples in each class of log CFU/swab values |
% ESCr E. coli prevalence (frequency)a | No. of samples in each class of log CFU/100 cm2 values |
|||||||||
| 1.70 to 1.99 | 2.00 to 2.99 | 3.00 to 3.99 | 4.00 to 4.99 | >4.99 | 1.30 to 1.99 | 2.00 to 2.99 | 3.00 to 3.99 | >3.99 | ||||
| Feedlot arrival | 153 | 3.9 (6) AB | 2 | 2 | 1 | 0 | 1 | 15.0 (23) A | 16 | 6 | 1 | 0 |
| Increased disease susceptibility | 128 | 5.5 (7) AB | 2 | 4 | 0 | 1 | 0 | 11.7 (15) A | 14 | 1 | 0 | 0 |
| December 2009 | 173 | 2.9 (5) B | 3 | 1 | 1 | 0 | 0 | 7.5 (13) AB | 3 | 8 | 2 | 0 |
| March 2010 | 178 | 1.7 (3) B | 1 | 1 | 1 | 0 | 0 | 1.7 (3) B | 3 | 0 | 0 | 0 |
| May 2010 | 178 | 2.2 (4) B | 3 | 1 | 0 | 0 | 0 | 17.4 (31) A | 28 | 2 | 1 | 0 |
| July 2010 | 178 | 11.2 (20) A | 12 | 8 | 0 | 0 | 0 | 8.4 (15) A | 15 | 0 | 0 | 0 |
| Total | 988 | 4.6 (45) | 23 | 17 | 3 | 1 | 1 | 10.1 (100) | 79 | 17 | 4 | 0 |
Prevalence values in the same column that do not have a common letter are statistically significantly different (P < 0.05).
When cattle arrived at the feedlot, the ESCr E. coli hide prevalence was 15.0%, which was not different (P ≥ 0.05) than subsequent hide prevalences except the 1.7% prevalence during March 2010, which was significantly lower (P < 0.05) than hide prevalences during all other periods except December 2009 (Table 2). Overall, ESCr E. coli bacteria were enumerated on 100 hides, but the hide concentrations of ESCr E. coli were <2.00 log CFU/100 cm2 for 79 (79.0%) of these samples (Table 2). The concentrations of ESCr E. coli on the hides were low in comparison to the concentrations of lactose-fermenting coliforms present on the cattle hides. Lactose-fermenting coliforms were enumerated from 980 of the 988 (99.2%) herd hide samples, with period mean concentrations of lactose-fermenting coliforms ranging from 3.11 to 4.66 log CFU/100 cm2 (data not shown).
Prevalences and concentrations of ESCr E. coli in feces and on hides of cattle injected with TIO during the period of increased disease susceptibility.
During the period of increased disease susceptibility, 50 cattle were injected with TIO (“IDS+TIO” cattle). Pre-TIO injection samples were recovered from 49 of these IDS+TIO cattle, and the fecal prevalence of ESCr E. coli was 8.2% (Table 3), which was not significantly different (P = 0.50) than the 5.5% herd fecal prevalence of ESCr E. coli during the period of increased disease susceptibility (see Table S1 in the supplemental material). Post-TIO injection samples were obtained from all 50 IDS+TIO cattle at 3 to 8 days following TIO injection, and the fecal prevalence of ESCr E. coli of 92.0% was significantly higher (P < 0.05) than the 8.2% prevalence pre-TIO injection (Table 3). The ESCr E. coli fecal prevalence post-TIO injection for IDS+TIO cattle was also significantly higher (P < 0.01) than the 5.5% herd fecal prevalence of ESCr E. coli during the period of increased disease susceptibility (see Table S1 in the supplemental material). For 11 additional samples obtained from five of these cattle during subsequent health examinations that occurred during the increased disease susceptibility period, the fecal prevalence of ESCr E. coli was 90.9%. During the December 2009, March 2010, May 2010, and July 2010 sampling periods, the ESCr E. coli fecal prevalence ranged from 0.0 to 6.1% (Table 3) and did not significantly differ (P ≥ 0.05) from the corresponding herd ESCr E. coli fecal prevalences during these periods (see Table S1 in the supplemental material). The fecal concentrations of ESCr E. coli were ≥2.00 log CFU/swab for 52 of the 61 (85.2%) post-TIO injection samples obtained during the period of increased disease susceptibility, but fecal concentrations of ESCr E. coli were ≥2.00 log CFU/swab for only 3 of the 193 (1.6%) samples obtained from these cattle during the subsequent December 2009, March 2010, May 2010, and July 2010 sampling periods (Table 3).
Table 3.
Qualitative and quantitative evaluation of ESCr E. coli for cattle injected with ceftiofur during the increased disease susceptibility period
| Period | No. sampled | Fecal samples |
Hide samples |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % ESCr E. coli prevalence (frequency)a | No. of samples in each class of log CFU/swab values |
% ESCr E. coli prevalence (frequency)a | No. of samples in each class of log CFU/100 cm2 values |
|||||||||
| 1.70 to 1.99 | 2.00 to 2.99 | 3.00 to 3.99 | 4.00 to 4.99 | >4.99 | 1.30 to 1.99 | 2.00 to 2.99 | 3.00 to 3.99 | >3.99 | ||||
| Increased disease susceptibility, pre-ceftiofur injection | 49 | 8.2 (4) B | 0 | 4 | 0 | 0 | 0 | 8.2 (4) AB | 2 | 2 | 0 | 0 |
| Increased disease susceptibility, 3 to 8 days post-ceftiofur injection | 50 | 92.0 (46) A | 3 | 19 | 17 | 6 | 1 | 26.0 (13) A | 10 | 1 | 2 | 0 |
| Increased disease susceptibility, additional post-ceftiofur injection samples | 11 | 90.9 (10) | 1 | 3 | 5 | 1 | 0 | 27.3 (3) | 1 | 2 | 0 | 0 |
| December 2009 | 49 | 6.1 (3) B | 1 | 1 | 0 | 1 | 0 | 24.5 (12) A | 7 | 5 | 0 | 0 |
| March 2010 | 48 | 0.0 (0) B | 0 | 0 | 0 | 0 | 0 | 0.0 (0) B | 0 | 0 | 0 | 0 |
| May 2010 | 48 | 2.1 (1) B | 0 | 1 | 0 | 0 | 0 | 20.8 (10) A | 9 | 1 | 0 | 0 |
| July 2010 | 48 | 2.1 (1) B | 1 | 0 | 0 | 0 | 0 | 0.0 (0) B | 0 | 0 | 0 | 0 |
| Total | 303 | 21.5 (65) | 6 | 28 | 22 | 8 | 1 | 13.9 (42) | 29 | 11 | 2 | 0 |
Prevalence values in the same column that do not have a common letter are statistically significantly different (P < 0.05). Prevalences for increased disease susceptibility with additional post-ceftiofur injection samples were not included in statistical comparisons.
The 26.0% hide prevalence of ESCr E. coli for IDS+TIO cattle sampled at 3 to 8 days post-TIO injection was higher than the 8.2% prevalence pre-TIO injection, but this difference was not statistically significant (Table 3). ESCr E. coli hide prevalences for IDS+TIO cattle were not significantly different than the pre-TIO injection hide prevalence during the December 2009, March 2010, May 2010, and July 2010 periods but ranged from 24.5 and 20.4% during December 2009 and May 2010 to 0.0% during March 2010 and July 2010 (Table 3). Hide concentrations of ESCr E. coli were ≥2.00 log CFU/100 cm2 for only 13 of the 303 (4.3%) hide samples obtained from IDS+TIO cattle (Table 3). Pre-TIO injection, March 2010, May 2010, and July 2010 hide ESCr E. coli prevalences for IDS+TIO were not significantly different (P ≥ 0.05) from their corresponding herd ESCr E. coli hide prevalences during these periods (see Table S1 in the supplemental material). However, the 26.0% hide prevalence of ESCr E. coli for IDS+TIO cattle sampled at 3 to 8 days post-TIO injection was significantly higher (P = 0.02) than the herd ESCr E. coli hide prevalence of 11.7% during the period of increased disease susceptibility (see Table S1 in the supplemental material). During December 2009, the 24.5% ESCr E. coli hide prevalence for IDS+TIO cattle was significantly higher (P < 0.01) than the herd ESCr E. coli hide prevalence of 7.5% (see Table S1 in the supplemental material).
Prevalences and concentrations of ESCr E. coli in feces and on hides of cattle injected with TIO at times other than during the period of increased disease susceptibility.
Cattle that had been injected with TIO at times other than during the period of increased disease susceptibility (“Other+TIO” cattle) were sampled during the feedlot arrival, December 2009, March 2010, May 2010, and July 2010 sampling periods. During the feedlot arrival period, samples were obtained from eight of the 19 cattle that had been injected with TIO while on pasture, and ESCr E. coli was isolated from one fecal sample and one hide sample, which were not from the same animal (Table 4). The number of Other+TIO cattle sampled increased during each subsequent period, from 33 during December 2009 to 51 during July 2010, since TIO injections continued to occur during the residence of the herd at the feedlot (Table 1). ESCr E. coli fecal prevalences for the samples obtained from December to July ranged from 0.0 to 9.8%. Fecal concentrations of ESCr E. coli were ≥2.00 log CFU/swab for only two of the 174 (1.1%) samples obtained from Other+TIO cattle (Table 4). Prevalences of ESCr E. coli on the hides of Other+TIO cattle were 9.1, 2.6, 25.6, and 3.9%, during December 2009, March 2010, May 2010, and July 2010, respectively (Table 4). Hide concentrations were ≥2.00 log CFU/100 cm2 for only 3 of the 174 (1.7%) hide samples obtained from these cattle. ESCr E. coli prevalences for Other+TIO cattle did not significantly differ (P > 0.05) from their corresponding herd prevalences during the December 2009, March 2010, May 2010, and July 2010 periods for both fecal and hide samples (see Table S2 in the supplemental material).
Table 4.
Qualitative and quantitative evaluation of ESCr E. coli for cattle injected with ceftiofur at times other than during the increased disease susceptibility period
| Period | No. sampled | Fecal samples |
Hide samples |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % ESCr E. coli prevalence (frequency)a | No. of samples in each class of log CFU/swab values |
% ESCr E. coli prevalence (frequency)a | No. of samples in each class of log CFU/100 cm2 values |
|||||||||
| 1.70 to 1.99 | 2.00 to 2.99 | 3.00 to 3.99 | 4.00 to 4.99 | >4.99 | 1.30 to 1.99 | 2.00 to 2.99 | 3.00 to 3.99 | >3.99 | ||||
| Feedlot arrival | 8 | 12.5 (1) | 1 | 0 | 0 | 0 | 0 | 12.5 (1) | 1 | 0 | 0 | 0 |
| December 2009 | 33 | 3.0 (1) A | 1 | 0 | 0 | 0 | 0 | 9.1 (3) AB | 1 | 2 | 0 | 0 |
| March 2010 | 39 | 0.0 (0) A | 0 | 0 | 0 | 0 | 0 | 2.6 (1) B | 1 | 0 | 0 | 0 |
| May 2010 | 43 | 0.0 (0) A | 0 | 0 | 0 | 0 | 0 | 25.6 (11) A | 10 | 1 | 0 | 0 |
| July 2010 | 51 | 9.8 (5) A | 3 | 2 | 0 | 0 | 0 | 3.9 (2) B | 2 | 0 | 0 | 0 |
| Total | 174 | 4.0 (7) | 5 | 2 | 0 | 0 | 0 | 10.3 (18) | 15 | 3 | 0 | 0 |
Prevalence values in the same column that do not have a common letter are statistically significantly different (P < 0.05). Feedlot arrival prevalences were not included in statistical comparisons.
PFGE genotypes of ESCr E. coli isolates.
XbaI PFGE genotypes were obtained for 383 ESCr E. coli isolates. Genotypes were obtained for 198 fecal isolates originating from 113 samples; thus, for 40 of the fecal samples more than one isolate was genotyped. Genotypes were obtained for 185 hide isolates originating from 158 samples; thus, more than one isolate was genotyped for 25 hide samples. Multiple XbaI PFGE banding patterns were identified from 39 of 65 samples that had more than one isolate genotyped (37 samples had 2 banding patterns, and 2 samples had 3 banding patterns). Seventy-one isolates had XbaI PFGE banding patterns identical to that of another isolate from the same sample and were considered redundant. Of the 312 nonredundant isolates, 133 originated from feces and 179 originated from hides. Overall, 26 unique XbaI PFGE banding patterns were identified, and these genotypes were assigned letters according to their prevalence (i.e., genotype A was most prevalent, followed by genotype B, etc.). Eight genotypes (S, T, U, V, W, X, Y, and Z) were identified from only one sample. Of the 18 genotypes identified from more than one sample, two (P and R) were identified exclusively from fecal samples, and two (I and L) were identified exclusively from hide samples (Table 5).
Table 5.
ESCr E. coli PFGE genotype prevalences
| Sample type and period | No. of isolates | PFGE genotype frequency |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | ||
| Fecal samples | |||||||||||||||||||||||||||
| Feedlot arrival | 7 | 1 | 6 | ||||||||||||||||||||||||
| Increased disease susceptibility | 78 | 36 | 17 | 1 | 1 | 6 | 2 | 2 | 3 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| December 2009 | 12 | 2 | 1 | 2 | 2 | 3 | 1 | 1 | |||||||||||||||||||
| March 2010 | 3 | 1 | 1 | 1 | |||||||||||||||||||||||
| May 2010 | 5 | 4 | 1 | ||||||||||||||||||||||||
| July 2010 | 28 | 24 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| Total | 133 | 66 | 2 | 18 | 6 | 4 | 1 | 7 | 3 | 4 | 5 | 4 | 2 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | |||||
| Hide samples | |||||||||||||||||||||||||||
| Feedlot arrival | 25 | 22 | 2 | 1 | |||||||||||||||||||||||
| Increased disease susceptibility | 42 | 13 | 14 | 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||
| December 2009 | 28 | 19 | 5 | 3 | 1 | ||||||||||||||||||||||
| March 2010 | 4 | 2 | 2 | ||||||||||||||||||||||||
| May 2010 | 63 | 8 | 39 | 1 | 6 | 9 | |||||||||||||||||||||
| July 2010 | 17 | 14 | 1 | 1 | 1 | ||||||||||||||||||||||
| Total | 179 | 35 | 40 | 15 | 22 | 19 | 13 | 2 | 6 | 9 | 3 | 1 | 5 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | |||||||
| Total (fecal and hide) | 312 | 101 | 42 | 33 | 28 | 23 | 14 | 9 | 9 | 9 | 7 | 6 | 5 | 5 | 5 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
The overall most prevalent genotype, genotype A, was identified from 101 of the 271 (37.3%) ESCr E. coli-positive samples. Genotype A was predominate in fecal samples, since it was identified in 66 of the 113 (58.4%) ESCr E. coli-positive fecal samples. The next most prevalent fecal genotype (genotype C) was isolated from only 18 (15.9%) of the ESCr E. coli-prevalent fecal samples. No single genotype predominated in ESCr E. coli-prevalent hide samples, since genotypes B, A, D, and E were prevalent in 40 (25.3%), 35 (22.2%), 22 (13.9%), and 19 (12.0%) of the 158 ESCr E. coli-positive hide samples, respectively. No genotype was isolated during all six sampling periods, but pattern A was prevalent during five of the six sampling periods, all except December 2009 (Table 5). Other genotypes prevalent during multiple sampling periods were C, E, L, and N, each being prevalent during three sampling periods. Genotypes B, F, G, H, J, K, and R were each prevalent during two sampling periods (Table 5).
Antibiotic resistance phenotypes of ESCr E. coli isolates.
Susceptibilities to 15 antibiotics were determined for the 312 nonredundant ESCr E. coli isolates. All isolates were MDR, since all were resistant to at least three classes of antibiotics. All isolates were susceptible to AMI and CIP. Seven resistance phenotypes were identified. Resistance to at least AMC, AMP, FOX, TIO, AXO, CHL, STR, FIS, and TET (ACSSuTAuCfCtCx+ phenotype) was detected with 99.7% (n = 311) of isolates (Table 6). One isolate was susceptible to CHL but resistant to AMC, AMP, FOX, TIO, AXO, STR, FIS, and TET. For 13 of 14 genotypes with more than four isolates examined, the predominate resistance phenotype was observed for >75% of the isolates; genotype F was the only exception. Notably, all eight genotype F isolates with the ACSSuTAuCfCtCx + NAL resistance phenotype were from samples from the increased disease susceptibility period, while all six genotype F isolates with the ACSSuTAuCfCtCx + KAN + NAL resistance phenotype were from May 2010 samples (data not shown).
Table 6.
Antibiotic resistance phenotypes of ESCr E. coli isolates
| PFGE genotype | No. of isolates | % of isolates with resistance to the indicated antibioticsa |
||||||
|---|---|---|---|---|---|---|---|---|
| AMC, AMP, FOX, AXO, TIO, STR, FIS, TET | AMC, AMP, FOX, AXO, TIO, CHL, STR, FIS, TET | AMC, AMP, FOX, AXO, TIO, CHL, NAL, STR, FIS, TET | AMC, AMP, FOX, AXO, TIO, CHL, STR, FIS, TET, COT | AMC, AMP, FOX, AXO, TIO, CHL, NAL, STR, FIS, TET, COT | AMC, AMP, FOX, AXO, TIO, CHL, KAN, NAL, STR, FIS, TET | AMC, AMP, FOX, AXO, TIO, CHL, GEN, KAN, STR, FIS, TET | ||
| A | 101 | 0.0 | 97.0 | 2.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| B | 42 | 0.0 | 0.0 | 97.6 | 0.0 | 0.0 | 2.4 | 0.0 |
| C | 33 | 0.0 | 3.0 | 0.0 | 90.9 | 6.1 | 0.0 | 0.0 |
| D | 28 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| E | 23 | 0.0 | 13.0 | 0.0 | 0.0 | 87.0 | 0.0 | 0.0 |
| F | 14 | 0.0 | 0.0 | 57.1 | 0.0 | 0.0 | 42.9 | 0.0 |
| G | 9 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| H | 9 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| I | 9 | 0.0 | 77.8 | 22.2 | 0.0 | 0.0 | 0.0 | 0.0 |
| J | 7 | 0.0 | 85.7 | 0.0 | 0.0 | 14.3 | 0.0 | 0.0 |
| K | 6 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| L | 5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 |
| M | 5 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| N | 5 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| O | 2 | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| P | 2 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Q | 2 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| R | 2 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| S | 1 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| T | 1 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| U | 1 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| V | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 |
| W | 1 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| X | 1 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Y | 1 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Z | 1 | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Total | 312 | 0.3 | 59.9 | 17.3 | 10.9 | 7.4 | 2.2 | 1.9 |
Antibiotics: AMC, amoxicillin-clavulanic acid; AMP, ampicillin; FOX, cefoxitin; AXO, ceftriaxone; TIO, ceftiofur; CHL, chloramphenicol; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; FIS, sulfisoxazole; TET, tetracycline; COT, trimethoprim-sulfamethoxazole.
Prevalence of the blaCMY-2 gene, plasmid replicons, and plasmids of >80 kbp in ESCr E. coli isolates.
The blaCMY-2 gene was present in all 312 nonredundant ESCr E. coli isolates. The plasmid replicons IncB/O, IncFIIA, IncFIC, IncHI1, IncHI2, IncK/B, IncL/M, IncP, IncT, IncW, and IncX were not detected from any isolate. The IncA/C replicon was the most prevalent replicon, being present in 69.2% (n = 216) of isolates (Table 7). Interestingly, all 97 isolates lacking the IncA/C plasmid replicon were genotype A (the IncA/C plasmid replicon was detected from only 5 of the 101 genotype A isolates). The next most prevalent plasmid replicon was IncY, present in 23.4% (n = 73) of the isolates. The IncFIB plasmid replicon was present in 20.2% (n = 63) of the isolates, and its presence was correlated with COT resistance, since 76.2% of the isolates with the IncFIB replicon were COT resistant and 82.8% of COT-resistant isolates possessed the IncFIB replicon (data not shown). One or more plasmids larger than 80 kbp were present in 68.9% (n = 215) of the 312 nonredundant ESCr E. coli isolates (Table 7). Of the 97 isolates lacking a plasmid larger than 80 kbp, 95 were genotype A isolates. A single 4-kbp plasmid was the only plasmid present in 89.1% (n = 90) of the genotype A isolates (data not shown).
Table 7.
Prevalence of plasmids >80 kbp and replicons in ESCr E. coli isolates
| PFGE genotype | No. of isolates | % of isolates with at least one plasmid >80 kbp | % of isolates with indicated replicon |
||||||
|---|---|---|---|---|---|---|---|---|---|
| IncA/C | IncY | IncFIB | IncFrep | IncFIA | IncN | IncI1 | |||
| A | 101 | 5.9 | 5.0 | 12.9 | 4.0 | 0.0 | 2.0 | 4.0 | 4.0 |
| B | 42 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| C | 33 | 100.0 | 100.0 | 39.4 | 84.8 | 48.5 | 3.0 | 6.1 | 3.0 |
| D | 28 | 100.0 | 100.0 | 100.0 | 0.0 | 3.6 | 100.0 | 0.0 | 0.0 |
| E | 23 | 100.0 | 100.0 | 4.3 | 78.3 | 0.0 | 0.0 | 0.0 | 0.0 |
| F | 14 | 100.0 | 100.0 | 0.0 | 0.0 | 85.7 | 0.0 | 0.0 | 0.0 |
| G | 9 | 100.0 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 11.1 | 0.0 |
| H | 9 | 88.9 | 100.0 | 11.1 | 44.4 | 22.2 | 0.0 | 0.0 | 0.0 |
| I | 9 | 100.0 | 100.0 | 0.0 | 0.0 | 66.7 | 0.0 | 100.0 | 0.0 |
| J | 7 | 100.0 | 100.0 | 0.0 | 28.6 | 14.3 | 0.0 | 0.0 | 0.0 |
| K | 6 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| L | 5 | 100.0 | 100.0 | 60.0 | 20.0 | 100.0 | 40.0 | 20.0 | 0.0 |
| M | 5 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| N | 5 | 100.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 0.0 |
| O | 2 | 100.0 | 100.0 | 50.0 | 0.0 | 0.0 | 50.0 | 0.0 | 0.0 |
| P | 2 | 100.0 | 100.0 | 0.0 | 0.0 | 50.0 | 0.0 | 0.0 | 0.0 |
| Q | 2 | 100.0 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| R | 2 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| S | 1 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| T | 1 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| U | 1 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| V | 1 | 100.0 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| W | 1 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 |
| X | 1 | 0.0 | 100.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Y | 1 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Z | 1 | 100.0 | 100.0 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Total | 312 | 68.9 | 69.2 | 23.4 | 20.2 | 14.4 | 12.5 | 5.8 | 1.6 |
DISCUSSION
A goal of this study was to determine the prevalence and levels of ESCr E. coli in a feedlot cattle population when TIO was the preferred therapeutic antibiotic. TIO injection is known to transiently increase the fecal concentrations of ESCr E. coli and blaCMY-2 in individual cattle before returning to levels observed prior to TIO injection (20–22). During the December 2009, March 2010, May 2010, and June 2010 sample periods, fecal prevalences of ESCr E. coli for IDS+TIO or Other+TIO cattle were not significantly higher (P ≥ 0.05) than the corresponding herd fecal prevalences (see Tables S1 and S2 in the supplemental material). This result demonstrates that once the transient increase of ESCr E. coli shedding that follows TIO injection abates, TIO-injected cattle are no more likely than untreated members of the same herd to shed ESCr E. coli. Two prior studies of the impact of TIO use on the prevalence of ESCr E. coli in dairy cow herds had conflicting results, with one study finding an association and the other finding no association between the extent of TIO use and herd prevalence of E. coli with reduced susceptibility to ESC (23, 24). In this study, herd fecal and hide ESCr E. coli prevalences when weaning calves arrived at the feedlot were not significantly lower (P < 0.05) than prevalences during any subsequent sampling period (Table 2). This suggests that the therapeutic use of TIO in feedlot cattle populations does not significantly increase the herd prevalence of ESCr E. coli. However, we note that differences exist between the practices of the USMARC feedlot and those of commercial feedlots. Specifically, the USMARC cattle populations are “closed” (i.e., all feedlot cattle are born on center and raised until weaned on USMARC pastures, and replacement heifers are selected from USMARC populations), and introduction of cattle to USMARC feedlot occurs only twice per year when weaning calves are transferred to the feedlot. Thus, patterns of therapeutic antibiotic use in some commercial feedlots may differ from those in the USMARC feedlot.
E. coli strains associated with cattle are known to harbor antibiotic resistance genes, including genes encoding ESCr (5, 32–36). These antibiotic resistance genes may be transmitted to human commensal and pathogenic bacteria, either by E. coli or when horizontally transferred to food-borne pathogenic bacteria, such as S. enterica (5, 37, 38). Therefore, fecal samples are typically examined to assess ESCr E. coli or blaCMY-2 presence in cattle environments (20–24, 39). Hide samples were cultured for ESCr E. coli, since hides have been demonstrated to harbor E. coli strains of fecal origin, including E. coli O157:H7 (40, 41). During this study, a total of 1,465 fecal and 1,465 hide samples were obtained, and blaCMY-2 positive ESCr E. coli was prevalent on 10.9% (n = 160) of hides, which is significantly higher (P < 0.01) than the 8.0% (n = 117) prevalence in feces (data not shown). The 92.0% (n = 46) fecal prevalence of ESCr E. coli for IDS+TIO at 3 to 8 days post-TIO was significantly higher (P < 0.01) than the 26.0% (n = 16) hide prevalence for these cattle (Table 3). Genotype prevalences also differed between fecal and hide samples; for example, genotype B was the most prevalent hide genotype during May 2010 (isolated from 39 hides) but was isolated from only one May 2010 fecal sample (Table 5). Prevalence and genotype distribution differences between fecal and hide samples were expected, since fecal samples recover the E. coli population shed by the animal at the moment of sampling, while the hide sample obtained from the same animal likely contains a more broad representation of the overall E. coli population present in the feedlot environment at the time of sampling. These results demonstrate that hide sampling is a useful complement to fecal sampling for examination of the antibiotic resistance status of commensal E. coli in cattle feedlot environments.
To our knowledge, ESCr E. coli isolated from a longitudinal study of a single cattle herd had not been subjected to genotypic analysis. In this study, we identified only 26 ESCr E. coli genotypes, 12 of which were isolated during multiple sampling periods, and we identified a predominant genotype (A) which was isolated during five of the six sampling periods (Table 5). Our results suggest that clonal expansion of feedlot-adapted E. coli strains containing blaCMY-2 contributes to the persistence of the ESCr E. coli in this cattle herd. We note that Daniels et al. identified 46 unique genotypes from 46 blaCMY-2 ESCr E. coli isolates obtained from 14 cattle herds, suggesting that blaCMY-2 ESCr E. coli populations between herds are diverse (7). Consideration of the results of this study, the study by Daniels et al., and the mathematical modeling of the persistence of blaCMY-2 ESCr E. coli in cattle environments performed by Volkova et al. (26), it is likely that both conjugal transfer and clonal expansion contribute to blaCMY-2 IncA/C plasmid persistence in commensal E. coli. We note that E. coli has been demonstrated to survive for at least 6 months in cattle fecal pats (42, 43) and for at least 6 weeks on feedlot surfaces after the removal of cattle (44). Thus, we theorize that in individual feedlots, persistence of blaCMY-2 ESCr E. coli occurs primarily by a cycle of ingested and shed clonal populations “adapted” to survive the specific stresses encountered in cattle and the environment at the individual feedlot level. We further theorize that conjugal transfer also occurs, ensuring that blaCMY-2 IncA/C plasmids are transferred to other receptive commensal E. coli strains, which are possibility more fit to survive when the environmental conditions change. Longitudinal studies of multiple cattle feedlots are required to test these theories of blaCMY-2 ESCr E. coli persistence in cattle feedlots.
All genotype A isolates possessed the ACSSuTAuCfCtCx+ phenotype, typically conferred by large blaCMY-2 IncA/C plasmids, but 89.1% of these isolates had only one 4-kbp plasmid, which is not large enough to carry all of the resistance genes (blaCMY-2, tetA, floR, aadA2, sul1, and sul2) typically harbored by blaCMY-2 IncA/C plasmids. Additionally, the IncA/C replicon was detected from only 5.0% of the genotype A isolates (Table 7). A few scenarios could explain these results. The integration of different genetic elements harboring the blaCMY-2 gene and the genes conferring the other antibiotic resistances could have occurred independently. Indeed, presence of blaCMY-2 in the bacterial chromosome is not unprecedented, since a blaCMY-2 gene is contained within a chromosomally integrated SXT/R391-like element in a human clinical isolate of Proteus mirabilis (45). However, since all genotype A isolates had the ACSSuTAuCfCtCx+ phenotype, a more likely explanation is integration of a blaCMY-2 IncA/C plasmid into the E. coli chromosome. Failure to detect the IncA/C replicon can be explained if the plasmid site of integration was located between the annealing sites of the primers used to amplify the IncA/C replicon. Alternatively, the plasmid site of integration could be located elsewhere, but one of the primer annealing sites may have been altered by mutation, insertion, or deletion.
Concerns have been raised that therapeutic use of TIO in animal agriculture, including its use in feedlot cattle, contributes significantly to the increased occurrence of ESCr infections in humans (2, 19, 46, 47). The isolation of blaCMY-2 ESCr E. coli from U.S. retail beef supports these concerns, but the source of the ESCr E. coli in these products is unknown (4). However, it is well established that the primary source of contamination of beef products with S. enterica and E. coli O157:H7 during processing are cattle hides with high levels of these pathogens (40, 41, 48, 49). The July 2010 hide samples were obtained within 2 weeks of harvest and approximate the population of ESCr E. coli likely to be present on the hides of these cattle during processing. Herd prevalence of ESCr E. coli on hides was 8.4% in July 2010, but ESCr E. coli concentrations were <2.00 log CFU/100 cm2 for all of these samples (Table 2). Thus, the ESCr E. coli likely present on these cattle hides when sent to harvest was a very small fraction of the overall lactose-fermenting coliform population on the hides, since the herd prevalence of lactose-fermenting coliforms on hides during July 2010 was 100% and the mean concentration was 4.60 log CFU/100 cm2 (data not shown). Complex factors, including contamination in lairage, can alter the microbial populations present on cattle hides at harvest (50, 51). Clearly, studies on the prevalence, concentrations, and genotypes of ESCr E. coli on cattle hides when processing begins and in final products from the same processing location are required for a complete understanding of frequency and sources of the ESCr E. coli contamination of beef products.
Supplementary Material
ACKNOWLEDGMENTS
We thank Julie Dyer, Bruce Jasch, Dee Kucera, Kim Kucera, Shannon Ostdiek, and Frank Reno for technical support. We thank Shuna A. Jones for providing data on patterns of therapeutic antibiotic use at the USMARC feeding operation. We thank the USMARC cattle operations staff for facilitating cattle sampling. We thank Jody Gallagher for administrative assistance. We thank Terrance M. Arthur and Tommy L. Wheeler for critical review of the manuscript.
Mention of trade names, proprietary products, or specified equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable.
Footnotes
Published ahead of print 25 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03592-12.
REFERENCES
- 1.Hohmann EL. 2001. Nontyphoidal salmonellosis. Clin. Infect. Dis. 32:263–269 [DOI] [PubMed] [Google Scholar]
- 2.Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. 2009. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 49:132–141 [DOI] [PubMed] [Google Scholar]
- 3.Hellerstein S. 2003. Antibiotic treatment for urinary tract infections in pediatric patients. Minerva Pediatr. 55:395–406 [PubMed] [Google Scholar]
- 4.Mollenkopf DF, Kleinhenz KE, Funk JA, Gebreyes WA, Wittum TE. 2011. Salmonella enterica and Escherichia coli harboring blaCMY in retail beef and pork products. Foodborne Pathog. Dis. 8:333–336 [DOI] [PubMed] [Google Scholar]
- 5.Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folster JP, Pecic G, McCullough A, Rickert R, Whichard JM. 2011. Characterization of blaCMY-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog. Dis. 8:1289–1294 [DOI] [PubMed] [Google Scholar]
- 7.Daniels JB, Call DR, Besser TE. 2007. Molecular epidemiology of blaCMY-2 plasmids carried by Salmonella enterica and Escherichia coli isolates from cattle in the Pacific Northwest. Appl. Environ. Microbiol. 73:8005–8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doublet B, Carattoli A, Whichard JM, White DG, Baucheron S, Chaslus-Dancla E, Cloeckaert A. 2004. Plasmid-mediated florfenicol and ceftriaxone resistance encoded by the floR and blaCMY-2 genes in Salmonella enterica serovars Typhimurium and Newport isolated in the United States. FEMS Microbiol. Lett. 233:301–305 [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Blickenstaff K, Glenn A, Ayers SL, Friedman SL, Abbott JW, McDermott PF. 2009. Beta-lactam resistance in Salmonella strains isolated from retail meats in the United States by the National Antimicrobial Resistance Monitoring System between 2002 and 2006. Appl. Environ. Microbiol. 75:7624–7630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli A, Tosini F, Giles WP, Rupp ME, Hinrichs SH, Angulo FJ, Barrett TJ, Fey PD. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donaldson SC, Straley BA, Hegde NV, Sawant AA, DebRoy C, Jayarao BM. 2006. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72:3940–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winokur PL, Brueggemann A, DeSalvo DL, Hoffmann L, Apley MD, Uhlenhopp EK, Pfaller MA, Doern GV. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44:2777–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. 1996. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40:221–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, White DG, Cebula TA, Ravel J. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376–386 [DOI] [PubMed] [Google Scholar]
- 16.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, Leclerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309 10.1371/journal.pone.0000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Alarcon C, Singer RS, Johnson TJ. 2011. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6:e23415.doi:10.1371/journal.pone.0023415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.European Food Safety Authority Panel on Biological Hazards 2011. Scientifc opinion on the public health risks of bacterial strains producing extended-spectrum beta-lactamases and/or AmpC beta-lactamases in food and food-producing animals. EFSA J. 9:2332 http://www.efsa.europa.eu/en/efsajournal/pub/2322.htm [Google Scholar]
- 20.Singer RS, Patterson SK, Wallace RL. 2008. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl. Environ. Microbiol. 74:6956–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowrance TC, Loneragan GH, Kunze DJ, Platt TM, Ives SE, Scott HM, Norby B, Echeverry A, Brashears MM. 2007. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am. J. Vet. Res. 68:501–507 [DOI] [PubMed] [Google Scholar]
- 22.Alali WQ, Scott HM, Norby B, Gebreyes W, Loneragan GH. 2009. Quantification of the bla(CMY-2) in feces from beef feedlot cattle administered three different doses of ceftiofur in a longitudinal controlled field trial. Foodborne Pathog. Dis. 6:917–924 [DOI] [PubMed] [Google Scholar]
- 23.Tragesser LA, Wittum TE, Funk JA, Winokur PL, Rajala-Schultz PJ. 2006. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67:1696–1700 [DOI] [PubMed] [Google Scholar]
- 24.Daniels JB, Call DR, Hancock D, Sischo WM, Baker K, Besser TE. 2009. Role of ceftiofur in selection and dissemination of blaCMY-2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl. Environ. Microbiol. 75:3648–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subbiah M, Top EM, Shah DH, Call DR. 2011. Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C plasmids. Appl. Environ. Microbiol. 77:4486–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volkova VV, Lanzas C, Lu Z, Grohn YT. 2012. Mathematical model of plasmid-mediated resistance to ceftiofur in commensal enteric Escherichia coli of cattle. PLoS One 7:e36738.doi: 10.1371/journal.pone.0036738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 28.Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C. 2009. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75:559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 73:1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abramson JH. 2011. WINPEPI updated: computer programs for epidemiologists, and their teaching potential. Epidemiol. Perspect. Innov. 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carattoli A. 2008. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14(Suppl. 1):117–123 [DOI] [PubMed] [Google Scholar]
- 33.Hammerum AM, Heuer OE. 2009. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin. Infect. Dis. 48:916–921 [DOI] [PubMed] [Google Scholar]
- 34.Karczmarczyk M, Walsh C, Slowey R, Leonard N, Fanning S. 2011. Molecular characterization of multidrug-resistant Escherichia coli isolates from Irish cattle farms. Appl. Environ. Microbiol. 77:7121–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khachatryan AR, Hancock DD, Besser TE, Call DR. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Bogaard AE, Stobberingh EE. 2000. Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 14:327–335 [DOI] [PubMed] [Google Scholar]
- 37.Sawant AA, Hegde NV, Straley BA, Donaldson SC, Love BC, Knabel SJ, Jayarao BM. 2007. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 73:156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Licht TR, Wilcks A. 2006. Conjugative gene transfer in the gastrointestinal environment. Adv. Appl. Microbiol. 58:77–95 [PubMed] [Google Scholar]
- 39.Jiang X, Yang H, Dettman B, Doyle MP. 2006. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog. Dis. 3:355–365 [DOI] [PubMed] [Google Scholar]
- 40.Arthur TM, Brichta-Harhay DM, Bosilevac JM, Kalchayanand N, Shackelford SD, Wheeler TL, Koohmaraie M. 2010. Super shedding of Escherichia coli O157:H7 by cattle and the impact on beef carcass contamination. Meat Sci. 86:32–37 [DOI] [PubMed] [Google Scholar]
- 41.Arthur TM, Keen JE, Bosilevac JM, Brichta-Harhay DM, Kalchayanand N, Shackelford SD, Wheeler TL, Nou X, Koohmaraie M. 2009. Longitudinal study of Escherichia coli O157:H7 in a beef cattle feedlot and role of high-level shedders in hide contamination. Appl. Environ. Microbiol. 75:6515–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avery SM, Moore A, Hutchison ML. 2004. Fate of Escherichia coli originating from livestock faeces deposited directly onto pasture. Lett. Appl. Microbiol. 38:355–359 [DOI] [PubMed] [Google Scholar]
- 43.Alexander TW, Reuter T, Sharma R, Yanke LJ, Topp E, McAllister TA. 2009. Longitudinal characterization of resistant Escherichia coli in fecal deposits from cattle fed subtherapeutic levels of antimicrobials. Appl. Environ. Microbiol. 75:7125–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berry ED, Wells JE, Arthur TM, Woodbury BL, Nienaber JA, Brown-Brandl TM, Eigenberg RA. 2010. Soil versus pond ash surfacing of feedlot pens: occurrence of Escherichia coli O157:H7 in cattle and persistence in manure. J. Food Prot. 73:1269–1277 [DOI] [PubMed] [Google Scholar]
- 45.Harada S, Ishii Y, Saga T, Tateda K, Yamaguchi K. 2010. Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob. Agents Chemother. 54:3545–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vieira AR, Collignon P, Aarestrup FM, McEwen SA, Hendriksen RS, Hald T, Wegener HC. 2011. Association between antimicrobial resistance in Escherichia coli isolates from food animals and blood stream isolates from humans in Europe: an ecological study. Foodborne Pathog. Dis. 8:1295–1301 [DOI] [PubMed] [Google Scholar]
- 47.Aarestrup FM, Wegener HC, Collignon P. 2008. Resistance in bacteria of the food chain: epidemiology and control strategies. Expert Rev. Anti Infect. Ther. 6:733–750 [DOI] [PubMed] [Google Scholar]
- 48.Brichta-Harhay DM, Guerini MN, Arthur TM, Bosilevac JM, Kalchayanand N, Shackelford SD, Wheeler TL, Koohmaraie M. 2008. Salmonella and Escherichia coli O157:H7 contamination on hides and carcasses of cull cattle presented for slaughter in the United States: an evaluation of prevalence and bacterial loads by immunomagnetic separation and direct plating methods. Appl. Environ. Microbiol. 74:6289–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nou X, Rivera-Betancourt M, Bosilevac JM, Wheeler TL, Shackelford SD, Gwartney BL, Reagan JO, Koohmaraie M. 2003. Effect of chemical dehairing on the prevalence of Escherichia coli O157:H7 and the levels of aerobic bacteria and enterobacteriaceae on carcasses in a commercial beef processing plant. J. Food Prot. 66:2005–2009 [DOI] [PubMed] [Google Scholar]
- 50.Arthur TM, Bosilevac JM, Brichta-Harhay DM, Guerini MN, Kalchayanand N, Shackelford SD, Wheeler TL, Koohmaraie M. 2007. Transportation and lairage environment effects on prevalence, numbers, and diversity of Escherichia coli O157:H7 on hides and carcasses of beef cattle at processing. J. Food Prot. 70:280–286 [DOI] [PubMed] [Google Scholar]
- 51.Arthur TM, Bosilevac JM, Brichta-Harhay DM, Kalchayanand N, King DA, Shackelford SD, Wheeler TL, Koohmaraie M. 2008. Source tracking of Escherichia coli O157:H7 and Salmonella contamination in the lairage environment at commercial U.S. beef processing plants and identification of an effective intervention. J. Food Prot. 71:1752–1760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.