Abstract
l-Homophenylalanine (l-Hph) is a useful chiral building block for synthesis of several drugs, including angiotensin-converting enzyme inhibitors and the novel proteasome inhibitor carfilzomib. While the chemoenzymatic route of synthesis is fully developed, we investigated microbial production of l-Hph to explore the possibility of a more efficient and sustainable approach to l-Hph production. We hypothesized that l-Hph is synthesized from l-Phe via a mechanism homologous to 3-methyl-2-oxobutanoic acid conversion to 4-methyl-2-oxopentanoic acid during leucine biosynthesis. Based on bioinformatics analysis, we found three putative homophenylalanine biosynthesis genes, hphA (Npun_F2464), hphB (Npun_F2457), and hphCD (Npun_F2458), in the cyanobacterium Nostoc punctiforme PCC73102, located around the gene cluster responsible for anabaenopeptin biosynthesis. We constructed Escherichia coli strains harboring hphABCD-expressing plasmids and achieved the fermentative production of l-Hph from l-Phe. To our knowledge, this is the first identification of the genes responsible for homophenylalanine synthesis in any organism. Furthermore, to improve the low conversion efficiency of the initial strain, we optimized the expression of hphA, hphB, and hphCD, which increased the yield to ∼630 mg/liter. The l-Hph biosynthesis and l-Leu biosynthesis genes from E. coli were also compared. This analysis revealed that HphB has comparatively relaxed substrate specificity and can perform the function of LeuB, but HphA and HphCD show tight substrate specificity and cannot complement the LeuA and LeuC/LeuD functions, and vice versa. Finally, the range of substrate tolerance of the l-Hph-producing strain was examined, which showed that m-fluorophenylalanine, o-fluorophenylalanine, and l-tyrosine were accepted as substrates and that the corresponding homoamino acids were generated.
INTRODUCTION
l-Homophenylalanine (l-Hph) is a nonproteinogenic amino acid and contains an integrated one-carbon-extended side chain compared to that of l-Phe. l-Hph is a useful chiral building block for the synthesis of several pharmaceutical drugs, such as angiotensin-converting enzyme (ACE) inhibitors and the novel proteasome inhibitor carfilzomib, which is used for treating multiple myeloma (1, 2). The majority of ACE inhibitors, including enalapril, lisinopril, and benazepril, contain a common l-Hph core in their structures. Furthermore, carfilzomib, which was recently approved by the U.S. Food and Drug Administration, is a synthetic tetrapeptide epoxyketone that contains an l-Hph residue. Because of the importance of l-Hph as a pharmaceutical ingredient, several processes for industrial-scale production of l-Hph have been developed (3). They include transamination from 2-oxo-phenylbutanoate using transaminase in a stereoselective manner and optical resolution of the hydantoin derivative by hydantoinase, followed by hydrolysis of the carbamoyl moiety, catalyzed by carbamoylase (4, 5, 6). Although chemoenzymatic methods seem to be quite efficient processes to supply l-Hph for pharmaceutical drug synthesis, the production cost depends on material derived from fossil fuel. To explore more efficient and sustainable production of l-Hph, we investigated microbial production of l-Hph from l-Phe and simple carbon and nitrogen sources, such as glucose and ammonia.
In nature, some secondary metabolites contain aromatic homoamino acids, for example, benzyl glucosinolate from mustard oil (7) and nonribosomally synthesized cyclic peptides from cyanobacteria (8). Biosynthetic studies on benzyl glucosinolate using radiolabeled precursors indicated that Hph exists as an intermediate during benzyl glucosinolate biosynthesis, although the genes responsible for glucosinolate synthesis have not been identified (9). Recently, the gene cluster for a protease inhibitor depsipeptide containing l-Hph residues, anabaenopeptin, produced by the cyanobacterium Anabaena sp. strain 90, was discovered (10). However, previous studies have focused on these pathways in respect to nonribosomal-peptide synthetases (11) and do not mention the homoamino acid biosynthetic genes.
In this study, we focused on the analogy of the biosynthetic pathway between l-Hph and l-Leu in regard to the side chain carbon elongation mechanism. Based on this speculation, we searched the homologous genes of leucine biosynthesis in the anabaenopeptin gene cluster of the cyanobacterium Nostoc punctiforme PCC73102 and found three putative genes responsible for l-Hph biosynthesis. We constructed Escherichia coli strains expressing these genes and successfully demonstrated the microbial production of l-Hph. Next, we performed complementary comparisons between l-Hph biosynthesis genes and the l-Leu biosynthetic genes, LeuA, LeuB, and LeuC/D. Additionally, we determined substrate specificity, which reveals that the l-Hph-producing strain can also produce fluorinated Hph analogs and homotyrosine (Hty). Finally, we suggest that microbial production of l-Hph using genetically engineered E. coli is a viable alternative process to existing chemoenzymatic syntheses.
MATERIALS AND METHODS
Chemicals.
l-Homophenylalanine was purchased from Tokyo Chemical Industry (Tokyo, Japan). Phenylalanine, tyrosine, and other amino acid derivatives were obtained from Wako Pure Chemical Industries (Osaka, Japan).
Cloning of genes and construction of l-Hph-producing strains.
The l-Hph biosynthesis genes hphA (Npun_F2464), hphB (Npun_F2457), and hphCD (Npun_F2458) were amplified from the genome of N. punctiforme PCC73102 using PrimeStar Max polymerase (TaKaRa Bio, Otsu, Japan) according to the manufacturer's instructions, with the oligonucleotide primer pairs listed in Table 1. The amplified hphCD fragment was digested with EcoRI and KpnI and cloned into pTrc99a (Invitrogen, Carlsbad, CA), which had been digested with the corresponding restriction enzymes, and subsequently dephosphorylated with shrimp alkaline phosphatase (TaKaRa Bio) to obtain pTrc99a-Ptrc-hphCD (named pHPH01). The PCR fragment containing hphA was digested with KpnI and BamHI and introduced into pHPH01 digested with KpnI and BamHI to construct pTrc99a-Ptrc-hphCD-hphA (pHPH02). The PCR fragment containing hphB was digested with BamHI and PstI and cloned into pSTV29 (TaKaRa Bio) digested with BamHI and PstI to produce pSTV29-Plac-hphB (pHPH03). E. coli strain W3110 (12) was transformed with pHPH02 and pHPH03 to generate the l-Hph-producing strain system 1. To improve Hph productivity, other production systems were also constructed using the oligonucleotide primers listed in Table 1, according to a procedure similar to that described above (Table 2).
Table 1.
Oligonucleotides used to construct plasmids for expression of hph genes and pheA and aroG genes
| Plasmid | Comment | Primer | Sequencea |
|---|---|---|---|
| pHPH01 (pTrc99a-Ptrc-hphCD) | hphCD_Fw_EcoRI | 5′-ATAGAATTCAGTCACGAAAATAACGTTTTG-3′ | |
| hphCD_Rv_KpnI | 5′-AGAGGTACCTTATTATTAATCAAAGCGATCGCTAATTTTTC-3′ | ||
| pHPH02 (pTrc99a-Ptrc-hphCD-hphA) | Derived from pHPH01 | hphA_Fw_KpnI | 5′-AGAGGTACCAAGAGGAGAAATTAACTATGGAAACCCTTCCTATC-3′ |
| hphA_Rv_BamHI | 5′-AGAGGATCCTGATGAATTATTTAAACACCCCATTTTTC-3′ | ||
| pHPH03 (pSTV29-Plac-hphB) | hphB_Fw_BamHI | 5′-AGAGGATCCAAGAGGAGAAATTAACTATGGAATCTTTAAGTAAGG-3′ | |
| hphB_Rv_PstI | 5′-AGACTGCAGTTATTATTATTTTCGTGACTCATGAAGTAC-3′ | ||
| pHPH04 (pSTV29-Ptrc-hphB) | Ptrc-hphB fragment was amplified from pHPH08 | Ptrc_Fw_SacI | 5′-ACAGAGCTCGCATAATTCGTGTCGCTCAAG-3′ |
| hphB_Rv_PstI | 5′-AGACTGCAGTTATTATTATTTTCGTGACTCATGAAGTAC-3′ | ||
| pHPH05 (pTrc99a-Ptrc-hphA) | hphA_Fw_NcoI | 5′-ACTACACCATGGAAACCCTTCCTATC-3′ | |
| hphA_Rv_BamHI | 5′-AGAGGATCCTGATGAATTATTTAAACACCCCATTTTTC-3′ | ||
| pHPH06 (pTrc99a-Ptrc-hphA-hphB) | Derived from pHPH05 | hphB_Fw_BamHI | 5′-AGAGGATCCAAGAGGAGAAATTAACTATGGAATCTTTAAGTAAGG-3′ |
| hphB_Rv_PstI | 5′-AGACTGCAGTTATTATTATTTTCGTGACTCATGAAGTAC-3′ | ||
| pHPH07 (pSTV29-Ptrc-hphCD) | Ptrc-hphCD fragment was amplified from pHPH01 | Ptrc_Fw_SacI | 5′-ACAGAGCTCGCATAATTCGTGTCGCTCAAG-3′ |
| hphCD_Rv_KpnI | 5′-AGAGGTACCTTATTATTAATCAAAGCGATCGCTAATTTTTC-3′ | ||
| pHPH08 (pTrc99a-Ptrc-hphB) | hphB_Fw_NcoI | 5′-ACTACACCATGGAATCTTTAAGTAAGG-3′ | |
| hphB_Rv_EcoRI | 5′-ATAGAATTCTTATTATTATTTTCGTGACTCATGAAGTAC-3′ | ||
| pHPH09 (pTrc99a-Ptrc-hphB-hphA) | Derived from pHPH08 | hphA_Fw_KpnI | 5′-AGAGGTACCAAGAGGAGAAATTAACTATGGAAACCCTTCCTATC-3′ |
| hphA_Rv_BamHI | 5′-AGAGGATCCTGATGAATTATTTAAACACCCCATTTTTC-3′ | ||
| pHPH10 (pTrc99a-Ptrc-hphA-hphCD) | Derived from pHPH05 | hphCD_Fw_BamHI | 5′-AGAGGATCCAAGAGGAGAAATTAACTATGAGTCACGAAAATAACG-3′ |
| hphCD_Rv_HindIII | 5′-AAAAGCTTTTATTATTAATCAAAGCGATCGCTAATTTTTC-3′ | ||
| pHPH11 (pTrc99a-Ptrc-hphB-hphCD) | Derived from pHPH08 | hphCD_Fw_EcoRI | 5′-ATAGAATTCAGTCACGAAAATAACGTTTTG-3′ |
| hphCD_Rv_KpnI | 5′-AGAGGTACCTTATTATTAATCAAAGCGATCGCTAATTTTTC-3′ | ||
| pHPH12 (pSTV29-Ptrc-hphA) | Ptrc-hphA fragment was amplified from pHPH05 | Ptrc_Fw_SacI | 5′-ACAGAGCTCGCATAATTCGTGTCGCTCAAG-3′ |
| hphA_Rv_BamHI | 5′-AGAGGATCCTGATGAATTATTTAAACACCCCATTTTTC-3′ | ||
| pMWF01 [pMW219-Ptrp-pheAfbr-aroG(P150L)] | pTrp_Fw_EcoRI | 5′-ATCGGAATTCTCATGTTTGACAGCTTATCATCG-3′ | |
| pTrp_Rv_BamHI | 5′-ATCGGGATCCGTGAACTTGCGTACTAGTTAAC-3′ | ||
| pheA_Fw_BamHI | 5′-ATCGGGATCCAAAAAGGCAACACTATGACATCGGA-3′ | ||
| pheA330_Rv_SalI | 5′-ATCGGTCGACTTATGATTCCAGACGGGTCATAATC-3′ | ||
| aroG_Fw_SalI | 5′-ATCGGTCGACAAAGGAACAGACATGAATTATCAGA-3′ | ||
| aroG_Rv_SphI | 5′-ATCGGCATGCATCCGACAATTAAACCTTACCCG-3′ | ||
| aroG_P150L_1 | 5′-CTTTCTCGATATGATCACCCtgCAATATCTC-3′ | ||
| aroG_P150L_2 | 5′-CAGGTCAGCGAGATATTGcaGGGTGATCAT-3′ |
Italics indicate sites digested by restriction enzymes; lowercase indicates the mutational site.
Table 2.
l-Hph-producing systems
| System | Description |
|---|---|
| 1 | E. coli W3110/pHPH02/pHPH03 |
| 2 | E. coli W3110/pHPH02/pHPH04 |
| 3 | E. coli W3110/pHPH06/pHPH07 |
| 4 | E. coli W3110/pHPH09/pHPH07 |
| 5 | E. coli W3110/pHPH10/pHPH04 |
| 6 | E. coli W3110/pHPH11/pHPH12 |
Production of l-Hph.
Single colonies from l-Hph-producing strains were inoculated into 5 ml Luria-Bertani (LB) medium containing 100 mg/liter ampicillin and 25 mg/liter chloramphenicol and incubated at 30°C overnight. An inoculum of the resulting overnight culture (100 μl) was transferred into production medium (5 ml) composed of 20 g/liter glucose, 2 g/liter MgSO4, 16 g/liter KH2PO4, 14 g/liter K2HPO4, 2 g/liter NH4SO4, 1 g/liter citric acid, 5 g/liter Casamino Acids (Difco, Franklin Lakes, NJ), 50 mg/liter FeSO4 · 7H2O, 10 mg/liter thiamine · HCl, and 10 mg/liter MnSO4 · 5H2O (pH 7.2). l-Phe was added to the medium at a final concentration of 1 g/liter (6.1 mM). Following 6 h of cultivation at 30°C, the cultures were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and further cultivated at 30°C for 30 or 48 h. l-Hph was produced from glucose and ammonia by direct fermentation, using an l-Hph-producing strain with an l-Phe production plasmid, Ptrp-pheAfbr-aroG(P150L)-pMW219 (named pMWF01), the genes of which are amplified from the genome of E. coli W3110 using the oligonucleotide primers listed in Table 1. The culture conditions were the same as those described above without the addition of l-Phe (13, 14). For Hph analog production, Phe analogs were added to the culture and cultivated as described above.
Product analysis.
Culture supernatants were diluted with water and analyzed by high-performance liquid chromatography (HPLC) on a LaChrom Elite HPLC system (Hitachi High-Technologies, Tokyo, Japan) equipped with an Inertsil ODS-3 column (GL Sciences; 4.6 by 250 mm; 5 μm) at a flow rate of 1.0 ml/min at 50°C, with spectrophotometric detection of substrates and products at 220 nm. Solvent gradients were as follows: for l-Hph and fluorinated derivatives, 0 to 15 min, 25 to 35% MeOH-0.1% trifluoroacetic acid (TFA); 15 to 16 min, 35 to 90% MeOH-0.1% TFA; 16 to 18 min, 90% MeOH-0.1% TFA; 18 to 19 min, 90% to 25% MeOH-0.1% TFA in H2O-0.1% TFA; for homotyrosine, 0 to 15 min, 5 to 15% MeOH-0.1% TFA; 15 to 16 min, 15 to 90% MeOH-0.1% TFA; 16 to 18 min, 90% MeOH-0.1% TFA; 18 to 19 min, 90% to 5% MeOH-0.1% TFA in H2O-0.1% TFA. Mass analyses were carried out using a 3200 QTrap liquid chromatography-tandem mass spectrometry (LC–MS-MS) system (AB Sciex, MA).
A complementation experiment between hphABCD and leuABCD.
Complementation of leuABCD function by hphABCD was examined by the following procedure. Strains BW25113 ΔleuA, BW25113 ΔleuB, BW25113 ΔleuC, and BW25113 ΔleuD were obtained from the KEIO library (15). pTrc99a plasmids harboring each hph gene were introduced into the corresponding leu gene deletion mutant as ΔleuA hphA, ΔleuB hphB, ΔleuC hphCD, and ΔleuD hphCD (Table 3, experiment A). The resulting strains were cultured in M9 medium (16) at 30°C for 24 h, and growth was checked by measurement of the optical density at 660 nm (OD660) using a UV-1800 spectrophotometer (Shimadzu, Kyoto, Japan). Complementation of hphABCD function by leuABCD from E. coli W3110 was examined as follows. Several plasmid systems that contained each hph gene exchanged with the corresponding leu gene were constructed to examine l-Hph production (Table 3, experiment B). Production and analysis were performed as described above.
Table 3.
Strain descriptions
| Strain | Description |
|---|---|
| Expt Aa | |
| Wild type | E. coli BW25113 |
| ΔleuA | E. coli BW25113 leuA::Kmr |
| ΔleuA hphA | E. coli BW25113 leuA::Kmr/pHPH05 |
| ΔleuB | E. coli BW25113 leuB::Kmr |
| ΔleuB hphB | E. coli BW25113 leuB::Kmr/pHPH08 |
| ΔleuC | E. coli BW25113 leuC::Kmr |
| ΔleuC hphCD | E. coli BW25113 leuC::Kmr/pHPH01 |
| ΔleuD | E. coli BW25113 leuD::Kmr |
| ΔleuD hphCD | E. coli BW25113 leuD::Kmr/pHPH01 |
| Expt Bb | |
| hphA | E. coli W3110/pHPH05 |
| hphD hphB | E. coli W3110/pHPH11 |
| hphA hphCD | E. coli W3110/pHPH10 |
| hphA hphB | E. coli W3110/pHPH06 |
| leuA hphCD hphB | E. coli W3110/pHPH11/pSTV29-Ptrc-leuA |
| hphA hphCD leuB | E. coli W3110/pHPH10/pSTV29-Ptrc-leuB |
| hphA leuC leuD hphB | E. coli W3110/pHPH06/pSTV29-Ptrc-leuC/D |
| hphA hphCD hphB | E. coli W3110/pHPH10/pHPH04 |
Strains constructed to evaluate cell growth by exchanging leu genes with corresponding hph genes.
Strains constructed to examine l-Hph production by removing hph or replacing it with a corresponding leu gene.
RESULTS
Discovery of l-Hph biosynthesis genes in N. punctiforme PCC73102.
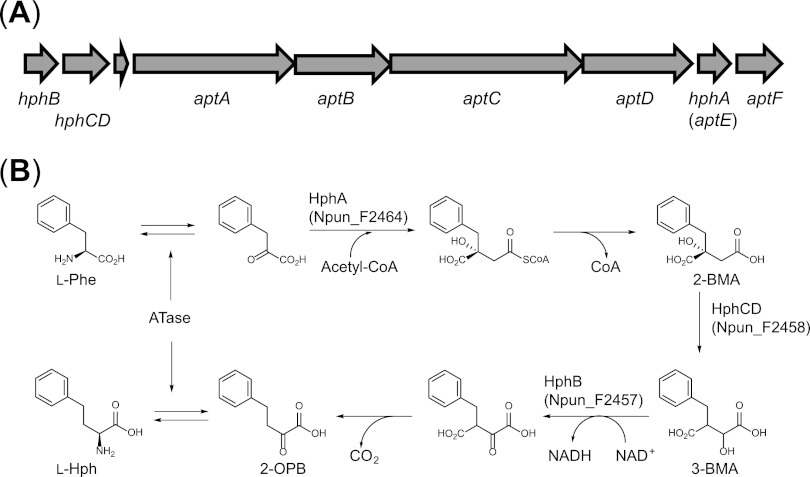
We hypothesized that if aromatic homoamino acids are synthesized via a one-carbon extension mechanism, similar to l-leucine biosynthesis from 3-methyl-2-oxobutanoic acid to 4-methyl-2-oxopentanoic acid (17), the homologous genes encoding isopropylmalate synthase (LeuA), 3-isopropylmalate dehydrogenase (LeuB), and 3-isopropylmalate isomerase (LeuC and LeuD) might exist within or around the anabaenopeptin biosynthetic gene cluster of the cyanobacterium Anabaena sp. strain 90 (10). However, we could not find the expected genes, because the sequence information for the gene cluster was thought not to be complete at this time. Instead, we searched the anabaenopeptin NZ 857 (nostamide) biosynthesis gene cluster from N. punctiforme PCC73102, the genome of which has been completely determined (GenBank accession number AAAY02000000) (18). Subsequently, we found three putative homophenylalanine biosynthetic genes, Npun_F2464 (the gene for benzylmalate synthase), Npun_F2457 (the gene for 3-benzylmalate dehydrogenase), and Npun_F2458 (the gene for 3-benzylmalate isomerase), named hphA, hphB, and hphCD, respectively, located around the anabaenopeptin gene cluster (Fig. 1A). Previously, Npun_F2464 was annotated as aptE, which is located downstream of a gene encoding nonribosomal peptide synthetases. Npun_F2457 and Npun_F2458 are adjacent to the cluster and annotated as encoding 3-isopropylmalate dehydrogenase and 3-isopropylmalate isomerase, respectively. Although hphB and hphCD could not be found in the anabaenopeptin gene cluster from Anabaena sp. strain 90 at this time, we used the BLAST server (19) to confirm the presence of the homologous genes in the anabaenopeptin gene cluster of Nodularia spumigena CCY9414. Also, we found the homologous genes in the genomes of other cyanobacterial species, Synechococcus sp. strain PCC 7335 and Oscillatoria sp. strain PCC 6506, from which secondary-metabolite-containing homoamino acid residues have not been isolated (Table 4). We compared the amino acid sequences of HphA, HphB, and HphCD from N. punctiforme to those of LeuA, LeuB, and LeuC/D from E. coli, respectively, using ClustalW (20). Alignment of HphA and LeuA indicated that HphA does not have a regulatory domain, which is located in the C-terminal region of bacterial LeuA and contains a leucine binding site (21). HphB shows 42% sequence identity with E. coli LeuB. HphCD appears to be a fused structure composed of a large and a small subunit, whereas bacterial isopropylmalate isomerase is a heterodimer composed of a large subunit, LeuC, and a small subunit, LeuD (22). Based on these analyses, we hypothesized the l-Hph biosynthetic pathway from l-Phe to be as shown in Fig. 1B.
Fig 1.
Locations of l-Hph biosynthesis genes in the anabaenopeptin synthetase cluster of N. punctiforme PCC73102 (A) and the proposed l-Hph biosynthetic pathway mediated by HphA, HphCD, HphB, and aromatic aminotransferase (ATase) in a similar route to leucine biosynthesis (B). The proposed Hph biosynthetic pathway is as follows. l-Phe is converted into phenylpyruvic acid via a transamination reaction mediated by an aminotransferase, such as tyrosine aminotransferase (TyrB). Phenylpyruvic acid is condensed with acetyl-coenzyme A (CoA) catalyzed by HphA, and the resulting thioester is spontaneously hydrolyzed, leading to 2-BMA. 2-BMA is converted into 3-BMA (3-benzylmalic acid) via isomerization of a hydroxyl group mediated by HphCD. The hydroxyl moiety of 3-BMA is oxidized by HphB, followed by spontaneous decarboxylation, producing 2-oxo-phenylbutanoic acid (2-OPB). Finally, 2-OPB is converted into l-Hph via a transamination reaction mediated by an aminotransferase, such as TyrB.
Table 4.
HphA, HphB, HphCD, and their homologues
| Protein | Organism | Length (aad) | Homology, similarity (%) | Accession no. |
|---|---|---|---|---|
| HphA and its homologuesa | ||||
| HphA | N. punctiforme PCC73102 | 392 | 100 | YP_001865968 |
| Putative HphA | Anabaena sp. 90 | 392 | 82, 92 | ACZ55947 |
| N. spumigena CCY9414 | 392 | 80, 90 | ZP_01629954 | |
| Oscillatoria sp. PCC 6506 | 394 | 60, 79 | ZP_07113410 | |
| Synechococcus sp. PCC 7335 | 411 | 56, 74 | ZP_05037378 | |
| LeuA | E. coli W3110 | 523 | 26, 44 | YP_488380 |
| HphB and its homologuesb | ||||
| HphB | N. punctiforme PCC73102 | 363 | 100 | YP_001865961 |
| Putative HphB | N. spumigena CCY9414 | 422 | 79, 90 | ZP_01629946 |
| Oscillatoria sp. PCC 6506 | 352 | 70, 88 | ZP_07113411 | |
| Synechococcus sp. PCC 7335 | 349 | 64, 76 | ZP_05038014 | |
| LeuB | E. coli W3110 | 363 | 42, 56 | YP_488379 |
| HphCD and its homologuesc | ||||
| HphCD | N. punctiforme PCC73102 | 569 | 100 | YP_001865962 |
| Putative HphCD | N. spumigena CCY9414 | 574 | 82, 91 | ZP_01629947 |
| Oscillatoria sp. PCC 6506 | 581 | 70, 83 | ZP_07113412 | |
| Synechococcus sp. PCC 7335 | 567 | 69, 83 | ZP_05037298 | |
| LeuC | E. coli W3110 | 466 | 33, 48 | YP_488378 |
| LeuD | E. coli W3110 | 201 | 59, 68 | YP_488377 |
Found in various cyanobacteria and E. coli LeuA.
Found in various cyanobacteria and E. coli LeuB.
Found in various cyanobacteria and E. coli LeuC/D.
aa, amino acids.
To demonstrate that these gene products synthesize l-Hph, we designed a system to produce l-Hph in a genetically engineered E. coli strain by expressing hphA, hphB, and hphCD. First, two plasmids, pHPH02 and pHPH03, were constructed and introduced into E. coli W3110. The resulting strain was cultivated in production medium containing 1 g/liter l-Phe at 30°C. After 48 h of incubation, the culture supernatant was analyzed by HPLC. As shown in Fig. 2, only the culture supernatant obtained from the strain containing all hph genes exhibited a peak that coincides with the peak of the l-Hph standard. Furthermore, LC–MS-MS analysis indicated that the molecular mass of this peak ([M + H]+ = 180.2) is identical to the theoretical mass of l-Hph ([M + H]+ = 180.1). The 14-unit mass increase compared to the molecular mass of l-Phe corresponds to the mass of a methylene carbon. Additionally, fragment ions at 134.2 and 117.2 clearly showed the presence of carboxyl and amino moieties, respectively. Thus, we demonstrated that HphA, HphB, and HphCD are responsible for the biosynthesis of l-Hph in N. punctiforme PCC73102. It is also noteworthy that an unknown peak was detected in the culture broth and also generated in the culture broth of the strain harboring the hphA gene only. LC–MS-MS analysis of the peak clearly showed that this unknown compound is 2-benzylmalic acid (2-BMA) (theoretical mass, [M − H]− = 223.1; observed mass, [M − H]− = 223.1), which is an intermediate produced by HphA (Fig. 1B).
Fig 2.
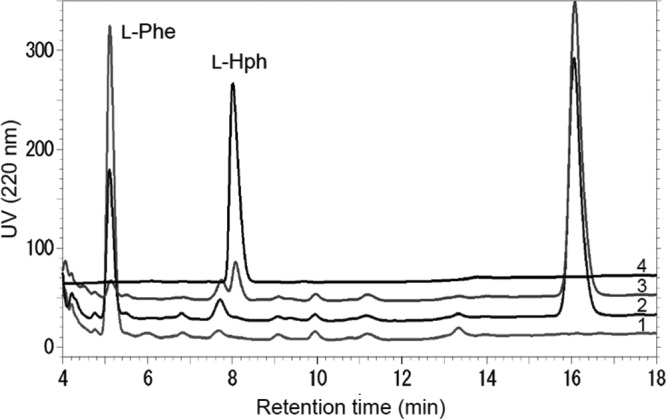
HPLC chromatogram of supernatants obtained from fermentative broths of E. coli W3110 expressing the corresponding Hph proteins. Line 1, HphCD expression only; line 2, HphA and HphCD expression; line 3, HphA, HphCD, and HphB expression; line 4, l-Hph standard (1 g/liter). l-Hph and l-Phe elute at 8.1 and 5.2 min, respectively. The peak at 16.1 min is 2-BMA.
Optimization of gene expression to increase l-Hph productivity.
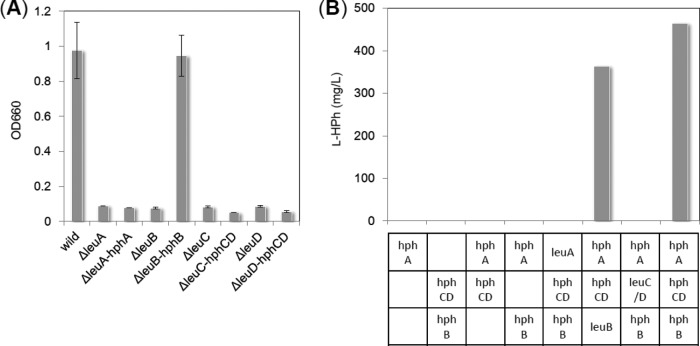
Although we identified the l-Hph biosynthesis genes, the efficiency of l-Hph production was relatively low (∼154 mg/liter at 48 h cultivation) at 1 g/liter l-Phe and 30°C, which we considered insufficient for industrial-scale production of l-Hph. It has been known that the optimal balance of the gene expression in the host cell resulted in an increase of the product yield, and several methods have been developed to fix the metabolic flow (23). Thus, we attempted the simple method to optimize expression of hphA, hphB, and hphCD to increase the yield of l-Hph; five new two-plasmid expression systems, with different arrays of genes, were constructed (Tables 1 and 2), and their l-Hph production was examined. As shown in Fig. 3, optimization dramatically increased l-Hph yields, especially of systems 5 and 6, which had yields of 630 mg/liter and 560 mg/liter, respectively, at 48 h cultivation. We also used these systems to investigate the influence of temperature on l-Hph production. Fermentation was carried out at 25, 30, and 35°C, and HPLC analyses showed yields of 900 mg/liter, 260 mg/liter, and 0 mg/liter, respectively, with 1 g/liter l-Phe. At 35°C, l-Hph production was completely abolished, while 2-BMA, which is a product of the HphA reaction, accumulated in the culture broth. This may indicate that either HphB or HphCD, or both, is thermosensitive.
Fig 3.
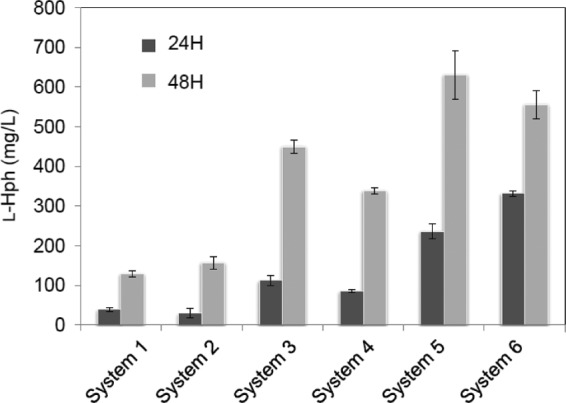
Improvement of l-Hph production by E. coli W3110 strains harboring the various plasmids listed in Table 2. The error bars represent standard deviations (SD) of the mean of three independent experiments.
Next, we examined production of l-Hph from glucose and ammonia, without the addition of l-Phe, by combining the HphABCD-expressing plasmids pHPH11 and pHPH12, with an l-Phe-producing strain as a host. This l-Phe-producing strain was constructed by introducing pMWF01, which contains the genes encoding feedback-resistant mutants of PheA and AroG, into E. coli W3110 (13, 14). The resulting strain produced a level of l-Hph (396 mg/liter at 30 h) from glucose and ammonia similar to that of the l-Phe-feeding culture (395 mg/liter at 30 h).
Complementation experiment between hphABCD and leuABCD.
Based on their respective reaction schemes, the catalytic properties of HphA, HphB, and HphCD are expected to be similar to those of LeuA, LeuB, and LeuC/LeuD, respectively. To evaluate whether these genes can compensate for each other's functions, we generated several strains with the corresponding hph and leu genes exchanged, as listed in Table 3. First, we examined whether leu gene-deficient mutants supplemented with the corresponding hph gene could grow in M9 medium (Fig. 4A). The leuB deletion mutant complemented with hphB grew as efficiently as the wild-type strain, whereas the ΔleuA hphA, ΔleuC hphCD, and ΔleuD hphCD strains did not grow. Second, we determined whether l-Hph-producing cells, in which one of the hph genes was complemented with the corresponding leu gene, could produce l-Hph (Fig. 4B). The strain in which hphB was exchanged with leuB produced amounts of l-Hph almost identical to those produced by the nonexchanged strain. However, the other strains did not produce a detectable amount of l-Hph. These results indicate that HphB has relatively relaxed substrate specificity and can perform the functions of LeuB, whereas HphA and HphCD have tight substrate specificity and cannot complement LeuA and LeuC/LeuD function, and vice versa.
Fig 4.
Complementation experiments with Hph and Leu biosynthesis genes. (A) Growth (OD660) of leu gene-deficient mutants and mutants complemented with the corresponding hph gene, as listed in Table 3, in M9 medium. The error bars represent SD of the mean of three independent experiments. (B) l-Hph production by strains containing hph and leu gene combinations as listed in Table 3. The yields of l-Hph are calculated as the average of two independent experiments, the values of which are within 10% error.
Substrate specificity of the l-Hph-producing system.
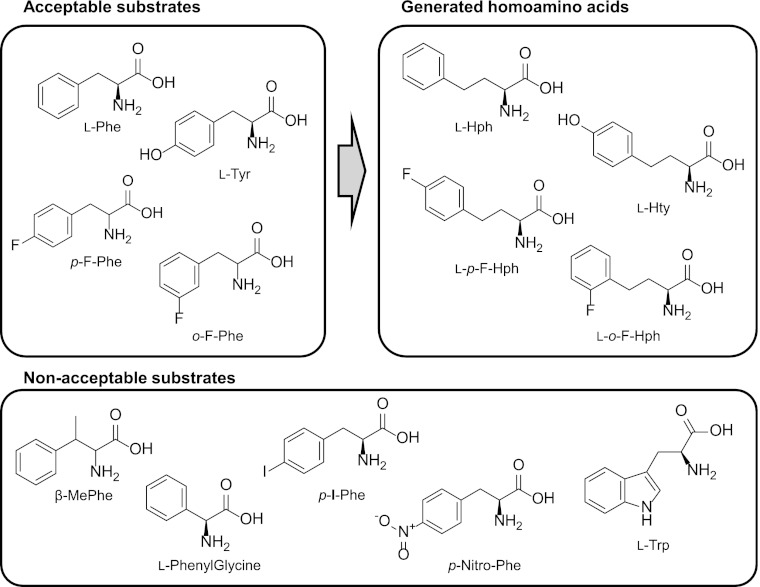
Finally, we explored the range of substrate specificity for homoamino acid synthesis by engineered E. coli with hph genes. l-Phe analogs, including l-Tyr, dl-m-fluoro-Phe, dl-o-fluoro-Phe, β-methyl-Phe, l-phenylglycine, l-p-iodo-Phe, l-p-nitro-Phe, and l-Trp, were added to the culture medium of the l-Hph-producing strain (Fig. 5). Following cultivation for 48 h, the culture supernatants were analyzed by HPLC and LC–MS-MS. dl-m-Fluoro-Phe, dl-o-fluoro-Phe, and l-Tyr were successfully converted into their corresponding homoamino acids, whereas β-methyl-Phe, l-phenylglycine, l-p-iodo-Phe, l-p-nitro-Phe, and l-Trp were not. The identities of the homoamino acids of dl-m-fluoro-Phe (theoretical [M + H]+ = 198.1; experimental [M + H]+ = 198.1), dl-o-fluoro-Phe (theoretical [M + H]+ = 198.1; experimental [M + H]+ = 198.1), and l-Tyr (theoretical [M + H]+ = 196.1, experimental [M + H]+ = 196.1) were verified by MS analysis. The 14-mass-unit increase of the homoamino acids compared to their amino acid substrates clearly indicated extension of the amino acid side chain by a methylene carbon. In the cases of β-methyl-Phe, l-phenylglycine, l-p-iodo-Phe, l-p-nitro-Phe, and l-Trp, the corresponding homoamino acids could not generated, and it is supposed that HphA, which is the first enzyme for Hph biosynthesis, could not accept these substrates. This is supported by the fact that both substrate consumption and HphA product generation could not be observed, although we did not determine whether the host amino acid aminotransferases (24, 25) could convert these amino acids to their corresponding keto acids.
Fig 5.
Classification of substrates used to examine the substrate specificity of the l-Hph-producing system. dl-m-Fluoro-Phe, dl-o-fluoro-Phe, and l-Tyr are converted into the corresponding homoamino acids. β-Methyl-Phe, l-phenylglycine, l-p-iodo-Phe, l-p-nitro-Phe, and l-Trp could not produce the homoamino acids or the biosynthetic intermediates.
DISCUSSION
In this study, we identified l-Hph biosynthesis genes hphA, hphB, and hphCD in N. punctiforme PCC73102 and demonstrated microbial production of l-Hph by genetically engineered E. coli strains employing hphA, hphB, and hphCD expression (Fig. 1 and 2). Genome projects for several cyanobacterial species are ongoing, and among the draft genome sequences, the homologous genes for hphA, hphB, and hphCD can be found in the genomes of N. spumigena CCY9414 (GenBank accession number AAVW00000000), Synechococcus sp. PCC 7335 (GenBank accession number ABRV00000000), and Oscillatoria sp. PCC 6506 (GenBank accession number NZ_CACA00000000) (26) (Table 4). N. spumigena CCY9414 produces the cyclic peptides nodulapeptin B and C, which contain both Hph and Hty residues in their structures (10). While Oscillatoria sp. PCC 6506 and Synechococcus sp. PCC 7335 are not known as cyclic-peptide producers, we propose that these organisms possess a capacity for producing nonribosomally synthesized peptides involving aromatic homoamino acid residues, which must be supplied by hphABCD-like genes. Interestingly, the cyclic peptides nostamide, from N. punctiforme PCC73102, and nodulapeptins and the linear nonribosomal peptide spumidins (27), from N. spumigena CCY9414, contain both Hph and Hty residues despite the fact that these organisms possess only one set of hphABCD genes. Our finding that Hph-producing strains could also generate Hty from Tyr clearly indicates that the cyanobacterial aromatic homoamino acid biosynthesis enzymes have dual substrate specificity, accepting both phenylpyruvic acid and 4-hydroxylphenyl pyruvic acid, and supply l-Hph and l-Hty as building blocks for nonribosomal-peptide synthesis. Apart from homoamino acid biosynthesis in cyanobacteria, there is some evidence that other plant and fungal organisms can also produce aromatic homoamino acids. Previous studies of benzyl glucosinolate biosynthesis using feeding experiments clearly indicated the presence of genes for Hph production in some specific plants, although the genes have not been isolated. Most recently, Cacho and coworkers identified the gene cluster for synthesis of Hty, which is a precursor of antifungal cyclic-peptide echinochandins from the fungus Emericella rugulosa NRRL 11440 (28). The Hty gene cluster is composed of genes for isopropyl malate synthase (htyA), transaminase (htyB), isopropyl malate dehydrogenase (htyC), and aconitase (htyD), along with two oxygenases (htyF and htyE) for further modification of Hty, although the last two are not involved in the heterologous production of Hty. Considering these bioinformatics analyses and the existence of various secondary metabolites containing aromatic homoamino acids, the gene sets responsible for aromatic homoamino acid biosynthesis appear to be distributed across a wide variety of organisms, such as fungi and plants, in addition to cyanobacteria.
Interestingly, cyclic peptides containing dihomotyrosine, named largamides, have been isolated from the cyanobacterium Oscillatoria sp. (29). Although genes involved in the synthesis of dihomotyrosine have not been identified, it is possible that an hph-like gene is involved, which should catalyze two rounds of methylene carbon extension in a manner similar to that for dihomomethionine synthesis catalyzed by leu-like genes from the plant Arabidopsis thaliana (30, 31). Discovery of largamide-producing enzymes and comparison of the crystal structures of enzymes for homoamino acid and dihomoamino acid synthesis will provide a good opportunity to study control of side chain elongation on homoamino acid and allow rational protein engineering to generate novel enzymes that synthesize unnatural homoamino acids, like the method described previously (32).
The remarkable accumulation of 2-BMA during fermentation of the l-Hph-producing strain implied that the HphA-catalyzed reaction is not inhibited by l-Hph, which is consistent with the absence of a C-terminal regulatory domain in HphA. Although it is possible that HphB or HphCD plays a key role in regulating this pathway, the high level of l-Hph production may indicate that the pathway does not contain critical regulatory machinery. The relatively high sequence identity of HphB and E. coli LeuB (42%) is consistent with the finding that HphB can function in place of LeuB, and vice versa (Fig. 4). HphCD is a fused structure composed of a large and a small subunit, as opposed to the bacterial 3-isopropylmalate isomerase, which is a heterodimer of a large subunit, LeuC, and a small subunit, LeuD. We could not determine whether the fusion of the two subunits has any particular role in HphCD. Like LeuC/D, HphCD requires an iron-sulfur cluster in its catalytic center for activity (33). We initially suspected that the existing iron-sulfur machinery of E. coli, such as the Icu or Suf pathway (34), could not deliver the iron-sulfur cluster to the cyanobacterial enzyme because of the structural differences between the HphCD and LeuC/LeuD complexes. However, l-Hph production by the engineered E. coli clearly indicates that the E. coli iron-sulfur machinery is sufficient to provide the iron-sulfur cluster for apo-HphCD, although we were not able to estimate the efficiency.
To improve the low yield of l-Hph produced by the initial strain, we constructed five strains harboring the plasmid set with altered gene arrangements. This successfully elevated the yield from 154 mg/liter to 630 mg/liter (Fig. 3). In a recent report on paclitaxel (originally named taxol) precursor overproduction in E. coli, optimization of the isoprenoid pathway was critical both for efficient production of the terpenoid and for decreasing formation of the by-product, indole (35). Like the optimization of the paclitaxel pathway, the optimization of the Hph pathway might lead to increased l-Hph production. SDS-PAGE analysis of enzyme expression indicated that lower expression of HphCD may improve yields, because higher expression of HphCD reduced growth (data not shown). We also carried out direct fermentative production of l-Hph from glucose and ammonia by introducing the plasmid (pMWF01) harboring feedback-resistant mutants of pheA and aroG, which are key enzymes for l-Phe biosynthesis. The constructed strain provided almost the same amount of l-Hph as fermentative production with feeding of 1 g/liter l-Phe. Thus, this technique may represent an alternative method for direct fermentative production of l-Hph.
Additionally, we investigated the substrate specificity of the system and revealed that fluorinated Phe analogs and l-Tyr were also accepted as substrates, whereas analogs with larger functional groups substituted in the aromatic ring were not (Fig. 5). d-Amino acids also could not be converted into homoamino acids, as half the amount of substrate remained when racemic amino acids were used as a substrate. However, it is possible that the d form could be utilized with the combination of the specific d-amino acid-oxidase or -racemase (36), leading to the efficient generation of various l-homoamino acid analogs from racemic materials.
In conclusion, we identified the l-Hph biosynthesis genes in N. punctiforme PCC73102 and applied them to microbial production of l-Hph in E. coli. To our knowledge, this is the first identification of the genes responsible for homophenylalanine synthesis in any organism. We also demonstrated that several aromatic homoamino acid analogs, including l-homotyrosine, could be produced using this system, even though the range of substrate specificity is moderately limited. We propose that various fine chemical materials, such as keto acids, hydroxy acids, and alcohols, derived from aromatic homoamino acids can be produced by combining the l-Hph-producing strain with various enzymes that have been developed for industrial production of useful chiral building blocks (37). Additionally, detailed structure-based comparison between enzymes for producing l-Hph and other homoamino acids will allow us to generate novel homoamino acid-producing systems through protein engineering in the future.
ACKNOWLEDGMENTS
We thank Satoshi Koizumi for revision of the manuscript and useful advice, and Nobuo Yokota and Kazuki Ishikawa for LC-MS analysis of homoamino acids. We also thank laboratory members for their advice.
Footnotes
Published ahead of print 25 January 2013
REFERENCES
- 1. Brown NJ, Vaughan DE. 1998. Angiotensin-converting enzyme inhibitors. Circulation 97: 1411–1420 [DOI] [PubMed] [Google Scholar]
- 2. Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FWB, Chanan-Khan AA, Orlowski RZ. 2007. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood 110: 3281–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmad AL, Oh PC, Shukor SRA. 2009. Sustainable biocatalytic synthesis of l-homophenylalanine as pharmaceutical drug precursor. Biotechnol. Adv. 27: 286–296 [DOI] [PubMed] [Google Scholar]
- 4. Asano Y, Yamada A, Kato Y, Yamaguchi K, Hibino Y, Hirai K, Kondo K. 1990. Enantioselective synthesis of (S)-amino acids by phenylalanine dehydrogenase from Bacillus sphaericus: use of natural and recombinant enzymes. J. Org. Chem. 55: 5567–5571 [Google Scholar]
- 5. Chen ST, Tseng MJ, Kao T, Sookkheo B, Surat T. November 2000. Facile synthesis of l-homophenylalanine by equilibrium shift enzymatic reaction using engineered tyrosine aminotransferase. US patent 6,146,–859. [Google Scholar]
- 6. Kao CH, Lo HH, Hsu SK, Hsu WH. 2008. A novel hydantoinase process using recombinant Escherichia coli cells with dihydropyrimidinase, and l-N-carbamoylase activities as biocatalyst for the production of l-homophenylalanine. J. Biotechnol. 134: 231–239 [DOI] [PubMed] [Google Scholar]
- 7. Dawson GW, Hick AJ, Bennett RN, Donald A, Pickett JA, Wallsgrove RM. 1993. Synthesis of glucosinolate precursors and investigations into the biosynthesis of phenylalkyl- and methylthioalkylglucosinolate. J. Biol. Chem. 268: 27154–27159 [PubMed] [Google Scholar]
- 8. Tan LT. 2007. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 68: 954–979 [DOI] [PubMed] [Google Scholar]
- 9. Underhill EW. 1967. Biosynthesis of mustard oil glucosides: 3-benzylmalic acid, a precursor of 2-amino-4-phenylbutyric acid and of gluconasturtiin. Can. J. Biochem. 46: 401–406 [DOI] [PubMed] [Google Scholar]
- 10. Rouhiainen L, Jokela J, Fewer DP, Urmann M, Sivonen K. 2010. Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in anabaena (cyanobacteria). Chem. Biol. 17: 265–273 [DOI] [PubMed] [Google Scholar]
- 11. Finking R, Marahiel MA. 2004. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58: 453–488 [DOI] [PubMed] [Google Scholar]
- 12. Bachmann BJ. 1987. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p 1191–1219 In Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC [Google Scholar]
- 13. Nelms J, Gonzalez DH, Yoshida T, Fotheringham I. 1992. Novel mutation in the pheA gene of Escherichia coli K-12 which result in highly feedback inhibition-resistant variants of chorismate mutase/prephenate dehydratase. Appl. Environ. Microbiol. 58: 2592–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kikuchi Y, Tsujimoto K, Kurahashi O. 1997. Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase of Escherichia coli. Appl. Environ. Microbiol. 63: 761–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 2006.0008 doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 17. Felnagle EA, Chaubey A, Noey EL, Houk KN, Liao JC. 2012. Engineering synthetic recursive pathways to generate non-natural small molecules. Nat. Chem. Biol. 8: 518–526 [DOI] [PubMed] [Google Scholar]
- 18. Meeks JC, Elhai J, Thiel T, Potts M, Larimer F, Lamerdin J, Predki Atlas PR. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynth. Res. 70: 85–106 [DOI] [PubMed] [Google Scholar]
- 19. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- 20. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinfomatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- 21. de Carvalho LPS, Frantom PA, Argyrou A, Blanchard JS. 2009. Kinetic evidence for interdomain communication in the allosteric regulation of R-isopropylmalate synthase from Mycobacterium tuberculosis. Biochemistry 48: 1996–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manikandan K, Geerlof A, Zozulya AV, Svergun DI, Weiss MS. 2011. Structural studies on the enzyme complex isopropylmalate isomerase (LeuCD) from Mycobacterium tuberculosis. Proteins 79: 35–49 [DOI] [PubMed] [Google Scholar]
- 23. Keasling JD. 2008. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 3: 64–76 [DOI] [PubMed] [Google Scholar]
- 24. Collier RH, Kohlhaw G. 1972. Nonidentity of the aspartate and the aromatic aminotransferase components of transaminase A in Escherichia coli. J. Bacteriol. 112: 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Powell JT, Morrison JF. 1978. The purification and properties of the aspartate aminotransferase and aromatic-amino-acid aminotransferase from Escherichia coli. Eur. J. Biochem. 87: 391–400 [DOI] [PubMed] [Google Scholar]
- 26. Méjean A, Mazmouz R, Mann S, Calteau A, Médigue C, Ploux O. 2010. The genome sequence of the cyanobacterium Oscillatoria sp. PCC 6506 reveals several gene clusters responsible for the biosynthesis of toxins and secondary metabolites. J. Bacteriol. 192: 5264–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fewer DP, Jokela J, Rouhiainen L, Wahlsten M, Koskenniemi K, Stal LJ, Sivonen K. 2009. The non-ribosomal assembly and frequent occurrence of the protease inhibitors spumigins in the bloom-forming cyanobacterium Nodularia spumigena. Mol. Microbiol. 73: 924–937 [DOI] [PubMed] [Google Scholar]
- 28. Cacho RA, Jiang W, Chooi Y-H, Walsh CT, Tang Y. 2012. Identification and characterization of the echinocandin B biosynthetic gene cluster from Emericella ruglosa NRRL11440. J. Am. Chem. Soc. 134: 16781–16790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Plaza A, Bewley CA. 2006. Largamides A-H, unusual cyclic peptides from the marine cyanobacterium Oscillatoria sp. J. Org. Chem. 71: 6898–6907 [DOI] [PubMed] [Google Scholar]
- 30. Falk FL, Vogel C, Textor S, Bartram S, Hick A, Pickett JA, Gershenzon J. 2004. Glucosinolate biosynthesis: demonstration and characterization of the condensing enzyme of the chain elongation cycle in Eruca sativa. Phytochemistry 65: 1073–1084 [DOI] [PubMed] [Google Scholar]
- 31. Knill T, Reichelt M, Paetz C, Gershenzon J, Binder S. 2009. Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. Plant Mol. Biol. 71: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marcheschi RJ, Li H, Zhang K, Noey EL, Kim S, Chaubey A, Houk KN, Liao JC. 2012. A synthetic recursive “+1” pathway for carbon chain elongation. ACS Chem. Biol. 7: 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Py B, Barras F. 2010. Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 8: 436–446 [DOI] [PubMed] [Google Scholar]
- 34. Fontecave Ollagnier de Choudens MS. 2008. Iron-sulfur cluster biosynthesis in bacteria: mechanisms of cluster assembly and transfer. Arch. Biochem. Biophys. 474: 226–237 [DOI] [PubMed] [Google Scholar]
- 35. Ajikumar PK, Xiao W-H, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. 2010. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330: 70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galkin A, Kulakova L, Yoshimura T, Soda K, Esaki N. 1997. Synthesis of optically active amino acids from α-keto acids with Escherichia coli cells expressing heterologous genes. Appl. Environ. Microbiol. 63: 4651–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koma D, Yamanaka H, Moriyoshi K, Ohmoto T, Sakai K. 2012. Production of aromatic compounds by metabolically engineered Escherichia coli with an expanded shikimate pathway. Appl. Environ. Microbiol. 78: 6203–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]