Abstract
Pseudomonas sp. strain HZN6 utilizes nicotine as its sole source of carbon, nitrogen, and energy. However, its catabolic mechanism has not been elucidated. In this study, self-formed adaptor PCR was performed to amplify the upstream sequence of the pseudooxynicotine amine oxidase gene. A 1,437-bp open reading frame (designated nox) was found to encode a nicotine oxidase (NOX) that shows 30% amino acid sequence identity with 6-hydroxy-l-nicotine oxidase from Arthrobacter nicotinovorans. The nox gene was cloned into a broad-host-range cloning vector and transferred into the non-nicotine-degrading bacteria Escherichia coli DH5α (DH-nox) and Pseudomonas putida KT2440 (KT-nox). The transconjugant KT-nox obtained nicotine degradation ability and yielded an equimolar amount of pseudooxynicotine, while DH-nox did not. Reverse transcription-PCR showed that the nox gene is expressed in both DH5α and KT2440, suggesting that additional factors required for nicotine degradation are present in a Pseudomonas strain(s), but not in E. coli. The mutant of strain HZN6 with nox disrupted lost the ability to degrade nicotine, but not pseudooxynicotine. These results suggested that the nox gene is responsible for the first step of nicotine degradation. The (RS)-nicotine degradation results showed that the two enantiomers were degraded at approximately the same rate, indicating that NOX does not show chiral selectivity. Site-directed mutagenesis revealed that both the conserved flavin adenine dinucleotide (FAD)-binding GXGXXG motif and His456 are essential for nicotine degradation activity.
INTRODUCTION
Nicotine, a major toxic alkaloid found in cigarettes, induces pleasure and reduces anxiety (1). However, a number of diseases are caused directly or indirectly by nicotine, such as cancer and pulmonary disease (2, 3). In addition, waste from the tobacco industry presents a serious threat to both the environment and human health (4–6). Several bacterial strains have the ability to mineralize nicotine by different degradation pathways (7–10). In the Gram-positive strain Arthrobacter nicotinovorans, the degradation pathway has been characterized and the related enzymes have been well elucidated (7, 11–16). In the Gram-negative Pseudomonas strains, there are at least four different degradation pathways (via N-methylmyosmine [NMM], cotinine, nicotyrine, and nornicotine, respectively) (8, 9, 17). In Pseudomonas putida S16, the pyrrolidine pathway (via NMM) has been fully elucidated (17). The genes were cloned, and the enzymes were fully characterized, except for the enzyme that catalyzes the conversion of 3-succinoyl-pyridine (SP) to 6-hydroxy-3-succinoyl-pyridine (HSP). The nicotine oxidoreductase (NicA) is responsible for the earlier steps by catalyzing the conversion of nicotine to SP via pseudooxynicotine (PN) (18). HSP can be cleaved to 2,5-dihydroxypyridine (DHP) and succinic acid by two different enzymes, HSP hydroxylase A and HSP hydroxylase B (19, 20). DHP is degraded to fumarate by four enzymes, 2,5-DHP dioxygenase, N-formylmaleamate deformylase, maleamate amidase, and maleate cis-trans isomerase, which is similar to the DHP-degrading mechanism in P. putida KT2440 (21, 22). The other three degradation pathways have been poorly studied (8), and no genes or enzymes have been reported.
Several nicotine-degrading strains have been isolated and characterized by our research group, e.g., Shinella sp. strain HZN1 (23) and Pseudomonas sp. strain HZN6 (9, 10). The strain HZN6 uses the pyrrolidine pathway, consisting of nicotine, NMM, PN, 3-succinoylsemialdehyde-pyridine (SAP), SP, and HSP. Shinella sp. HZN1 uses the other three pathways. Previous studies showed that strain HZN6 carries genetic information that is different from that of any reported nicotine-degrading strains (9). Transposon mutagenesis of Pseudomonas sp. HZN6 identified a sulfurtransferase homologue (SirA2) that is involved in SP hydroxylation. Further studies identified two novel genes (pao and sap) that encode pseudooxynicotine amine oxidase (PNAO) and 3-succinoylsemialdehyde-pyridine dehydrogenase (SAPD), respectively. The former enzyme catalyzes the conversion of PN to SAP, while the latter catalyzes the conversion of SAP to SP. However, the enzyme that converts the nicotine to NMM and PN is unknown.
In this study, we cloned the upstream sequences of the pao gene using the self-formed adaptor PCR (SEFA-PCR) method (24). A novel nicotine oxidase gene (nox) was analyzed and identified (Fig. 1). The nox gene product, NOX, was responsible for the first step of nicotine degradation in Pseudomonas sp. strain HZN6, which catalyzed the conversion of nicotine to NMM, which then hydrolyzed spontaneously into PN. The enantioselectivity of the nox gene product was also determined.
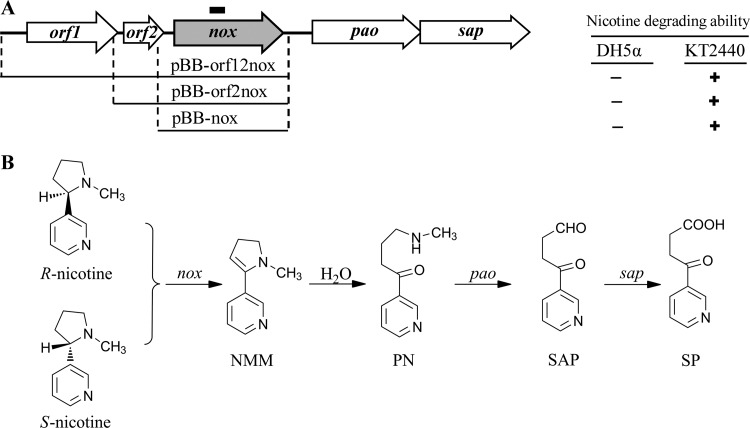
Fig 1.
Schematic representation of the genes responsible for the initial steps of nicotine degradation from Pseudomonas sp. strain HZN6. (A) (Left) Organization of the cloned genes. The short black line above the nox gene represents the region for RT-PCR analysis. (Right) Nicotine-degrading abilities of E. coli DH5α and P. putida KT2440 harboring different plasmids. +, ability; −, no ability. (B) Proposed initial steps of nicotine catabolism.
MATERIALS AND METHODS
Chemicals.
(S)-(−)-Nicotine (>99%) and (RS)-(±)-nicotine (>99%) were purchased from Sigma-Aldrich. PN (98%) was obtained from Toronto Research Chemicals, Inc. (Canada). The enzymes used in the DNA manipulations were obtained from TaKaRa Biotechnology (Dalian, China). All analytical and high-performance liquid chromatography (HPLC) grade reagents were from the Shanghai Chemical Reagent Co. Ltd. (Shanghai, China).
Bacterial strains and culture conditions.
The wild-type (WT) strain Pseudomonas sp. HZN6 can utilize nicotine as its the sole source of carbon, nitrogen, and energy (Table 1) and was deposited in the China Center for Type Culture Collection (CCTCC 2010196) (10). P. putida strain KT2440 is a non-nicotine-degrading bacterium (25). All of the Pseudomonas strains were cultured aerobically at 30°C in LB medium or mineral salt medium (MSM), as described previously (10). Escherichia coli strains were routinely grown in LB medium at 37°C. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 mg/liter; chloramphenicol (Cm), 34 mg/liter; kanamycin (Km), 50 mg/liter; and gentamicin (Gm), 50 mg/liter.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference(s) |
|---|---|---|
| Strains | ||
| Pseudomonas | ||
| Pseudomonas sp. HZN6 | Apr; Wild type, nicotine degrader; G− | 9, 10 |
| P. putida KT2440 | Apr; non-nicotine-degrading strain | 25 |
| N6Δnox | Apr Kmr; nox::Kmr mutant of HZN6 | This study |
| N6ΔnoxC | Apr Kmr Gmr; N6Δnox containing pBB-nox | This study |
| KT-nox | Apr Gmr; KT2440 containing pBB-nox | This study |
| E. coli | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | TaKaRa |
| HB101(pRK2013) | Conjugation helper strain | Laboratory stock |
| SM10λpir | thi thr leu tonA lacY supE recA::RP-4-Tc::Mu (λ pir) | Laboratory stock |
| DH-nox | Gmr; DH5α containing pBB-nox | This study |
| Plasmids | ||
| pMD18-T | Apr; T-A clone vector | TaKaRa |
| pJQ200SK | Gmr mob+ orip15A lacZα+ sacB; suicide vector | 29 |
| pBBR1-MCS5 | Gmr; broad-host-range cloning vector | 27 |
| pJQΔnox | Gmr; ApaI-SacI fragment containing nox inserted into pJQ200SK where nox was disrupted by kanamycin resistance gene | This study |
| pBB-orf12nox | Gmr; KpnI-SacI fragment containing orf1, orf2, and nox inserted into pBBR1-MCS5 | This study |
| pBB-orf2nox | Gmr; KpnI-SacI fragment containing orf2 and nox inserted into pBBR1-MCS5 | This study |
| pBB-nox | Gmr; KpnI-SacI fragment containing nox inserted into pBBR1-MCS5 | This study |
| pBB-orf12noxH456R | Gmr; pBB-orf12nox with a mutation of His456 to Arg | This study |
| pBB-orf2noxH456R, | Gmr; pBB-orf2nox with a mutation of His456 to Arg | This study |
| pBB-noxH456R, | Gmr; pBB-nox with a mutation of His456 to Arg | This study |
| pBB-Δmotif | Gmr; pBB-nox with deletion of FAD-binding motif GxGxxG | This study |
Gene cloning and sequence analysis.
The genomic DNA of the HZN6 strain was extracted by a high-salt-concentration precipitation method (26). Three primers, UPpao1 (5′-CATCAAGATGTTCTACGGTCCAGGTGT-3′), UPpao2 (5′-CCCGACTTACGCAGTGCGGAAGAAA-3′), and UPpao3 (5′-GGTACTTTGCCAACCNNNNNNNNNACCACA-3′), were designed to amplify the upstream sequences of the pao gene by SEFA-PCR. The final amplified PCR products were purified and cloned into the pMD18-T plasmid. Nucleotide sequences were determined by the Invitrogen Technologies Co. (Shanghai, China). Open reading frames (ORFs) were identified using the ORF Finder program, and comparisons of the amino acid (or nucleotide) sequences were performed with the BLAST programs on the National Center for Biotechnology Information (NCBI) website.
Construction of plasmids and transconjugants.
A DNA fragment containing the nox gene and its upstream sequences (250 bp) was amplified from the genomic DNA of HZN6 using the primers noxF (5′-TAAGAGCTCGACTTCAGTAACGTTGTTAG-3′; the SacI site is underlined) and noxR (5′-TAAGGTACCAGGCACAACAACACGGCTTG-3′; the KpnI site is underlined). The PCR product was excised by restriction digestion with both SacI and KpnI and inserted into the same sites in the broad-host-range plasmid pBBR1-MCS5 (27), yielding the plasmid pBB-nox (Fig. 1). Similarly, the DNA fragments carrying two ORFs (orf2 and the nox gene) and three ORFs (orf1, orf2, and the nox gene) were amplified with the primer pairs orf2F (5′-TAAGAGCTCGGATCAGGTCCGTCACCAAG-3′; SacI underlined)/noxR and orf1F (5′-TAAGAGCTCCGTTCCCATCTAATCTCATC-3′; SacI underlined)/noxR, respectively. The fragments were inserted into the pBBR1-MCS5 plasmid, yielding pBB-orf2nox and pBB-orf12nox, respectively (Fig. 1). These three plasmids were transferred into E. coli DH5α cells. The plasmids were then introduced from the DH5α strains into P. putida KT2440 with the help of the pRK2013 plasmid from E. coli HB101. The recombinant DH5α and KT2440 strains containing the pBB-nox plasmid were designated DH-nox and KT-nox, respectively.
Construction of the nox deletion mutant strain.
Two oligonucleotide primers, noxUF (5′-ATAGAGCTCCGTAATAGCACCAAGTGCAT-3′; the SacI site is underlined) and noxUR (5′-AGCAGACAGTTTTATTGTGTAGCGTTGCATCTCAGAC-3′; sequence complementary to KmF is underlined), were designed to amplify the 5′-terminal region of the nox gene from the genomic DNA of the HZN6 strain using PCR. Two other oligonucleotide primers, noxDF (5′-ACGCTGACTTGACGGGACTGGGTCCAGACTCACGACTA-3′; sequence complementary to KmR is underlined) and noxDR (5′-TATGGGCCCTACCATTATCTGGATTGCAA-3′; the ApaI site is underlined), were designed to amplify the 3′-terminal region of the nox gene. The kanamycin resistance gene was amplified from the pSC123 plasmid using the primers KmF and KmR, as previously described (10). Three DNA fragments were fused using the overlap extension PCR method (28). This fusion DNA fragment was digested with both ApaI and SacI and inserted into the pJQ200SK vector (29). The resulting plasmid, pJQ-Δnox, contained a deleted nox gene with a 720-bp fragment instead of the 918-bp kanamycin resistance gene as a selectable marker.
A two-step recombination was performed to delete the nox gene from the chromosome of Pseudomonas sp. HZN6, as described previously (10). The pJQ-Δnox plasmid was introduced into HZN6 cells via SM10λpir. The single-crossover mutants were screened on an LB plate containing Ap, Km, and Gm. The gentamicin-resistant strains were then subjected to repeated cultivation in LB medium containing 10% sucrose but no gentamicin. The double-crossover mutants, which lost the vector backbone and were sensitive to gentamicin, were selected on LB plates with Ap and Km. The deletion of the nox gene was confirmed by PCR using the noxUF and noxDR primers. This procedure resulted in the deletion of the mutant N6Δnox. The pBB-nox plasmid was transferred into the N6Δnox strain to generate the complementation strain N6ΔnoxC.
Degradation assays and analysis methods.
Pseudomonas sp. strain HZN6, P. putida strain KT2440, E. coli DH5α, and their derivative strains were cultured in LB medium for 12 h, harvested by centrifugation (6,000 × g for 5 min), washed twice with MSM, and resuspended in MSM (the cell density was adjusted to an approximate optical density of 1.5 at 600 nm [OD600]). An aliquot of the cells (30% [vol/vol] for degradation; 5% [vol/vol] for cell growth) was inoculated in 100 ml of MSM supplemented with 1 mM (S)-nicotine in a 250-ml Erlenmeyer flask. The cultures were incubated at 30°C and shaken at 180 rpm on a rotary shaker. Samples were taken at regular intervals. Each treatment was performed in three replicates. Concentrations of nicotine and PN were determined by HPLC analysis, as described previously (10).
Isolation of total RNA and RT-PCR.
During the nicotine degradation process, samples were taken at 2 h (for HZN6 and DH-nox) and 12 h (for KT-nox) after incubation in MSM containing 1 mM nicotine. Total RNA was extracted from the cell pellets using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Contaminating DNA was removed by a 30-min digestion at 37°C with 20 units of RNase-free DNase I (MBI Fermentas). Synthesis of total cDNA was carried out with 20 μl of the reverse transcription (RT) reaction mixtures containing 1 μg of RNA, 2.5 mM each dNTP, 200 U of SuperScript II reverse transcriptase (Invitrogen), and 2.5 mM random primers in the buffer recommended by the manufacturer. Samples were initially heated at 42°C for 45 min and then incubated at 70°C for 15 min. The cDNA obtained was stored at −20°C. The cDNA products were then amplified in 25-μl PCR mixtures with 2.5 μl of the RT reaction mixture as the template. The thermocycler program was as follows: 94°C for 2 min; 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s; and then 72°C for 2 min. A 295-bp fragment of the nox gene was amplified with the primers RTnoxF (5′-AGACAAAATGTCCGTCTT-3′) and RTnoxR (5′-GCCATTTAGCTGTTCGAC-3′). The primer pairs used in the RT-PCR were also used in standard PCRs with genomic DNA (positive control), as well as with RNA isolated from strains (HZN6, DH-nox, or KT-nox) grown in the presence of nicotine and treated with DNase (negative control).
Isomer selectivity in nicotine degradation.
(RS)-Nicotine was used to determine the isomer selectivity of the HZN6 strain and recombinants containing the nox gene. Samples (1 ml) were removed periodically and extracted three times with 1 ml dichloromethane. The organic phase was dehydrated by passing it through a filter paper filled with anhydrous sodium sulfate, evaporated to dryness under nitrogen flow, and redissolved in 1 ml n-hexane for HPLC analyses. Similar to the method reported by Tang et al. (30), the nicotine enantiomers were separated and quantitatively characterized with the JASCO 2000 plus HPLC (Japan) with a UV detector set at 259 nm, which was equipped with a Chiralpak AD-H column (4.6 by 250 mm; 5 μm). The mobile phase was the n-hexane–methanol mixture with trifluoroacetic acid (95:4.98:0.02 [vol/vol]; flow rate, 1.0 ml/min), and the column temperature was 25°C. Enantioselectivity in nicotine biodegradation was determined by evaluating the changes in the enantiomer fraction (EF) as follows: EF = E1/(E1 + E2), where E1 and E2 correspond to the peak areas of (R)- and (S)-nicotine.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out by the overlap extension PCR method (28). His456Arg was made by replacement of the codon CAC with AGA using the primers H456RF (5′-AGTAACGGCTGGAGAGCCAACATCGAT-3′) and H456RR (5′-ATCGATGTTGGCTCTCCAGCCGTTACT-3′) (mutated codons are underlined). The external primers for overlap extensions were noxF and noxR, orf2F and noxR, and orf1F and noxR. PCR products from overlap extensions were digested with SacI and KpnI and inserted into the same sites in plasmid pBBR1-MCS5, yielding pBB-noxH456R, pBB-orf2noxH456R, and pBB-orf12noxH456R, respectively. The constructed mutants were confirmed by sequencing. The conserved GXGXXG motif was deleted using the primers DMF (5′-TACGATGTAATCGTGGTTGCGACTGCCGCGCGTGAA-3′) and DMR (5′-TTCACGCGCGGCAGTCGCAACCACGATTACATCGTA-3′). The two external primers for overlap extensions were noxF and noxR. Similarly, overlap PCR products were cloned into the plasmid pBBR1-MCS5, yielding pBB-Δmotif. All of the constructed plasmids were introduced into P. putida KT2440 to detect the nicotine-degrading activity.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in GenBank under accession number JN391188.
RESULTS
Cloning the upstream DNA fragments of the pao gene and sequence analysis.
Our previous study revealed that two consecutive genes, pao and sap, are responsible for the second and third enzymatic steps of nicotine degradation, respectively (9). Because degrading genes are frequently located in a cluster (21, 31), we hypothesized that the gene responsible for the first step is located adjacent to the pao gene. Then, SEFA-PCR was performed to amplify the upstream sequences of the pao gene. A 3,640-bp DNA fragment was amplified and cloned into the pMD18-T plasmid. The nucleotide sequence of the insert DNA was determined, and three ORFs (>500 bp) consisting of 1,005, 573, and 1,437 bp were found and designated orf1, orf2, and orf3, respectively (Fig. 1A). Sequence comparisons revealed that the 3 ORFs did not show any similarity to any gene with a definite function at the nucleotide level.
A homology search of the NCBI database revealed that the deduced amino acid sequence of ORF1 shared 33% and 28% identity with the amino acid sequences of the AraC family transcriptional regulators from Pseudomonas entomophila L48 (YP_608148.1) and P. putida KT2440 (NP_746618.1), respectively. ORF1 encodes a 334-residue protein with a predicted molecular mass of 38.01 kDa. The G+C content is 50.35%. The deduced amino acid sequence of ORF2 is similar to the amino acid sequence of the YjgF family proteins, which do not have definite functions.
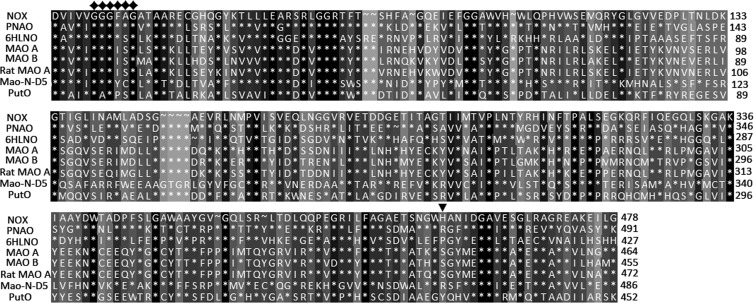
The orf3 gene, with a length of 1,437 bp, encodes a 478-amino acid protein with a calculated molecular mass of 52.225 kDa. The G+C content is 52.89%. The deduced amino acid sequence was compared with the known enzymes available in the GenBank database. The results indicated that the enzyme shares 26 to 40% identity with several amine oxidases, including PNAO (40% identity) (9), 6-hydroxy-l-nicotine oxidase (6HLNO) (Protein Data Bank [PDB] 3K7M_X) from A. nicotinovorans (30% identity) (16), monoamine oxidase (Mao-N-D5) (PDB 2VVM_A) from Aspergillus niger (30% identity) (32), human monoamine oxidase A (MAO A) (PDB 2BXR_A) (26% identity), human MAO B (PDB 1OJ9_A) (25% identity) (33), rat MAO A (PDB 1O5W_A) (26% identity) (34), l-amino acid oxidase (PDB 1F8R_A) from Calloselasma rhodostoma (24% identity) (35), and putrescine oxidase (PutO) (PDB 2YG5_A) from Rhodococcus erythropolis (25% identity) (36). The alignment of protein sequences from these amine oxidases is shown in Fig. 2. Three conserved partial sequences are proposed to encode flavin adenine dinucleotide (FAD)-binding domain (pfam01593). The canonical GXGXXG (amino acids 56 to 61) motif was found in the N-terminal sequence. Based on both sequence similarity and the nicotine degradation pathway of the HZN6 strain (9), the orf3 gene was considered to be a probable candidate for the transformation of nicotine to pseudooxynicotine and was designated nox (nicotine oxidase) (Fig. 1A).
Fig 2.
Alignment of amino acid sequences of NOX and related amine oxidases. 6HLNO, 3K7M_X; Mao-N-D5, 2VVM_A; MAO A, 2BXR_A; MAO B, 1OJ9_A; Rat MAO A, 1O5W_A; PutO, 2YG5_A. ⧫, the conserved FAD-binding GXGXXG motif; ▼, His456 of NOX; *, amino acids are the same as the first line; ∼, gaps. Conserved residues are shaded, with the highest identity level ranging from black (100% identity) to light gray (0% identity).
Identification of a nicotine oxidase gene.
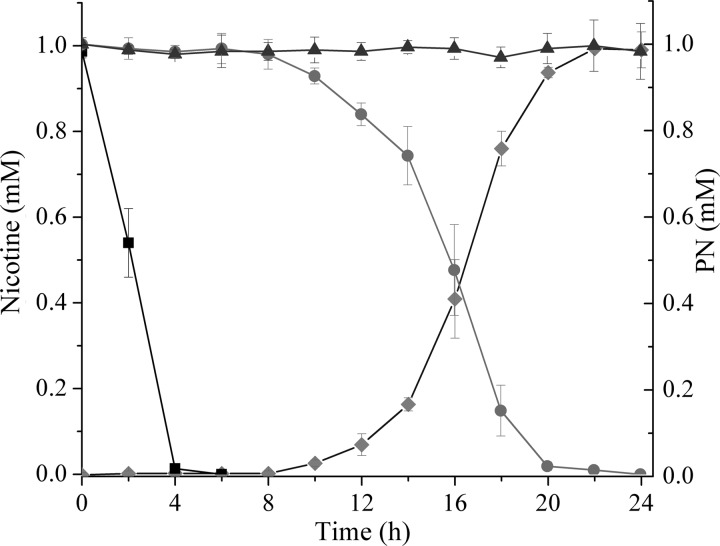
To confirm the assumption that the nox gene is responsible for the first step of nicotine degradation, several plasmids and transconjugants were constructed to detect its ability to degrade nicotine. First, three plasmids, pBB-orf12nox, pBB-orf2nox, and pBB-nox, were constructed and transformed into E. coli DH5α cells. The results indicated that none of these recombinant DH5α strains possessed any nicotine degradation activity (Fig. 1A). However, when the three plasmids were transferred into the non-nicotine-degrading bacterium P. putida strain KT2440, all of the recombinant strains acquired the ability to convert nicotine (1 mM) into a stoichiometric amount of PN (0.98 ± 0.03 mM) (Fig. 1A and 3). The KT2440 strain that contains the plasmid pBB-nox (KT-nox) was used for further study. The HPLC results showed that KT-nox needed 24 h to degrade the nicotine completely, while the WT strain HZN6 only needed 4 h (Fig. 3). Therefore, we conclude that this ORF is the target gene responsible for the dehydrogenation of the pyrrolidine moiety of nicotine (Fig. 1B).
Fig 3.
Degradation dynamics of nicotine by strains HZN6 and KT-nox in MSM. Left axis, nicotine concentrations in strain HZN6 (■), strain KT-nox (●), and the uninoculated control (▲). Right axis, PN concentration in strain KT-nox (⧫). The error bars represent the standard deviations of three replicates.
Transcription of the nox gene.
To confirm whether expression of the nox gene can be induced by nicotine, specific primers were designed for RT-PCR analyses using the RNA of Pseudomonas sp. HZN6 as a template. Total RNA was isolated from cultures grown either on MSM containing nicotine as the sole carbon source or on MSM containing glucose. RT-PCR of the nox gene resulted in amplified fragments of the expected size (295 bp) when nicotine was added to the MSM (Fig. 4). In contrast, PCR products were not detected with the MSM containing glucose, suggesting that expression of the nox gene is induced in the presence of nicotine.
Fig 4.
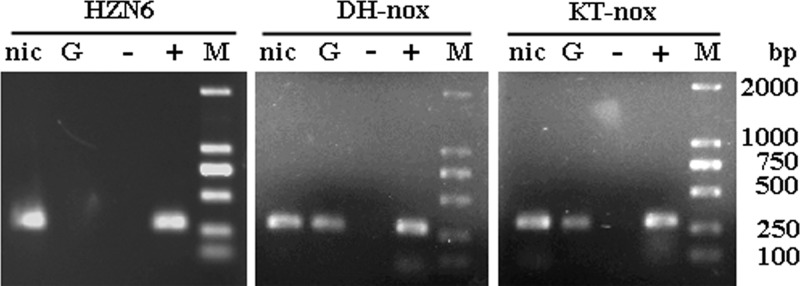
Agarose gel electrophoresis of RT-PCR products. The analysis was performed using total RNA isolated from strain HZN6, DH-nox, or KT-nox grown in MSM with nicotine (lane nic) or glucose (lane G). Control reactions: lane −, negative controls, DNase-treated RNAs from strain HZN6, DH-nox, or KT-nox grown in the presence of nicotine and used as templates; lane +, positive controls, genomic DNAs from strain HZN6, DH-nox, or KT-nox used as templates. The sizes of the amplified fragments were confirmed by comparison with the DL2000 marker (lane M).
RT-PCR analysis was also performed on RNA samples isolated from the cultures of DH-nox and KT-nox grown in MSM with 1 mM nicotine or 1 mM glucose. As shown in Fig. 4, the 295-bp PCR products that indicated the transcription of the nox gene were detected with both DH-nox and KT-nox cultured in MSM with either nicotine or glucose.
Nonenantioselective degradation of nicotine by the nox gene product.
One millimolar (RS)-nicotine was used as a substrate to determine the chiral selectivity of nicotine by the WT strain HZN6. Separation of the two isomers of nicotine in the culture was achieved on a Chiralpak AD-H column. Racemic standard EFs of (RS)-nicotine were 0.498 ± 0.010 (n = 10). With these criteria, an EF of >0.508 or <0.488 of (RS)-nicotine was considered to be significantly nonracemic. The results showed that the EFs had no significant deviation during biodegradation (see Table S1 in the supplemental material), indicating that both (R)- and (S)-nicotine could be degraded by strain HZN6 and that there were no significant differences in the degradation rates. Furthermore, the chiral selectivity of nicotine by the transconjugant KT-nox was determined. As with the WT strain, the EFs of (RS)-nicotine were constant during the degradation, indicating that two nicotine enantiomers can be catalyzed by one gene product, i.e., the nox gene, with the same degradation rates (see Table S1 in the supplemental material). Thus, based on the above data, we concluded that nicotine degradation by the nox gene product showed an absence of chiral selectivity (Fig. 1B).
Phenotypic characterization of the mutant with nox disrupted.
To examine the actual role of the nox gene in nicotine degradation in vivo, the strain HZN6 was inactivated by inserting the kanamycin resistance gene using a gene replacement technique based on homologous recombination. The double-recombination event was confirmed by PCR and sequencing analysis (data not shown). The degrading activities of the WT strain and mutant N6Δnox were investigated. The resulting nox mutant, N6Δnox, was unable to grow on nicotine plates, but it was able to grow on PN as the sole carbon source. Both in liquid form and on plates, N6Δnox could not degrade nicotine but could use PN for cell growth (Fig. 5). As shown in Fig. 5A, both the WT and N6ΔnoxC completely degraded 1 mM nicotine within 12 h of incubation, and cell growth displayed no significant differences (approximate growth rate, 0.012 ± 0.002 h−1). Figure 5B shows that 1 mM PN was completely degraded within 10 h by both N6Δnox and N6ΔnoxC without significant differences. The growth rates of N6Δnox and N6ΔnoxC were both approximately 0.011 ± 0.002 h−1, similar to that of the WT strain (9).
Fig 5.
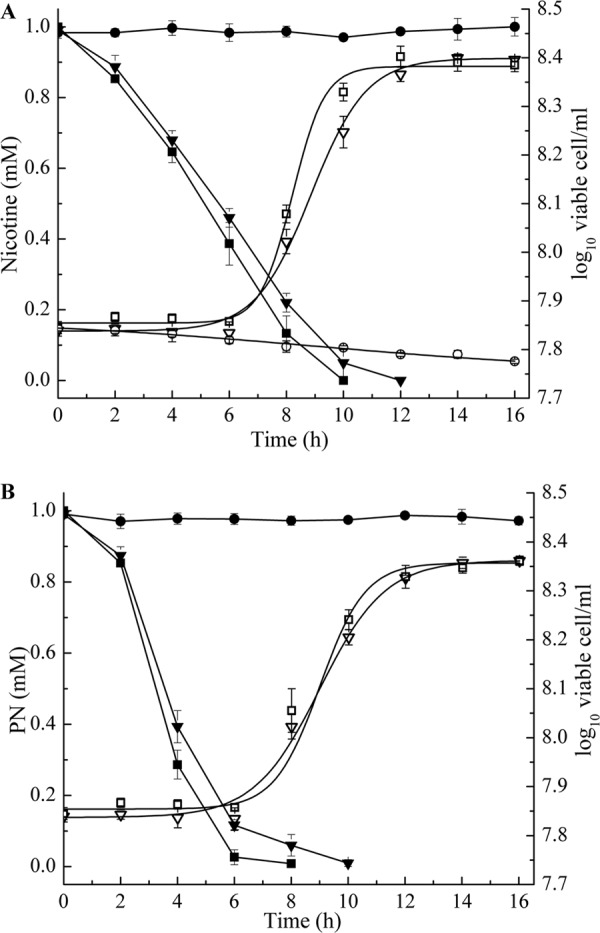
Time course of nicotine and PN degradation and cell growth in cultures of strain HZN6 and its derivatives. (A) Left axis, nicotine concentrations in strains HZN6 (■), N6Δnox (●), and N6ΔnoxC (▼). Right axis, cell growth of strains HZN6 (□), N6Δnox (○), and N6ΔnoxC (▽). (B) Left axis, PN concentrations in strains N6Δnox (■), N6ΔnoxC (▼), and an uninoculated control (●). Right axis, cell growth of strains N6Δnox (□) and N6ΔnoxC (▽). The error bars represent the standard deviations of three replicates.
Identification of the essential residues for NOX catalysis.
The nox gene, which encodes the amino oxidase NOX, shows some homology to known amino oxidase genes, such as the 6-hydroxy-l-nicotine oxidase gene. Based on known proteins, the tertiary structure of NOX was predicted (see the supplemental material). The conserved FAD-binding motif (GXGXXG) is proposed to interact with FAD, and His456 is proposed to bind nicotine. To confirm the importance of the GXGXXG motif and His456, P. putida KT2440 containing the mutant plasmids pBB-noxH456R, pBB-orf2noxH456R, pBB-orf12noxH456R, and pBB-Δmotif was used for the degradation assay of nicotine. After 7 days of incubation, all concentrations of nicotine in MSM were constant, and no product was produced. These results showed that strain KT2440 containing the mutant plasmids had no nicotine-degrading activities, indicating that both the GXGXXG motif and His456 are essential in the nicotine-degrading process.
DISCUSSION
Pseudomonas sp. strain HZN6 is able to utilize nicotine as its sole source of carbon, nitrogen, and energy (10). Our previous study revealed that the nicotine-degrading mechanism used by the HZN6 strain is different from those described in any other known reports. Two novel genes, pao and sap, were cloned and identified as responsible for the second and third enzymatic steps of nicotine degradation, respectively (9). In this study, a putative amine oxidase gene (nox) was cloned and considered a candidate for the enzyme that catalyzes nicotine to PN. After transformation of the nox gene into the non-nicotine-degrading strain KT2440, a recombinant was obtained with nicotine-degrading activity. The mutant N6Δnox with nox disrupted lost the ability to utilize nicotine, but not PN, as a sole carbon source. Based on these results, we concluded that the nox gene is responsible for the transformation of nicotine into PN in Pseudomonas sp. strain HZN6 (Fig. 1).
Many gene products have activities in E. coli DH5α, such as two genes cloned by the shotgun method, the nicA gene for nicotine oxidoreduction from P. putida S16 (18) and the pytH gene for the hydrolyzation of pyrethroid from Sphingobium sp. strain JZ-1 (37). Whole DH5α cells containing the exogenous gene can degrade the corresponding substrates. However, in our study, the transconjugant of DH5α that contained the nox gene did not show nicotine-degrading activity. RT-PCR results showed that mRNAs from the nox gene were produced in both DH5α and KT2440. These results indicated that additional factors that are present in the Pseudomonas strains but not in E. coli are involved in the nicotine degradation process. In addition, nicotine-degrading assays showed that KT-nox exhibited a long lag phase (about 8 h), which was much longer than that of the WT strain HZN6 (Fig. 3). These results might suggest that the additional factors in P. putida KT2440 show low activities and require a long time to generate active NOX. Further studies are required before we can identify the additional factors and demonstrate the mechanism of action between these factors and the nox gene.
Chiral selectivity is a common phenomenon in the biodegradation of both natural and man-made compounds by organisms (38, 39). Nicotine is a naturally chiral compound, and the enantiomeric (S-isomer/R-isomer) ratio is approximately 200:1 (40). The biodegradation of (RS)-nicotine in the strain A. nicotinovorans showed chiral selectivity that has been well elucidated (16, 40, 41). However, selective degradation of (RS)-nicotine in Pseudomonas strains has been poorly studied. In the strain P. putida S16, NicA converts nicotine to SP via PN, but the stereospecificity of the enzyme is unknown (18). Nicotine is first hydroxylated at position 6 in A. nicotinovorans. This reaction is catalyzed by nicotine dehydrogenase, which accepts both enantiomers and retains their configurations. The chiral carbons (C-2′ of the pyrrolidine ring) of the two 6-hydroxy-nicotine enantiomers are both oxidized and dehydrogenated, generating 6-hydroxy-N-methylmyosmine, followed by the formation of 6-hydroxy-pseudooxynicotine. The enantiomers of 6-hydroxy-nicotine are catalyzed by two distinct amine oxidases, 6HLNO and the 6-hydroxy-d-nicotine oxidase (6HDNO), with absolute stereospecificity. By comparing the two different nicotine degradation processes with both the HZN6 and A. nicotinovorans strains, we can conclude that the first nicotine-degrading step by strain HZN6 has the same catalytic process as the step catalyzed by the 6-hydroxy-l/d-nicotine oxidase. Unexpectedly, in this study, we found that the strain HZN6 could degrade both (R)- and (S)-nicotine at nearly the same rate. Furthermore, the KT2440 strain containing the nox gene displayed approximately equal oxidative activities toward the two enantiomers of nicotine, i.e., NOX showed no isomer selectivity. There are other known enzymes that do not show isomer selectivity toward chiral substrates. In Sphingobium sp. strain JZ-1, the degradation of pyrethroids is catalyzed by a purified esterase, PytH, which hydrolyzes the ester bond of pyrethroids (37). In the above-mentioned case, the enzyme showed nonenantioselectivity, possibly due to the enzyme acting on the achiral carbons. In contrast, our results indicated that, although the product of the nox gene acts on the chiral carbon, the degradation does not show chiral selectivity, which is interesting.
The initial steps of nicotine metabolism (nicotine, PN, and SP) by Pseudomonas strains were confirmed 60 years ago (42). The degrading mechanism at the gene and enzyme levels had not been demonstrated until NicA was found in P. putida strain S16 (18). NicA converts nicotine directly to SP through PN. In contrast to strain S16, our findings clearly showed that these initial degradation processes were catalyzed by the products of three consecutive genes, nox, pao, and sap, in Pseudomonas sp. strain HZN6. This study provides us with a better understanding of the genetic and biochemical diversity of nicotine catabolic mechanisms in Pseudomonas spp. In addition, following the third wave of biocatalysis (43), our study provides a new candidate for genetic engineering of nicotine into PN for use in medical research and bioremediation of nicotine-contaminated environments.
Supplementary Material
ACKNOWLEDGMENTS
We thank three anonymous reviewers who helped us make critical improvements to the manuscript. We are also grateful to Eva Top (Department of Biological Sciences, University of Idaho) for her kind provision of plasmids pGemT7cat and pMaT7cat.
This work was supported by the National Basic Research Program of China (no. 2009CB421603) and the National Natural Science Foundation of China (no. 20837002, 21007058, 21177112, and 21277122).
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03824-12.
REFERENCES
- 1. Benowitz NL. 2010. Nicotine addiction. N. Engl. J. Med. 362: 2295–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hukkanen J, Jacob P, III, Benowitz NL. 2005. Metabolism and disposition kinetics of nicotine. Pharmacol. Rev. 57: 79–115 [DOI] [PubMed] [Google Scholar]
- 3. Hecht SS. 1999. Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst. 91: 1194–1210 [DOI] [PubMed] [Google Scholar]
- 4. Seckar JA, Stavanja MS, Harp PR, Yi Y, Garner CD, Doi J. 2008. Environmental fate and effects of nicotine released during cigarette production. Environ. Toxicol. Chem. 27: 1505–1514 [DOI] [PubMed] [Google Scholar]
- 5. Zarrelli A, DellaGreca M, Parolisi A, Iesce MR, Cermola F, Temussi F, Isidori M, Lavorgna M, Passananti M, Previtera L. 2012. Chemical fate and genotoxic risk associated with hypochlorite treatment of nicotine. Sci. Total Environ. 426: 132–138 [DOI] [PubMed] [Google Scholar]
- 6. Civilini M, Domenis C, Sebastianutto N, de Bertoldi M. 1997. Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Manage. Res. 15: 349–358 [Google Scholar]
- 7. Brandsch R. 2006. Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69: 493–498 [DOI] [PubMed] [Google Scholar]
- 8. Li HJ, Li XM, Duan YQ, Zhang KQ, Yang JK. 2010. Biotransformation of nicotine by microorganism: the case of Pseudomonas spp. Appl. Microbiol. Biotechnol. 86: 11–17 [DOI] [PubMed] [Google Scholar]
- 9. Qiu J, Ma Y, Wen Y, Chen L, Wu L, Liu W. 2012. Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl. Environ. Microbiol. 78: 2154–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qiu J, Ma Y, Chen L, Wu L, Wen Y, Liu W. 2011. A sirA-like gene, sirA2, is essential for 3-succinoyl-pyridine metabolism in the newly isolated nicotine-degrading Pseudomonas sp. HZN6 strain. Appl. Microbiol. Biotechnol. 92: 1023–1032 [DOI] [PubMed] [Google Scholar]
- 11. Baitsch D, Sandu C, Brandsch R, Igloi GL. 2001. Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J. Bacteriol. 183: 5262–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiribau CB, Mihasan M, Ganas P, Igloi GL, Artenie V, Brandsch R. 2006. Final steps in the catabolism of nicotine—deamination versus demethylation of gamma-N-methylaminobutyrate. FEBS J. 273: 1528–1536 [DOI] [PubMed] [Google Scholar]
- 13. Chiribau CB, Sandu C, Fraaije M, Schiltz E, Brandsch R. 2004. A novel gamma-N-methylaminobutyrate demethylating oxidase involved in catabolism of the tobacco alkaloid nicotine by Arthrobacter nicotinovorans pAO1. Eur. J. Biochem. 271: 4677–4684 [DOI] [PubMed] [Google Scholar]
- 14. Chiribau CB, Sandu C, Igloi GL, Brandsch R. 2005. Characterization of PmfR, the transcriptional activator of the pAO1-borne purU-mabO-folD operon of Arthrobacter nicotinovorans. J. Bacteriol. 187: 3062–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Igloi GL, Brandsch R. 2003. Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J. Bacteriol. 185: 1976–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kachalova GS, Decker K, Holt A, Bartunik HD. 2011. Crystallographic snapshots of the complete reaction cycle of nicotine degradation by an amine oxidase of the monoamine oxidase (MAO) family. Proc. Natl. Acad. Sci. U. S. A. 108: 4800–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang SN, Liu Z, Tang HZ, Meng J, Xu P. 2007. Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology 153: 1556–1565 [DOI] [PubMed] [Google Scholar]
- 18. Tang HZ, Wang LJ, Meng XZ, Ma LY, Wang SN, He XF, Wu G, Xu P. 2009. Novel nicotine oxidoreductase-encoding gene involved in nicotine degradation by Pseudomonas putida strain S16. Appl. Environ. Microbiol. 75: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang H, Yao Y, Zhang D, Meng X, Wang L, Yu H, Ma L, Xu P. 2011. A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J. Biol. Chem. 286: 39179–39187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang HZ, Wang SN, Ma LY, Meng XZ, Deng ZX, Zhang D, Ma CQ, Xu P. 2008. A novel gene, encoding 6-hydroxy-3-succinoylpyridine hydroxylase, involved in nicotine degradation by Pseudomonas putida strain S16. Appl. Environ. Microbiol. 74: 1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiménez JI, Canales Á Jiménez-Barbero J, Ginalski K, Rychlewski L, García JL, Díaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. U. S. A. 105: 11329–11334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang H, Yao Y, Wang L, Yu H, Ren Y, Wu G, Xu P. 2012. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci. Rep. 2: 377 doi:10.1038/srep00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang JH, Ma Y, Qiu JG, Wu LF, Chen LS. 2011. Biodegradation of nicotine by a novel strain Shinella sp. HZN1 isolated from activated sludge. J. Environ. Sci. Health B 46: 703–708 [DOI] [PubMed] [Google Scholar]
- 24. Wang SM, He J, Cui ZL, Li SP. 2007. Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl. Environ. Microbiol. 73: 5048–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nelson K, Weinel C, Paulsen I, Dodson R, Hilbert H, Martins dos Santos V, Fouts D, Gill S, Pop M, Holmes M. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4: 799–808 [DOI] [PubMed] [Google Scholar]
- 26. Miller S, Dykes D, Polesky H. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176 [DOI] [PubMed] [Google Scholar]
- 28. Horton RM, Cai Z, Ho S, Pease L. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8: 528–535 [PubMed] [Google Scholar]
- 29. Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127: 15–21 [DOI] [PubMed] [Google Scholar]
- 30. Tang Y, Zielinski WL, Bigott HM. 1998. Separation of nicotine and nonicotine enantiomers via normal phase HPLC on derivatized cellulose chiral stationary phases. Chirality 10: 364–369 [Google Scholar]
- 31. Liu H, Wang SJ, Zhang JJ, Dai H, Tang H, Zhou NY. 2011. Patchwork assembly of nag-like nitroarene dioxygenase genes and the 3-chlorocatechol degradation cluster for evolution of the 2-chloronitrobenzene catabolism pathway in Pseudomonas stutzeri ZWLR2-1. Appl. Environ. Microbiol. 77: 4547–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Atkin KE, Reiss R, Koehler V, Bailey KR, Hart S, Turkenburg JP, Turner NJ, Brzozowski AM, Grogan G. 2008. The structure of monoamine oxidase from Aspergillus niger provides a molecular context for improvements in activity obtained by directed evolution. J. Mol. Biol. 384: 1218–1231 [DOI] [PubMed] [Google Scholar]
- 33. De Colibus L, Li M, Binda C, Lustig A, Edmondson DE, Mattevi A. 2005. Three-dimensional structure of human monoamine oxidase A (MAO A): relation to the structures of rat MAO A and human MAO B. Proc. Natl. Acad. Sci. U. S. A. 102: 12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma J, Yoshimura M, Yamashita E, Nakagawa A, Ito A, Tsukihara T. 2004. Structure of rat monoamine oxidase A and its specific recognitions for substrates and inhibitors. J. Mol. Biol. 338: 103–114 [DOI] [PubMed] [Google Scholar]
- 35. Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, Vrielink A. 2000. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 19: 4204–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kopacz MM, Rovida S, van Duijn E, Fraaije MW, Mattevi A. 2011. Structure-based redesign of cofactor binding in putrescine oxidase. Biochemistry 50: 4209–4217 [DOI] [PubMed] [Google Scholar]
- 37. Wang B, Guo P, Hang B, Li L, He J, Li S. 2009. Cloning of a novel pyrethroid-hydrolyzing carboxylesterase gene from Sphingobium sp. strain JZ-1 and characterization of the gene product. Appl. Environ. Microbiol. 75: 5496–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garrison AW, Avants JK, Jones WJ. 2011. Microbial transformation of triadimefon to triadimenol in soils: selective production rates of triadimenol stereoisomers affect exposure and risk. Environ. Sci. Technol. 45: 2186–2193 [DOI] [PubMed] [Google Scholar]
- 39. Liu W, Gan J, Schlenk D, Jury WA. 2005. Enantioselectivity in environmental safety of current chiral insecticides. Proc. Natl. Acad. Sci. U. S. A. 102: 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koetter JWA, Schulz GE. 2005. Crystal structure of 6-hydroxy-d-nicotine oxidase from Arthrobacter nicotinovorans. J. Mol. Biol. 352: 418–428 [DOI] [PubMed] [Google Scholar]
- 41. Kachalova GS, Bourenkov GP, Mengesdorf T, Schenk S, Maun HR, Burghammer M, Riekel C, Decker K, Bartunik HD. 2010. Crystal structure analysis of free and substrate-bound 6-hydroxy-l-nicotine oxidase from Arthrobacter nicotinovorans. J. Mol. Biol. 396: 785–799 [DOI] [PubMed] [Google Scholar]
- 42. Wada E, Yamasaki K. 1953. Mechanism of microbial degradation of nicotine. Science 117: 152–153 [DOI] [PubMed] [Google Scholar]
- 43. Bornscheuer U, Huisman G, Kazlauskas R, Lutz S, Moore J, Robins K. 2012. Engineering the third wave of biocatalysis. Nature 485: 185–194 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.