Abstract
Manganese peroxidases (MnPs) are key players in the ligninolytic system of white rot fungi. In Pleurotus ostreatus (the oyster mushroom) these enzymes are encoded by a gene family comprising nine members, mnp1 to -9 (mnp genes). Mn2+ amendment to P. ostreatus cultures results in enhanced degradation of recalcitrant compounds (such as the azo dye orange II) and lignin. In Mn2+-amended glucose-peptone medium, mnp3, mnp4, and mnp9 were the most highly expressed mnp genes. After 7 days of incubation, the time point at which the greatest capacity for orange II decolorization was observed, mnp3 expression and the presence of MnP3 in the extracellular culture fluids were predominant. To determine the significance of MnP3 for ligninolytic functionality in Mn2+-sufficient cultures, mnp3 was inactivated via the Δku80 strain-based P. ostreatus gene-targeting system. In Mn2+-sufficient medium, inactivation of mnp3 did not significantly affect expression of nontargeted MnPs or their genes, nor did it considerably diminish the fungal Mn2+-mediated orange II decolorization capacity, despite the significant reduction in total MnP activity. Similarly, inactivation of either mnp4 or mnp9 did not affect orange II decolorization ability. These results indicate functional redundancy within the P. ostreatus MnP gene family, enabling compensation upon deficiency of one of its members.
INTRODUCTION
Manganese peroxidases (MnPs) are the most common extracellular ligninolytic peroxidases produced by white rot fungi (WRF); MnPs, along with laccases and lignin peroxidases, are considered to play an active role in the lignin depolymerization process (1–5). This class of peroxidases includes two types of enzymes: Mn2+-dependent peroxidase (MnP) and Mn2+-independent peroxidase (versatile peroxidase [VP]). MnP oxidizes Mn2+ to Mn3+, the former being an obligatory cosubstrate for this enzyme, as it is required to complete the catalytic cycle, whereas VP, in addition to preferentially having MnP activity and being structurally related to MnP at the gene and protein levels, can directly oxidize aromatic substrates (2–4, 6–13). Hence, to date, many VP-encoding genes are classified as part of the MnP gene family (mnp genes) (3, 9–13). Accordingly, we have used this nomenclature in the current report.
Many WRF typically possess a set of multiple manganese peroxidase isoenzymes, encoded by apparently redundant structurally related genes, composing a gene family. Studies on the catalytic properties of the various isoenzymes demonstrated distinct differences in both the culture conditions and substrate specificity associated with their transcription and kinetic constants (1, 3, 4, 9, 14). This diversity, along with the broad range of substrate specificity and their remarkable degradative potential, makes these peroxidases attractive versatile biocatalysts for biotechnological and environmental applications, e.g., in pulping and bleaching, in removal of hazardous wastes, and in synthesis of certain organic compounds. Effective application of these enzymes is dependent on establishing a comprehensive understanding of their properties and mode of action (2, 5, 13, 14).
Pleurotus ostreatus is a commercially important edible WRF known as the oyster mushroom (15–17). Based on the ongoing P. ostreatus genome deciphering project (http://genome.jgi-psf.org/PleosPC9_1/PleosPC9_1.home.html), it was recently found that this fungus expresses nine mnp genes, designated mnp1 to -9, composing its MnP gene family. P. ostreatus MnPs correspond to the subfamily of “short” MnPs, in comparison to the typical “long” MnPs (including an extra C-terminal extension in their amino acid sequences) from the model ligninolytic fungus Phanerochaete chrysosporium. Long MnPs are specific for Mn2+, whereas short MnPs have a wider substrate specificity and are capable of oxidizing phenols, amines, and 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) in the absence of Mn2+ (9). The peroxidases encoded by mnp3, -6, -7, -8, and -9 have been characterized as short MnPs (MnP3, -6, -7, -8, and -9, respectively), while mnp1, -2, -4, and -5 encode VPs (VP1, -2, -4, and -5, respectively) (9, 10). The large number of genes encoding MnP isoenzymes raised the questions of what is the significance of this multiplicity and whether or not potential redundancy may exist among the members of this gene family (10).
Previous studies have shown that Mn2+ amendment to P. ostreatus growth media enables Mn2+-mediated decolorization of the azo dye orange II (OII) and increases fungal lignin degradation capacity (10, 13, 18–21). In parallel, gene expression analysis showed that Mn2+ amendment results in a drastic increase in the transcript levels of the predominantly expressed genes mnp3 and mnp9 (while transcription of mnp4 was reduced), suggesting that the two corresponding short MnPs play significant roles in these processes (10, 12, 13, 18–20).
In order to obtain direct evidence for the significance of mnp3, a highly expressed member of the MnP gene family, for the functionality of the P. ostreatus ligninolytic system, we implemented a reverse genetics strategy based on silencing the mnp3 gene by RNA interference (RNAi). Knockdown of mnp3 resulted in marked inhibition of its expression, which was colinear with a reduction in OII decolorization (10). Nevertheless, silencing mnp3 as a target via this approach also resulted in inhibition of expression of the other, structurally related mnp genes. While this phenomenon may have been to our advantage, since it possibly diminished the compensatory effects originating from the other mnp genes, it also did not allow us to focus on the significance of a specific mnp (10–12). Whether or not various mnp genes truly have redundant or compensatory functions remained to be determined. Directly answering these questions requires the production of homokaryon knockout strains in which specific mnp genes have been inactivated.
Gene targeting in Pleurotus was made possible by the recent production of a Δku80 strain, which exhibits exclusive homologous recombination (HR) during the integration of transforming DNA, in conjunction with the development of a protocol for isolation of specific gene-targeted homokaryon strains. The first implementation of this procedure was for the inactivation of the VP encoded by mnp4 (12). Enzymatic activity assays of the Δmnp4 strain provided direct proof that it encodes the predominant VP under Mn2+-deficient culture conditions, supporting our previous gene expression-based conclusions (12).
However, direct evidence linking a specific mnp to the various enhanced lignocellulose and xenobiotic degradative activities observed under Mn2+-amended culture conditions (10, 13, 18–22) has yet to be provided. Here, we have studied the functionality of the predominantly expressed MnPs, focusing on characterization of Δmnp3 strains. We have determined that inactivation of either mnp3, mnp4, or mnp9 did not affect the Mn2+-mediated decolorization ability of the fungus, providing direct evidence for functional redundancy within the P. ostreatus MnP gene family.
MATERIALS AND METHODS
Fungal and bacterial strains and growth conditions.
Pleurotus ostreatus monokaryon strain PC9 (Spanish Type Culture Collection accession number CECT20311), which is a protoclone derived by dedikaryotization of the commercial dikaryon strain N001 (Spanish Type Culture Collection accession number CECT20600) (23), and its derivative homokaryon Δku80 strain (carboxin resistant, designated strain 20b) (12) were used throughout this study. Fungal strains were grown and maintained in YMG medium (1% [wt/vol] glucose, 1% [wt/vol] malt extract [Difco], 0.4% [wt/vol] yeast extract [Difco]) (10, 24) or in GP medium (2% [wt/vol] glucose, 0.5% [wt/vol] peptone [Difco], 0.2% [wt/vol] yeast extract [Difco], 0.1% [wt/vol] K2HPO4, 0.05% [wt/vol] MgSO4 · 7H2O); Mn2+ was added as MnSO4 (10, 25). When required, 1.5% [wt/vol] agar was added to the appropriate medium. Liquid cultures were maintained in stationary 100-ml Erlenmeyer flasks containing 10 ml media. Cultures were incubated at 28°C in the dark. The inoculum for all growth conditions was one disk (5 mm in diameter) of mycelium obtained from the edge of a young colony grown on solid medium and positioned at the center of the petri dish or flask. The azo dye OII [4-(2-hydroxy-1-naphthylazo)benzenesulfonic acid sodium salt], the fungicide carboxin (Sigma-Aldrich), and the antibiotic hygromycin B (Alexis Biochemicals) were added to final concentrations of 100 mg/liter, 2 mg/liter (50% lethal dose [LD50] = 0.16 mg/liter), and 100 mg/liter (LD50 = 7 mg/liter), respectively. Escherichia coli JM109 cells (Promega) were used for standard cloning procedures according to the manufacturer's protocol. Culture biomass production was measured as dry weight (oven dried to a constant weight at 65°C) in liquid GP culture containing OII. The mycelial linear growth rate was determined by measuring the position of the advancing mycelial front (leading hyphae) in solid GP culture containing OII.
Nucleic acid manipulation and analyses.
Molecular manipulations were carried out on the basis of standard protocols described by Sambrook et al. (26). Genomic DNA was extracted from culture biomass first ground with dry ice in a cryogenic tissue grinder (BioSpec Products) with the DNeasy plant minikit (Qiagen). Nucleic acid concentration and purity measurements were performed using a NanoDrop-2000 apparatus (Thermo Scientific). PCR was performed in an Eppendorf Mastercycler gradient thermocycler using Phusion high-fidelity PCR master mix (Finnzymes) with the primers listed in Table 1. Isolation and purification of DNA fragments from agarose gel or PCR amplification were performed using the Wizard SV gel and PCR clean-up system (Promega). Cloning into plasmids was performed using the pGEM-T Vector System II (Promega). Plasmid DNA was purified using the QIAprep spin miniprep kit (Qiagen). DNA endonuclease restriction was performed with restriction enzymes from Fermentas. Digoxigenin (DIG)-labeled DNA probes were used for Southern blotting (Table 1) according to the DIG system procedures (Roche Applied Science). Total RNA was extracted from culture biomass first ground with dry ice in a cryogenic tissue grinder (BioSpec Products), then homogenized with QIAshredder spin columns (Qiagen). RNA was purified from the lysate using the RNeasy Plus minikit (Qiagen). cDNA was synthesized using the SuperScript III first-strand synthesis system for reverse transcription-PCR (RT-PCR; Invitrogen). Gene expression analyses were performed with an ABI StepOnePlus real-time PCR sequence detection system and software (Applied Biosystems) using Power SYBR green PCR master mix (Applied Biosystems), the primers listed in Table 1, and an annealing temperature of 63°C according to the manufacturer's default operating procedures. The endogenous internal control gene used was the β-tubulin gene identified in the JGI genome database of PC9 v1.0 (http://genome.jgi-psf.org/PleosPC9_1/PleosPC9_1.home.html); the corresponding protein identification (ID) is 117235 (Table 1), which corresponds to the previously identified protein ID 16119 in PC15 v1.0 (http://genome.jgi-psf.org/PleosPC15_1/PleosPC15_1.home.html) (10, 12, 13, 18). To determine the amplification threshold cycle (CT) of each target gene, the automatically determined threshold level of mnp7 (Table 1) was used for all the target genes within a tested sample. Target gene transcript abundance, in arbitrary units, was expressed relative to levels of β-tubulin according to their CT values with the formula 2CT for β-tubulin −CT for target gene × β-tubulin reaction efficiency/target gene reaction efficiency. DNA fragments, plasmid inserts, and RT-PCR amplicons were fully sequenced at the Center for Genomic Technologies of the Hebrew University of Jerusalem.
Table 1.
Oligonucleotides used in this study
| Purpose and template | Target | Primer designation | Sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|---|---|
| Cassette construction | ||||
| mnp3 replacement cassette (TMS8) | ||||
| PC9 genomic DNA | mnp3 | mnp3PF | CACGACATGCACGTTTGAGCTTAGAAGC | 2,031 |
| mnp3 | btubPF-mnp3PR | ATTTAGTTTCCTCCCAACAGCATTGCTGGAGTTGAAGAGACGA | ||
| pTMS14 (12) | Hygr | mnp3PR-btubPF | TCGTCTCTTCAACTCCAGCAATGCTGTTGGGAGGAAACTAAAT | 2,749 |
| Hygr | mnp3TF-hygR | GTGTCAACAGGAGAACTTGAGACCTATTCCTTTGCCCTCGGA | ||
| PC9 genomic DNA | mnp3 | hygR-mnp3TF | TCCGAGGGCAAAGGAATAGGTCTCAAGTTCTCCTGTTGACAC | 2,028 |
| mnp3 | mnp3TR | GTCTTCGATACTGGTATCCCTCCTTTATGG | ||
| mnp9replacement cassette (TMS11) | ||||
| PC9 genomic DNA | mnp9 | mnp9PF | CGGTTGTTGACAACTCTCTTTCTT | 2,023 |
| mnp9 | btubPF-mnp9PR | ATTTAGTTTCCTCCCAACAGCATGGCGAGTTCGGTGGAGTAAG | ||
| pTMS14 (12) | Hygr | mnp9PR-btubPF | CTTACTCCACCGAACTCGCCATGCTGTTGGGAGGAAACTAAAT | 2,748 |
| Hygr | mnp9TF-hygR | CTGAATGAGCGTTACGAGGTACCTATTCCTTTGCCCTCGGA | ||
| PC9 genomic DNA | mnp9 | hygR-mnp9TF | TCCGAGGGCAAAGGAATAGGTACCTCGTAACGCTCATTCAG | 2,019 |
| mnp9 | mnp9TR | CAAGCAGACTACTAACAAGTTGTAGGTT | ||
| Analysis of construct integration: mnp3 replacement cassette (TMS8)a | ||||
| Recombination probe (strain-specific genomic DNA) | Hygr | hph+118 | GATGTAGGAGGGCGTGGATA | 3,738 |
| mnp3 | mnp3+4376 | TTTTGGAAGCATGCTGATTG | ||
| Homokaryon probe (strain-specific genomic DNA) | mnp3 | mnp3+255 | CAAACTTCGCATCTTCGTGA | 1,009 |
| mnp3 | mnp3+1264 | TTGGCCTGGTTGTCTAGAGG | ||
| Southern probe (pTMS8) | mnp3 | mnp3−1677 | ACCACCCCCATTCCAATTAT | 1,332 |
| mnp3 | mnp3−345 | GCTCCCGATTTGATGGTAGA | ||
| Amplicona (12) | ||||
| Strain-specific genomic DNA | mnp2 | mnp2+947 | TTGACCCCTCCGTAAGTGAC | 94 |
| mnp2+1041 | CGAGCGAGAACACCTTTACC | |||
| Strain-specific genomic DNA | mnp3 | mnp3+1103 | GCCCGTGGTATGTATTCAGC | 165 |
| mnp3+1268 | AAGCTTGGCCTGGTTGTCTA | |||
| Strain-specific genomic DNA | mnp4 | mnp4+1084 | CCCGGAGTTTTTGATTCTCA | 282 |
| mnp4+1366 | ATCAAAGGCTGCAAGGAAGA | |||
| Strain-specific genomic DNA | mnp9 | mnp9+216 | ATACCCTGCGTTTTCTGTGG | 496 |
| mnp9+712 | TACCAAGGGGAAAGCACTTG | |||
| Gene expressiona (10) | ||||
| Strain-specific total cDNA | β-tubulin | tubF543 | GTGCGTAAGGAAGCTGAGGG | 201 |
| tubR777 | TGTGGCATTGTACGGCTCAAC | |||
| Strain-specific total cDNA | mnp1 | MnP1F1334 | GTTCGCTCGAGACGATAGGACAT | 187 |
| MnP1R1619 | GGGGAGATGTGGTTTGGTTACA | |||
| Strain-specific total cDNA | mnp2 | MnP2F737 | GCCTTTCGATAGCGTGGATAAG | |
| MnP2R1123 | GGTCCCTTGCAACATTGTCTC | |||
| Strain-specific total cDNA | mnp3 | MnP3F30 | CCTCCTGACTTTGGCATCTCA | 199 |
| MnP3R240 | CACCTCCTCCACCTTTGGTT | |||
| Strain-specific total cDNA | mnp4 | MnP4F1269 | TTGTTGGCTAGAGACCCCCAGA | 230 |
| MnP4R1609 | CAAGTGGGCCGCTCCGAC | |||
| Strain-specific total cDNA | mnp5 | MnP5F52 | CAGGCCGTCAATGCCGTA | |
| MnP5R433 | CGTAAGGACGGAACCATCA | |||
| Strain-specific total cDNA | mnp6 | MnP6F1133 | TTCCCAGGTACTGCCGGA | 186 |
| MnP6R1515 | CCAAAGACAGTTTCAACATCGC | |||
| Strain-specific total cDNA | mnp7 | MnP7F1293 | CTTCATCTCCAACCCCAACTC | 237 |
| MnP7R1519 | GCTTTGCGGCAGGATG | |||
| Strain-specific total cDNA | mnp8 | MnP8F1091 | CTTCGACTCTACTCCCAACAGC | |
| MnP8R1335 | GAGGCGTGGTCGGTGAT | |||
| Strain-specific total cDNA | mnp9 | MnP9F1359 | ATTGCTAATGAAAGGCTCATGGTT | 181 |
| MnP9R1588 | GGTGGCAGCACAAGCAG |
Numbers indicate genomic coordinates from the relevant target gene transcription start site (ATG) coding sequences.
Construction of transforming DNA: mnp3 and mnp9 replacement cassettes.
The flanking DNA (2 kb 5′ and 3′) of mnp3 (9, 10) was amplified from genomic DNA and the hygromycin B resistance gene (Hygr) from plasmid pTMS14 (12) using primers mnp3PF and btubPF-mnp3PR, hygR-mnp3TF and mnp3TR, and mnp3PR-btubPF and mnp3TF-hygR for the 5′ flank, the 3′ flank, and Hygr, respectively (Table 1). The resulting amplicons were fused together using the double-joint PCR technique (12, 27, 28) to produce the mnp3 replacement cassette (TMS8), which was cloned to produce plasmid pTMS8 (Fig. 1A). Similarly, the flanking DNA (2 kb 5′ and 3′) of mnp9 (9, 10) was amplified from genomic DNA and Hygr from plasmid pTMS14 (12) using primers mnp9PF and btubPF-mnp9PR, hygR-mnp9TF and mnp9TR, and mnp9PR-btubPF and mnp9TF-hygR for the 5′ flank, the 3′ flank, and Hygr, respectively (Table 1). The resulting amplicons were fused together using the double-joint PCR technique (12, 27, 28) to produce the mnp9 replacement cassette (TMS11), which was cloned to produce plasmid pTMS11 (Fig. 2A).
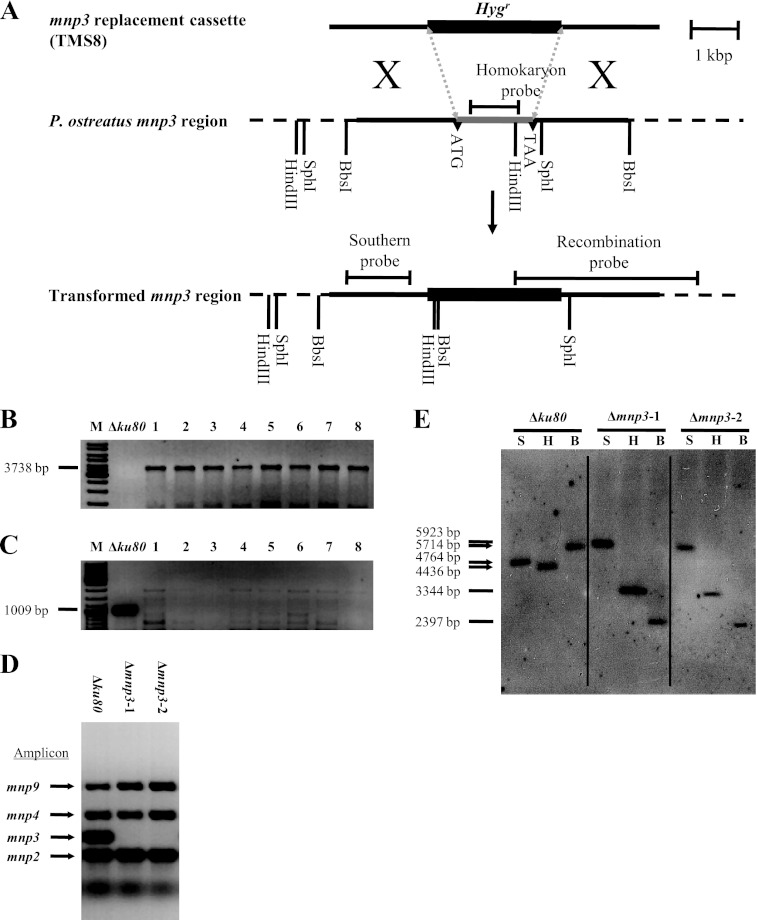
Fig 1.
(A) Strategy for replacement of mnp3 in P. ostreatus (20b). The hygromycin B resistance gene (Hygr) fused to 2 kb of 5′ and 3′ DNA flanking mnp3 (TMS8) was used for transformation. (B) PCR screening for transformants that have undergone homologous recombination using primers targeting the indicated mnp3 recombination probe amplicon (Table 1) (lanes 1 to 8). (C) PCR screening for homokaryon transformants that have undergone homologous recombination using primers targeting the indicated mnp3 homokaryon probe amplicon (Table 1) (lanes 1 to 8). (D) PCR screening for homokaryon Δmnp3 transformants, while showing that mnp2, -4, and 9 were not targeted, using primers targeting the indicated amplicons (Table 1) (12). (E) Southern blot analysis of DNA from Δku80 (20b) (restriction patterns marked with arrows) and two Δmnp3 transformant strains (Δmnp3-1 and Δmnp3-2) (restriction patterns marked with lines) digested with either SphI (S), HindIII (H), or BbsI (B) and probed with a DIG-labeled DNA probe corresponding to the indicated Southern probe amplicons (Table 1).
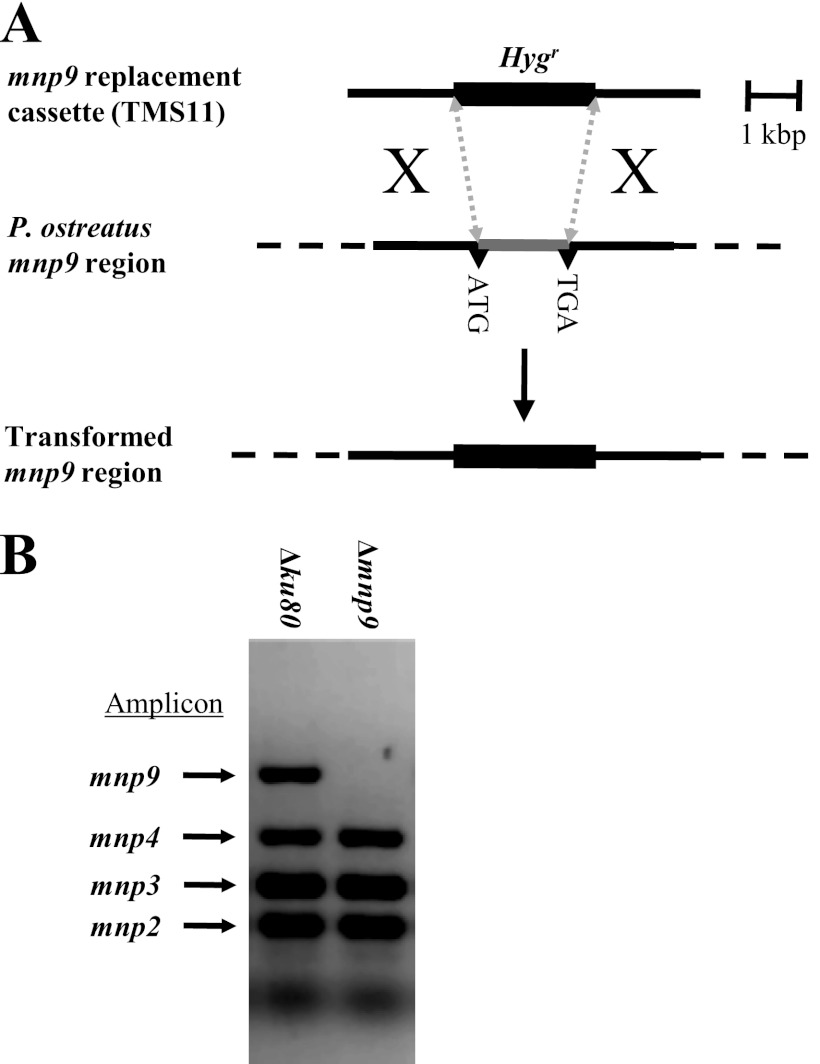
Fig 2.
(A) Strategy for replacement of mnp9 in P. ostreatus (20b). The hygromycin B resistance gene (Hygr) fused to 2 kb of 5′ and 3′ DNA flanking mnp9 (TMS11) was used for transformation. (B) PCR screening for homokaryon Δmnp9 transformants, while showing that mnp2, -3, and -4 were not targeted, using primers targeting the indicated amplicons (Table 1) (12).
Fungal transformation.
Transformation was performed based on the polyethylene glycol (PEG)-CaCl2 protocol previously adapted for P. ostreatus (10, 12, 13, 24, 29). Hygromycin B was used as a selection marker, and resistance was conferred via introduction of the hygromycin B resistance cassette (Hygr) (12). Competent protoplasts were produced by digestion of the vegetative mycelium of P. ostreatus from YMG liquid culture with lytic enzymes. The lytic enzyme solution consisted of 2% (wt/vol) lysing enzymes from Trichoderma harzianum (Sigma-Aldrich; product number L1412) and 0.05% (wt/vol) chitinase from Trichoderma viride (Sigma-Aldrich; product number C8241) in 0.5 M sucrose as an osmotic stabilizer. The protoplasts were washed (by centrifugation at 450 × g, 8 min, 4°C) in STC solution (18.2% [wt/vol] sorbitol, 50 mM Tris-HCl [pH 8.0], 50 mM CaCl2, 0.5 M sucrose) and adjusted to a final concentration of 5 × 107 protoplasts/ml. Then, 2 ml protoplasts was mixed with 100 μl transforming DNA (300 ng/μl), 150 μl heparin solution (Sigma-Aldrich; product number H4784) (5 mg dissolved in 1 ml STC solution), and 300 μl single-strand λ phage carrier DNA (Fermentas; product number SD0011) (500 μg/ml after denaturation at 95°C for 5 min and immediate transfer to ice). After 40 min of incubation on ice, 10 ml PTC solution (40% [wt/vol] PEG 4000, 50 mM Tris-HCl [pH 8.0], 50 mM CaCl2, 0.5 M sucrose) was added, and the mixture was incubated for 20 min at room temperature. The mixture was then plated on solid YMG regeneration medium containing 0.5 M sucrose and left to regenerate overnight. Subsequently a medium overlay containing hygromycin B was applied, and a final concentration of 150 mg/liter was obtained. Transformants were isolated after 10 days of incubation at 28°C. Transformant stability was verified by three successive transfers (inoculated from the edge of a 10-day-old colony) to solid medium without the selection drug and then return of the transformant to solid culture conditions in which the selective drug was present.
Protein expression profiles.
Culture fluids were filtered through Whatman no. 1 filter paper followed by filtration through a 0.45-μm mixed cellulose ester filter (Whatman). The sample was then concentrated 60-fold using a 10-kDa-cutoff PM-10 membrane (Amicon Division) and treated with cOmplete, EDTA-free protease inhibitor cocktail tablets (Roche Applied Science). The concentrated eluate was separated on a NuPAGE 4 to 12% bis-Tris gel in MES (morpholineethanesulfonic acid)-SDS running buffer (Invitrogen), and the 40- to 45-kDa gel fraction was excised and maintained at 4°C. The sample was subsequently analyzed using a label-free quantitative (LFQ) proteomics approach (30). This method entails three steps: separation, identification, and quantification. Separation was carried out by high-pressure liquid chromatography (HPLC)/tandem mass spectrometry (LC-MS/MS) in an Orbitrap (Thermo Scientific) mass spectrometer, and identification was carried out with Sequest 3.31 software against the JGI genome database of P. ostreatus PC9 v1.0 (http://genome.jgi-psf.org/PleosPC9_1/PleosPC9_1.home.html). Analysis of quantitative changes in protein abundance was based on measuring peptide ion peak intensities. The LFQ analyses were carried out at Smoler Proteomics Center of The Israel Institute of Technology (Technion).
Enzymatic activity assays.
Samples of culture fluids were collected, centrifuged (4,720 × g, 10 min, 4°C), and maintained at 4°C. Enzymatic activity assays were conducted in a volume of 200 μl in microtiter plates at 32°C using the Synergy 2 multimode microplate reader (BioTek). Mn2+-dependent and Mn2+-independent activities were determined using phenol red (Sigma-Aldrich) as the substrate (31). Oxidation of phenol red was measured by monitoring the A610 (ε = 22.0 mM−1 cm−1). The reaction mixture contained 0.1 mM MnSO4, 0.1 mM H2O2, 0.01% (wt/vol) phenol red, 25 mM lactate, 0.1% (wt/vol) bovine serum albumin, and 20 mM sodium succinate buffer (pH 4.5). After incubation, the reaction was terminated by addition of NaOH to a final concentration of 80 mM. Activity in the absence of either MnSO4 or H2O2, was measured to establish specific (either Mn2+-dependent or Mn2+-independent) peroxidase activity. Accordingly, Mn2+-independent activity was deduced from the reaction in the absence of Mn2+ after subtraction of its corresponding reaction activity in the absence of H2O2, and Mn2+-dependent activity was deduced from the reaction mixture containing Mn2+ by subtraction of the corresponding Mn2+-independent activity. Corresponding boiled samples served as blanks. One unit of enzymatic activity was defined as the amount of enzyme that catalyzes the formation of 1.0 μmol of product per minute per milliliter of culture filtrate.
Analysis of OII decolorization.
OII decolorization capacity was estimated in GP cultures amended with OII. In solid culture, decolorization capacity was estimated according to the visually decolorized area, as measured from the center of the inoculation point. In liquid culture, 100 μl of the media was centrifuged (4,720 × g, 10 min, room temperature) and mixed with 900 μl phosphate buffer (0.1 M, pH 7.0) and the OII concentration in the media was quantified according to the absorption reading of the solution at a λmax of 483 nm using a BioMate 3 spectrophotometer (Thermo Spectronic), according to a standard curve. Noninoculated medium amended with OII was used as a control (10, 12, 13).
RESULTS
P. ostreatus MnP3 is predominantly expressed in Mn2+-amended GP medium.
In order to study the significance of specific MnP gene family members for the functionality of the P. ostreatus ligninolytic system, the protein expression profile in the extracellular fluids was analyzed. The 40- to 45-kDa fraction of the fungal secretome, which contains the peroxidases (based on in silico genomic analyses [9–13]), was analyzed for MnP/VP isoenzymes in extracellular fluids collected from the wild-type PC9 strain liquid GP culture amended with 27 μM Mn2+ after 7 days of incubation. This day was selected since at this time point the greatest amount of OII decolorization was observed.
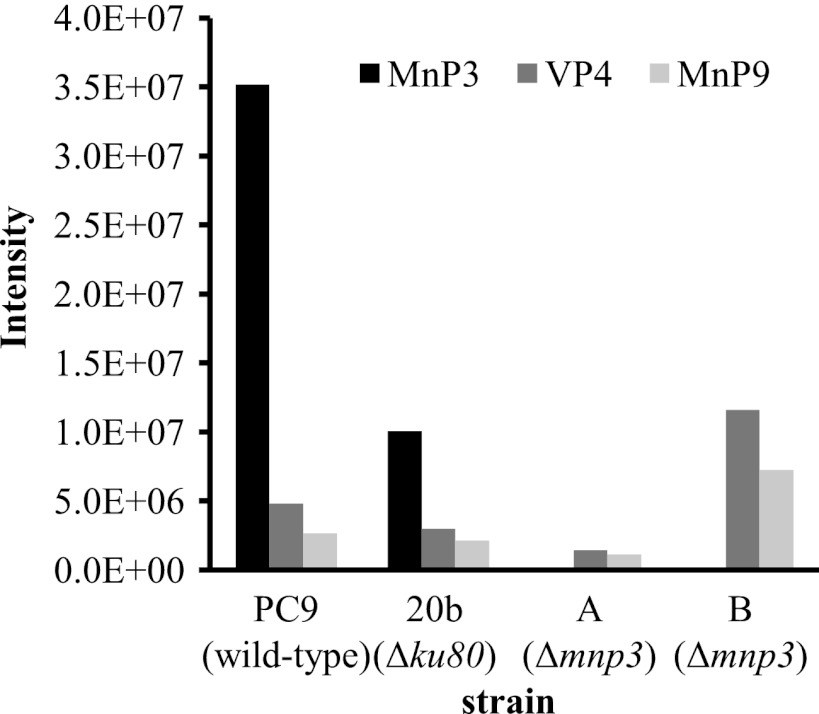
The identification and quantification of proteins were based on the presence of unique peptides corresponding to MnP/VP isoenzymes and on the presence of at least 2 identified peptides per protein in each sample (Table 2). MnP3, VP4, and MnP9, encoded by mnp3, mnp4, and mnp9, respectively (9, 10), were detected. MnP3 was found to be the most abundant (approximately 10-fold more than VP4 or MnP9) (Fig. 3). The MnP/VP isoenzyme abundance profiles were in line with the measured MnP gene family transcript abundance on day 7, showing the predominance of mnp3 (being at least 3-fold more than other mnp genes) (Fig. 4). On the basis of these results, mnp3 was targeted for gene disruption in order to elucidate its contribution to ligninolytic functionality.
Table 2.
Identified manganese-dependent peroxidase (MnP) and versatile peroxidase (VP) isoenzyme peptides
| MnP/VP isoenzyme | Sequence | Length (aaa) | No. of identified ions from each sample from strain: |
|||
|---|---|---|---|---|---|---|
| PC9 (wild type) | 20b (Δku80) | A (Δmnp3) | B (Δmnp3) | |||
| MnP3 | GTAFPGVGGNQGEVESPIAGEIR | 23 | 2 | 2 | 0 | 0 |
| IIDCSDVIPVPKPIQSK | 17 | 2 | 2 | 0 | 0 | |
| INFIIGRPPATAASPNGIIPEPFDTVTDIIAR | 32 | 1 | 0 | 0 | 0 | |
| IQSDHDIAR | 9 | 1 | 2 | 0 | 0 | |
| ITFHDAIGFSPTK | 13 | 1 | 0 | 0 | 0 | |
| SKIIDCSDVIPVPKPIQSK | 19 | 1 | 0 | 0 | 0 | |
| TACEWQSFVNNQAK | 14 | 1 | 1 | 0 | 0 | |
| Total | 9 | 7 | 0 | 0 | ||
| VP4 | DPQTACEWQSMVNNQPK | 17 | 1 | 0 | 0 | 0 |
| IFPGTPDNKGEVQSPIQGEIR | 21 | 2 | 1 | 1 | 2 | |
| IPFFIGRPDAVAASPDHIVPEPFDSVDTIIAR | 32 | 0 | 0 | 0 | 1 | |
| IQSDHIIAR | 9 | 1 | 1 | 1 | 1 | |
| MAIIGQDK | 8 | 0 | 0 | 0 | 1 | |
| Total | 4 | 2 | 2 | 5 | ||
| MnP9 | IATIGQVR | 8 | 0 | 0 | 0 | 1 |
| IEFIAGR | 7 | 1 | 0 | 0 | 1 | |
| ITDCSDVIPVPQVR | 14 | 1 | 1 | 1 | 1 | |
| ITKPVTIPAGR | 11 | 1 | 1 | 1 | 1 | |
| WQSFIANER | 9 | 1 | 0 | 0 | 1 | |
| Total | 4 | 2 | 2 | 4 | ||
aa, amino acids.
Fig 3.
Quantitative profile of manganese-dependent peroxidase (MnP) and versatile peroxidase (VP) isoenzymes in extracellular medium of P. ostreatus wild-type (PC9), Δku80 (20b), and Δmnp3 strains using a label-free quantitative approach. The strains were grown in liquid GP medium amended with 27 μM Mn2+ for 7 days. Data represent a combined sample of 10 biological replicates.
Fig 4.
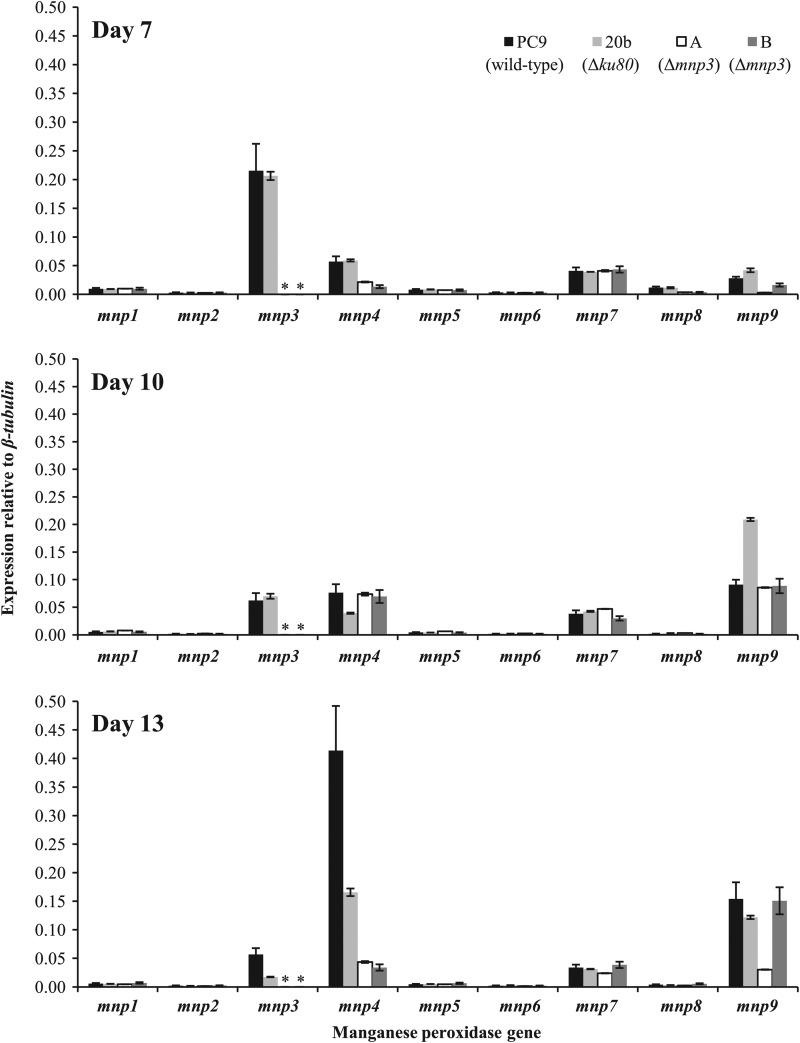
Relative transcript abundance of MnP gene family members in P. ostreatus wild-type (PC9), Δku80 (20b), and two Δmnp3 (A and B) strains, grown for 7, 10, and 13 days in liquid GP medium amended with 27 μM Mn2+. The abundances of mnp1 to -9 transcripts, relative to that for β-tubulin, were measured by real-time PCR and quantified using the standard curve method. Asterisks indicate no detection of transcript. Data represent the averages of two biological replicates. Bars denote the standard deviations.
Production and characterization of P. ostreatus PC9 homokaryon Δmnp3 strains.
MnP3 is identified in the JGI genome database of PC9 v1.0 (http://genome.jgi-psf.org/PleosPC9_1/PleosPC9_1.home.html) as protein ID 137740, which corresponds to the previously identified protein ID 185959 in PC15 v1.0 (http://genome.jgi-psf.org/PleosPC15_1/PleosPC15_1.home.html) (9, 10).
Based on the P. ostreatus gene-targeting system methodology (12), the TMS8 gene replacement cassette, targeted for HR with the mnp3 coding sequence (Fig. 1A; Table 1), was used to transform the P. ostreatus PC9 homokaryon Δku80 strain. After the initial isolation step the transformants were grown without selection.
Genomic DNA analyses of the selected transformants confirmed that mnp3 was replaced by Hygr and that they are homokaryons (Fig. 1B and C). In parallel, it was shown that the nontargeted mnp genes (i.e., mnp2, mnp4, and mnp9) were not disrupted due to the recombination event (Fig. 1D). Southern blot hybridization analysis of genomic DNA digested with either SphI, HindIII, or BbsI and probed with a DIG-labeled DNA probe corresponding to the Southern probe amplicon confirmed that replacement of mnp3 was the result of a single HR and that these strains are homokaryons (Fig. 1E).
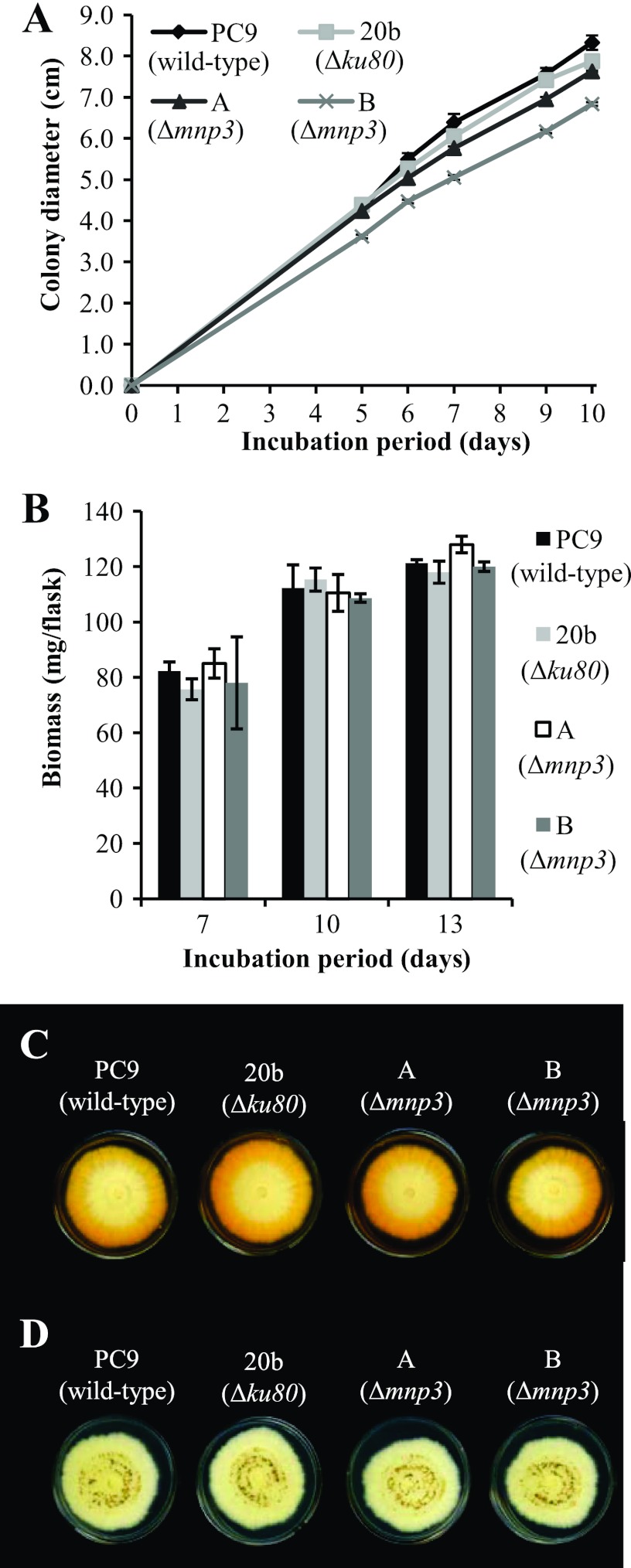
The Δmnp3 transformants showed growth parameters similar to those of the wild-type strain PC9 and the Δku80 strain both in solid GP culture (linear growth rate of 7.6 ± 0.8 mm/24 h) (Fig. 5A) and in liquid GP culture (biomass production of 121.8 ± 6.2 mg/flask after 13 days of incubation) (Fig. 5B). Unexpectedly, they also exhibited similar capabilities for OII decolorization in GP agar medium amended with 27 μM Mn2+ (Fig. 5C). Two transformants, designated Δmnp3A and Δmnp3B, were chosen for further analyses.
Fig 5.
(A) Linear mycelial growth rate of P. ostreatus wild-type (PC9), Δku80 (20b), and Δmnp3 (A and B) strains grown on solid GP medium amended with 27 μM Mn2+ and OII at a concentration of 100 mg/liter, during 10 days of incubation. Data represent the averages of three biological replicates. Bars denote the standard deviations. (B) Biomass production of PC9, 20b, A, and B strains grown in liquid GP medium amended with 27 μM Mn2+ and OII at a concentration of 100 mg/liter, after 7, 10, and 13 days of incubation. Data represent the averages of three biological replicates. Bars denote the standard deviations. (C) OII decolorization by PC9, 20b, A, and B strains grown on solid GP medium amended with 27 μM Mn2+ and OII at a concentration of 100 mg/liter after 10 days of incubation. (D) Mn2+ oxidation (apparent as dark brown to black MnO2 precipitates) by PC9, 20b, A, and B strains grown on solid GP medium amended with 108 μM Mn2+ after 10 days of incubation.
Gene transcript, peroxidase activity, and MnP protein expression profiles of P. ostreatus Δmnp3 strains.
To determine the effect of mnp3 disruption on the MnP gene family expression profiles, their transcript abundances were quantitatively evaluated in total RNA extracted from the Δmnp3, PC9, and Δku80 strains grown in liquid GP medium amended with 27 μM Mn2+ and OII, following 7, 10, and 13 days of incubation (Fig. 4). While PC9 and the Δku80 strains showed typical mnp3 expression profiles, no mnp3 transcripts were detected in the Δmnp3 strains, confirming absolute inactivation of mnp3 and the homokaryon nature of the strains. The results also clearly show that the nontargeted mnp genes were expressed at levels comparable to those for the wild type throughout the incubation period, indicating that inactivation of mnp3 does not result in compensatory effects (i.e., higher expression of other mnp genes) at the transcript level (Fig. 4).
An additional observation made by this time course analysis was that the culture age is another factor differentially affecting mnp gene expression. Generally, the expression of mnp3 peaked at 7 days, while mnp4 and mnp9 transcription increased throughout the incubation period (Fig. 4).
Analysis of the fungal secretome supported the gene expression data (Fig. 4). In the wild-type PC9 and Δku80 strains, MnP3, VP4, and MnP9 were detected, with no indication of the presence of the other MnPs. MnP3 was the predominant isoenzyme. However, in the two Δmnp3 strains MnP3 was not detected, while the abundances of VP4 and MnP9 were similar to those for the PC9 and Δku80 strains (Fig. 3).
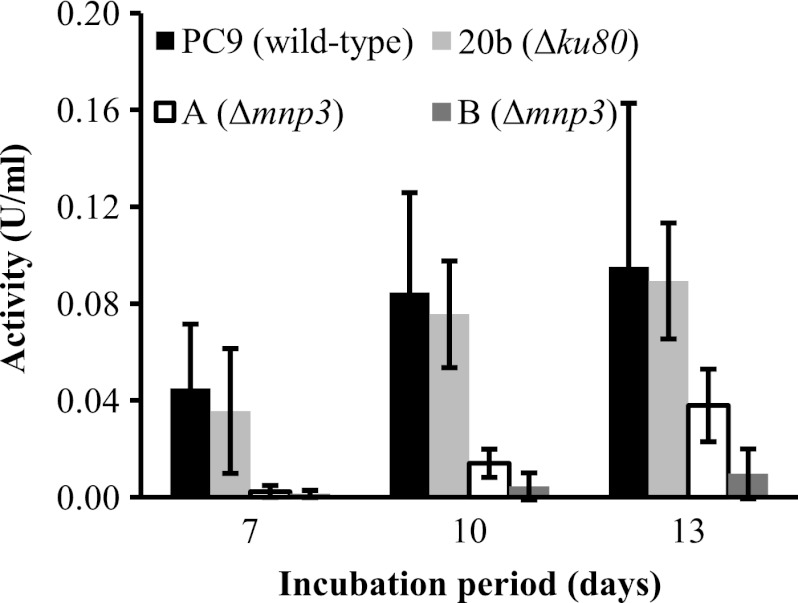
In order to evaluate the effect of mnp3 inactivation on Mn2+-dependent peroxidase activity, a time course assay was carried out using phenol red as a substrate. In general, oxidation activity increased in all strains as the incubation period progressed. In the wild-type and Δku80 strains similar activity levels were measured and were generally higher than those in the Δmnp3 strains (Fig. 6).
Fig 6.
Time course assay of Mn2+-dependent peroxidase activity of P. ostreatus wild-type (PC9), Δku80 (20b), and Δmnp3 (A and B) strains. The strains were grown in liquid GP medium amended with 27 μM Mn2+ and OII at a concentration of 100 mg/liter during 13 days of incubation. One unit of enzymatic activity was defined as the amount of enzyme that catalyzes the formation of 1.0 μmol of oxidized phenol red per minute per milliliter of culture filtrate. Data represent the averages of three biological replicates. Bars denote the standard deviations.
Inactivation of mnp3 does not decrease Mn2+-mediated OII decolorization capacity.
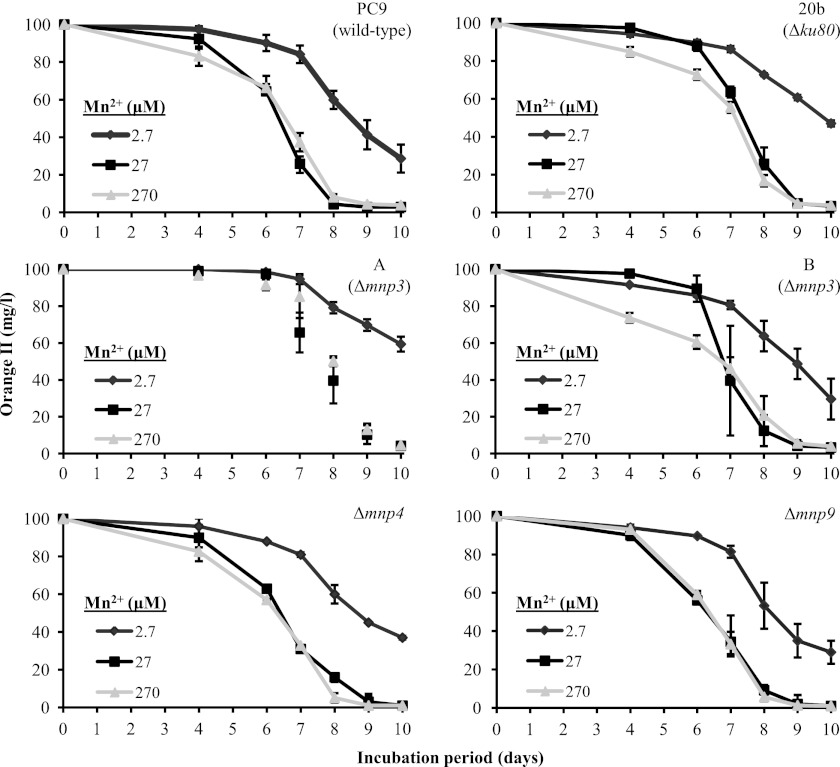
To evaluate the effect of mnp3 inactivation on the Mn2+-mediated functionality of the ligninolytic system, the in vivo capacities of the Δmnp3, PC9, and Δku80 strains to decolorize the azo dye OII were determined in liquid GP cultures amended with either 2.7, 27, or 270 μM Mn2+ during 10 days of incubation (Fig. 7). Examination of the decolorization capacity in this range of Mn2+ concentrations was based on our previous study showing a positive correlation between these factors (10) and the assumption that the transformants will react differently under the various Mn2+ concentrations. The differences observed in decolorization between the mnp3 knockout strains and the PC9 and Δku80 controls were within the experimental error (Fig. 7).
Fig 7.
Time course assay of OII decolorization by P. ostreatus wild-type (PC9), Δku80 (20b), Δmnp3 (A and B), Δmnp4, and Δmnp9 strains in liquid GP media amended with either 2.7, 27, or 270 μM Mn2+ during 10 days of incubation. Data represent the averages of four biological replicates. Bars denote the standard deviations.
An additional screen consisted of monitoring the appearance of dark brown to black MnO2 precipitates typically observed in solid GP cultures containing Mn2+ concentrations higher than 54 μM (10, 13). The tested strains formed similar MnO2 precipitate patterns, indicative of similar Mn2+ oxidation activities (Fig. 5D).
Taken together, our results indicate that, even though expression analyses suggested that MnP3 may play a key role, no significant differences were found in decolorization capacity between the examined strains in either of the tested Mn2+ concentrations (Fig. 7). These data indicate that mnp3 gene function may be redundant and may be compensated for by the standard expression levels of other mnp genes (Fig. 4).
Production of a P. ostreatus PC9 Δmnp9 strain.
To further study the phenomenon of compensation in the MnP gene family, mnp9 was selected for targeted inactivation. This gene was selected since, as detailed above, MnP9 was detected along with MnP3 and VP4 in the fungal secretome (Fig. 3). MnP9 and MnP3 are both Mn2+-dependent peroxidases, and mnp3 and mnp9 are both upregulated in the presence of Mn2+ (10). Thus, MnP9 and MnP3 are also the most likely candidates to substitute for each other under conditions limiting the expression of one of them.
MnP9 is identified in the JGI genome database of PC9 v1.0 (http://genome.jgi-psf.org/PleosPC9_1/PleosPC9_1.home.html) as protein ID 137764, which corresponds to the previously identified protein ID 23172 in PC15 v1.0 (http://genome.jgi-psf.org/PleosPC15_1/PleosPC15_1.home.html) (9, 10). The P. ostreatus gene-targeting system (12) and the TMS11 gene replacement cassette, targeted for HR at the mnp9 gene locus (Fig. 2A, Table 1), were used to transform the P. ostreatus PC9 homokaryon Δku80 strain.
Genomic DNA analyses of the selected transformants confirmed that mnp9 was disrupted and that the transformants are homokaryons, while showing that the nontargeted mnp genes (i.e., mnp2, mnp3, and mnp4) were not disrupted due to the recombination event (see representative PCR results in Fig. 2B). All the disruptants exhibited growth parameters similar to those of wild-type strain PC9 both in solid GP culture and in liquid GP culture. One was selected and designated the P. ostreatus PC9 homokaryon Δmnp9 strain.
Inactivation of mnp9 or mnp4 did does decrease Mn2+-mediated OII decolorization capacity.
To evaluate the effect of mnp9 inactivation on the functionality of the ligninolytic system, the in vivo Mn2+-mediated OII decolorization capacity was determined using the same experimental setup described above. Furthermore, this assay was expanded to also include the third isoenzyme detected in the fungal secretome, VP4, using the previously described Δmnp4 strain (12).
Similar to the phenotype observed with the Δmnp3 strains, only a minor delay in decolorization was observed in the mnp9 and mnp4 knockout strains relative to the PC9 and Δku80 strains (Fig. 7). Additionally, the OII decolorization and MnO2 precipitation phenotypes in solid GP culture were similar among all the tested strains.
Conclusively, these results further substantiate a general phenomenon of functional redundancy among the members of the MnP gene family in Mn2+-amended GP culture.
DISCUSSION
Whether predominantly expressed members of the P. ostreatus MnP gene family have a specific role is a question addressed in this study.
Even though there is structural resemblance among mnp genes, the marked upregulation of mnp3, combined with the enhanced capabilities to degrade lignin and recalcitrant anthropogenic compounds, observed due to amendment of Mn2+ to the P. ostreatus culture, suggested a central role for MnP3 in substrate utilization by this fungus. It has therefore been the center of attention of studies concerning the ligninolytic system of P. ostreatus (7, 10, 15, 18–22, 25).
In a previous study Salame et al. (10) showed that gene silencing targeted at mnp3 resulted in the cosilencing of the entire MnP gene family, which correlated to reduced OII decolorization capacity in Mn2+-amended GP culture (10, 11). Thus, the purpose of this study was to dissect the specific significance of the predominantly expressed MnP3 by isolation and characterization of a homokaryon Δmnp3 strain using a gene knockout strategy.
Inactivation of mnp3 did not significantly affect the expression of nontargeted members of the MnP gene family and their encoded isoenzymes, nor did it considerably affect the Mn2+-mediated OII decolorization capacity compared to that of the control strains (PC9 and the Δku80 strains, used as background in transformation). However, total MnP activity (as evaluated by Mn2+-dependent oxidation of phenol red) was reduced in the Δmnp3 strains. It seems that the relatively low activity level was sufficient for maintaining the fungal decolorization capacity. This is in line with our previous observation showing that overexpression of mnp4 and the corresponding increase in activity resulted in earlier but not accelerated decolorization (13).
In light of these results, the other strongly upregulated MnP-encoding gene, mnp9, a probable candidate for functional compensation of mnp3 (and vice versa), was inactivated. As with the Δmnp3 strains, the Δmnp9 strain exhibited Mn2+-mediated decolorization ability similar to that of the control strains. Similarly, the Δmnp4 strain, encoding the predominant VP (also possessing Mn2+ peroxidase activity), exhibited a similar phenotype under these conditions.
Conclusively, these outcomes point to functional redundancy within the P. ostreatus MnP gene family, enabling compensation (“buffering”) for the deficiency of one of its members in the tested Mn2+-amended GP culture conditions. Particularly, the observed redundancy may be explained by the concomitant high expression of three mnp genes under these conditions. Also, the dye-decolorizing peroxidases whose genes were detected in the P. ostreatus genome could be involved in the observed Mn2+-mediated decolorization (10, 13). However, they lack a Mn2+ oxidation site (9). This is in utter contrast to the situation observed in Mn2+-deficient GP culture conditions, where only one mnp, mnp4, is predominant; its inactivation resulted in a clear phenotype of a drastic decrease in phenol red oxidation activity (12).
In addition, membrane-bound MnP/VP isoenzymes may contribute to in vivo activity, while being “transparent” to extracellular activity assays and secretome analyses. Specifically, this may be the case with MnP7, identified in the JGI genome database of PC9 v1.0 (http://genome.jgi-psf.org/PleosPC9__1/PleosPC9_1.home.html) as protein ID 137755 (Table 1), which corresponds to the previously identified protein ID 153232 in PC15 v1.0 (10, 12). Although mnp7 was found to be transcribed at a significant level, it was not identified in the fungal secretome profile obtained by LFQ analysis. This could be explained by bioinformatics analysis indicating MnP7 to be potentially membrane bound. While the N termini of all nine MnP/VP isoenzymes were found to have typical signal peptides (9) predicted to direct them for secretion (Predisi Server; http://www.predisi.de), MnP7 was the only isoenzyme predicted to also contain a transmembrane domain overlapping and transcending the cleavage site of its signal peptide (TMHMM Server 2.0; http://www.cbs.dtu.dk/services/TMHMM). Hence, MnP7 may be both secreted and anchored to the membrane.
A strain in which several mnp genes have been inactivated may impair decolorization of OII and reveal the extent of redundancy within this gene family. Thus far, our attempts to produce mnp double-knockout strains by using a mixture of gene replacement cassettes, targeting mnp3, -4, and 9, have been unsuccessful, as only single-knockout strains were obtained.
Comparison of the mnp3 knockout effects to those seen by targeting mnp3 for knockdown via RNAi (10, 11) demonstrates the distinct difference in the outcomes of employing these methods of genetic manipulation. While the former was utilized here for specific and complete targeted inactivation of a single mnp, the latter resulted in temporal cosuppression of multiple mnp genes. Based on the results presented in the current report, the direct association between the level of MnP gene family expression and Mn2+-mediated OII decolorization could not be established without the use of an RNAi-based strategy (unless multiple knockouts could be produced in this fungus).
Functional redundancy is a characteristic feature of many biological systems, especially when gene families are present. In these cases, deletion or mutation of a gene has minimal or no impact on the phenotype or fitness of the organism because of functional compensation conferred by one or more genes capable of performing similar functions. This trait can provide organisms with robustness in terms of functioning in frequently harsh and variable environments, buffering internal molecular noise, genetic variation, and unpredictable environmental fluctuations (1, 32).
The exact contribution of genetic redundancy is not clear. On the one hand, since fully functional redundant genes have identical functions, their maintenance throughout evolution should not be stable. On the other hand, if these genes have similar, overlapping functions, they can enable “fine-tuning” for adaption to different environmental conditions, in parallel to having a role in providing functional redundancy, thus justifying their preservation (32).
The fact that P. ostreatus has a large number of distinct genes encoding transcripts of various isoenzymes belonging to the MnP gene family is a classic indication of potential redundancy and/or diversity of these enzymes and may imply that complex and versatile strategies are employed by this fungus for the degradation of aromatic and recalcitrant compounds such as amorphic lignin and azo dyes.
In line with these hypotheses, the recently available genomic data from various wood decay fungi, such as the WRF P. chrysosporium, Ceriporiopsis subvermispora, and Trametes versicolor and the brown rot fungi Postia placenta and Ganoderma lucidum, substantiate a trend of multiplicity in genes encoding enzymes considered to be involved in ligninolysis-related processes, which are typically grouped together in large gene families, such as those encoding laccases, lignin peroxidases, and cytochrome P450s. Overall, this extensive and seemingly highly redundant array of genes is consistent with the role of WRF within the ecosystem, specifically, interaction with highly heterogeneous compounds and support in maintaining important metabolic roles (1, 3, 4, 9, 14, 33–38).
In this study, we have shown that either mnp3, -4, or -9, the three predominantly expressed genes of the nine P. ostreatus MnP gene family members in Mn2+-amended GP culture, is functionally redundant for Mn2+-mediated OII decolorization. Nevertheless, the extent and role of the functional redundancy exhibited by the MnP gene family under other circumstances could be different. From our previous report, showing predominance of a single mnp (mnp4) in Mn2+-deficient GP culture (12), and this work we deduce that under conditions in which more than one mnp is highly expressed inactivation of a single mnp is “buffered,” whereas, when only one gene is predominant, its inactivation incapacitates its corresponding functionally.
To date, most of the insight into the mechanism of lignin degradation by P. ostreatus has been based on biochemical and physiological analyses (15–17). The above-mentioned gene function studies were accomplished by implementation of our recently developed gene silencing (10, 11) and specific gene-targeting (12) system enabling, among others, production of homokaryon knockout strains. Application of these versatile genetic modification tools markedly advances our understanding of the genetic basis of the ligninolytic system.
ACKNOWLEDGMENTS
We are deeply grateful to Takashi Watanabe and Yoichi Honda, Research Institute for Sustainable Humanosphere, Kyoto University, Uji City, Japan, for generously providing plasmid pTM1. We are also grateful to Assaf Eybishtz, Adi Moshe, Dagan Sade, Aurelia Zemach, and Carmit Ziv for their advice and comments. We thank the Joint Genome Institute (U.S. Department of Energy) and the Pleurotus Genome Consortium for access to the P. ostreatus genome database.
This research was partially supported by the Ministry of Science and Technology, Israel, and by grant no. 2011505 from the U.S.-Israel Binational Science Foundation.
Footnotes
Published ahead of print 1 February 2013
REFERENCES
- 1.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TK, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Dueñas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, St John F, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719 [DOI] [PubMed] [Google Scholar]
- 2.Hammel KE, Cullen D. 2008. Role of fungal peroxidases in biological ligninolysis. Curr. Opin. Plant Biol. 11:349–355 [DOI] [PubMed] [Google Scholar]
- 3.Hofrichter M, Ullrich R, Pecyna MJ, Liers C, Lundell T. 2010. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 87:871–897 [DOI] [PubMed] [Google Scholar]
- 4.Morgenstern I, Klopman S, Hibbett DS. 2008. Molecular evolution and diversity of lignin degrading heme peroxidases in the Agaricomycetes. J. Mol. Evol. 66:243–257 [DOI] [PubMed] [Google Scholar]
- 5.Ruiz-Dueñas FJ, Martínez AT. 2009. Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb. Biotechnol. 2:164–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Ruiz E, Gonzalez-Perez D, Ruiz-Dueñas FJ, Martínez AT, Alcalde M. 2012. Directed evolution of a temperature-, peroxide- and alkaline pH-tolerant versatile peroxidase. Biochem. J. 441:487–498 [DOI] [PubMed] [Google Scholar]
- 7.Kamitsuji H, Honda Y, Watanabe T, Kuwahara M. 2004. Production and induction of manganese peroxidase isozymes in a white-rot fungus Pleurotus ostreatus. Appl. Microbiol. Biotechnol. 65:287–294 [DOI] [PubMed] [Google Scholar]
- 8.Morales M, Mate MJ, Romero A, Martínez MJ, Martínez AT, Ruiz-Dueñas FJ. 2012. Two oxidation sites for low redox-potential substrates: a directed mutagenesis, kinetic and crystallographic study on Pleurotus eryngii versatile peroxidase. J. Biol. Chem. 287:41053–41067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz-Dueñas FJ, Fernández E, Martínez MJ, Martínez AT. 2011. Pleurotus ostreatus heme peroxidases: an in silico analysis from the genome sequence to the enzyme molecular structure. C. R. Biol. 334:795–805 [DOI] [PubMed] [Google Scholar]
- 10.Salame TM, Yarden O, Hadar Y. 2010. Pleurotus ostreatus manganese-dependent peroxidase silencing impairs decolourization of Orange II. Microb. Biotechnol. 3:93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salame TM, Ziv C, Hadar Y, Yarden O. 2011. RNAi as a potential tool for biotechnological applications in fungi. Appl. Microbiol. Biotechnol. 89:501–512 [DOI] [PubMed] [Google Scholar]
- 12.Salame TM, Knop D, Tal D, Levinson D, Yarden O, Hadar Y. 2012. Predominance of a versatile-peroxidase-encoding gene, mnp4, as demonstrated by gene replacement via a gene targeting system for Pleurotus ostreatus. Appl. Environ. Microbiol. 78:5341–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salame TM, Knop D, Levinson D, Mabjeesh SJ, Yarden O, Hadar Y. 2012. Release of Pleurotus ostreatus versatile peroxidase from Mn2+ repression enhances anthropogenic and natural substrate degradation. PLoS One 7:e52446 doi:10.1371/journal.pone.0052446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP, Kuske C, Magnuson JK, Martin F, Spatafora JW, Tsang A, Baker SE. 2011. Fueling the future with fungal genomics. Mycology 2:192–209 [Google Scholar]
- 15.Cohen R, Persky L, Hadar Y. 2002. Biotechnological applications and potential of wood-degrading mushrooms of the genus Pleurotus. Appl. Microbiol. Biotechnol. 58:582–594 [DOI] [PubMed] [Google Scholar]
- 16.Sánchez C. 2010. Cultivation of Pleurotus ostreatus and other edible mushrooms. Appl. Microbiol. Biotechnol. 85:1321–1337 [DOI] [PubMed] [Google Scholar]
- 17.Stajić M, Vukojević J, Duletić-Laušević S. 2009. Biology of Pleurotus eryngii and role in biotechnological processes: a review. Crit. Rev. Biotechnol. 29:55–66 [DOI] [PubMed] [Google Scholar]
- 18.Cohen R, Hadar Y, Yarden O. 2001. Transcript and activity levels of different Pleurotus ostreatus peroxidases are differentially affected by Mn2+. Environ. Microbiol. 3:312–322 [DOI] [PubMed] [Google Scholar]
- 19.Cohen R, Persky L, Hazan-Eitan Z, Yarden O, Hadar Y. 2002. Mn2+ alters peroxidase profiles and lignin degradation by the white-rot fungus Pleurotus ostreatus under different nutritional and growth conditions. Appl. Biochem. Biotechnol. 102:415–429 [DOI] [PubMed] [Google Scholar]
- 20.Cohen R, Yarden O, Hadar Y. 2002. Lignocellulose affects Mn2+ regulation of peroxidase transcript levels in solid-state cultures of Pleurotus ostreatus. Appl. Environ. Microbiol. 68:3156–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerem Z, Hadar Y. 1995. Effect of manganese on preferential lignin degradation by Pleurotus ostreatus during solid-state fermentation. Appl. Environ. Microbiol. 61:3057–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golan-Rozen N, Chefetz B, Ben-Ari J, Geva J, Hadar Y. 2011. Transformation of the recalcitrant pharmaceutical compound carbamazepine by Pleurotus ostreatus: role of cytochrome P450 monooxygenase and manganese peroxidase. Environ. Sci. Technol. 45:6800–6805 [DOI] [PubMed] [Google Scholar]
- 23.Larraya LM, Pérez G, Peñas MM, Baars JJ, Mikosch TS, Pisabarro AG, Ramírez L. 1999. Molecular karyotype of the white rot fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 65:3413–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irie T, Honda Y, Watanabe T, Kuwahara M. 2001. Efficient transformation of filamentous fungus Pleurotus ostreatus using single-strand carrier DNA. Appl. Microbiol. Biotechnol. 55:563–565 [DOI] [PubMed] [Google Scholar]
- 25.Irie T, Honda Y, Watanabe T, Kuwahara M. 2001. Homologous expression of recombinant manganese peroxidase genes in ligninolytic fungus Pleurotus ostreatus. Appl. Microbiol. Biotechnol. 55:566–570 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Ninomiya Y, Suzuki K, Ishii C, Inoue H. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101:12248–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973–981 [DOI] [PubMed] [Google Scholar]
- 29.Honda Y, Matsuyama T, Irie T, Watanabe T, Kuwahara M. 2000. Carboxin resistance transformation of the homobasidiomycete fungus Pleurotus ostreatus. Curr. Genet. 37:209–212 [DOI] [PubMed] [Google Scholar]
- 30.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, Scrivens JH. 2009. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome Res. 8:3752–3759 [DOI] [PubMed] [Google Scholar]
- 31.Grinhut T, Salame TM, Chen Y, Hadar Y. 2011. Involvement of the ligninolytic enzymes and Fenton like reaction during humic acid degradation by Trametes sp. M23. Appl. Microbiol. Biotechnol. 91:1131–1140 [DOI] [PubMed] [Google Scholar]
- 32.Zhang J. 2012. Genetic redundancies and their evolutionary maintenance. Adv. Exp. Med. Biol. 751:279–300 [DOI] [PubMed] [Google Scholar]
- 33.Kersten P, Cullen D. 2007. Extracellular oxidative systems of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Fungal Genet. Biol. 44:77–87 [DOI] [PubMed] [Google Scholar]
- 34.Kilaru S, Hoegger PJ, Kües U. 2006. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 50:45–60 [DOI] [PubMed] [Google Scholar]
- 35.MacDonald J, Suzuki H, Master ER. 2012. Expression and regulation of genes encoding lignocellulose-degrading activity in the genus Phanerochaete. Appl. Microbiol. Biotechnol. 94:339–351 [DOI] [PubMed] [Google Scholar]
- 36.Pezzella C, Autore F, Giardina P, Piscitelli A, Sannia G, Faraco V. 2009. The Pleurotus ostreatus laccase multi-gene family: isolation and heterologous expression of new family members. Curr. Genet. 55:45–57 [DOI] [PubMed] [Google Scholar]
- 37.Syed K, Yadav JS. 2012. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit. Rev. Microbiol. 38:339–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanden Wymelenberg A, Gaskell J, Mozuch M, BonDurant SS, Sabat G, Ralph J, Skyba O, Mansfield SD, Blanchette RA, Grigoriev IV, Kersten PJ, Cullen D. 2011. Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species. Appl. Environ. Microbiol. 77:4499–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]