Abstract
The phenylacetic acid (PAA) degradation pathway is a widely distributed funneling pathway for the catabolism of aromatic compounds, including the environmental pollutants styrene and ethylbenzene. However, bacterial chemotaxis to PAA has not been studied. The chemotactic strain Pseudomonas putida F1 has the ability to utilize PAA as a sole carbon and energy source. We identified a putative PAA degradation gene cluster (paa) in P. putida F1 and demonstrated that PAA serves as a chemoattractant. The chemotactic response was induced during growth with PAA and was dependent on PAA metabolism. A functional cheA gene was required for the response, indicating that PAA is sensed through the conserved chemotaxis signal transduction system. A P. putida F1 mutant lacking the energy taxis receptor Aer2 was deficient in PAA taxis, indicating that Aer2 is responsible for mediating the response to PAA. The requirement for metabolism and the role of Aer2 in the response indicate that P. putida F1 uses energy taxis to detect PAA. We also revealed that PAA is an attractant for Escherichia coli; however, a mutant lacking a functional Aer energy receptor had a wild-type response to PAA in swim plate assays, suggesting that PAA is detected through a different mechanism in E. coli. The role of Aer2 as an energy taxis receptor provides the potential to sense a broad range of aromatic growth substrates as chemoattractants. Since chemotaxis has been shown to enhance the biodegradation of toxic pollutants, the ability to sense PAA gradients may have implications for the bioremediation of aromatic hydrocarbons that are degraded via the PAA pathway.
INTRODUCTION
Aromatic hydrocarbons have been a major source of environmental contamination, and because of their toxicity and persistence in the environment, these pollutants remain a critical problem (reviewed in references 1 and 2). Understanding the catabolic pathways by which microorganisms can assimilate and mineralize these compounds has been important in the development of bioremediation strategies. The phenylacetic acid (PAA) degradation pathway has been studied as a funneling pathway for degradation of the toxic environmental contaminants styrene and ethylbenzene (3) and is present in 16% of completely sequenced bacterial genomes, many of which are members of the Proteobacteria, Actinobacteria, and Thermus/Deinococcus phylogenetic groups (4). PAA is also a naturally occurring plant metabolite that serves as a plant growth hormone (5), so it is likely to be common in the environment. In the first step of the aerobic bacterial PAA degradation pathway, phenylacetyl-coenzyme A (CoA) ligase converts PAA to phenylacetyl-CoA, which is a substrate for further degradation (Fig. 1) (4, 6–9). Interestingly, all of the intermediates in this pathway are CoA derivatives, which is more typical of anaerobic degradation strategies, and thus, the pathway is regarded as an aerobic/anaerobic hybrid pathway (4, 6, 10, 11).
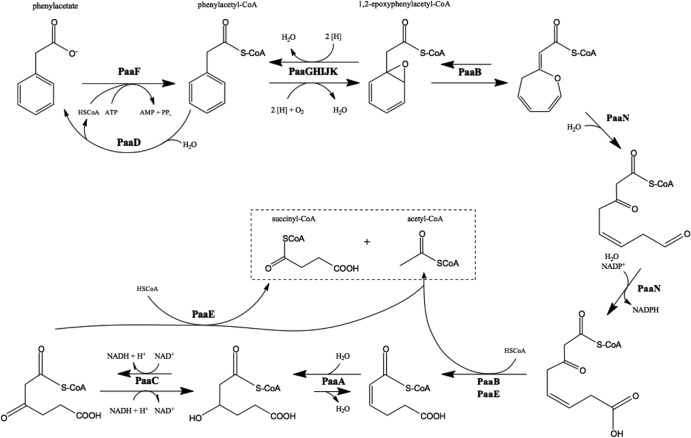
Fig 1.
PAA degradation pathway (4, 6). PaaF, phenylacetyl-CoA ligase; PaaD, acyl-CoA thioesterase; PaaGHIJK, ring 1,2-phenylacetyl-CoA epoxidase; PaaB, ring 1,2-epoxyphenylacetyl-CoA isomerase; PaaN, oxepin-CoA hydrolase/3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase; PaaE, 3-oxoadipyl-CoA/3-oxo-5,6-dehydrosuberyl-CoA thiolase; PaaA, 2,3-dehydroadipyl-CoA hydratase; PaaC, 3-hydroxyadipyl-CoA dehydrogenase.
The gene clusters necessary for PAA catabolism have been characterized in Escherichia coli and in several Pseudomonas strains, including Pseudomonas putida U and Pseudomonas sp. strain Y2 (6, 11–13). In this study, we identified a putative PAA degradation gene cluster (paa) in P. putida F1 with a gene organization similar to those of the paa2 gene cluster in Pseudomonas sp. Y2 and the pha gene cluster in Pseudomonas putida U. Although PAA degradation has been widely studied, no examples of chemotaxis have been reported for this compound. Since P. putida F1 has been studied for its ability to both degrade environmental pollutants such as aromatic hydrocarbons and exhibit chemotaxis toward these toxic chemicals (14), we were interested to determine whether P. putida F1 could sense PAA as an attractant.
Chemotaxis, the ability of motile bacteria to travel toward or away from specific chemicals (chemoeffectors), allows bacteria to actively seek an environment of optimal growth and survival. Chemoeffectors can be detected directly through physically binding to a chemoreceptor or indirectly by responding to changes in the intracellular energy levels created by that chemoeffector (reviewed in references 15, 16, 17). Chemotaxis has been shown to increase the biodegradation rate of pollutants, as it can improve bioavailability (18–20). Therefore, chemotaxis can enhance the removal of toxic pollutants and improve on current bioremediation strategies. The purpose of this study was to investigate the ability of Pseudomonas putida F1 to sense PAA via chemotaxis, which may have implications for the bioremediation of toxic aromatic compounds that are degraded by pathways that funnel into the PAA degradation pathway.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α λpir (21), which was used for cloning and plasmid propagation, and E. coli HB101(pRK2013) (22), which was used for plasmid mobilization in triparental matings, were grown at 37°C in lysogeny broth (LB) or on LB agar (23). For chemotaxis assays, P. putida F1 and its derivatives were grown in minimal medium (MSB) (24) at 30°C with 10 mM succinate, 5 mM succinate plus 5 mM PAA, or 5 mM PAA as a carbon source. E. coli RP437 and its derivatives (Table 1) were grown in MSB containing 5 mM PAA as the carbon source; 0.5 mM (each) methionine, leucine, histidine, and threonine and 1 μg/ml thiamine were provided to satisfy auxotrophic requirements. For plasmid selection and maintenance in E. coli and P. putida, antibiotics were added at the following concentrations: tetracycline at either 10 μg/ml or 20 μg/ml, kanamycin at 50 μg/ml, and gentamicin at 15 μg/ml.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference(s) |
|---|---|---|
| E. coli strains | ||
| DH5α λpir | Cloning host | 21 |
| HB101 | Host for mobilization plasmid pRK2013 | 23 |
| RP437 | Wild type for chemotaxis | 41 |
| UU1117 | RP437 Δaer1 | 42 |
| P. putida strains | ||
| F1 | Wild type | 61, 62 |
| F1 cheA mutant | F1 cheA::mini-Tn5; Kmr | 37 |
| XLF016 | F1 Δaer1 (locus tag Pput_3481) | This study |
| XLF019 | F1 Δaer2 (locus tag Pput_3628) | This study |
| RLF1 | F1 ΔpaaF (locus tag Pput_2480) | This study |
| RLF2 | F1 ΔpaaI (locus tag Pput_2483) | This study |
| Plasmids | ||
| pAW19 | sacB containing cloning vector, Apr, Kmr | 21 |
| pEX18Gm | sacB containing cloning vector, Gmr | 63 |
| pRK2013 | ColE1 ori, RP4 mobilization function, Kmr | 22 |
| pRK415 | Broad-host-range cloning vector, Tcr | 26 |
| pRK415Km | Broad-host-range cloning vector, Kmr | This study |
| pXLF219 | pRK415 containing aer2 | This study |
| pJPF1 | paaF deletion construct: 1-kb PCR fragments from upstream and downstream of paaF fused and cloned into pEX18Gm | This study |
| pJPF2 | paaI deletion construct: 1-kb PCR fragments from upstream and downstream of paaI fused and cloned into pEX18Gm | This study |
| pRLF1 | pRK415Km carrying paaF | This study |
| pRLF2 | pRK415Km carrying paaI | This study |
| pHRP309 | Broad-host-range lacZ transcriptional fusion vector, Gmr | 29 |
| pHRP310 | pK19 with Ω Smr/Spr cassette | 29 |
| pHRP311 | pHRP309 with Ω Smr/Spr cassette | 29 |
| pXLF016 | 1-kb PCR fragments from upstream and downstream of Gene Pput_3481 (aer1) fused and cloned into SpeI-SacI sites of pAW19 | This study |
| pXLF019 | 1-kb PCR fragments from upstream and downstream of Gene Pput_3628 (aer2) fused and cloned into SpeI-SacI sites of pAW19 | This study |
| pXLF219 | Gene Pput_3628 (aer2) from strain F1 cloned into HindIII-BamHI sites of pRK415, Tcr | This study |
| pVNF1 | pHRP310 containing cloned paaA promoter region | This study |
| pVNF2 | pHRP309 containing cloned paaA promoter region upstream of promoterless lacZ | This study |
Kmr, kanamycin resistance; Gmr, gentamicin resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance; Smr, streptomycin resistance; Spr, spectinomycin resistance.
DNA manipulations.
Standard methods were used for the manipulation of plasmids and DNA fragments (23). E. coli strains were transformed with plasmid DNA following standard procedures (23). Restriction endonucleases and DNA modification enzymes were purchased from New England BioLabs (Beverly, MA). Plasmids were purified using commercial kits purchased from Fermentas (Glen Burnie, MD), and genomic DNA was isolated using a 5′ ArchivePure DNA kit (5 Prime, Gaithersburg, MD). DNA fragments were purified by gel extraction using a Gene JET gel extraction kit from Fermentas.
The primers used in this study are shown in Table S1 in the supplemental material. Pfu high-fidelity polymerase with Pfu reaction buffer [200 mM Tris-Cl (pH 8.8), 100 mM (NH4)2PO4, 100 mM KCl, 1% Triton X-100, bovine serum albumin (BSA) (1 mg/ml), 20 mM Mg2SO4] and standard conditions (95°C denaturing, 55°C annealing, and 72°C elongation temperatures and an elongation time of approximately 1 min per kb of PCR product) were used for all reactions. All cloned PCR products were verified by fluorescent automated DNA sequencing at the University of California Davis Sequencing Facility with an Applied Biosystems 3730 automated sequencer.
E. coli HB101(pRK2013) was used to mobilize plasmids into P. putida F1 strains in triparental matings as previously described (25). Exconjugants were selected and isolated on MSB plates containing 10 mM succinate and the appropriate antibiotic. To select for deletion mutants that arose from double-crossover events, cells were grown on MSB containing 10 mM succinate and 20% sucrose. Individual colonies were then screened for loss of the plasmid based on antibiotic susceptibility, and deletions were verified by PCR using the appropriate primers (see Table S1 in the supplemental material).
pRK415Km construction.
In order to generate a variant of pRK415 (26) that encodes kanamycin resistance, the kanamycin resistance gene from pET-28b (EMD4Biosciences, San Diego, CA) was PCR amplified with primers pETKm_RsrII_for and pETKm_ApaI_Rev (see Table S1 in the supplemental material). The PCR product was digested with RsrII and ApaI and then ligated with similarly digested pRK415. The resulting plasmid was used to transform DH5α, and transformants were selected on LB plates containing 100 μg/ml kanamycin and screened for loss of tetracycline resistance. The presence of the kanamycin resistance gene was verified by restriction digestion.
Construction of PAA pathway mutants and complementation plasmids.
To construct the paaF in-frame deletion mutant RLF1, 1-kb regions upstream and downstream of the paaF gene (locus tag Pput_2480) were PCR amplified using primers listed in Table S1 in the supplemental material. The resulting PCR fragments were gel purified and were directionally cloned into HindIII- and EcoRI-digested pEX18Gm using an In-Fusion HD cloning kit (Clonetech, Mountain View, CA). The correct construct was verified by restriction digestion and DNA sequence analysis. Isolation of exconjugants and verification of the deletion mutant were carried out as described above. To complement the mutation, the wild-type paaF gene was amplified by PCR using primers paaF_For_HindIII and paaF_Rev_BamHI and then directionally cloned into dephosphorylated BamHI- and HindIII-digested pRK415Km to generate plasmid pRLF1. Construction of an in-frame paaI deletion mutant, RLF2, was carried out as described above, using primer pairs listed in Table S1 in the supplemental material to amplify 1-kb upstream and downstream fragments of the paaI gene (locus tag Pput_2483). To complement the mutant, the wild-type paaI gene was amplified by PCR using primers paaI_For_HindIII and paaI_Rev_BamHI and then directionally cloned into dephosphorylated BamHI- and HindIII-digested pRK415Km to generate plasmid pRLF2.
Construction of the energy taxis mutant XLF019.
Mutants lacking the putative energy taxis receptors aer1 and aer2 (locus tags Pput_3481 and Pput_3628, respectively), designated as they are in P. putida strain KT2440 (27), were constructed. To generate deletion constructs, 1-kb regions upstream and downstream of each gene were amplified by PCR using primers listed in Table S1 in the supplemental material. The resulting PCR fragments were fused by blunt-end ligation (28) and further amplified by PCR, to generate a 2-kb fragment with an in-frame deletion of the gene. Each fragment was cloned into pAW19 and sequenced. The resulting plasmids (Table 1) were introduced into E. coli DH5α λpir and mated into P. putida F1. Isolation of exconjugants and verification of the mutation were carried out as described above. The aer2 gene was cloned into pRK415 after amplification with 3628 HindIII-F and 3628 BamHI-R primers (see Table S1 in the supplemental material), forming plasmid pXLF219.
Construction of a paaA-lacZ transcriptional fusion.
A paaA-lacZ transcriptional fusion was constructed using primers paa_Prom_For and paa_Prom_Rev (see Table S1 in the supplemental material) to amplify the promoter region between the divergently transcribed paaY and paaA genes (the amplified fragment includes the first 16 bp of paaY and the first 173 bp of paaA). The resulting PCR fragment was digested and cloned into the KpnI-EcoRI sites of the cohort vector pHRP310 containing a Ω cassette to facilitate cloning (29), generating plasmid pVNF1. The correct construct was verified by restriction digestion and DNA sequence analysis. The plasmid was purified and digested with SalI and EcoRI, and the resulting fragment was cloned into the SalI-EcoRI site of the lacZ transcriptional fusion vector pHRP309 (29) to generate plasmid pVNF2. E. coli strain DH5α was transformed with pVNF2, and the resulting strain was mated with wild-type P. putida F1 as described above.
Soft-agar swim plate assays.
Soft-agar swim plates were used to measure chemotaxis in MSB soft-agar medium (30) containing 0.3% Noble agar and 5 mM succinate, 5 mM aspartate, or 1 mM PAA. In assays involving E. coli, the medium also contained 0.1 mM (each) methionine, leucine, histidine, and threonine and 1 μg/ml thiamine to satisfy auxotrophic requirements. Strains were harvested during the exponential phase (optical density at 660 nm [OD660], 0.3 to 0.4) after growth in MSB containing 5 mM PAA. Pellets were washed and resuspended in chemotaxis buffer (CB; 50 mM potassium phosphate buffer [pH 7.0], 0.05% glycerol, 10 μM EDTA) to an OD660 of approximately 0.4. Plates were inoculated by pipetting 2 μl of resuspended cells into the agar, and the cultures were incubated at 30°C. A ring of growth, which is indicative of a positive response, was observed with backlighting (31) approximately 24 h after inoculation. Photographs were taken using a Canon EOS Rebel T2i camera. The colony diameters were measured for quantitative analysis.
Gradient swim plate assays.
The gradient plate assay was adapted from an assay described by Pham and Parkinson (2011) (32). MSB soft-agar plates contained 0.3% Noble agar and 1 mM glycerol. Agar plugs containing 5 mM PAA in MSB and 2% Noble agar were made by pouring the medium into a sterile petri dish to a depth of 5 mm. After the medium solidified, the large end of a sterile 1,000-μl pipette tip was used to excise an agar plug, which was gently placed onto the center of the solidified MSB soft-agar plate. P. putida cell suspensions were generated as described for soft-agar swim plate assays. Plates were inoculated by pipetting 2 μl of resuspended cells 2 cm from the center of the agar plug. Plates were incubated at 30°C for 20 to 24 h, and photographs were taken when under observation using backlighting (31). A positive response is indicated by an oblong colony migrating toward the agar plug.
Chemical-in-plug assays.
Chemical-in-plug assays were performed as described by Storch et al. (33). P. putida strains were grown to the exponential phase (OD660, 0.4 to 0.5) in MSB containing either 10 mM succinate (uninduced) or 5 mM succinate plus 5 mM PAA (induced). Tetracycline was added to reach a concentration of 20 μg/ml when appropriate. Cells were harvested and resuspended to an OD660 of 0.4 in 2× chemotaxis buffer. The resuspended cells were mixed with an equal volume of a 0.5% (wt/vol) Noble agar solution at 40°C, and the cell suspension was poured into sterile petri dishes (35 by 10 mm). Agar plugs (described above) containing 5 mM PAA, 10 mM succinate, or no carbon source were placed into the center of the cell suspension. The plates were incubated at room temperature, and the formation of a ring of cells around the agar plug, indicating a positive chemotactic response, was observed using backlighting (31) after 1 h.
β-Galactosidase enzyme assays.
Cells were grown to an OD660 of between 0.45 and 0.55 in MSB containing 10 mM succinate (uninduced) or 5 mM succinate plus 5 mM PAA (induced), and assays were carried out as described by Miller (34).
RESULTS
Identification of a gene cluster for PAA degradation in P. putida F1.
The complete genome of P. putida F1 has been sequenced (GenBank accession number NC 009512). Orthologs of PAA degradation pathway genes were identified in P. putida F1 (locus tags Pput_2473 to Pput_2489) and annotated according to the experimentally characterized PAA degradation genes in P. putida U, Pseudomonas sp. Y2, and E. coli W (4, 6, 11–13). Organization of the paa gene cluster in P. putida F1 (paaXYABCDEFGHIJKPLMN) is similar to those in other Pseudomonas strains. However, orthologs of P. putida F1 genes paaD (locus tag Pput_2478) and paaP (locus tag Pput_2486), which encode an acyl-CoA thioesterase (35) and a putative membrane protein, respectively, are present in the second functional paa2 gene cluster of Pseudomonas sp. Y2 but not in P. putida U. Calculated percent amino acid identities of paa gene products from P. putida F1 are more similar to those from P. putida U (78% to 98% amino acid sequence identity). The predicted PAA pathway appears to be functional in P. putida F1, as this strain was able to utilize PAA as a sole source of carbon and energy, growing with a doubling time of 67 min.
P. putida F1 is attracted to PAA, and the response is inducible.
To test whether PAA is an attractant for P. putida F1 and whether or not the response requires induction, chemical-in-plug assays were used. Cells were pregrown with 10 mM succinate (uninduced) or with 5 mM succinate plus 5 mM PAA (induced). Both cultures exhibited a strong positive response to the positive-control attractant succinate, as shown by the accumulation of a ring of cells around the attractant-containing agar plug (Fig. 2). In contrast, no response was seen to the negative CB control. With PAA as the attractant, only cells that had been pregrown with PAA showed a positive response (Fig. 2). These results indicate that P. putida F1 exhibits taxis toward PAA and that the response is induced in the presence of PAA.
Fig 2.
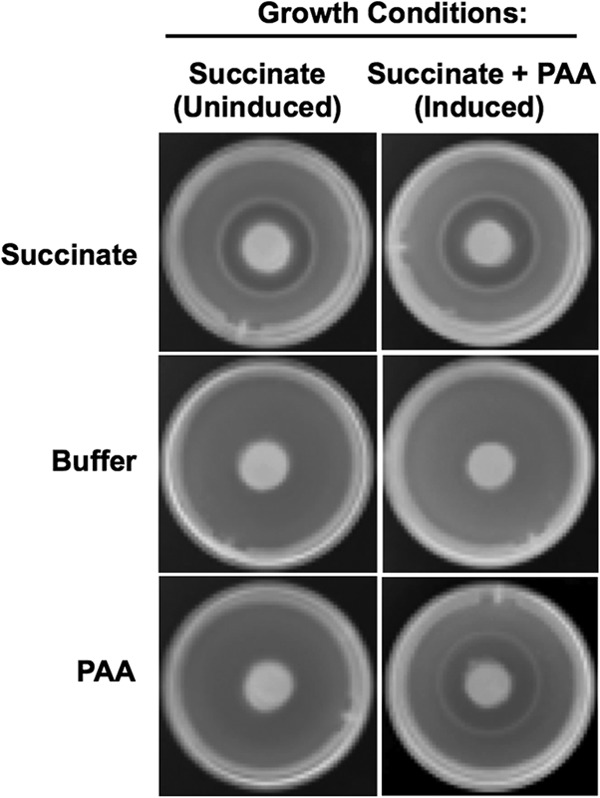
P. putida F1 response to PAA in chemical-in-plug assays. Wild-type P. putida F1 was pregrown in 10 mM succinate (uninduced) or in 5 mM succinate and 5 mM PAA (induced). Responses to 10 mM succinate (positive control), buffer (negative control), and 5 mM PAA are shown.
The PAA degradation genes in P. putida U were previously shown to be induced in response to PAA (8, 9). We verified that the paa gene cluster is inducible by PAA in P. putida F1 by monitoring expression from a paaA-lacZ transcriptional fusion. As expected, cells that were pregrown with PAA (induced) showed higher β-galactosidase activity than uninduced cells grown with succinate only (2,723.5 ± 328.3 Miller units in induced cells and 1,426.3 ± 343.6 Miller units in uninduced cells). These results show that degradation of and chemotaxis to PAA require induction by PAA in P. putida F1.
CheA is required for signal transduction in response to PAA.
To determine whether PAA is sensed through the conventional chemotaxis signaling pathway, we tested the response of a cheA mutant in soft-agar swim plates. CheA is a histidine kinase that initiates the phosphorylation of cytoplasmic chemotaxis proteins that control the direction of flagellar rotation in response to chemoeffectors (reviewed in reference 36). A P. putida F1 cheA mutant [strain F1(cheA)] that displays constant smooth swimming behavior and is unable to form chemotactic rings on LB swim agar plates (37) did not respond to PAA (Fig. 3A), indicating that taxis to PAA is mediated through the conserved chemotaxis signal transduction pathway.
Fig 3.
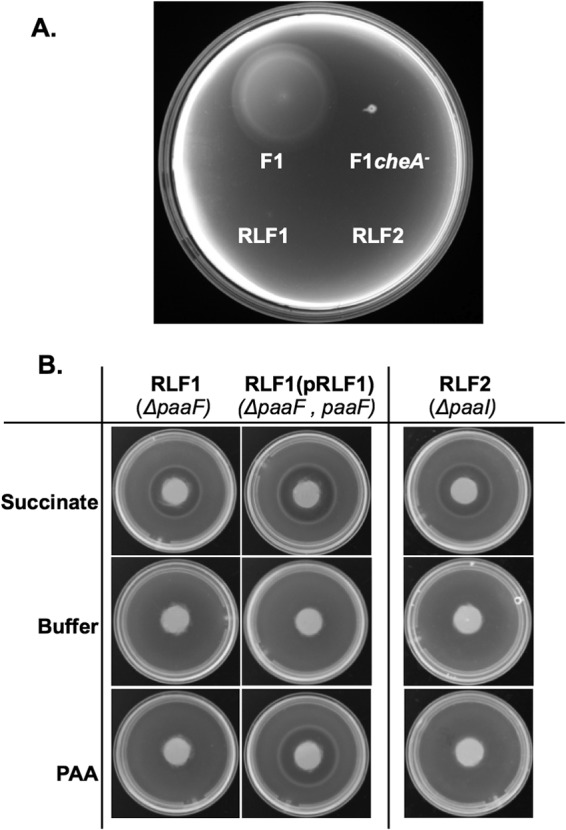
Responses to PAA by wild-type P. putida F1, a F1 cheA mutant, and PAA catabolic mutants. (A) Responses of wild-type P. putida F1, a F1 cheA mutant, RLF1 (ΔpaaF), and RLF2 (ΔpaaI) in MSB soft-agar swim plates containing 1 mM PAA. Plates were incubated at 30°C for approximately 24 h. (B) Chemical-in-plug assays showing responses of the PAA catabolic mutants RLF1(pRK415Km) (ΔpaaF), the complemented strain RLF1(pRLF1), and RLF2 (ΔpaaI) to 10 mM succinate (positive control), buffer (negative control), and 5 mM PAA. Photographs were taken after 1 h.
PAA catabolic mutants are deficient in taxis to PAA.
In order to examine the role of PAA metabolism in the chemotactic response, we generated mutants that were blocked at different steps of the PAA degradation pathway. P. putida RLF1 lacks a functional paaF gene, which encodes the phenylacetyl-CoA ligase that catalyzes the first step in the degradation pathway, converting PAA to phenylacetyl-CoA (4, 6–9). P. putida RLF2 lacks a functional paaI gene, which encodes one of the major subunits of the ring-hydroxylating complex (ring 1,2-phenylacetyl-CoA epoxidase) that catalyzes the conversion of phenylacetyl-CoA to 1,2-epoxyphenylacetyl-CoA (4, 11, 13). Mutant strains RLF1 and RLF2 were no longer able to grow on PAA as a sole carbon and energy source (Fig. 3A and data not shown). To evaluate the chemotaxis phenotypes of the catabolic mutants, we tested the responses of the P. putida ΔpaaF (RLF1) and ΔpaaI (RLF2) mutants in chemical-in-plug assays, which do not require growth of the cells on the attractant to visualize the chemotactic response. Strain RLF1(pRK415Km) was unable to respond to PAA, as indicated by the absence of a ring of cells around the PAA agar plug (Fig. 3B). Complementation of the ΔpaaF mutant [strain RLF1(pRLF1)] restored both the ability to grow on PAA (data not shown) and the chemotactic response to PAA (Fig. 3B). As expected, the RLF1(pRK415Km) and RLF1(pRLF1) strains responded equally well to succinate, and no response to the negative CB control was seen. Strain RLF2 (ΔpaaI) also did not respond to PAA (Fig. 3B); however, we were unable to complement RLF2 (data not shown). Because paaI encodes one subunit of a large enzyme complex, it is possible that providing paaI on a multicopy plasmid affected the stoichiometry of the components and prevented the formation of an active complex. Nevertheless, these results provide further evidence that PAA metabolism is required for the response to PAA.
aer2 (locus tag Pput_3628) encodes an aerotaxis/energy taxis receptor.
The observation that PAA taxis is metabolism dependent in P. putida F1 suggested that the response to PAA might be a form of energy taxis. To test this possibility, we needed to identify the energy taxis receptor in P. putida F1. Two methyl-accepting chemotaxis protein (MCP)-like receptors encoded in the P. putida F1 genome have high amino acid sequence identity to known aerotaxis receptors in other P. putida strains. The product of locus tag Pput_3481 (aer1) shares 94% amino acid sequence identity with the aerotaxis receptor in P. putida PRS2000 (38), and that of locus tag Pput_3628 (aer2) shares 99% amino acid sequence identity with the Aer2 aerotaxis/energy taxis receptor in P. putida KT2701 (27). To identify the energy taxis receptor in P. putida F1, the wild-type strain and the single-deletion mutants XLF016 (Δaer1) and XLF019 (Δaer2) were tested using MSB soft-agar swim plates containing succinate. The wild type exhibited a cone-shaped growth pattern with the largest ring at the bottom of the plate where oxygen conditions are expected to be most limiting (Fig. 4A). Strain XLF016 (Δaer1) had a wild-type phenotype; however, strain XLF019 (Δaer2) consistently formed a smaller ring and failed to form the cone-shaped growth pattern (Fig. 4A), which is consistent with the colony morphology of E. coli aer mutants (39). Introduction of plasmid pXLF219 carrying the wild-type aer2 gene from P. putida F1 into strain XLF019 restored the wild-type phenotype (Fig. 4B). Aerotaxis capillary assays (39) were carried out to confirm the phenotypes of the wild-type, mutant, and complemented strains (data not shown and reference 40). These results indicate that Aer2 is required for aerotaxis in P. putida F1.
Fig 4.
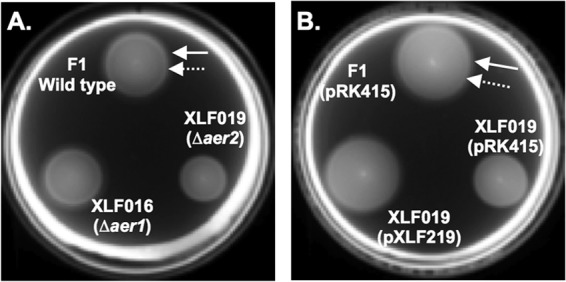
Soft-agar swim plate assays to identify the aerotaxis/energy taxis receptor in P. putida F1. (A) Wild-type P. putida F1 and mutant strains XLF016 (Δaer1) and XLF019 (Δaer2) in a MSB soft-agar plate containing 5 mM succinate. (B) Complementation of the Δaer2 defect. The images shows wild-type P. putida F1(pRK415), vector control strain XLF019(pRK415), and the complemented strain XLF019(pXLF219) in a MSB plate containing 5 mM succinate plus tetracycline. In both panels, F1 (wild type) is marked by solid arrows to indicate the cone-shaped growth pattern indicative of an energy taxis response (outer ring) and dashed arrows indicate the normal chemotaxis response phenotype (inner ring). Plates were incubated at 30°C for approximately 8 h. Growth studies demonstrated that strains had similar growth rates on succinate (data not shown), indicating that the defect in the response of strain XLF019 is solely due to a taxis defect.
The Aer2 energy taxis receptor in P. putida F1 mediates taxis to PAA.
To examine whether Aer2 mediated the metabolism-dependent response to PAA, strain XLF019 (Δaer2 mutant) was tested for its response to PAA in gradient plate, soft-agar swim plate, and chemical-in-plug assays. In gradient plate assays and soft-agar swim plate assays, the XLF019 aer2 deletion strain showed a markedly reduced response to PAA compared to the wild type (Fig. 5A and B). In addition, the characteristic cone-shaped growth pattern indicative of an energy taxis response (39) was absent for the XLF019 aer2 deletion strain. In chemical-in-plug assays, no visibly detectable response was observed (Fig. 5C). Complementation of XLF019 with aer2 cloned on a multicopy plasmid [strain XLF019(pXLF219)] restored the wild-type response in all three assays (Fig. 5). The results presented here demonstrate a key role of Aer2 in PAA chemotaxis and provide evidence that the response to PAA in P. putida F1 is mediated through energy taxis.
Fig 5.
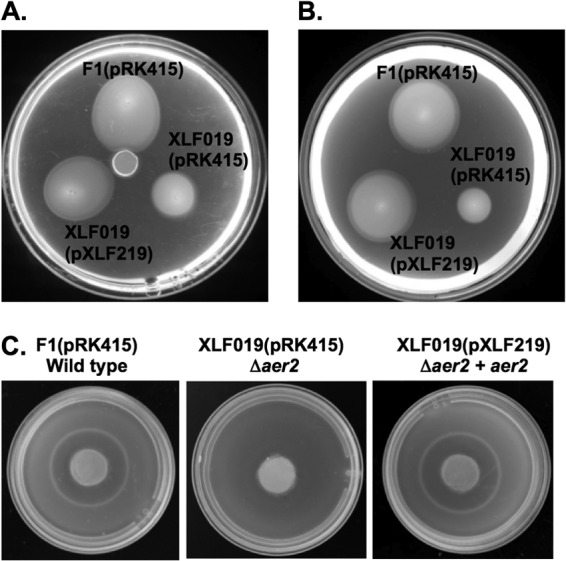
Responses of wild-type P. putida F1(pRK415), the Δaer2 mutant XLF019(pRK415), and the aer2 complemented strain XLF019(pXLF219) in (A) gradient plate, (B) soft-agar swim plate, and (C) chemical-in-plug assays. PAA was tested at a concentration of 1 mM in soft-agar swim plates; agar plugs containing 5 mM PAA were used in the gradient plate and chemical-in-plug assays. Tetracycline was included in the gradient plate and soft-agar swim plate assays. All experiments were replicated at least three times, and representative results are shown. Responses of strains F1 and XLF019 (Δaer2) lacking plasmids were also tested using all three assays, and the results were similar (data not shown). Growth studies with wild-type strain F1 and XLF019 on PAA demonstrated similar growth rates (data not shown), indicating that the defects in the response of strain XLF019 were solely due to a taxis defect.
E. coli is chemotactic to PAA, but Aer is not required for the response.
E. coli W is capable of PAA catabolism, and although the organization of the PAA gene cluster differs from that in P. putida strains, the catabolism of PAA proceeds through the same pathway (11, 13). We were interested in determining if E. coli exhibits chemotaxis to PAA and, if so, whether the response is also mediated through energy taxis. For this experiment, we tested the wild-type chemotaxis E. coli RP437 reference strain (41) and confirmed its ability to utilize PAA as the sole carbon and energy source (data not shown). We carried out quantitative soft-agar swim plate assays and measured colony diameters in 24 replicates from at least 3 independent experiments. RP437 displayed a positive taxis response to PAA (data not shown). To determine whether the response was mediated through energy taxis in E. coli, the E. coli UU1117 aer mutant strain (42) was tested. Measurements were normalized to 100% of the wild-type (RP437) response. No significant differences were seen in the responses of the aer mutant to PAA (100.8% ± 3.8%) or the positive-control attractant aspartate (99.6% ± 2.7%) compared to the wild type. These data suggest that Aer is not required for the response to PAA in E. coli.
DISCUSSION
Metabolism-dependent behavior is a characteristic of energy taxis, and the Aer protein in E. coli is the most well-studied energy taxis receptor (42, 43). The P. putida F1 Aer2 protein is 31% identical in amino acid sequence to the E. coli Aer protein, and their PAS (Per-Arnt-Sim) domains are 55% identical. The PAS domain in Aer contains a binding site for flavin adenine dinucleotide (FAD). During energy taxis, oxidation and reduction of FAD in response to changes in internal energy (i.e., rate of electron transport, redox, proton motive force, etc.) modulate signaling and cell behavior (reviewed in references 16, 17, 44, 45). The PAS domain of Aer2 in P. putida F1 contains 13 of the 17 amino acid residues identified to be important for signal transduction in E. coli Aer. Furthermore, 3 of the 4 residues responsible for FAD binding in E. coli Aer are conserved (45, 46). Aer homologs have been shown to function in energy taxis in diverse bacteria (43, 47, 48). The inducible metabolism-dependent response to PAA and the requirement for the PAS-domain-containing receptor Aer2 are consistent with an energy taxis response to this compound by P. putida F1. The ability of energy taxis receptors such as Aer2 to detect changes in intracellular energy levels and drive the movement of the cell in response is dependent on the metabolism of the chemoeffector (reviewed in references 16, 17, 44). Thus, it is expected that the degradation of PAA and, in effect, the resulting energy taxis response require a functional and induced paa degradation pathway.
The PAA degradation pathways in E. coli W and P. putida U are repressed in the absence of the inducer phenylacetyl-CoA, which is the first intermediate in the pathway (11, 13). The paa gene cluster in P. putida F1 encodes a putative repressor (PaaX) which is 37.3% and 88.6% identical to the functionally characterized E. coli W and P. putida U repressor proteins PaaX and PhaN, respectively. We showed that the paa genes in P. putida F1 are induced in the presence of PAA, and it is expected that PaaX controls their expression in P. putida F1.
Although bacterial taxis to PAA has not been previously reported, there is precedent for energy taxis in the response to certain aromatic compounds. The response to methylphenols by P. putida was shown to be mediated by energy taxis (27), as was a portion of the response to 2-nitrotoluene by Acidovorax sp. strain JS42 (49). However, chemotaxis to many aromatic compounds does not involve energy taxis. For example, although the receptor for toluene chemotaxis in P. putida F1 has not been identified, based on the analysis of catabolic mutants, the response is metabolism independent (14). In some cases, specific receptors dedicated to the detection of aromatic compounds have been identified. For example, the MCPs NahY, NbaY, and McpT detect naphthalene in P. putida G7 (50), 2-nitrobenzoate in P. fluorescens KU-7 (51), and toluene and other related chemicals in P. putida DOT-T1E (52), respectively. The mechanisms of 4-hydroxybenzoate detection in P. putida PRS2000 (30, 53–55) and 2,4-dichlorophenoxyacetate detection in Ralstonia eutropha JMP134(pJP4) (56) appear to be different, as chemotaxis to these compounds requires participation of the major facilitator superfamily (MFS) protein transporters PcaK and TfdK, respectively. Finally, recent studies have demonstrated that although phenol sensing in E. coli requires a functional MCP, the chemical is not sensed through the periplasmic ligand binding domain but rather through the transmembrane bundles and HAMP domain (32).
Unlike the results seen with specific receptors for aromatic compounds, the detection of PAA by Aer2 is indirect and nonspecific, as it senses the change in intracellular energy generated by catabolism rather than through direct binding of PAA. This may provide an advantage for the detection of other aromatic growth substrates that funnel into the PAA degradation pathway. The toxic pollutant styrene, for example, is degraded by a number of Pseudomonas strains such as Pseudomonas sp. strain Y2, P. fluorescens ST, and P. putida CA-3 (57–60). In these strains, styrene is converted into PAA through the styrene upper catabolic pathway (57–60). It would be interesting to examine whether styrene also serves as a substrate for energy taxis in chemotactic strains containing styrene and PAA catabolic pathways.
Our results indicate that PAA is an attractant for E. coli; however, we were unable to detect a role for Aer in the response to PAA. It remains unclear whether this was due to the sensitivity of the assay or if a different receptor is involved in E. coli chemotaxis to PAA. Currently, we are attempting to identify the receptor(s) responsible for PAA taxis in E. coli. We have not yet ruled out energy taxis as the sensory mechanism, since the Tsr chemoreceptor has also been shown to sense energy changes in E. coli (43). Differences in the mechanism of PAA sensing in E. coli and P. putida are interesting because comparisons of the PAA degradation gene clusters of E. coli and P. putida also show some functional and evolutionary differences (11, 13). The products of the paa genes in E. coli share 40% to 65% amino acid sequence identity with homologs in P. putida U (11). In addition, the gene organizations within the cluster differ between these two species. A major difference is the absence of a PAA transport system encoded by the paa gene cluster in E. coli. In order to grow on PAA, P. putida U requires phaJ and phaK, which encode a MFS permease and a specific outer membrane channel-forming protein, respectively (13). Homologs of these genes are also present in other Pseudomonas strains with a PAA degradation pathway, including Pseudomonas sp. strain Y2 and P. putida F1. The variations in PAA taxis mechanisms provide an interesting aspect of diversity in distantly related bacteria that carry a conserved and widely distributed catabolic pathway.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Science Foundation to R.E.P. and J.L.D. (MCB0919930).
We thank the Department of Energy, the Joint Genome Institute, and the IMG Informatics, especially Cheryl Kerfeld and Seth Axen, for their support of the IMG-ACT interface for student annotations. Finally, we thank the University of St. Thomas undergraduates enrolled in the 2010 BIOL464 bioinformatics class for identifying PAA as a chemoattractant and annotating the paa genes of P. putida F1.
Footnotes
Published ahead of print 1 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03895-12.
REFERENCES
- 1.Samanta SK, Singh OV, Jain RK. 2002. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol. 20:243–248 [DOI] [PubMed] [Google Scholar]
- 2.Haritash AK, Kaushik CP. 2009. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J. Hazard. Mater. 169:1–15 [DOI] [PubMed] [Google Scholar]
- 3.Luengo JM, García JL, Olivera ER. 2001. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol. Microbiol. 39:1434–1442 [DOI] [PubMed] [Google Scholar]
- 4.Teufel A, Mascaraque V, Ismail W, Voss M, Perera J, Eisentreich W, Haehnel W, Fuchs G. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. U. S. A. 107:14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrightman F, Lighty DL. 1982. Identification of phenylacetic acid as a natural auxin in the shoots of higher plants. Physiol. Plant. 55:17–24 [Google Scholar]
- 6.Ismail W, El-Said Mohamed M, Wanner BL, Datsenko KA, Eisenreich W, Rohdich F, Bacher A, Fuchs G. 2003. Functional genomics by NMR spectroscopy. Phenylacetate catabolism in Escherichia coli. Eur. J. Biochem. 270:3047–3054 [DOI] [PubMed] [Google Scholar]
- 7.Miñambres B, Martinez-Blanco H, Olivera ER, García B, Diez B, Barredo JL, Moreno MA, Schleissner C, Salto F, Luengo JM. 1996. Molecular cloning and expression in different microbes of the DNA encoding Pseudomonas putida U phenylacetyl-CoA ligase. J. Biol. Chem. 271:33531–33538 [DOI] [PubMed] [Google Scholar]
- 8.Schleissner C, Olivera ER, Fernández-Valverde M, Luengo JM. 1994. Aerobic catabolism of phenylacetic acid in Pseudomonas putida U: biochemical characterization of a specific phenylacetic acid transport system and formal demonstration that phenylacetyl-coenzyme A is a catabolic intermediate. J. Bacteriol. 176:7667–7676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez-Blanco H, Reglero A, Rodriquez-Aparicio LB, Luengo JM. 1990. Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. J. Biol. Chem. 265:7084–7090 [PubMed] [Google Scholar]
- 10.Vitovski S. 1993. Phenylacetate-coenzyme A ligase is induced during growth on phenylacetic acid in different bacteria of several genera. FEMS Microbiol. Lett. 108:1–5 [DOI] [PubMed] [Google Scholar]
- 11.Ferrández A, Miñambres B, García B, Olivera ER, Luengo JM, García JL, Díaz E. 1998. Catabolism of phenylacetic acid in Eschericia coli. J. Biol. Chem. 273:25974–25986 [DOI] [PubMed] [Google Scholar]
- 12.Bartolomé-Martín D, Martínez-García E, Mascaraque V, Rubio J, Perera J, Alonso S. 2004. Characterization of a second functional gene cluster for the catabolism of phenylacetic acid in Pseudomonas sp. strain Y2. Gene 341:167–179 [DOI] [PubMed] [Google Scholar]
- 13.Olivera ER, Miñambres B, García B, Muñiz C, Moreno MA, Ferrández A, Díaz E, García JL, Luengo JM. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. U. S. A. 95:6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parales RE, Ditty JL, Harwood CS. 2000. Toluene-degrading bacteria are chemotactic to the environmental pollutants benzene, toluene, and trichloroethylene. Appl. Environ. Microbiol. 66:4098–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazelbauer GI, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors; high-performance signalling in networked arrays. Trends Biochem. Sci. 33:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandre G, Zhulin IB. 2001. More than one way to sense chemicals. J. Bacteriol. 183:4681–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law AM, Aitken MD. 2003. Bacterial chemotaxis to naphthalene desorbing from a nonaqueous liquid. Appl. Environ. Microbiol. 69:5968–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marx RB, Aitken MD. 2000. Bacterial chemotaxis enhances naphthalene degradation in a heterogeneous aqueous system. Environ. Sci. Technol. 34:3379–3383 [Google Scholar]
- 20.Pandey G, Jain RK. 2002. Bacterial chemotaxis toward environmental pollutants: role in bioremediation. Appl. Environ. Microbiol. 68:5789–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White AK, Metcalf WW. 2004. The htx and ptx operons of Pseudomonas stutzeri WM88 are new members of the Pho regulon. J. Bacteriol. 186:5876–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Nat. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159–271 [DOI] [PubMed] [Google Scholar]
- 25.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (NY) 1:784–791 [Google Scholar]
- 26.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191–197 [DOI] [PubMed] [Google Scholar]
- 27.Sarand I, Osterberg S, Holmqvist S, Holmfeldt P, Skarfstad E, Parales RE, Shingler V. 2008. Metabolism-dependent taxis towards (methyl)phenols is coupled through the most abundant of three polar localized Aer-like proteins of Pseudomonas putida. Environ. Microbiol. 10:1320–1334 [DOI] [PubMed] [Google Scholar]
- 28.Adereth Y, Champion KJ, Hsu T, Dammai V. 2005. Site-directed mutagenesis using Pfu DNA polymerase and T4 DNA ligase. Biotechniques 38:864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parales RE, Harwood CS. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram− bacteria. Gene 133:23–30 [DOI] [PubMed] [Google Scholar]
- 30.Harwood CS, Nichols NN, Kim MK, Ditty JL, Parales RE. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkinson JS. 2007. A “bucket of light” for viewing bacterial colonies in soft agar. Methods Enzymol. 423:432–435 [DOI] [PubMed] [Google Scholar]
- 32.Pham HT, Parkinson JS. 2011. Phenol sensing by Escherichia coli chemoreceptors: a nonclassical mechanism. J. Bacteriol. 193:6597–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storch KF, Rudolph J, Oesterhelt D. 1999. Car: a cytoplasmic sensor responsible for arginine chemotaxis in the archaeon Halobacterium salinarum. EMBO J. 18:1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JH. 1975. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 35.Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Bugliano JA, Solorzano V, Wu J, Lima CD. 2006. Structure, function, and mechanism of the phenylacetate pathway hog dog-fold thioesterase PaaI. J. Biol. Chem. 281:11028–11038 [DOI] [PubMed] [Google Scholar]
- 36.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024–1037 [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Wood PL, Parales JV, Parales RE. 2009. Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J. Bacteriol. 191:2909–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols NN, Harwood CS. 2000. An aerotaxis transducer gene from Pseudomonas putida. FEMS Microbiol. Lett. 182:177–183 [DOI] [PubMed] [Google Scholar]
- 39.Taylor BL, Watts KJ, Johnson MS. 2007. Oxygen and redox sensing by two-component systems that regulate behavioral responses: behavioral assays and structural studies of aer using in vivo disulfide cross-linking. Methods Enzymol. 422:190–232 [DOI] [PubMed] [Google Scholar]
- 40.Liu X. 2009. Chemotaxis to pyrimidines and s-triazines in Pseudomonas and Escherichia coli. Ph.D. dissertation. University of California, Davis, Davis, CA [Google Scholar]
- 41.Parkinson JS, Houts SE. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bibikov SI, Biran R, Rudd KE, Parkinson JS. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. U. S. A. 94:10541–10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexandre G, Greer-Phillips S, Zhulin IB. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 28:113–126 [DOI] [PubMed] [Google Scholar]
- 45.Taylor BL. 2007. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 65:1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Repik A, Rebbapragada A, Johnson MS, Haznedar JÖ, Zhulin IB, Taylor BL. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweinitzer T, Josenhans C. 2010. Bacterial energy taxis: a global strategy? Arch. Microbiol. 192:507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexandre G. 2010. Coupling metabolism and chemotaxis-dependent behaviours by energy taxis receptors. Microbiology 156:2283–2293 [DOI] [PubMed] [Google Scholar]
- 49.Rabinovitch-Deere CA, Parales RE. 2012. Three types of taxis used in the response of Acidovorax sp. strain JS42 to 2-nitrotoluene. Appl. Environ. Microbiol. 78:2308–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grimm AC, Harwood CS. 1999. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 181:3310–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwaki H, Muraki T, Ishihara S, Hasegawa Y, Rankin KN, Sulea T, Boyd J, Lau PCK. 2007. Characterization of a pseudomonad 2-nitrobenzoate nitroreductase and its catabolic pathway-associated 2-hydroxylaminobenzoate mutase and a chemoreceptor involved in 2-nitrobenzoate chemotaxis. J. Bacteriol. 189:3502–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lacal J, Muñoz-Martínez F, Reyes-Darías JA, Duque E, Matilla M, Segura A, Calvo JJ, Jímenez-Sánchez C, Krell T, Ramos JL. 2011. Bacterial chemotaxis towards aromatic hydrocarbons in Pseudomonas. Environ. Microbiol. 13:1733–1744 [DOI] [PubMed] [Google Scholar]
- 53.Ditty JL, Harwood CS. 1999. Conserved cytoplasmic loops are important for both the transport and chemotaxis functions of PcaK, a protein from Pseudomonas putida with 12 membrane-spanning regions. J. Bacteriol. 181:5068–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ditty JL, Harwood CS. 2002. Charged amino acids conserved in the aromatic acid/H+ symporter family of permeases are required for 4-hydroxybenzoate transport by PcaK from Pseudomonas putida. J. Bacteriol. 184:1444–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nichols NN, Harwood CS. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179:5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hawkins AC, Harwood CS. 2002. Chemotaxis of Ralstonia eutropha JMP134(pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Appl. Environ. Microbiol. 68:968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beltrametti F, Marconi AM, Bestetti G, Colombo C, Galli E, Ruzzi M, Zennaro E. 1997. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl. Environ. Microbiol. 63:2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Conner K, Buckley CM, Hartmans S, Dobson ADW. 1995. Possible regulatory role for nonaromatic carbon sources in styrene degradation by Pseudomonas putida CA-3. Appl. Environ. Microbiol. 61:544–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Leary ND, O'Conner KE, Duetz W, Dobson ADW. 2001. Transcriptional regulation of styrene degradation in Pseudomonas putida CA-3. Microbiology 147:973–979 [DOI] [PubMed] [Google Scholar]
- 60.Velasco A, Alonso S, García JL, Perera J, Díaz E. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 180:1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finette BA, Subramanian V, Gibson DT. 1984. Isolation and characterization of Pseudomonas putida PpF1 mutants defective in the toluene dioxygenase enzyme system. J. Bacteriol. 160:1003–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibson DT, Hensley M, Yoshioka H, Mabry TJ. 1970. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry 9:1626–1630 [DOI] [PubMed] [Google Scholar]
- 63.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.