Abstract
Although molecular mechanisms promoting adherence of enterohemorrhagic Escherichia coli (EHEC) O157:H7 on epithelial cells are well characterized, regulatory mechanisms controlling biofilm formation are not fully understood. In this study, we demonstrate that biofilm formation in EHEC O157:H7 strain 86-24 is highly repressed compared to that in an isogenic hha mutant. The hha mutant produced large quantities of biofilm compared to the wild-type strain at 30°C and 37°C. Complementation of the hha mutant reduced the level of biofilm formation to that of the wild-type strain, indicating that Hha is a negative regulator of biofilm production. While swimming motility and expression of the flagellar gene fliC were significantly reduced, the expression of csgA (encoding curlin of curli fimbriae) and the ability to bind Congo red were significantly enhanced. The expression of both fliC and csgA and the phenotypes of motility and curli production affected by these two genes, respectively, were restored to wild-type levels in the complemented hha mutant. The csgA deletion abolished biofilm formation in the hha mutant and wild-type strain, and csgA complementation restored biofilm formation to these strains, indicating the importance of csgA and curli in biofilm formation. The regulatory effects of Hha on flagellar and curli gene expression appear to occur via the induction and repression of FlhDC and CsgD, as demonstrated by reduced flhD and increased csgD transcription in the hha mutant, respectively. In gel shift assays Hha interacted with flhDC and csgD promoters. In conclusion, Hha regulates biofilm formation in EHEC O157:H7 by differential regulation of FlhDC and CsgD, the global regulators of motility and curli production, respectively.
INTRODUCTION
Escherichia coli O157:H7 (referred to herein as O157) is the most frequent cause of sporadic and multiperson outbreaks of hemorrhagic colitis and hemolytic uremic syndrome (HUS) in humans (1). O157 uses diverse sets of adherence mechanisms to colonize intestinal epithelial cells and to produce biofilms on abiotic and biotic surfaces (2–5). Intimate adherence of O157 to bovine and human intestinal epithelial cells results in the formation of characteristic attaching and effacing (A/E) lesions and requires the LEE (locus of enterocyte effacement)-encoded type III secretion system and adherence factors (6).
On the other hand, formation of biofilms in both pathogenic and nonpathogenic E. coli strains requires differential and temporal expression of extracellular matrix components that facilitate a transition from an independent planktonic state to an organized multicellular community living within a highly structured extracellular matrix (7). One prominent component of this matrix is the adhesive fimbrial structures referred to as curli, which are thin, coiled, highly aggregative fibers of various lengths that protrude from the bacterial cell surface (8). Besides binding Congo red dye, the adhesive properties of curli enable E. coli to bind various host proteins, colonize animal tissues, activate the immune system, and produce biofilms on inanimate surfaces (8–13). Curli fimbriae are composed primarily of the protein curlin, encoded by the csgA gene of the csgBAC operon (14).
In addition to curli, flagella are required for the initial stages of biofilm formation in E. coli, as demonstrated by the inability of motility-deficient mutants to produce biofilms (15). However, motility is not required for biofilm formation by all E. coli strains, such as nonflagellated enteroaggregative E. coli strains that are as capable of producing biofilms like the flagellated wild-type strains on glass and plastic surfaces (16). The flagellar requirement is also bypassed in E. coli strains that overproduce curli or harbor conjugative F plasmids (17, 18).
In E. coli, flagellum-dependent swimming motility and curli-mediated adherence to solid surfaces during biofilm formation are regulated by a complex network of regulatory cascades. The csgD gene of the csgDEFG is a FixJ/LuxR family of transcriptional regulators that along with sigma factors RpoS-RpoE forms a regulatory cascade for activation of curli gene expression and biofilm production (19). The regulation of flagellar gene expression in E. coli is controlled by the master regulator FlhDC (class I flagellar operon) and RpoF cascade for activating the expression of class II and III flagellar genes (20–22). FliC, which represents the major structural protein of the E. coli flagellum, is encoded by the class III flagellar gene fliC (21, 22).
The expression of flhDC and csgD is controlled by a variety of physiological and environmental cues that activate or repress transcriptional regulators of the flhDC and csgD operons (19, 23–25). For example, flagellar genes are expressed under conditions favoring rapid growth and expression of curli is enhanced during slow growth or stationary phase, nutritional deprivation, low temperature, low osmolarity, and oxidative stress (26–30). Temperature was one of the first environmental cues recognized to affect the expression of global regulatory cascades that redirect bacterial cells to form biofilms by upregulating production of curli and extracellular polysaccharides along with bacterial cell surface-associated proteins and nucleic acids (19, 24, 30–36). The selective expression of desired sets of genes in response to temperature cues is a common theme among commensal and pathogenic bacterial species for conserving energy and ensuring their survival and persistence in a specific niche as well as transmission to a new host or locale (28, 29, 37). A recent study comparing the global gene expression profiles of E. coli K-12 grown at 37°C or 30°C showed elevated expression of metabolic pathways, such as uptake and utilization of amino acids, carbohydrates, and iron needed for rapid growth at 37°C (29). On the other hand, growth of E. coli at low temperatures (≤32°C) resulted in reduced expression of genes affecting energy metabolism and amino acid biosynthesis and uptake, gene expression changes linked to slow growth and biofilm formation (30, 33). A recent review summarized how bacterial pathogens use the mammalian host body temperature of 37°C as a cue for growth and to regulate the expression of virulence genes and to cause disease in infected hosts (37).
Biofilms have been shown to enhance persistence of food-borne bacterial pathogens, including O157, on meat-processing equipment and surfaces in animal slaughter and meat packing plants due to their ability to produce biofilms or be recruited into mixed-community biofilms in mammalian intestines (38, 39). The importance of curli as important virulence determinants in human disease has been confirmed by reports demonstrating that many clinical isolates of E. coli, including those isolated from sepsis patients showing anti-curli antibodies in their blood and from patients who had developed HUS due to infection with nonmotile O157, expressed curli at 28°C and 37°C and showed enhanced adherence to epithelial cells (9, 40). However, the regulatory networks enabling expression of curli and production of biofilms by O157 in response to environmental factors, such as temperature, and host factors are not fully understood. In a recent study, we reported that SdiA, a LuxR homologue, acts as a repressor of flagellar and curli gene expression, while Hha represses curli but induces flagellar gene expression (41). The hha mutant showed enhanced biofilm formation relative to that of the sdiA mutant, indicating that the reduced motility and increased curli production are essential for biofilm formation at 30°C. Hha, originally identified as a negative regulator of hemolysin expression in E. coli strains, has also been demonstrated to act as a negative regulator of LEE expression and LEE-mediated adherence of O157 to epithelial cells (42–44).
Since low temperatures (≤30°C) are considered the norm for bacterial biofilm production (8, 28, 30), the major objectives of the present study were to determine whether the hha mutant that produces increased amounts of biofilms at 30°C (41) would do the same at 37°C through increased transcriptional levels of csgA resulting in the increased production of curli fimbriae. In addition, we wanted to determine the importance of curli fimbriae to and correlate relative effects of reduced motility of the hha mutant due to reduced fliC expression (41) on biofilm formation at 37°C and 30°C. And finally, we were interested in determining if hha-mediated differential regulation of csgA and fliC, which determines the quantity of biofilm being produced, occurs through the direct transcriptional regulation by Hha of FlhDC and CsgD, the master regulators of fliC and csgA expression, respectively.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
Table 1 lists the bacterial strains and plasmids used in this study. Escherichia coli O157:H7 strain 86-24 (46) was used as the wild-type parent strain for constructing isogenic mutants. All E. coli strains were cultivated in Luria-Bertani broth (LB) or LB (Becton, Dickinson Companies, Sparks, MD) containing 1.5% agar (Sigma-Aldrich, St. Louis, MO). LB used contained 1% NaCl. YESCA broth (47, 48) was used for growth of bacterial strains to isolate RNA and for biofilm assays. YESCA agar (1.5% agar) was used to assess Congo red binding of bacterial strains. Swimming motility was determined on motility agar prepared by adding Noble agar (Becton, Dickinson) to YESCA broth at a final concentration of 0.33%. Media were supplemented with antibiotics at the following final concentrations: streptomycin, 100 mg liter−1; kanamycin, 50 mg liter−1; and carbenicillin, 100 mg liter−1. The dyes Congo red and Coomassie brilliant blue G-250 were used at 40 μg ml−1 and 15 μg ml−1, respectively.
Table 1.
Bacterial strains and plasmidsa
| E. coli strain or plasmid | Genotype and description | Source or reference(s) |
|---|---|---|
| Strains | ||
| E. coli O157:H7 strain 86–24 | stx2+ and streptomycin resistant | 46, 77 |
| 86–24 Δhha | 86–24 deleted of hha | 44 |
| 86–24 ΔcsgA | 86–24 deleted of csgA | This study |
| 86–24 Δhha ΔcsgA | 86–24 deleted of hha csgA | This study |
| TOP 10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG λ | Invitrogen |
| BL21 | F− ompT hsdS(rB− mB−) gal dcm | Amersham Biosciences |
| Plasmids | ||
| pKD46 | Recombineering vector | 49 |
| pCP20 | FLP recombinase vector | 51 |
| pCR2.1 | Cloning vector | Invitrogen |
| pSM197R | 44 | |
| pSM569 | csgA-complementing plasmid (pCR2.1 containing a cloned copy of csgBAC) | This study |
| pSM138 | Hha expression plasmid (pGEX4T-3 containing hha ORF) | 44 |
See Materials and Methods for a detailed description of bacterial strains and plasmids listed in this table.
Determination of bacterial generation times.
Bacterial strains were grown overnight with shaking (200 rpm) at 37°C in YESCA broth containing carbenicillin at 100 μg−1 ml (YESCA-Carb). The overnight cultures were diluted 1:100 in YESCA-Carb, and after the addition of 300 μl of the diluted suspension to the wells of a 120-well Honeycomb 2 plate, the plate was incubated at 37°C or 30°C in an automated growth curve reader programmed for continuous shaking and collecting optical density readings at 600 nm (OD600) every 30 min (Growth Curves USA, Piscataway, NJ). The optical density data were analyzed by using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA). The generation times of bacterial strains were estimated from the exponential portions of the growth curves and equaled the time required for the doubling of the OD600.
Recombinant DNA procedures.
The construction of the hha deletion mutant and the hha-complementing plasmid (pSM197R) has been described previously (44). A previously reported recombineering method was used for deleting the csgA gene in the wild-type and hha mutant strains to construct the csgA and hha csgA deletion mutants, respectively (49). Briefly, a 1.2-kb fragment containing the neomycin resistance gene (neo), which was flanked at its 5′ and 3′ ends with 45 and 46 bases of csgA upstream and downstream nucleotide sequences, respectively, was generated by PCR using a FailSafe PCR kit (Epicenter, Madison, WI). The nucleotides sequences of 5′ and 3′ primers used in this PCR were CGTGACACAACGTTAATTTCCATTCGACTTTTAAATCAATCCGATAGCTGAATGAGTGACGTGC and GTTACCAAAGCCAACCTGAGTTACGTTGACGGTGGAATTAGATGCATAGAGCAGTGACGTAGTCGC, respectively. The nucleotides in bold type represent sequences complementary to the nucleotide sequences located immediately at the 5′ and 3′ ends, respectively, of the csgA gene. The underlined nucleotides represent nucleotides complementary to the 5′ and 3′ ends, respectively, of the oBBI 92/93 neo cassette containing FRT (flippase recognition target) sequences for enabling a FLP (flippase recombination enzyme)-catalyzed deletion of neo (50). The PCR fragment was gel purified and electroporated into the arabinose-induced competent cells of the wild-type and isogenic hha mutant strains of O157 containing the plasmid pKD46 (49). Kanamycin-resistant transformants were screened by PCR for the replacement of csgA with the neo gene. The kanamycin-resistant csgA deletion mutant (csgA mutant) was transformed with plasmid pCP20 encoding FLP recombinase (51) to facilitate deletion of the neo gene when the csgA mutant was grown at 37°C. For complementation of the csgA deletion mutation, the 1.5-kb DNA fragment containing the csgBAC operon was isolated by PCR using primers csgBACF and csgBACR (Table 2). The PCR-amplified DNA was resolved in a 1% agarose gel by a standard DNA gel electrophoresis technique, and the 1.5-kb DNA fragment was excised from the gel and purified with a gel extraction kit according to the manufacturer's recommended protocol (Qiagen Inc., Valencia, CA). The purified 1.5-kb fragment was cloned in pCR2.1 TOPO cloning system using E. coli TOP10 electrocompetent cells and the manufacturers' recommended protocol (Invitrogen, Carlsbad, CA). The pCR2.1 plasmid carrying csgBAC was named pSM569 (Table 1).
Table 2.
Primers used for PCR and QRT-PCR
| Primerc | Nucleotide sequencea | Locationb |
|---|---|---|
| fliCF (RT-PCR) | TTAGCTGCCACCCTTCATG | 2700535–2700517 |
| fliCR (RT-PCR) | TCGTCAAGTTGCCTGCATC | 2700385–2700403 |
| fliCP (RT-PCR) | TCTACGTATGCCTGGCTTCCACCG | 2700405–2700428 |
| csgAF (RT-PCR) | TACTATTACCCAGCATGGTGG | 1548878–1548898 |
| csgAR (RT-PCR) | CAAGAGTGGCGCTGTTACC | 1548984–1548966 |
| csgAP (RT-PCR) | CCACGTTGGGTCAGATCGATTGAG | 1548962–1548938 |
| flhDCF(RT-PCR) | ACAACATTAGCGGCACTGAC | 2655207–2655188 |
| flhDCR (RT-PCR) | AGAGTAATCGTCTGGTGGCTG | 2655107–2655124 |
| flhDCP (RT-PCR) | AAACGGAAGTGACAAACCAGCTGATTG | 2655131–2655158 |
| csgDF(RT-PCR) | CGCTGGCAATTACAGGAAAATTAC | 1547324–1547301 |
| csgDR (RT-PCR) | CTTTTTATCCGCTTCCATCATATCC | 1547224–1547248 |
| csgDP (RT-PCR) | ATATTCAACGTTCTCTGGACGATATCTCTTCAGGCT | 1547297–1547262 |
| rpoAF (RT-PCR) | GGCTTGACGATTTCGACATC | 4242887–4242906 |
| rpoAR (RT-PCR) | GGTGAGAGTTCAGGGCAAAG | 4242997–4242978 |
| rpoAP (RT-PCR) | TGAAGTTATTCTTACCTTGAATAAATCTGGCATTG | 4242976–4242942 |
| csgBACF | AACAAAAAAAGAAAAATACAACGC | 1548016–1548039 |
| csgBACR | GATCTCTAGATCTGAAGAGGGCGGCCATTG | 1549511–1549492 |
| flhDCF(promoter) | GATCGTCGACGTGCGCAACATCCCATTTCG | 5655748- 5655729 |
| flhDCR(promoter) | GATCAGATCTGTATGCATTATTCCCACCCAG | 5655349–5655329 |
| csgDF (promoter) | GATCAGATCTTGAAACCCCGCTTTTTTTATTG | 1547424–1547445 |
| csgDR (promoter) | GATCGTCGACAATGTACAACTTTTCTATCATTTC | 1547694–1547671 |
Nucleotide sequence of E. coli primers used in this study were selected from the published genome sequence of E. coli O157:H7 strain EDL933 with the accession number AE005174.2.
Location refers to the position of primer sequence in the genome of EDL933.
Subscripts F, R, and P indicate forward primers, reverse primers, and TaqMan probes; “RT-PCR” denotes primers used in reverse transcriptase-based quantitative PCR.
Quantitative reverse transcriptase PCR (QRT-PCR).
Bacterial strains grown overnight with shaking (200 rpm) at 37°C in LB broth containing carbenicillin at 100 μg ml−1 (LB-Carb) were diluted 1:100 into fresh YESCA-Carb and incubated at 30°C or 37°C until the culture A600 was 1.3 to 1.4 (5 to 5.5 h). One milliliter of bacterial culture was mixed with 2 ml of RNA Protect solution and processed for RNA isolation using the RNeasy minikit (Qiagen Inc., Valencia, CA). RNA was treated with Turbo DNase for DNA removal (Applied Biosystems/Ambion, Austin, TX). QRT-PCR was performed by adding 50 ng of RNA, 0.75 μM (each) antisense and sense primers, 0.25 μM TaqMan probe (labeled at the 5′ and 3′ends with FAM reporter and TAMRA quencher dyes, respectively) (Table 2) to QRT-PCR Master Mix (Agilent Technologies, Inc., Santa Clara, CA). The reaction mixtures were incubated in a Mx3005P (Agilent Technologies, Inc., Santa Clara, CA) for cDNA synthesis (50°C for 30 min), amplification, and real-time detection of amplified products (95°C for 10 min; 35 cycles of 95°C for 30 s, 55°C for 60 s, 72°C for 30 s) specific for fliC, csgA, flhD, and csgD. The expression of each of the four genes in the mutant strain was compared to the expression of the same genes in the wild type and the mutant strain complemented for the deleted gene using the Mx3005 software. The expression data were normalized to endogenous levels of rpoA in order to account for any minor variations in the amounts of RNA across samples. The expression data for each target gene in the mutant strains were plotted as relative expression by adjusting the expression of the same gene in the wild-type calibrator strain to 1.
Quantification of biofilm formation.
The bacterial cultures grown overnight at 37°C with shaking (200 rpm) in YESCA-Carb were diluted 1:100 in fresh YESCA-Carb. To quantify total biofilm biomass, 200 μl of the diluted culture was added into the wells of 96-well, round-bottom microtiter plates (Immulon, 2HB; Thermo Scientific, Milford, MA). Inoculating four independently grown cultures into 6 wells each resulted in a total of 24 wells being analyzed for the production of biofilms per strain. To determine the proportion of viable cells present in biofilms, 2 ml of 1:100 diluted overnight cultures prepared in YESCA-Carb from three independently grown cultures was added to 13-ml disposable plastic tubes with snap caps. After 48 h of incubation at 30°C or 37°C, the growth medium was aspirated, and plates or tubes were rinsed with phosphate-buffered saline (PBS, pH 7.3). The biofilms produced in the plates were heat fixed (30 min at 80°C), stained with 0.1% crystal violet for 30 min at 25°C, and washed three times with PBS. The wells were filled with 200 μl of 95% ethanol, and absorbance at 590 nm was measured to quantify the total biofilm mass (48). The biofilms produced in 13-ml tubes were not subjected to heat fixation and were scraped off into 1 ml of PBS with a sterile wooden applicator, vortexed vigorously, and diluted in 10-fold serial dilutions in PBS, and 100-μl aliquots of these dilutions were plated on LB agar containing carbenicillin (100 μg ml−1). The agar plates were incubated at 37°C for 24 h, and bacterial colonies produced on these plates were enumerated.
Determination of Congo red binding by bacterial cells.
Overnight cultures of bacterial strains grown in LB-Carb were streaked on Congo red agar plates. After incubation for 36 to 48 h at 30°C or 18 to 24 h at 37°C, plates were photographed.
Determination of bacterial motility.
For swimming motility assays, overnight bacterial cultures grown in LB-Carb were centrifuged at 10,000 × g for 2 min to pellet bacterial cells. The cell pellets were resuspended in YESCA broth at one-tenth the original volume of the culture, and 2 μl of this suspension was spotted on motility plates (YESCA broth containing 0.33% Noble agar and carbenicillin at 100 μg ml−1). The diameters of motility halos were determined after incubation for 24 to 36 h at 30°C and 12 to 16 h at 37°C.
Gel shift assays.
For gel shift assays, DNA fragments containing promoter sequences for flhDC and csgD were amplified by PCR using primers (Table 2) and an AmpliTaq Gold DNA polymerase-containing kit according to the manufacturer's instructions (Life Technologies, Grand Island, NY). The PCR amplified fragments were resolved on 2% agarose gels by electrophoresis and extracted from the gel using a Qiagen gel extraction kit according to the manufacturer's recommended protocol (Qiagen Inc., Valencia, CA). Each gel-purified fragment (100 ng) was incubated in a 20-μl reaction volume containing excess of poly(dI-dC) and 2-fold-increasing amounts (0 to 4 μg) of Hha and a 1× reaction buffer (44). After 20 min at 25°C, the promoter-Hha mixtures were analyzed on a 6% nondenaturing polyacrylamide gel (44). The gel was stained for 20 min in Sybr green diluted to 1× per the instructions of the manufacturer (Molecular Probes). The stained gel was visualized using a charge-coupled device (CCD) camera equipped with a 510-nm filter to capture Sybr green fluorescence.
Statistics.
Statistical analysis of the data for the biofilm production, motility zones, and gene expression was performed by using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). The statistical significance of the differences in the sample means of the mutant and wild-type strain was calculated by using the unpaired Student's t test. Results were considered significant at a P value of <0.05.
RESULTS
The generation times of mutant bacterial strains were similar to those of the wild-type bacterial strain.
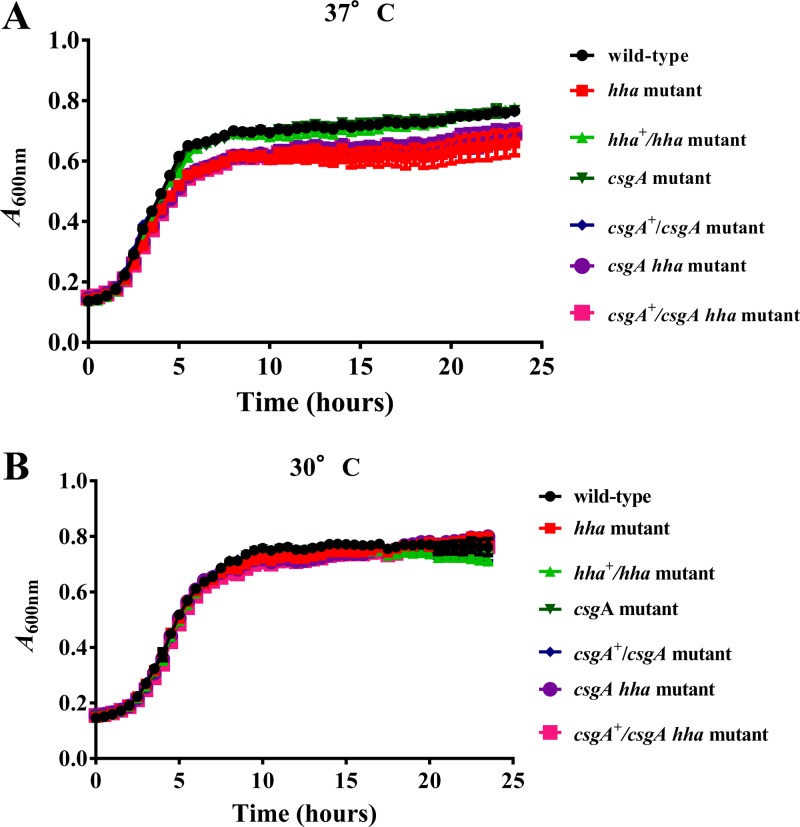
Reduced bacterial growth in response to specific environmental and nutritional factors, such as suboptimal growth temperatures and limiting nutrients, and due to genetic aberrations has been linked to increased biofilm formation (23, 26, 27, 36). Although temperatures between 26°C and 30°C are considered conducive to biofilm formation, many E. coli and E. coli O157 strains are capable of producing biofilms at 37°C (9, 12, 40). Thus, in order to determine the molecular basis of hha regulation of biofilm formation at 37°C (body temperature of the human host) and 30°C, we first ascertained that wild-type O157 and its isogenic hha, csgA, and hha csgA mutants would have similar growth rates at 37°C and 30°C. For this, 1:100 dilutions of overnight (37°C) bacterial cultures were cultivated in YESCA-Carb at 37°C or 30°C for 24 h with continuous shaking in an automated growth reader. This instrument is suitable for analyzing growth kinetics of large numbers of bacterial strains and has excellent sensitivity to detect differences in bacterial growth kinetics. However, the major limitation of this growth analyzer is that the generation times computed from the growth curves produced by this system cannot be compared to the generation times calculated from growth curves resulting from bacterial growth in large-volume flasks subjected to vigorous shaking. In other words, the OD600 of bacterial cultures in the microtiter plate wells of the growth reader increases by much smaller increments than that of cultures grown in larger vessels with a several-times air-to-culture volume ratio. In addition, small culture volumes that are confined to the wells of a microtiter plate are not conducive to achieving the high cell densities that are easily attained in large-volume containers over a shorter time (data not shown). The main purpose, therefore, of using this instrument was to determine if there are substantial differences in the growth kinetics of the strains that we used in this study, and the data that we obtained clearly showed that the generation times for each of the seven bacterial strains were fairly similar to each other. The generation times at 37°C ranged from 82 to 97 min and were shorter than the generation times for these strains at 30°C (112 to 128 min) (Fig. 1). The generation times of bacterial strains carrying the hha deletion were slightly longer at both 37°C (97 min) and 30°C (128 min) than those of the hha-positive strains (Fig. 1).
Fig 1.
Determination of bacterial generation times at 37°C and 30°C in YESCA broth. Bacterial strains were grown for 24 h in YESCA broth, and optical density at 600 nm (OD600) was recorded every 30 min. Three independent cultures were tested in triplicate, and each point on the growth curve represents an average of nine readings. The generation times were determined by computing the time required for doubling of the optical density of the bacterial culture during the exponential phase of growth.
Biofilm formation was enhanced and bacterial motility was reduced in hha mutant strains at 37°C and 30°C, but the expression of the flagellar gene fliC decreased only at 37°C.
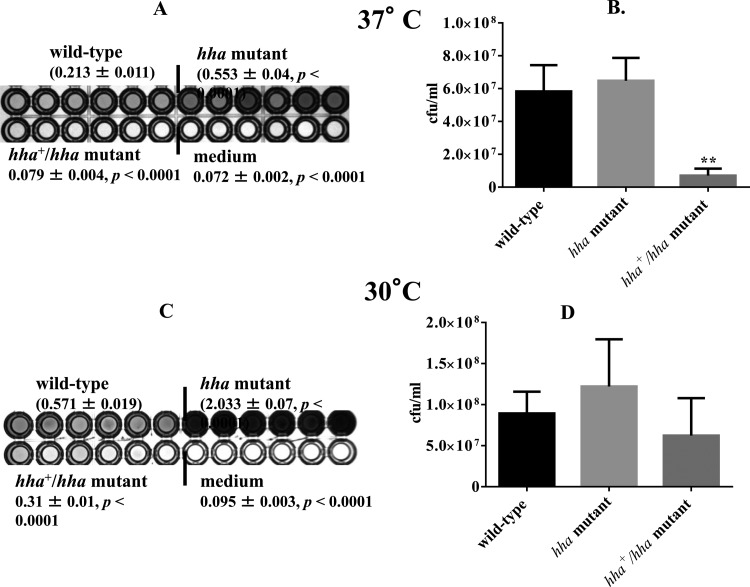
The total biofilm mass produced by the hha mutant was 2.5-fold higher (P < 0.0001) than that of the wild-type strain at 37°C, as indicated by the higher intensity of the blue color produced in the wells inoculated with the mutant strain (Fig. 2A). The mean intensity of the blue (0.079) color for the hha mutant complemented with a plasmid-cloned hha (pSM197R) was equivalent to that produced in the medium control wells, indicating that increased expression of hha from pSM197R (a multicopy plasmid) exerts a negative effect on biofilm formation (Fig. 2A). The proportions of viable cells recovered from the biofilms produced by the hha mutant were about 1.1-fold higher (P = 0.305) than those recovered from the biofilms produced by the wild-type strain (Fig. 2B). The numbers of viable bacterial cells in the biofilms produced by the hha-complemented strain (hha+/hha mutant) were 8-fold (P < 0.0022) and 9.1-fold (P < 0.0022) lower than those of the wild-type and hha mutant strains, respectively (Fig. 2B). On the other hand, the largest amount of total biofilm mass was produced by the hha mutant after 48 h of growth at 30°C, as indicated by the development of the dark blue color in the wells inoculated with this strain (Fig. 2C). The biofilm mass produced by the hha mutant at 30°C was 3.5-fold and 6.5-fold (P < 0.0001) higher than those of the wild-type and hha+/hha mutant strains, respectively (Fig. 2C). The hha+/hha mutant was a faint shade of blue whose intensity was approximately 2-fold lower (P < 0.0001) than that of the wild-type strain (Fig. 2C). The numbers of viable cells recovered from the biofilms produced by the hha mutant at 30°C were about 1.4-fold higher (P = 0.387) than those recovered from the wild-type strain (Fig. 2D). The hha+/hha mutant showed recovery of 0.7-fold (P = 0.238) and 2-fold (P = 0.132) fewer viable cells than the wild-type and hha mutant strains, respectively (Fig. 2D). The medium control wells appeared white.
Fig 2.
Effect of hha on the biofilm formation and on the proportion of viable bacterial cells recovered from biofilms at 37°C and 30°C after 48 h of growth in YESCA broth. Total biofilm masses produced by the wild type, hha mutant, and the hha-complemented mutant (hha+/hha mutant) at 37°C (A) and 30°C (C) are given as averages ± standard errors of the means (SEM) for 24 replicate wells from four independent cultures. Only 6 of 24 wells are shown for each strain. Significant (P < 0.05) differences in biofilm biomass between the wild-type and the mutant were determined using an unpaired Student's t test (Mann-Whitney test). The proportion of viable bacterial cells recovered from biofilms are shown as bars representing average CFU ml−1 of three independent cultures of the wild-type, hha mutant, and hha+/hha mutant grown at 37°C (B) and 30°C (D). Error bars represent the SEM. Significant differences between the wild-type and mutant strains were calculated as described above. **, P < 0.005.
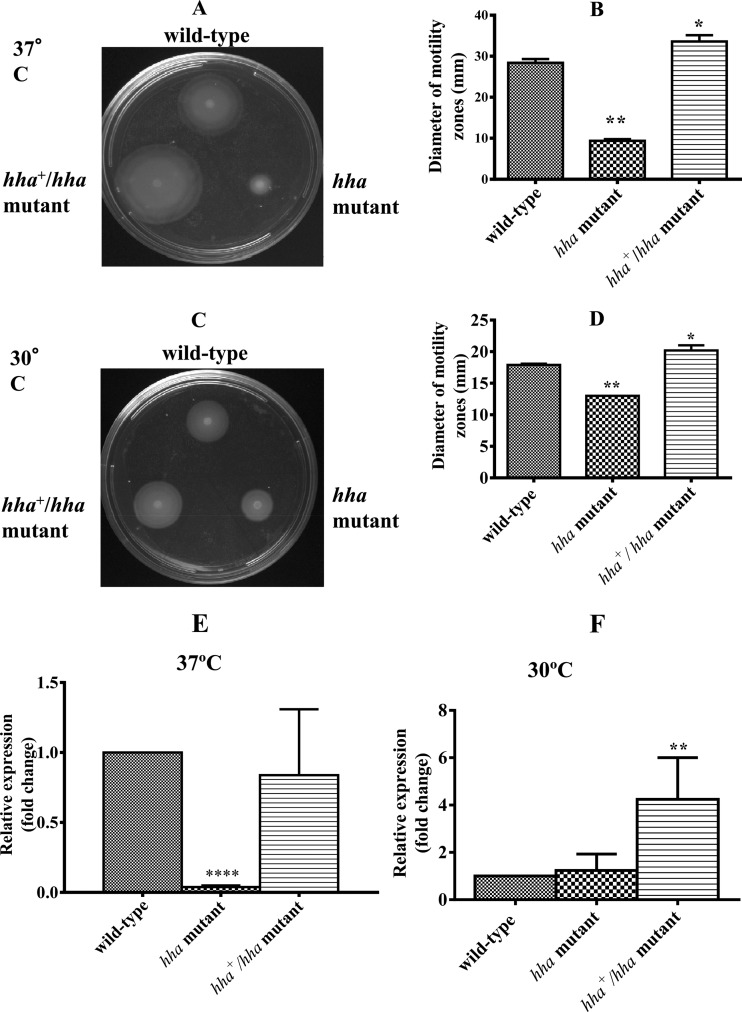
Bacterial motility has also been shown to be an important factor in the transition from a planktonic state to a sedentary state, as a prerequisite for biofilm formation, and in segregation and dispersal of mature biofilms (15). Determination of the relative motility of bacterial strains on motility plates showed that the hha mutant produced much smaller motility zones than the wild-type strain, and the motility zones produced by the hha-complemented hha+/hha strain were larger than those of the wild-type strain (Fig. 3A and C). Based on the measurements of the diameters of the motility zones produced at 37°C, motility zones of the hha mutant were 3.0-fold (P = 0.0079) smaller than that of the wild-type strain. The motility zones of the complemented hha+/hha mutant were 1.2-fold larger than that of the wild-type strain, presumably due to the hha dosage effect from pSM197R (Fig. 3B). The complementation of the motility deficiency in the hha mutant by pSM197R confirmed that hha is a positive regulator of motility in O157 (41). The mean diameter of the motility zones produced at 30°C by the hha mutant was about 1.4-fold smaller (P = 0.0079) than that of zones produced by the wild-type strain (Fig. 3D). The motility zones produced by the complemented hha+/hha mutant were about 1.1-fold (P = 0.0317) and 1.5-fold (P = 0.0079) larger in diameter than those of the wild-type and hha mutant strains, respectively (Fig. 3D).
Fig 3.
Effect of hha on bacterial motility and flagellar gene (fliC) expression at 37°C and 30°C. (A and C) The motility of wild-type, hha mutant, and hha+/hha mutant strains was determined by spotting 10-fold-concentrated overnight bacterial cultures on 0.33% soft agar plates. The diameter of the motility zone produced around the point of bacterial inoculation was measured after incubation for 12 to 16 h at 37°C (A) or 24 to 36 h at 30°C (C). (B and D) Diameters of swimming or motility zones of five independent bacterial cultures grown overnight at 37°C (B) and 30°C (D). (E and F) The expression of fliC was determined by QRT-PCR, and DNA-free RNA was prepared from the wild-type, hha mutant, and hha+/hha mutant strains grown at 37°C (E) and 30°C (F). Bars represent the average relative expression of fliC determined using RNA from three independent bacterial cultures. Error bars represent the SEM. Significant (P < 0.05) differences in motility between the wild-type and mutant strains were determined using an unpaired Student's t test (Mann-Whitney test). *, P = 0.0317; **, P = 0.0079. For fliC gene expression, significant (P < 0.05) differences between the wild-type and mutant strains were determined using an unpaired Student's t test and Welch's correction. **, P = 0.0063; ****, P < 0.0001.
Since increased biofilm formation correlated with reduced motility in the hha mutant strain at both 37°C and 30°C, and complementation of hha mutant with pSM197R restored its motility to the wild-type levels, we examined whether the expression of fliC was equally affected at 37°C and 30°C. The QRT-PCR analysis of RNA prepared from bacterial strains grown at 37°C showed a 26-fold (P < 0.0001) reduction in the fliC gene expression in the hha mutant compared to the wild-type strain (Fig. 3E). The complementation with pSM197R restored the expression of fliC in the hha mutant to the wild-type levels (P = 0.611) (Fig. 3E). The QRT-PCR analysis of bacterial strains grown at 30°C showed no significant difference in the fliC gene expression between the hha mutant and the wild-type strain (P = 0.436) (Fig. 3F). However, complementation with pSM197R increased fliC expression in the hha mutant strain 4-fold compared to expression in the wild-type strain (P = 0.0063) (Fig. 3F). Thus, the presence of multiple copies of hha enhanced fliC expression at 37°C and 30°C.
Increased biofilm formation by the hha mutant resulted from increased expression of curli independent of growth temperature.
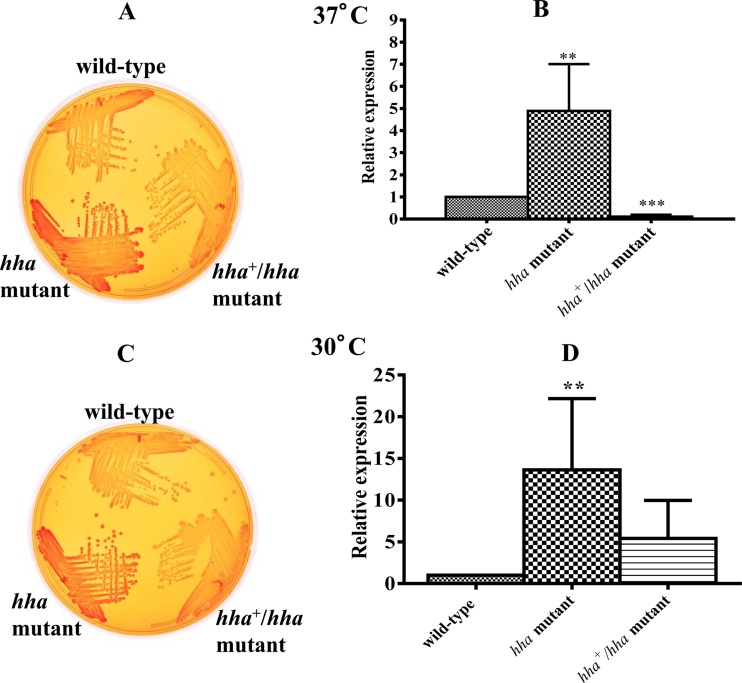
Among the cell surface components, curli fimbriae play an important role in the formation of biofilms (8, 28). Although the temperature range of 26°C to 30°C is considered optimal for the expression of curli fibers (28), many commensal and pathogenic strains of E. coli, including O157:H7 and O157:NM serotypes, are capable of expressing and assembling curli fibers on their cell surfaces at 37°C (9, 12, 40). Phenotypic expression of curli fimbriae on bacterial cell surfaces can be inferred from the intensity of the red color of the colonies produced on Congo red indicator plates (47, 48). The bacterial colonies produced by the hha mutant were dark red following 24 h of growth at 37°C (Fig. 4A) or 48 h of growth at 30°C (Fig. 4C), compared to the light red colonies of the wild-type strain. The hha mutant complemented with pSM197R produced colonies that had slightly weaker red coloration than the wild-type strain at both temperatures, presumably due to the increased production of hha from pSM197R (Fig. 4A and C). Since Congo red binding provides a qualitative assessment of the expression of curli fimbriae at the bacterial cell surface, we next determined by QRT-PCR whether the increased Congo red binding by the hha mutant strain correlated with the increased expression of csgA, the gene encoding curlin of curli fimbriae. QRT-PCR analysis showed that in the hha mutant, the expression of csgA increased about 5-fold when the strain was grown at 37°C (Fig. 4B) and about 14-fold (Fig. 4D) (P = 0.002) when the strain was grown at 30°C, compared to expression in the wild-type strain. The complementation of the hha mutant with pSM197R caused significant reduction (about 8-fold) (P = 0.0098) in csgA expression in cultures grown at 37°C (Fig. 4B). In the hha+/hha mutant grown at 30°C, the expression of csgA was reduced about 5-fold (P = 0.0022) compared to that in the hha mutant strain (Fig. 4D) but did not decrease completely to the wild-type level (Fig. 4D).
Fig 4.
Correlation between Congo red binding by and curli gene (csgA) expression in the wild-type and hha mutant at 37°C and 30°C. Congo red binding was assessed based on the intensity of the red color of the colonies produced on YESCA agar containing Congo red after incubation for 24 h at 37°C (A) or after incubation for 48 h at 30°C (C). Coloration of the colonies was interpreted as light red (low), dark red (intermediate), or darker red (high). The expression of csgA, the gene encoding curlin of curli fimbriae, was determined by QRT-PCR using DNA-free RNA prepared from the wild-type, hha mutant, and hha+/hha mutant strains grown at 37°C (B) and 30°C (D). Bars with different hatching patterns represent the average relative expression of csgA determined using RNA from three independent bacterial cultures grown at 37°C (B) and 30°C (D). Error bars represent the SEM. Significant (P < 0.05) differences between the wild-type and mutant strains were determined using an unpaired Student's t test and Welch's correction. **, P = 0.004; ***, P < 0.0001.
Curli fimbriae are important for biofilm formation in O157.
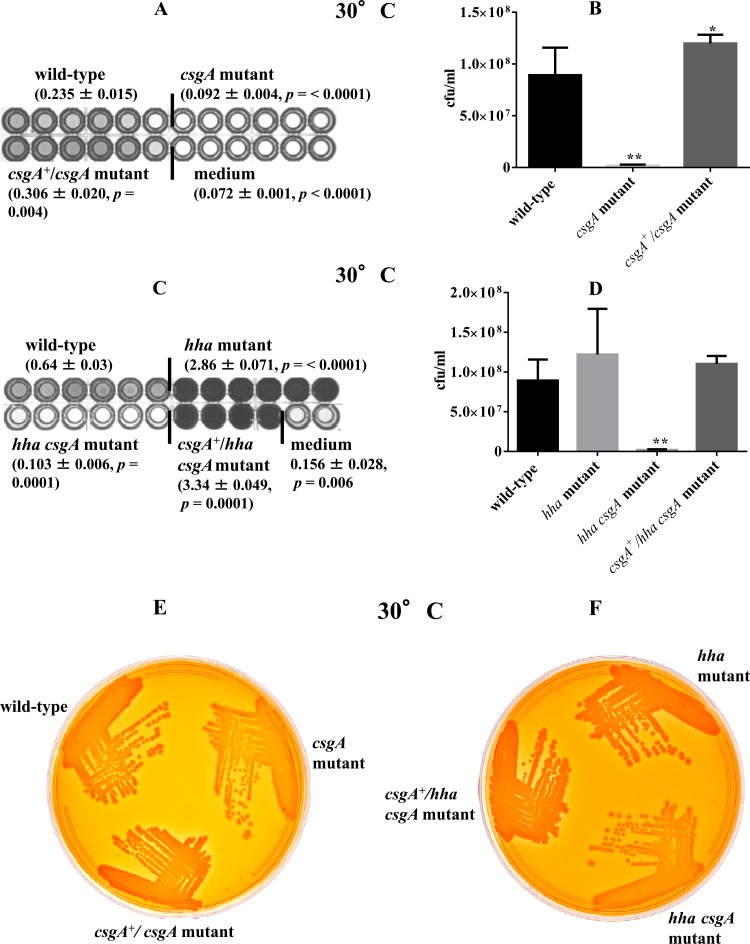
Curli fimbriae are among the most important cell surface structures for biofilm formation in E. coli, and expression of curli fimbriae is turned on in the early stationary phase (8, 23). In order to assess whether the increased biofilm formation and Congo red binding by the hha mutant were due to the increased production of curli fimbriae, which are encoded by the csgA gene, we examined the effect of csgA deletion on production of biofilm and Congo red binding in the wild type and the hha mutant at 30°C (a temperature that promotes enhanced Congo red binding and biofilm formation). The total biofilm mass produced by the csgA mutant was 2.5-fold lower (P < 0.0001) than that produced by the wild-type strain, and the intensity of the blue color produced by this mutant in the plate wells was similar to that in the wells inoculated with YESCA broth (medium control), indicating that the csgA deletion compromises the ability of csgA mutant to produce biofilms (Fig. 5A). The ability of the csgA mutant to produce biofilm was restored to wild-type levels by complementation with a plasmid-cloned csgA (pSM569) (Fig. 5A). The csgA mutant also showed significantly lower recovery of viable cells from the biofilms (1.90 × 106 CFU ml−1; P = 0.0022) than the wild-type strain (8.9 × 107 CFU ml−1) (Fig. 5B). The complementation of the csgA mutant with pSM569 resulted in recovery of viable cells (1.19 × 108, P = 0.041) that was only slightly but significantly higher than that in the wild-type strain (8.9 × 107 CFU ml−1), indicating a gene-dosage effect of pSM569 (Fig. 5B). Since the hha mutant produced higher levels of biofilm than the wild-type strain, we also determined whether csgA deletion would abolish the biofilm formation in the hha mutant. The hha mutant produced total biofilm mass that was 4.5-fold higher (P < 0.0001) than that produced by the wild-type strain, but the biofilm mass produced by the hha csgA mutant was reduced 22-fold compared to that of the hha mutant and 6-fold compared to that of the wild-type strain (Fig. 5C). The complementation of the hha csgA mutant strain by pSM569 increased its biofilm mass production to levels that were about 1.2-fold higher than those of the hha mutant (Fig. 5C). These effects of csgA and hha on biofilm production correlated with the proportions of viable bacterial cells recovered from the biofilms. As shown in Fig. 5D, the hha csgA mutant had 65-fold and 48-fold fewer viable cells (1.85 × 106 CFU ml−1; P = 0.0022) recovered from its biofilms than the hha mutant (1.22 × 108 CFU ml−1) and wild-type strain (8.9 × 107 CFU ml−1), respectively. The complementation of hha csgA mutant with pSM569 restored the level of viable bacterial cells in its biofilms (1.1 × 108 CFU ml−1) to levels similar to those recovered from the wild type (8.9 × 107 CFU ml−1) and the hha mutant strain (1.22 × 108 CFU ml−1) (Fig. 5D). These results indicated that csgA is the key gene conferring on O157 the ability to produce biofilms and that Hha is the negative regulator of csgA expression.
Fig 5.
Requirement of csgA (A and B) and effect of csgA expression (C and D) on biofilm formation, recovery of viable bacterial cells from biofilms, and Congo red binding (E and F) after 48 h of growth at 30°C. Values for total biofilm mass produced are averages ± standard errors of the means (SEM) for 24 replicate wells (A) and 22 replicate wells (C) from four independent cultures. Only 6 of 24 wells are shown for the each strain, except for the csgA+/hha csgA mutant strain, for which 4 of 22 wells are shown. Significant (P < 0.05) differences in biofilm biomass comparing the wild-type and mutant strains were determined using an unpaired Student's t test (Mann-Whitney test). (B and D) Numbers of viable bacterial cells recovered from biofilms, shown as average CFU ml−1 for three independent cultures of the wild-type, csgA mutant, and csgA-complemented (csgA+/csgA) mutant (B) and wild-type, hha mutant, hha csgA mutant, and csgA-complemented (csgA+/hha csgA) mutant (D). Error bars represent the SEM. Significant (P < 0.05) differences between the wild-type and mutant strains were determined using an unpaired Student's t test (Mann-Whitney). *, P = 0.041; **, P = 0.0022. (E and F) Congo red binding was assessed based on the intensity of the red color of the colonies produced on YESCA agar containing Congo red after incubation for 48 h at 30°C. Coloration of the colonies was interpreted as light red (low), dark red (intermediate), or darker red (high).
The effects of the differential expression of csgA on the formation of biofilms in the wild-type, hha, and hha csgA strains with or without complementation for csgA were also assessed in terms of the degree of Congo red binding. The csgA mutant produced lighter red colonies on Congo red plates after 48 h of incubation at 30°C, in contrast to the dark red colonies of the wild type and the csgA mutant complemented with pSM569 (Fig. 5E). The hha mutant produced darker red colonies in the presence of the csgA gene, because the colonies produced by the hha csgA mutant on the Congo red medium were a lighter red (Fig. 5F). The complementation of hha csgA mutant with pSM569 restored a dark-red colony phenotype (Fig. 5F). The results of the Congo red binding assay corroborated the data generated by quantitative biofilm and QRT-PCR assays and demonstrated that hha regulates biofilm formation in O157 by the negative regulation of csgA.
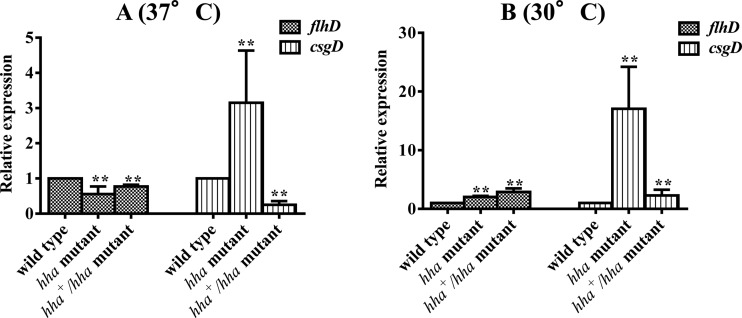
Reduced motility and increased Congo red binding correlated with reduced flhD and increased csgD transcriptional levels.
We showed previously that hha deletion resulted in reduced motility and enhanced curli expression due to reduced fliC and increased csgA transcriptional levels, respectively (41). Since global regulators FlhDC and CsgD control fliC and csgA expression, respectively (21, 23, 24), we examined if transcriptional levels of flhD and csgD would correlate with the transcriptional levels of fliC and csgA in the hha mutant strain. The transcriptional analysis showed that the flhD level was reduced about 2-fold in the hha mutant strain, and complementation of this mutant with pSM197R restored flhD expression similar to that in the wild-type strain at 37°C (Fig. 6A). On the other hand, expression of csgD was increased 3-fold in the hha mutant strain, while the expression of csgD was reduced 4-fold in the hha mutant complemented with pSM197R compared to that in the wild-type strain at 37°C (Fig. 6A). At 30°C, however, the expression of flhD was 2-fold and 3-fold higher in the hha and the hha+/hha mutant strains, respectively (Fig. 6B). The expression of csgD at 30°C was 17-fold higher in the hha mutant than in the wild-type strain, but the complementation of hha mutant with pSM197R reduced the expression of csgD from 17-fold to only 2.3-fold higher than the wild-type strain.
Fig 6.
Differential effects of hha on flhD and csgD expression. DNA-free RNAs were subjected to real-time QRT-PCR. Relative transcriptional levels of flhD and csgD were determined by QRT-PCR and DNA-free RNA prepared from three independent bacterial cultures of the wild-type, hha mutant, and hha+/hha mutant at 37°C (A) and 30°C (B). Relative expressions of flhD and csgD were normalized to the transcriptional levels of the rpoA gene. Values are expression of flhD and csgD in the mutant and complemented mutant strains relative to the expression of these genes in the wild-type strain. Error bars represent the SEM. Significant (P < 0.05) differences between the wild-type and mutant strains were determined using an unpaired Student's t test (Mann-Whitney). **, P = 0.0022.
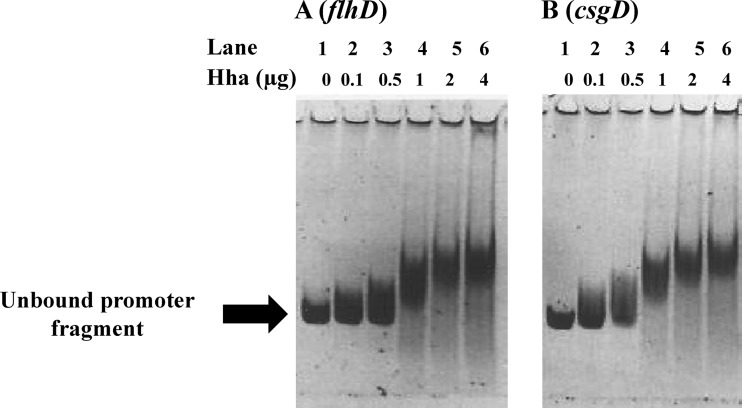
Purified Hha binds to flhDC and csgD promoters in gel shift assays.
Differential expression of fliC and csgA genes is essential for E. coli to switch from the planktonic to the sedentary life style in response to environmental cues (24). Because the expression of flagellar and curli genes is controlled by master regulators FlhDC and CsgD, and the expression of flhD and csgD was differentially affected in the hha mutant, especially at 37°C, we examined whether a purified Hha would bind to the promoter regions of flhDC and csgDEFG operons encoding the master regulators FlhDC and CsgD, which regulate transcription of fliC and csgA genes, respectively. Incubation of PCR-amplified fragments containing the promoter regions for flhDC (Fig. 7A) and csgD (Fig. 7B) with increasing concentrations of Hha and in the presence of a 10-fold molar excess of nonspecific competing DNA poly(dI-dC) showed reduction in the mobility of these fragments in a concentration-dependent manner relative to the same fragments incubated in the absence of Hha. The maximal shift in the mobility of the two promoter fragments was achieved at Hha concentrations of 2 to 4 μg (Fig. 7). These results indicated that Hha presumably regulates both flhD and csgD expression by its direct interactions with the flhDC and csgDEFG promoter regions in bacterial cells.
Fig 7.
Gel shift assays for assessing the binding of Hha with promoter regions controlling transcription of flhDC and csgD. One hundred nanograms of PCR-amplified and gel-purified DNA fragments containing promoter regions for flhDC (A) and csgD (B) were incubated in a binding buffer with or without purified Hha protein for 20 min at room temperature. The samples were analyzed on a 6% nondenaturing polyacrylamide gel stained with SYBR green and visualized using a filter with a bandwidth of 520 nm. The promoter fragments were incubated with 0 μg (lane 1), 0.1 μg (lane 2), 0.5 μg (lane 3), 1 μg (lane 4), 2 μg (lane 5), and 4 μg (lane 6) of Hha. The arrow indicates the unbound flhDC and csgDEFG promoter fragments.
DISCUSSION
E. coli biofilms are complex sedentary communities, and their morphogenesis is highly coordinated and is controlled by several genetic factors and environmental and nutritional cues (3, 7, 15, 21, 23–25, 27, 52, 53). A critical step in transitioning from a planktonic state to biofilm formation is cessation or reduction in the expression of the flagellar system in order to reduce bacterial motility and promote expression of adhesive fimbriae, such as curli fimbriae, and other extracellular adhesins that allow adherence of bacterial cells to solid surfaces (8, 23). In the current study, we have demonstrated that Hha represses biofilm formation in O157 by induction of flagellar and repression of curli gene expression. In comparison to the wild-type and hha+/hha mutant strains, the hha mutant showed significant reduction in bacterial motility but enhanced binding of Congo red, the phenotype indicative of increased curli production at both 37°C and 30°C (47, 48). The differentially affected motility and curli phenotypes in the hha mutant were restored to wild-type levels by complementation with pSM197R, indicating that Hha is responsible for regulating the expression of these two phenotypes. The transcriptional analysis revealed that csgA was expressed at significantly higher levels in the hha mutant than in the wild-type and hha+/hha mutant strains at both 37°C and 30°C. The repressor effects of Hha that we observed with respect to curli gene expression have also been reported for several other bacterial virulence genes (42, 44, 54, 55). For example, Hha, a member of the Hha-YmoA family of small transcriptional regulatory proteins, was initially identified as a repressor of α-hemolysin expression in E. coli (42). Other virulence factors that are negatively regulated by Hha include genes required for adherence of pathogenic E. coli O157:H7 and Salmonella strains to animal tissues and tissue culture cells (44, 54). A recent study showed that Hha acts as a repressor of biofilm formation in nonpathogenic E. coli K-12 strains through the repression of type I fimbriae (56). Although type I fimbriae are not expressed by E. coli O157:H7 due to the locked-on promoter configuration of this operon (57), the negative effect of Hha on biofilm formation by the repression of curli genes in O157 suggests a broader role for Hha in regulation of different clusters of fimbrial genes and in the complex regulatory scheme controlling the transition from planktonic to biofilm formation.
Several studies have reported that production of flagella and curli fimbriae promotes adherence to and colonization of epithelial cells in animal models and tissue cultures (13, 40, 58, 59). Flagella, for example, are essential for bacterial motility and for maintaining bacterial cells in a planktonic state when conditions are favorable for rapid growth (60, 61). On the other hand, curli expression is increased under conditions that are less favorable for rapid growth but conducive to adherence to solid surfaces (8, 23). Low temperature is conducive to biofilm formation in environmental and pathogenic E. coli strains (8, 62). Our quantitative analysis of biofilm formation showed that the hha mutant produced significantly larger amounts of biofilm and contained higher counts of viable bacterial cells in biofilms than the wild-type and hha+/hha mutant strains at both 37°C and 30°C. Overall, the hha mutant produced the largest amount of biofilm at 30°C compared to that at 37°C, and these results were in agreement with studies showing that low temperatures are conducive to biofilm formation in environmental and pathogenic E. coli strains (8, 62). Although the hha mutant produced larger amounts of biofilm than the wild-type strain at 37°C, these amounts were much lower than the amount of biofilm produced at 30°C. The differences in the amounts of biofilms produced at these two temperatures correlated with the highest transcriptional levels of csgA and csgD in the hha mutant grown at 30°C, in contrast those obtained at 37°C. The importance of csgA in biofilm formation, as has been shown by other studies (8, 63–66), was confirmed by demonstrating the inability of the csgA and hha csgA mutants to produce biofilms.
Since growth rates of wild-type and isogenic hha and hha+/hha mutant strains were higher at 37°C than at 30°C, the lower level of biofilm production by the hha mutant at 37°C compared to at 30°C might in part be due to differences in growth physiology at these two temperatures. For example, it is conceivable that higher growth of the hha mutant strains at 37°C would prevent csgD expression from reaching levels that were expressed at 30°C. Since increased expression of CsgD is critical for increased expression of csgA and biofilm formation (67), lower levels of CsgD would reduce biofilm production at 37°C compared to that at 30°C. It has also been shown that at growth temperatures of <32°C, E. coli exhibits a global-stress response resulting in upregulation of many RpoS-dependent regulons, including those that are involved in biofilm formation (30). A recent study has demonstrated that the production of indole, which inhibits biofilm formation, is enhanced when cultures are shifted from 30°C to 37°C (68).
Contrary to the enhanced expression of curli as indicated by the increased Congo red binding and biofilm formation, motility of hha mutant was significantly reduced at 37°C and 30°C. Although the motility halos produced by the hha and hha+/hha mutants at 37°C were much bigger (presumably due to rapid cellular metabolism) than those produced at 30°C, the motility of hha mutant was reduced at both temperatures relative to the motilities of the wild-type and hha+/hha mutant strains, though the reduction was more pronounced at 37°C than at 30°C. These results implied that a reduction in bacterial motility is essential for increased biofilm formation by the hha mutant strains. This reciprocal correlation between reduced motility or lack of motility and increased curli formation resulting in increased biofilm formation has been observed in the nonmotile O157 strains isolated from HUS patients (40). Although motility and transcriptional levels of flhD and fliC in the hha mutant strain were reduced at 37°C and reduced motility of the hha mutant was restored to levels even higher than those of the wild-type strain at 37°C and 30°C, the transcriptional levels of fliC were almost similar to and that of the flhD strain about 2-fold higher than that of the wild-type strain at 30°C. The latter results are not surprising given that the motility differences between the hha mutant and the wild-type strain were much less substantial at 30°C, and the reduced motility of hha mutant at 30°C could potentially be due either to the high levels of curli expression promoting bacterial aggregation and biofilm formation without affecting flagellar gene expression or to compromised flagellar motor functions (15, 17).
In vitro gel shift analysis revealed positive interactions of Hha with the promoter sequences of flhDC and csgDEFG, suggesting that similar interactions in vivo, which could be direct or indirect, regulate the expression of FlhDC and CsgD, two global regulators affecting the expression of not only the flagellar and curli gene regulons, respectively, but also many other regulons reported to directly or indirectly influence motility and biofilm formation in O157. Several factors, including bacterial growth rate, temperature, osmolarity, oxygen tension, and physiological stress due to nutrient limitation, affect relative expression of FlhDC and CsgD (23, 24, 53). Hence, diverse signal transduction mechanisms presumably work in a specific hierarchical order to respond to physical and physiological alterations in bacterial growth environments to affect and fine tune the expression of flhDC and csgD (36, 69–73). FlhDC and CsgD are considered to have hierarchical superiority in terms of their importance in regulation of bacterial motility and curli expression, respectively (21, 24). For example, FlhDC expression is regulated during the exponential and early post-exponential growth phases, while CsgD levels become higher during the stationary growth phase (24). In addition to FlhDC, which activate class II and class III flagellar and chemotaxis genes, several other transcriptional factors activate flagellar gene expression mostly by affecting the expression of flhDC (24, 52, 74). Similarly, expression of csgD is regulated by multiple transcriptional regulators (23, 24, 28, 75, 76). Based on the present study, which demonstrates Hha-mediated differential regulation of flhDC and csgD, and several other studies describing the negative regulation by Hha of many virulence-associated genes and regulons (41, 42, 44, 54), Hha could also qualify as a global regulator having a hierarchical superiority in the overall network of transcriptional regulators controlling bacterial motility, biofilm formation, and virulence gene expression.
ACKNOWLEDGMENTS
We thank Lindsay Smith, Robert Morgan, and Kellie Winter for technical assistance. We also thank Shawn Bearson and Randy Sacco for the critical review of the manuscript.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print 1 February 2013
REFERENCES
- 1.Karmali MA. 2004. Infection by Shiga toxin-producing Escherichia coli: an overview. Mol. Biotechnol. 26:117–122 [DOI] [PubMed] [Google Scholar]
- 2.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79 [DOI] [PubMed] [Google Scholar]
- 4.Saldana Z, Erdem AL, Schuller S, Okeke IN, Lucas M, Sivananthan A, Phillips AD, Kaper JB, Puente JL, Giron JA. 2009. The Escherichia coli common pilus and the bundle-forming pilus act in concert during the formation of localized adherence by enteropathogenic E. coli. J. Bacteriol. 191:3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres AG, Jeter C, Langley W, Matthysse AG. 2005. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl. Environ. Microbiol. 71:8008–8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott SJ, Yu J, Kaper JB. 1999. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67:4260–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 8.Olsen A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655 [DOI] [PubMed] [Google Scholar]
- 9.Bian Z, Brauner A, Li Y, Normark S. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181:602–612 [DOI] [PubMed] [Google Scholar]
- 10.La Ragione RM, Collighan RJ, Woodward MJ. 1999. Non-curliation of Escherichia coli O78:K80 isolates associated with IS1 insertion in csgB and reduced persistence in poultry infection. FEMS Microbiol. Lett. 175:247–253 [DOI] [PubMed] [Google Scholar]
- 11.Olsen A, Wick MJ, Morgelin M, Bjorck L. 1998. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect. Immun. 66:944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saldana Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, Giron JA. 2009. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 11:992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlich GA, Keen JE, Elder RO. 2002. Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect. Immun. 70:395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661–670 [DOI] [PubMed] [Google Scholar]
- 15.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285–293 [DOI] [PubMed] [Google Scholar]
- 16.Sheikh J, Hicks S, Dall'Agnol M, Phillips AD, Nataro JP. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41:983–997 [DOI] [PubMed] [Google Scholar]
- 17.Barrios AF, Zuo R, Ren D, Wood TK. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K-12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188–200 [DOI] [PubMed] [Google Scholar]
- 18.Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933–946 [DOI] [PubMed] [Google Scholar]
- 19.Gualdi L, Tagliabue L, Landini P. 2007. Biofilm formation-gene expression relay system in Escherichia coli: modulation of σS-dependent gene expression by the CsgD regulatory protein via σS protein stabilization. J. Bacteriol. 189:8034–8043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claret L, Hughes C. 2002. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J. Mol. Biol. 321:185–199 [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Matsumura P. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798–808 [DOI] [PubMed] [Google Scholar]
- 23.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruss BM, Besemann C, Denton A, Wolfe AJ. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 188:3731–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnhart MM, Lynem J, Chapman MR. 2006. GlcNAc-6P levels modulate the expression of curli fibers by Escherichia coli. J. Bacteriol. 188:5212–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen A, Arnqvist A, Hammar M, Normark S. 1993. Environmental regulation of curli production in Escherichia coli. Infect. Agents Dis. 2:272–274 [PubMed] [Google Scholar]
- 29.White-Ziegler CA, Malhowski AJ, Young S. 2007. Human body temperature (37°C) increases the expression of iron, carbohydrate, and amino acid utilization genes in Escherichia coli K-12. J. Bacteriol. 189:5429–5440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White-Ziegler CA, Um S, Perez NM, Berns AL, Malhowski AJ, Young S. 2008. Low temperature (23°C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154:148–166 [DOI] [PubMed] [Google Scholar]
- 31.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JA, Molin S, Prensier G, Arbeille B, Ghigo JM. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659–674 [DOI] [PubMed] [Google Scholar]
- 32.Domka J, Lee J, Bansal T, Wood TK. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9:332–346 [DOI] [PubMed] [Google Scholar]
- 33.Gadgil M, Kapur V, Hu WS. 2005. Transcriptional response of Escherichia coli to temperature shift. Biotechnol. Prog. 21:689–699 [DOI] [PubMed] [Google Scholar]
- 34.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515–524 [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Preston JF, III, Romeo T. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto K, Silhavy TJ. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 99:2287–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konkel ME, Tilly K. 2000. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2:157–166 [DOI] [PubMed] [Google Scholar]
- 38.Cassels FJ, Wolf MK. 1995. Colonization factors of diarrheagenic E. coli and their intestinal receptors. J. Ind. Microbiol. 15:214–226 [DOI] [PubMed] [Google Scholar]
- 39.Kumar CG, Anand SK. 1998. Significance of microbial biofilms in food industry: a review. Int. J. Food Microbiol. 42:9–27 [DOI] [PubMed] [Google Scholar]
- 40.Rosser T, Dransfield T, Allison L, Hanson M, Holden N, Evans J, Naylor S, La Ragione R, Low JC, Gally DL. 2008. Pathogenic potential of emergent sorbitol-fermenting Escherichia coli O157:NM. Infect. Immun. 76:5598–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma VK, Bearson SM, Bearson BL. 2010. Evaluation of the effects of sdiA, a luxR homologue, on adherence and motility of Escherichia coli O157:H7. Microbiology 156:1303–1312 [DOI] [PubMed] [Google Scholar]
- 42.Nieto JM, Carmona M, Bolland S, Jubete Y, de la Cruz F, Juarez A. 1991. The hha gene modulates haemolysin expression in Escherichia coli. Mol. Microbiol. 5:1285–1293 [DOI] [PubMed] [Google Scholar]
- 43.Sharma VK, Carlson SA, Casey TA. 2005. Hyperadherence of an hha mutant of Escherichia coli O157:H7 is correlated with enhanced expression of LEE-encoded adherence genes. FEMS Microbiol. Lett. 243:189–196 [DOI] [PubMed] [Google Scholar]
- 44.Sharma VK, Zuerner RL. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186:7290–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reference deleted.
- 46.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109:705–712 [DOI] [PubMed] [Google Scholar]
- 47.Hammar M, Bian Z, Normark S. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93:6562–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlich GA, Cooke PH, Solomon EB. 2006. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 72:2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bearson BL, Bearson SM, Uthe JJ, Dowd SE, Houghton JO, Lee I, Toscano MJ, Lay DC., Jr 2008. Iron regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microbes Infect. 10:807–816 [DOI] [PubMed] [Google Scholar]
- 51.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 52.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hengge-Aronis R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148–152 [DOI] [PubMed] [Google Scholar]
- 54.Fahlen TF, Wilson RL, Boddicker JD, Jones BD. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620–6629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mourino M, Madrid C, Balsalobre C, Prenafeta A, Munoa F, Blanco J, Blanco M, Blanco JE, Juarez A. 1996. The Hha protein as a modulator of expression of virulence factors in Escherichia coli. Infect. Immun. 64:2881–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Contreras R, Zhang XS, Kim Y, Wood TK. 2008. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS One 3:e2394 doi:10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roe AJ, Currie C, Smith DG, Gally DL. 2001. Analysis of type 1 fimbriae expression in verotoxigenic Escherichia coli: a comparison between serotypes O157 and O26. Microbiology 147:145–152 [DOI] [PubMed] [Google Scholar]
- 58.Erdem AL, Avelino F, Xicohtencatl-Cortes J, Giron JA. 2007. Host protein binding and adhesive properties of H6 and H7 flagella of attaching and effacing Escherichia coli. J. Bacteriol. 189:7426–7435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres AG, Milflores-Flores L, Garcia-Gallegos JG, Patel SD, Best A, La Ragione RM, Martinez-Laguna Y, Woodward MJ. 2007. Environmental regulation and colonization attributes of the long polar fimbriae (LPF) of Escherichia coli O157:H7. Int. J. Med. Microbiol. 297:177–185 [DOI] [PubMed] [Google Scholar]
- 60.Amsler CD, Cho M, Matsumura P. 1993. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J. Bacteriol. 175:6238–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao K, Liu M, Burgess RR. 2007. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 35:4441–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arnqvist A, Olsen A, Pfeifer J, Russell DG, Normark S. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol. Microbiol. 6:2443–2452 [DOI] [PubMed] [Google Scholar]
- 63.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 49:875–884 [DOI] [PubMed] [Google Scholar]
- 64.Kim SH, Kim YH. 2004. Escherichia coli O157:H7 adherence to HEp-2 cells is implicated with curli expression and outer membrane integrity. J. Vet. Sci. 5:119–124 [PubMed] [Google Scholar]
- 65.Sukupolvi S, Lorenz RG, Gordon JI, Bian Z, Pfeifer JD, Normark SJ, Rhen M. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 65:5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uhlich GA, Keen JE, Elder RO. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. 2010. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156:2470–2483 [DOI] [PubMed] [Google Scholar]
- 68.Han TH, Lee JH, Cho MH, Wood TK, Lee J. 2011. Environmental factors affecting indole production in Escherichia coli. Res. Microbiol. 162:108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17–23 [DOI] [PubMed] [Google Scholar]
- 70.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298 [DOI] [PubMed] [Google Scholar]
- 71.Dorel C, Vidal O, Prigent-Combaret C, Vallet I, Lejeune P. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169–175 [DOI] [PubMed] [Google Scholar]
- 72.Martino PD, Fursy R, Bret L, Sundararaju B, Phillips RS. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443–449 [DOI] [PubMed] [Google Scholar]
- 73.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 62:1014–1034 [DOI] [PubMed] [Google Scholar]
- 74.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521–532 [DOI] [PubMed] [Google Scholar]
- 75.Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R., III 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 41:349–363 [DOI] [PubMed] [Google Scholar]
- 76.Hengge-Aronis R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341–346 [PubMed] [Google Scholar]
- 77.Melton-Celsa AR, Rogers JE, Schmitt CK, Darnell SC, O'Brien AD. 1998. Virulence of Shiga toxin-producing Escherichia coli (STEC) in orally-infected mice correlates with the type of toxin produced by the infecting strain. Jpn. J. Med. Sci. Biol. 51(Suppl):S108–S114 [DOI] [PubMed] [Google Scholar]