Abstract
Thaumarchaeota are abundant and active in marine waters, where they contribute to aerobic ammonia oxidation and light-independent carbon fixation. The ecological function of thaumarchaeota in marine sediments, however, has rarely been investigated, even though marine sediments constitute the majority of the Earth's surface. Thaumarchaeota in the upper layer of sediments may contribute significantly to the reservoir of nitrogen oxides in ocean waters and thus to productivity, including the assimilation of carbon. We tested this hypothesis in the northern South China Sea (nSCS), a section of a large oligotrophic marginal sea with limited influx of nutrients, including nitrogen, by investigating the diversity, abundance, community structure, and spatial distribution of thaumarchaeotal signatures in surface sediments. Quantitative real-time PCR using primers designed to detect 16S rRNA and amoA genes in sediment community DNA revealed a significantly higher abundance of pertinent thaumarchaeotal than betaproteobacterial genes. This finding correlates with high levels of hcd genes, a signature of thaumarchaeotal autotrophic carbon fixation. Thaumarchaeol, a signature lipid biomarker for thaumarchaeota, constituted the majority of archaeal lipids in marine sediments. Sediment temperature and organic P and silt contents were identified as key environmental factors shaping the community structure and distribution of the monitored thaumarchaeotal amoA genes. When the pore water PO43− concentration was controlled for via partial-correlation analysis, thaumarchaeotal amoA gene abundance significantly correlated with the sediment pore water NO2− concentration, suggesting that the amoA-bearing thaumarchaeota contribute to nitrite production. Statistical analyses also suggest that thaumarchaeotal metabolism could serve as a pivotal intersection of the carbon, nitrogen, and phosphorus cycles in marine sediments.
INTRODUCTION
Nitrification, the microbial process of ammonia oxidation to nitrate via nitrite (NH3→NO2−→NO3−), is a key component of the global N cycle. Similar to chemolithoautotrophic ammonia-oxidizing bacteria (AOB), ammonia-oxidizing archaea (AOA) in the phylum Thaumarchaeota (references 1 and 2 and references therein) use aerobic ammonia oxidation to support their chemolithoautotrophic lifestyle (3) and thus contribute to nitrification and autotrophic carbon fixation (reference 4 and references therein). AOA are affiliated with several groups of the Thaumarchaeota, including marine group I (MGI), soil group I.1b, thermophilic AOA (ThAOA), and putatively marine group pSL12 (references 2 and 5 and references therein). Thaumarchaeotal amoA genes encoding the alpha subunit of ammonia monooxygenase (AMO) are abundant in marine environments and are usually present at higher concentrations than homologous proteobacterial amoA genes (references 2 and 6 and references therein). However, there is evidence that not all thaumarchaeota carrying the amoA gene are chemolithoautotrophic ammonia-oxidizing microbes, which led to the designation amoA-encoding archaea (AEA) in the literature (5, 7, 8). Ecophysiological studies of AOA suggest adaptation to low ammonia concentrations, and the finding that AOA dominate the nitrifying microbial community in oligotrophic marine environments has led to the proposal of niche separation between AOA and AOB (9, 10).
Thaumarchaeota use the 3-hydroxypropionate/4-hydroxybutyrate pathway for autotrophic carbon fixation, which employs acetyl-coenzyme A (CoA) carboxylase (encoded by accA) and 4-hydroxybutyryl-CoA dehydratase (encoded by hcd) (11, 12). The activity or potential of autotrophic carbon fixation by AOA communities in marine waters has recently been reported (13–17). In contrast, only a few studies have investigated the potential thaumarchaeotal contributions to ammonia oxidation and autotrophic carbon fixation in marine sediments (18, 19), despite the fact that marine surface sediments cover the majority (>70%) of the Earth's surface and constitute suitable habitats for thaumarchaeotal ammonia oxidizers (20). Marginal seas are critical regions of the world's oceans, playing important roles in nutrient cycling, marine production and fishery, and climate-related ecological functions (21). It is thus important to systematically investigate the biogeography, biogeochemistry, and ecological functions of marine sediment thaumarchaeota in these regions.
Membrane lipids, as specific biomarkers, can assist in the investigation of microbial abundance, distribution, and source environment (22). Archaea and bacteria have different membrane core lipid glycerol dialkyl glycerol tetraethers (GDGTs). In marine environments, branched GDGTs (bGDGTs) are specific to terrestrial source bacteria, whereas thaumarchaeol (originally called crenarchaeol) and its isomers are specific to marine thaumarchaeota (23). This observation has been employed to calculate the relative proportion of terrestrial bacterial membrane lipids to marine thaumarchaeotal membrane lipids, called the branched versus isoprenoid tetraether (BIT) index, which has been proposed to serve as a proxy for terrestrial organic matter in marine sediments (23). The BIT index may prove particularly useful for tracing terrestrial inputs in marginal sea environments.
The South China Sea (SCS) is the largest marginal sea (∼3.5 × 106 km2) in the western Pacific Ocean, characterized as a huge oligotrophic subtropical and tropical water body with usually undetectable nitrate and phosphate in the euphotic zone (24). The bioproductivity of the SCS is likely limited by N nutrient supply characterized by a surface water average N/P ratio well below the Redfield ratio of 16:1 (25). Although riverine inputs, such as those from the Pearl and Mekong Rivers, may provide nutrients for aquatic bioproduction in the estuarine and coastal areas, these supplies are not adequate to support productivity in the entire oligotrophic SCS (26). Deep waters may be another source of nutrient supply to the oligotrophic surface water in the SCS, which can occur via monsoonal wind driving eddies and other upwelling and vertical-mixing processes (27). The northern SCS (nSCS) includes several identified and prospective deep-sea methane hydrate reservoirs, and these areas may sustain productive methane seep-specific ecosystems, requiring a large supply or rapid recycling of nitrogenous nutrients (28, 29). It is an important ecological question to identify whether and, if so, how the methane seep environments influence the community structure, spatial distribution, and biogeochemical function of the thaumarchaeota in deep-sea sediments (6). Although sediments have been investigated in specific areas of the nSCS (30), no investigation of the methane hydrate reservoirs or their prospective areas has been reported. A recent study indicated the potential of increased thaumarchaeotal autotrophic carbon fixation in the bathypelagic zone of the SCS (13). Due to the close connection between the bathypelagic zone and the benthic environment, a similar autotrophic carbon fixation potential of thaumarchaeota may also exist in the surface sediments of the SCS; however, this hypothesis has not yet been tested.
In the current study, we investigated the ecology and biogeography of the marine thaumarchaeota and their genetic potential for ammonia oxidation and autotrophic carbon fixation in the surface sediments of the nSCS, including prospective gas hydrate reservoirs. In addition to molecular analyses of the thaumarchaeotal amoA and hcd functional marker genes, a study of thaumarchaeotal membrane core lipids was employed to analyze the relationship of the sediment fossil thaumarchaeotal community with environmental conditions. Key environmental factors likely shaping the diversity, abundance, community structure, spatial distribution, and biogeochemical potentials of the surface sediment thaumarchaeota were identified.
MATERIALS AND METHODS
Sample collection and environmental-factor measurements.
Surface sediment samples were collected from 12 sites of the nSCS during a cruise in August 2007 (reference 29 provides details). These sampling sites represent numerous typical sediment environments of the nSCS, including estuarine, coastal, and offshore sites close to the Pearl River estuary and Hainan Island and deep-water sites (>1,000-m water depth) close to Taiwan, Luzon, the Dongsha Islands, the Xisha Islands, and the Xisha trough (see Fig. S1 in the supplemental material). Several of our sampling sites are at or near the identified deep-sea gas hydrate environment in the Shenhu area southwest of the Dongsha Islands and gas hydrate prospective areas in the Xisha trough, the Jiulong methane reef southwest of Taiwan, and the Bijia'nan basin northwest of Luzon (29). Replicate surface sediment subcore samples down to a 5-cm depth from undisturbed core sediments were collected for microbiological, membrane lipid, and environmental-factor analyses. Environmental-factor measurement methods and results were reported previously (29) and are provided in Table S1 in the supplemental material.
DNA extraction and thaumarchaeotal amoA gene clone library analyses.
Sediment DNA extraction, DNA concentration measurement, and thaumarchaeotal amoA gene clone library construction and analysis followed previously described procedures (7, 28, 31). To test the reproducibility of our experimental procedure and to identify any potential small-scale (∼20-cm) spatial variability of the sediment AEA community, three separate amoA gene clone libraries (A3-I, A3-II, and A3-III) were constructed for sampling station A3, each from a distinct subcore DNA sample. The amoA gene sequences obtained were grouped into operational taxonomic units (OTUs) based on a 0.05 sequence distance cutoff, calculated using the DOTUR program (32), to facilitate comparison with previous studies (7, 20, 28, 31). The amoA gene sequences were translated into conceptual AmoA protein sequences, and the BLASTp program was used for retrieval of the top hit sequences from GenBank (33). Phylogenetic analysis followed a previous procedure (7, 28, 31), with CLUSTAL X (version 2.0) for sequence alignment and PHYLIP (version 3.69) for phylogenetic-tree construction using distance neighbor-joining inference (34, 35).
Quantification of major microbial groups.
The abundances of surface sediment thaumarchaeota in groups MGI and pSL12 were determined with real-time fluorescent quantitative PCR (qPCR) using community DNA and group-specific 16S rRNA gene primers listed in Table S2 in the supplemental material (36). The abundances of putative betaproteobacterial AOB and amoA-bearing and autotrophically carbon-fixing thaumarchaeota were estimated by using qPCR and primers targeting the betaproteobacterial amoA and the thaumarchaeotal amoA and hcd genes, respectively, as listed in Table S2 in the supplemental material (19, 20, 28, 37). All qPCR assays were carried out as described previously, including experiments for optimization, quality control, and the generation of standard curves, which employed linearized reference plasmids with target 16S rRNA, amoA, or hcd gene fragments as inserts (28). The ranges of the reference plasmid copy numbers used for standard-curve construction are shown in Table S3 in the supplemental material, along with data documenting the efficiency and sensitivity of each individual qPCR assay.
Membrane core lipid analyses.
Sediments from site E801 were analyzed for microbial-membrane core lipids, including both archaeal isoprenoidal GDGTs (iGDGTs) and bacterial bGDGTs, by using previous experimental procedures (38). These lipid data, along with the data from other sampling stations reported previously (38), were used for in-depth analyses of the relationship between sediment core lipid composition and environmental factors.
Statistical analyses.
The coverage (C) of each clone library was calculated as follows: C = [1 − (n1/N)] × 100, where n1 is the number of unique OTUs and N is the total number of clones in a library (39). Indices of gene diversity (Shannon-Wiener H and Simpson D) and evenness (J) were calculated using the OTU data (31). Rarefaction analysis and two nonparametric richness estimators, abundance-based coverage estimator (SACE) and bias-corrected Chao1 (SChao1), were calculated using DOTUR (32).
Community classification of sediment AEA assemblages using the thaumarchaeotal amoA gene sequences was determined with Fast UniFrac environmental-clustering and principal-coordinates analyses (PCoA) as detailed previously (7, 28, 31, 40). Correlations between the AEA assemblages and environmental factors were analyzed with canonical correspondence analysis (CCA) using the software Canoco (version 4.5; Microcomputer Power, Ithaca, NY) following previously described procedures (7, 28, 31). Pearson correlation analyses of the abundances of sediment 16S rRNA, amoA, and hcd genes with environmental factors were performed with the statistical software MINITAB (release 13.32; Minitab Inc., State College, PA) as detailed previously (28). Partial-correlation analyses of the abundances of the 16S rRNA, amoA, and hcd genes with major environmental factors were performed with the statistical software SPSS (release 16.0; SPSS Inc., Chicago, IL).
Cluster analysis was also performed using the relative abundances of iGDGTs to evaluate the variations of archaeal membrane core lipid composition and spatial distribution with MINITAB using a Euclidean distance measure and Ward linkage (41). Pearson correlation analyses were performed to investigate the relationship of the prevalent environmental factors in the nSCS with the abundance of the sediment thaumarchaeol and the calculated BIT values.
Nucleotide sequence accession numbers.
The determined thaumarchaeotal amoA gene sequences have been deposited in GenBank under accession numbers JX537480 to JX537762, and the determined thaumarchaeotal pSL12 and MGI 16S rRNA and hcd gene sequences (for creating qPCR standard curves) have been deposited under accession numbers KC433538 to KC433540, respectively.
RESULTS
Molecular diversity of thaumarchaeal amoA genes in sediments.
Community classification using Fast UniFrac environmental clustering (see Fig. S2 in the supplemental material) and PCoA (see Fig. S3 in the supplemental material) analyses revealed that the three parallel thaumarchaeotal amoA gene clone libraries (A3-I, A3-II, and A3-III) constructed from separate sediment subcore samples from station A3 were similar. This result confirmed the reproducibility of our experimental procedures, illustrated negligible within-site variability of the sediment AEA community, and justified pooling of the three amoA gene clone libraries into a single A3 library for subsequent analysis (see Table S4 in the supplemental material).
Of 12 amoA gene clone libraries, 1,457 clones contained a thaumarchaeotal amoA gene fragment insert, resulting in 283 unique DNA sequences and 131 OTUs. The values for library coverage (C) ranged from 87.8% to 97.0% (see Table S4 in the supplemental material), which, together with rarefaction analysis (see Fig. S4 in the supplemental material), indicated a high probability that the clone libraries produced represented the diversity of respective in situ AEA communities. Based on the diversity indices (H, 1/D, and J), sampling sites E422 and E501 exhibited the lowest and highest diversity of OTUs, respectively, while site CF14 exhibited the highest OTU richness based on the SACE and SChao1 estimators (see Table S4 in the supplemental material).
The 283 distinct thaumarchaeotal amoA sequences obtained were 68.8 to 99.8% identical with one another and 84.7 to 99.8% identical to the closest matches among amoA sequences deposited in GenBank. The corresponding deduced AmoA protein sequences were 77.7 to 100.0% identical with one another and 91.9 to 100.0% identical to the closest matches among AmoA sequences deposited in GenBank that were associated with a variety of terrestrial, estuarine, coastal, deep-sea sediment, and seawater source environments. Except for two sequences that were closely related to sequences in GenBank with an origin in soils from China, the AmoA sequences matched top-hit GenBank sequences that were originally obtained from marine or estuarine environments.
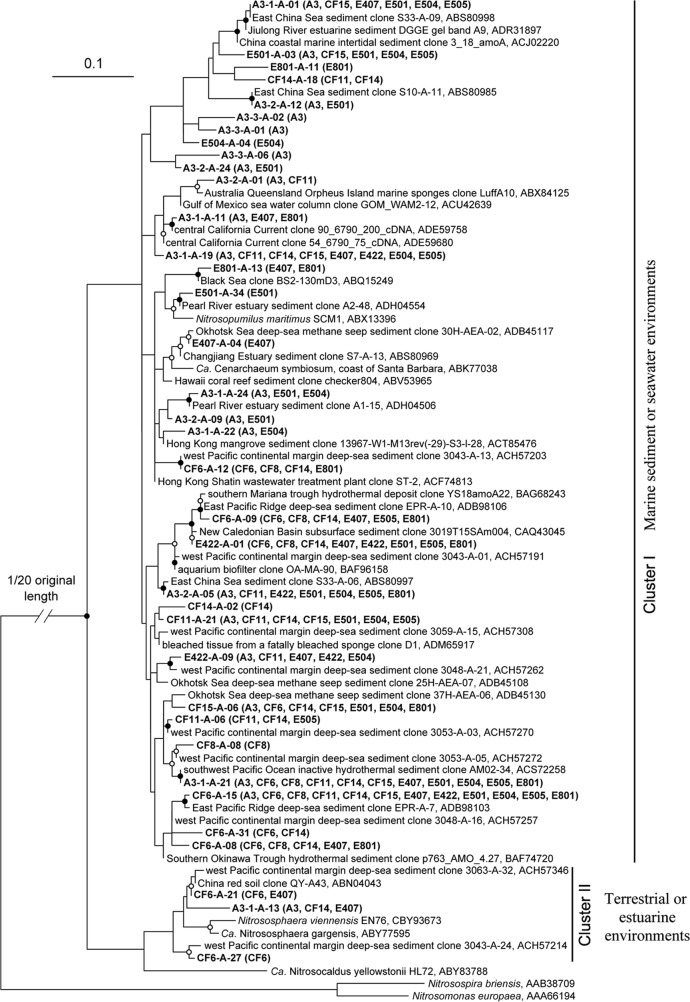
The phylogenetic tree of thaumarchaeotal AmoA sequences revealed two sequence clusters with >20% between-cluster distance, as determined using DOTUR (Fig. 1). The AmoA sequences in cluster I were mainly associated with marine sediments, seawater, or related environments, while the sources of the AmoA sequences in cluster II were soils or estuarine and marine environments with a high degree of terrestrial inputs (7, 31). The sequences in cluster I (98.2% of the unique AmoA sequences and 99.5% of the total clones) were much more abundantly represented in our experimental clone libraries than the sequences associated with cluster II, indicating that marine-environment-related AEA predominate in sediment environments in the nSCS.
Fig 1.
Consensus phylogenetic tree constructed from the AmoA protein sequences deduced from the amoA genes recovered from the surface sediments of the nSCS. The unique archaeal AmoA protein sequences obtained in this study were grouped at 0.05 distance cutoff using the DOTUR program. Bacterial AmoA sequences from Nitrosomonas europaea and Nitrosospira briensis were used as the outgroup. Bootstrap values of no less than 70% of 100 resamplings are marked with solid circles, and those less than 70% but no less than 50% are marked with open circles on the corresponding nodes. The archaeal AmoA sequences obtained in this study are shown in boldface, along with their distributions in each clone library in parentheses.
Two of the sequence types identified in the current study, A3-1-A-21 and CF6-A-15, were detected in almost all the sampling sites of the nSCS (Fig. 1). The clones that were associated with these two sequence types accounted for 37.4% of all the clones in our libraries, potentially representing the most abundant and prevalent AEA in the surface sediments of the nSCS.
AEA community classification and spatial distribution.
The results of both fast UniFrac all-environment P test significance (P = 0.00) and UniFrac significance (P < 0.01) statistics indicated that the different nSCS sediment environments harbored distinct AEA assemblages. The heterogeneous distribution of the AEA communities was further confirmed via fast UniFrac PCoA (Fig. 2) and clustering analyses (see Fig. S5 in the supplemental material), both showing that the AEA assemblages of the estuarine and coastal sites, A3 and E501, were significantly different from those of the offshore and deep-water sites. This classification pattern of the sediment AEA assemblages was strongly supported by high jackknife values of the community clustering analysis (see Fig. S5 in the supplemental material) and high PCoA resolving power, showing that the first PCoA principal coordinate (P1) explained more than half (61.03%) of the total AEA community variability and clearly distinguished the AEA assemblages of sites A3 and E501 from those of the other sites (Fig. 2).
Fig 2.
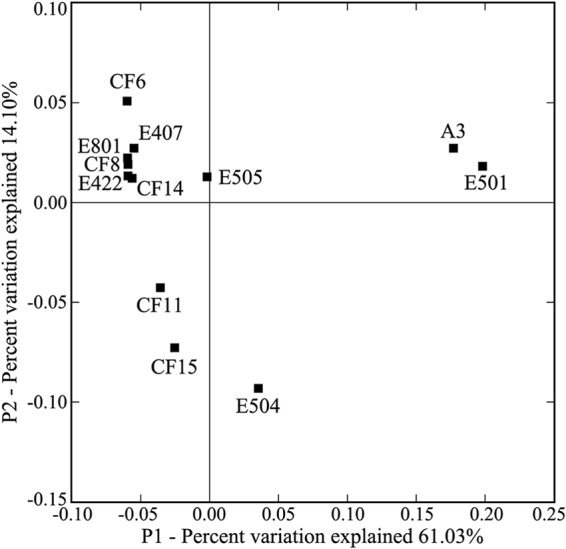
Ordination diagram of the surface sediment AEA assemblages from the nSCS calculated with weighted UniFrac PCoA analysis. Shown is the plot of the first two principal-coordinate axes (P1 and P2) for PCoA and the distributions of the AEA assemblages (designated by the sampling station names) in response to these axes.
CCA analysis was used to identify the key environmental factors that contributed to the heterogeneous distribution of the sediment AEA assemblages in the nSCS and showed that the environmental variables in the first two CCA dimensions (CCA1 and CCA2) explained 38.7% of the total variance in the AEA community composition and 39.7% of the cumulative variance of the AEA-environment relationship. The CCA analysis result confirmed that the AEA assemblages of the estuarine and coastal sites A3 and E501 were distinctly different from those of the offshore and deep-water sites (Fig. 3). CCA1 clearly distinguished the AEA assemblages of sites A3 and E501 from those of the other sites, and sediment temperature was identified as the most significant environmental factor (P = 0.001; 1,000 Monte Carlo permutations), contributing the most to this distinction (Fig. 3). Sediment OrgP was identified as another significant environmental factor (P = 0.039) and sediment silt content as a marginally significant environmental factor (P = 0.082) contributing to the heterogeneous distribution of the AEA assemblages in the nSCS (Fig. 3). These three environmental factors together provided 42.1% of the total CCA explanatory power. All the other environmental factors analyzed were not significant (P > 0.100) in their contributions to the heterogeneous distribution of the AEA assemblages in the surface sediments of the nSCS.
Fig 3.
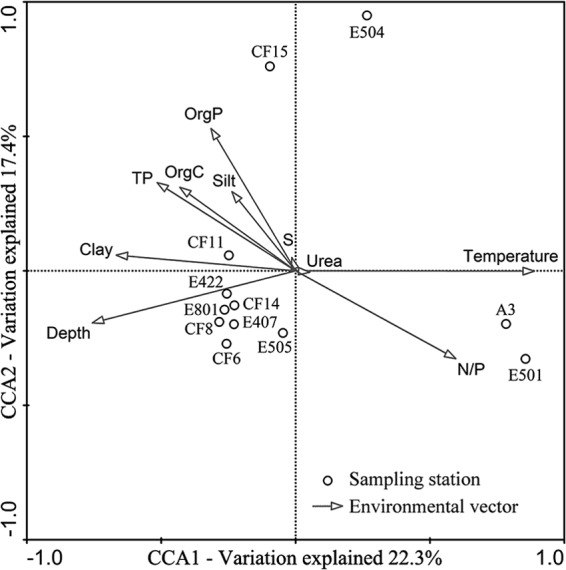
CCA ordination plot for the first two dimensions (CCA1 and CCA2) showing the relationship between the surface sediment AEA assemblages from the nSCS and environmental parameters analyzed using the weighted OTU data. Correlations between environmental variables and CCA axes are represented by the lengths and angles of arrows (environmental-factor vectors).
Quantitation of major microbial groups in sediments.
Quantitation of 16S rRNA gene copies using qPCR identified MGI as the dominant thaumarchaeotal phylotype in surface sediments of the nSCS (ranging from 1.18 × 107 to 1.51 × 108 gene copies g sediment−1). In contrast, the abundance of the pSL12 thaumarchaeotal 16S rRNA gene copies (ranging from 1.10 × 104 to 1.00 × 105 g sediment−1) was quite low (Table 1). The thaumarchaeotal amoA gene abundance varied quite widely in the nSCS, ranging from 3.62 × 106 to 4.86 × 108 copies g sediment−1, while the betaproteobacterial amoA gene abundance was more stable, ranging from 9.77 × 105 to 5.84 × 106 copies g sediment−1 (Table 1). The ratio of thaumarchaeotal to betaproteobacterial amoA gene abundance ranged from 2.7:1 to 232.3:1, with an average of 43.5:1, suggesting that thaumarchaeotal AEA outnumbered betaproteobacterial AOB in nSCS surface sediments. The thaumarchaeotal hcd gene abundance in surface sediments ranged from 2.81 × 107 to 4.26 × 108 copies g sediment−1 (Table 1). The ratio of thaumarchaeotal amoA to 16S rRNA gene abundance ranged from 0.1:1 to 3.2:1, with an average of 1.1:1. The ratio of thaumarchaeotal hcd to 16S rRNA gene abundance ranged from 0.6:1 to 2.9:1, with an average of 1.6:1. The ratio of thaumarchaeotal hcd to amoA gene abundance ranged from 0.4:1 to 13.6:1, with an average of 3.4:1. Although hcd gene abundances were generally slightly higher than the 16S rRNA and amoA gene abundances in the surface sediments of the nSCS, the 3 thaumarchaeotal genes showed significant positive correlations (amoA versus 16S rRNA, r = 0.741, P = 0.006; hcd versus 16S rRNA, r = 0.721, P = 0.008; hcd versus amoA, r = 0.913, P = 0.000), indicating that the majority of the nSCS surface sediment thaumarchaeotal communities possessed the genetic potential for ammonia oxidation and autotrophic carbon fixation.
Table 1.
Abundances of target genes in sediments from the 12 sampling stations in the nSCS
| Sampling station | Target gene (no. g sediment−1)a |
||||
|---|---|---|---|---|---|
| 16S rRNA |
Thaumarchaeotal hcd |
amoA |
|||
| MGI | pSL12 | Thaumarchaeotal | Betaproteobacterial | ||
| A3 | 5.50 × 107 (1.43 × 107) | 2.84 × 104 (1.39 × 103) | 1.45 × 108 (2.27 × 106) | 5.57 × 107 8.15 × 105 | 5.84 × 106 (8.14 × 104) |
| CF6 | 1.18 × 107 (1.07 × 107) | 3.68 × 104 (3.16 × 103) | 2.81 × 107 (2.78 × 106) | 8.32 × 106 6.49 × 104 | 1.53 × 106 (3.20 × 104) |
| CF8 | 7.00 × 107 (3.02 × 107) | 5.55 × 104 (2.38 × 103) | 3.87 × 107 (3.06 × 106) | 6.24 × 107 7.74 × 104 | 1.46 × 106 (8.73 × 104) |
| CF11 | 1.02 × 108 (3.37 × 107) | 2.21 × 104 (6.64 × 102) | 7.95 × 107 (4.84 × 106) | 1.99 × 107 3.05 × 105 | 2.49 × 106 (8.13 × 104) |
| CF14 | 1.05 × 108 (3.23 × 107) | 1.00 × 105 (5.67 × 102) | 7.76 × 107 (3.26 × 106) | 2.17 × 107 2.08 × 105 | 2.81 × 106 (2.08 × 105) |
| CF15 | 6.82 × 107 (5.90 × 107) | 8.14 × 104 (5.76 × 103) | 7.36 × 107 (6.90 × 106) | 1.73 × 108 1.59 × 105 | 3.37 × 106 (1.89 × 105) |
| E407 | 2.74 × 107 (5.49 × 106) | 1.16 × 104 (6.52 × 102) | 4.93 × 107 (2.31 × 106) | 3.62 × 106 4.43 × 104 | 1.23 × 106 (8.89 × 104) |
| E422 | 2.75 × 107 (3.78 × 106) | 1.10 × 104 (4.68 × 102) | 3.01 × 107 (2.05 × 106) | 4.36 × 106 4.02 × 104 | 1.63 × 106 (5.81 × 104) |
| E501 | 1.76 × 107 (1.27 × 107) | 1.41 × 104 (7.12 × 102) | 5.07 × 107 (2.73 × 106) | 3.35 × 107 2.70 × 105 | 1.25 × 106 (8.72 × 104) |
| E504 | 1.51 × 108 (4.63 × 107) | 2.25 × 104 (5.22 × 101) | 4.26 × 108 (1.32 × 106) | 4.86 × 108 1.25 × 106 | 3.88 × 106 (4.43 × 105) |
| E505 | 1.27 × 108 (1.01 × 108) | 1.40 × 104 (1.30 × 103) | 1.30 × 108 (1.04 × 106) | 2.27 × 108 2.11 × 106 | 9.77 × 105 (5.45 × 104) |
| E801 | 2.29 × 107 (2.57 × 107) | 5.40 × 104 (3.71 × 103) | 3.55 × 107 (1.60 × 106) | 1.44 × 107 1.66 × 105 | 2.03 × 106 (5.95 × 104) |
Means and standard errors (indicated in parentheses) were calculated based on 3 replicate measurements.
The abundance of MGI thaumarchaeotal 16S rRNA genes was significantly positively correlated with the sediment silt content, whereas the abundance of 16S rRNA genes of pSL12 thaumarchaeota exhibited a significant positive correlation with sediment OrgC and OrgN. The abundance of the thaumarchaeotal hcd gene showed a significant negative correlation with water depth. The abundance of the thaumarchaeotal amoA gene showed a significant negative correlation with sediment pore water DO, while the abundance of the betaproteobacterial amoA gene showed a significant positive correlation with sediment water content (WC) (see Table S5 in the supplemental material). The ratio of thaumarchaeotal to betaproteobacterial amoA gene abundance showed significant positive correlations with sediment pore water salinity and sediment silt content (see Table S5 in the supplemental material).
The ratio of the thaumarchaeotal amoA gene abundance to the MGI 16S rRNA gene abundance showed significant negative correlation with water depth and sediment pore water DO and NO3−, and the sum of NO2− and NO3− (NOx−) (see Table S5 in the supplemental material). However, partial-correlation analyses showed that the correlations of this ratio with sediment pore water NO3− (P = 0.356, controlling for DO) and NOx− (P = 0.498, controlling for DO) were due to its correlation with sediment pore water DO. The ratio of the thaumarchaeotal hcd gene abundance to the MGI 16S rRNA gene abundance showed a significant positive correlation with sediment temperature and negative correlations with sediment OrgC and OrgN (see Table S5 in the supplemental material). The ratio of the thaumarchaeotal hcd gene abundance to the amoA gene abundance showed significant positive correlations with sediment pore water DO, NO3−, NOx−, sediment OrgC/OrgN, and clay content and a significant negative correlation with sediment skewness (see Table S5 in the supplemental material). However, partial-correlation analysis showed that the correlation of this ratio with sediment pore water NOx− (r = 0.147, P = 0.666, controlling for NO3−) was due to the correlation with sediment pore water NO3−.
Microbial-membrane core lipids in sediments.
In general, the total amounts of archaeal membrane core lipid iGDGTs increased with water depth, with thaumarchaeol being the major component (56.5 to 68.8%) in the surface sediments of the nSCS (see Table S6 in the supplemental material). The levels of terrestrial bacterial membrane lipids (bGDGTs) were low at all the nSCS stations (see Table S6 in the supplemental material). The calculated BIT values ranged from 0.03 to 0.13 across the nSCS (see Fig. S6 in the supplemental material). The deep-water site E407 had the highest BIT value (0.13), while the BIT values calculated for all the other sites were quite low (<0.1).
Clustering analysis of membrane core lipid data showed two distinct major groups of archaeal iGDGT compositions, with the shallow-water sites (A3, E501, E504, and E505) forming one group and the deep-water sites the other group (see Fig. S7 in the supplemental material), indicating that the fossil archaeal assemblages of the shallow-water surface sediments were distinctly different from those of the deep-water surface sediments in the nSCS. CCA analysis using the archaeal iGDGT data further confirmed this community classification pattern (Fig. 4). The environmental variables in the first two CCA dimensions (CCA1 and CCA2) explained 85.2% of the total variance in the iGDGT composition and 98.1% of the cumulative variance of the iGDGT-environment relationship. CCA1 clearly distinguished the iGDGT compositions of the shallow-water sites from those of the deep-water sites. Of all the environmental factors analyzed (see Table S1 in the supplemental material), only sediment temperature correlated significantly (P = 0.001; 1,000 Monte Carlo permutations) with the composition and distribution of the sediment fossil archaeal community. This environmental factor alone contributed 100.0% of the total CCA explanatory power.
Fig 4.
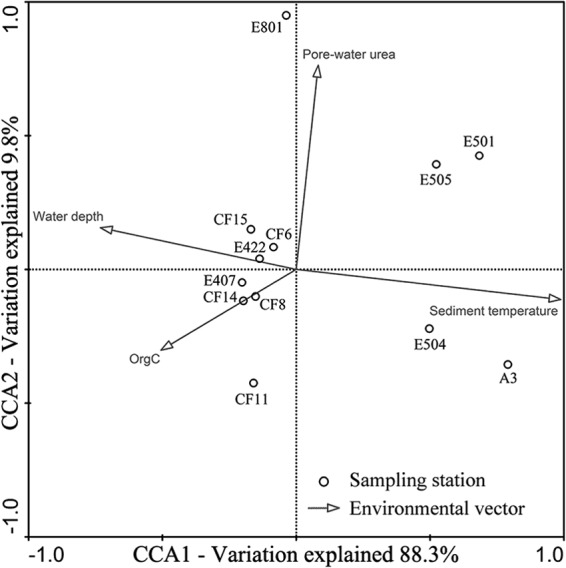
CCA ordination plot for the first two dimensions (CCA1 and CCA2) showing the relationship between the surface sediment fossil archaeal assemblages from the nSCS and environmental parameters analyzed using the archaeal membrane core lipid data.
The abundance of thaumarchaeol in sediments exhibited significant positive correlations with sediment OrgC and OrgN and correlated negatively with sediment temperature (see Table S5 in the supplemental material). The calculated BIT index was significantly positively correlated with sediment OrgC/OrgN but exhibited a negative correlation with sediment OrgN (see Table S5 in the supplemental material).
DISCUSSION
Environment-dependent distribution of sediment thaumarchaeotal microbiota.
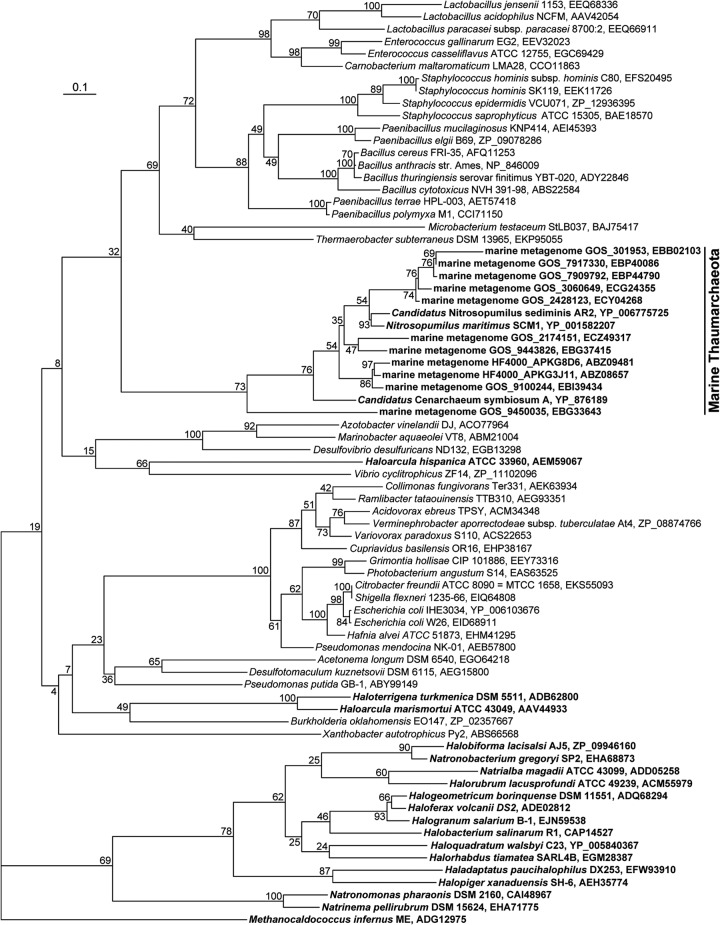
While prior studies suggested ecophysiological adaptation of AOA to low-ammonia conditions (9), numerous other environmental factors may also shape the niches of amoA-bearing thaumarchaeota, including the AOA (references 4 to 6 and references therein). In marine waters, the community structure and biogeochemical contribution of planktonic AOA may change with changing water chemistry and depth (13, 14, 42). Our previous studies indicated that the estuarine and marine sediment AEA community changes under the influence of terrestrial input and sedimentological conditions (7, 28, 31). The current study extended these findings by showing that the estuarine and coastal sites (A3 and E501) had surface sediment AEA assemblages that are distinct from the offshore and deep-water sites in the nSCS (Fig. 2 and 3; see Fig. S5 in the supplemental material). CCA analysis identified sediment temperature, OrgP, and silt content as the key environmental factors that shaped this AEA distribution pattern (Fig. 3). While both temperature and sedimentological conditions have previously been identified as major environmental factors influencing the ecophysiology, community structure, diversity, and spatial distribution of marine AOA or AEA (28, 30, 31, 43), our finding that sediment OrgP is a key environmental factor influencing the composition and distribution of surface sediment AEA is without precedent. Marine thaumarchaeota, including AOA, such as Nitrosopumilus maritimus and “Candidatus Nitrosopumilus sediminis” (12, 44), as well as AEA, such as “Candidatus Cenarchaeum symbiosum” (45), harbor the genes encoding the Phn phosphonate-specific uptake transporter. Homologues of phn genes have also been found in marine metagenomes of putative thaumarchaeotal origin (46, 47). Phosphonates comprise 20 to 30% of the ocean's OrgP and thus are an important P source (reference 4 and references therein). In the domain Archaea, phosphonate transporter genes have been found only in marine thaumarchaeota, some halophilic euryarchaeota, and one extremely thermophilic methanogen (Fig. 5). The unique phosphonate uptake mechanism may indicate the genetic and ecophysiological adaptability of thaumarchaeota to P-limited conditions in marine environments, such as the nSCS (48, 49).
Fig 5.
Consensus phylogenetic tree constructed by using distance neighbor-joining inference of the permease inner membrane protein subunit PhnE sequences of the phosphonate ABC transporter systems. The archaeal PhnE sequences are shown in boldface. The PhnE sequence of the extremely thermophilic methanogen Methanocaldococcus infernus was used as the outgroup.
No significant correlation was found between the abundance of thaumarchaeol and any of the thaumarchaeotal amoA, hcd, or MGI 16S rRNA gene abundances in nSCS surface sediments (data not shown). The thaumarchaeol detected in sediments may represent only fossil (dead) thaumarchaeotal populations (50), the majority of which may originate from the water column (51, 52). In contrast, the measured abundances of signature genes likely reflect the in situ (vital and dead cells with nondegraded DNA) abundance of thaumarchaeotal cells in the sediments. Clustering and CCA analyses of the surface sediment archaeal core lipids documented a difference between the shallow-water and deep-water sites (Fig. 4; see Fig. S7 in the supplemental material). Our CCA analysis also suggests that sediment temperature and the distribution pattern of core lipids are correlated (Fig. 4). Archaeal membrane lipid composition and abundance are responsive to temperature variations, and thus, they have been used to develop paleotemperature proxies (51). Previous studies have also indicated that the sediment diagenesis process does not appear to affect the relative proportions of the GDGTs (53). It is unclear whether the correlation between sediment temperature and archaeal core lipid composition identified by the CCA analysis (Fig. 4) represents a cause-effect relationship.
Low terrestrial nutrient input is the major cause of the oligotrophic conditions in the nSCS, which is supported by gene- and membrane lipid-based analyses of this and other studies (41). Soil-related thaumarchaeotal amoA sequences were only occasionally detected in the constructed clone libraries of the nSCS (cluster II in Fig. 1), which is in stark contrast to the eutrophic Changjiang estuary and the adjacent East China Sea, which receive tremendous inputs of terrestrial materials and thaumarchaeotal microorganisms (31). The low BIT values indicate low terrestrial input in most of the nSCS sampling sites (see Fig. S6 in the supplemental material). This was further supported by the finding that the BIT values correlated positively with sediment OrgC/OrgN and negatively with sediment OrgN in the nSCS (see Table S5 in the supplemental material). High OrgC/OrgN and low OrgN values are usually associated with old and recalcitrant organic matter that may originate from terrestrial input. The nSCS BIT values in general were lower in deep-water sites, except for site E407 (see Fig. S6 in the supplemental material). The relatively high BIT value at site E407 indicated a likely greater terrestrial input. Detection of soil-related thaumarchaeotal amoA sequences in the clone library of site E407 provides another line of evidence of terrestrial input (cluster II in Fig. 1). This seemingly puzzling observation that deep-sea environments may receive stronger terrestrial inputs than estuarine and coastal areas (i.e., A3 and E501) has been reported previously (7).
Our data showed that the surface sediment AEA assemblages responded to the variation of certain key environmental factors in the nSCS (Fig. 3). Sediment physical and geochemical conditions, rather than localized dispersal, play a key role in controlling the spatial distribution of the AEA assemblages in the nSCS surface sediments, consistent with a previous finding about marine water column AOA (42). Our current analyses also indicated that the deep-sea methane hydrate reservoirs or their prospective areas harbored sediment AEA assemblages similar to those of the nonhydrate deep-sea environments in the nSCS (Fig. 2 and 3; see Fig. S5 in the supplemental material). As discussed in a previous publication (29), the nSCS methane hydrates are buried very deeply in the sediments, and the seeping activity may be very low and localized, therefore not causing significant changes to the deep-sea sediment geochemistry.
Ecophysiological potential of sediment MGI thaumarchaeota.
MGI thaumarchaeota constitute a significant cohort of the resident archaea in marine sediments (54, 55). Quantitation of thaumarchaeotal signature genes suggests that the surface sediments in the nSCS also harbor high levels of MGI thaumarchaeota, the majority of which appeared to carry amoA genes (Fig. 1 and Table 1). We found that the abundance of thaumarchaeotal amoA genes was higher than that of betaproteobacterial amoA genes in nSCS surface sediments (Table 1). The thaumarchaeotal-to-betaproteobacterial amoA abundance ratio showed a significant positive correlation with sediment pore water salinity (see Table S5 in the supplemental material), consistent with the results of studies in the water column (42). A direct correlation of the thaumarchaeotal amoA gene abundance with sediment pore water NH4+ was not found (see Table S5 in the supplemental material). Urea was detected in pore water of the nSCS surface sediments (see Table S1 in the supplemental material), and it is possible that some thaumarchaeota utilize urea as an alternative source of ammonia for assimilation, as well as for nitrite production (17, 56, 57). On the other hand, partial-correlation analysis showed that the thaumarchaeotal amoA gene abundance in nSCS surface sediments is significantly positively correlated (r = 0.738, P = 0.009) with sediment pore water NO2− when pore water PO43− is controlled for. This suggests that the surface sediment amoA-bearing thaumarchaeota contribute to in situ nitrite production controlled by PO43− availability. Thaumarchaeota, including marine or estuarine AOA, such as Nitrosopumilus maritimus and “Candidatus Nitrosoarchaeum limnia” SFB1 (12, 58), as well as AEA, such as “Ca. Cenarchaeum symbiosum” (45), harbor genes encoding the Pst phosphate ABC transporter (see Fig. S8 in the supplemental material), a high-affinity, high-activity, but energetically costly phosphate transport system usually activated in P-deficient environments. Homologues of pst genes have also been found in marine metagenomes of putative thaumarchaeotal origin (46). The presence of this P-scavenging genetic inventory indicates the importance of inorganic-P acquisition for thaumarchaeotal ecophysiology. Recent studies have suggested that the nutrient regime of the nSCS ecosystem has been changing from N-limited to P-limited conditions, especially in its estuarine and coastal areas (48, 49). The importance of P availability (including both PO43− and OrgP) to the amoA-bearing thaumarchaeota potentially identifies a previously overlooked linkage of N, C, and P cycles in marine environments.
The average ratio of thaumarchaeotal amoA to MGI 16S rRNA gene abundance is 1.1:1, close to unity; however, this ratio ranged widely (0.1:1 to 3.2:1) in the different sampling sites of the nSCS. Fully sequenced marine thaumarchaeotal species, such as N. maritimus, “Ca. Cenarchaeum symbiosum,” “Ca. Nitrosoarchaeum limnia,” and “Candidatus Nitrosopumilus salaria,” harbor only one amoA gene per genome (12, 45, 58, 59). Nearly equal abundances of amoA and 16S rRNA genes from environmental MGI thaumarchaeota have been reported from most studied marine environments (reference 60 and references therein). However, a recent study indicated that the commonly used amoA primers might miss a significant fraction of the amoA-bearing thaumarchaeota (61). This potentially incomplete coverage of thaumarchaeotal amoA gene diversity by PCR primers may explain some of the amoA/16S rRNA gene ratios below 1 obtained in many studies, including ours (5). A recent report of some MGI thaumarchaeota that likely do not carry amo genes (62) contributes another explanation for the observed unequal abundances of thaumarchaeotal amoA and 16S rRNA genes. Previous studies also showed that the MGI thaumarchaeotal diversity is quite high and that MGI 16S rRNA gene clusters specific to marine sediment environments exist (55). However, commonly used archaeal 16S rRNA gene PCR primers may miss some of the thaumarchaeotal diversity included in marine sediment environments (63). This may be part of the reason why some of the nSCS sediment sites had a thaumarchaeotal amoA/16S rRNA gene ratio greater than unity (Table 1). Some studies have reported that MGI thaumarchaeota can take up amino acids and other organic compounds to live chemo-organotrophically or mixotrophically (64, 65). Hence, the mere detection of amoA genes is insufficient for inferring ammonia-dependent chemolithotrophy (2, 8, 28, 66).
Thaumarchaeotal hcd genes were also found in the surface sediments of the nSCS (Table 1). In general, the abundances of all three targeted thaumarchaeotal genes, hcd, amoA, and 16S rRNA, were significantly positively correlated, suggesting that the majority, if not all, of the amoA-bearing thaumarchaeota in marine sediments fix carbon autotrophically.
MGI thaumarchaeotal amoA abundance exhibited a significant negative correlation with sediment pore water DO in the nSCS (see Table S5 in the supplemental material), likely indicating active oxygen consumption by sediment thaumarchaeotal ammonia oxidation. The amoA-bearing thaumarchaeota, including AOA, are abundant and well adapted to hypoxic marine environments, including the water column and sediments (references 18, 20, 28, 55, and 67–69 and references therein). In our current study, sediment pore water NO3− showed a significant positive correlation with the ratio of thaumarchaeotal hcd to amoA gene abundances in the surface sediments (see Table S5 in the supplemental material). Partial-correlation analysis indicated that this correlation is real (P = 0.016, controlling for DO; P = 0.008, controlling for OrgC/OrgN; P = 0.008, controlling for sediment clay content; and P = 0.025, controlling for sediment skewness). Because specific MGI thaumarchaeotal clusters have been identified recently in nitrate-reducing marine sediments (55), it is possible that thaumarchaeota cooperate metabolically with diverse N-transforming microbes, including nitrate reducers, in marine sediments (66–69).
The molecular basis for catabolism of MGI thaumarchaeota has not yet been elucidated (5). The sole report so far that correlated genetic potential with evidence of a chemo-organoheterotrophic lifestyle investigated nonmarine AEA in wastewater (8), and the mode of carbon fixation in the putatively amoA-lacking thaumarchaeon Giganthauma has not yet been reported (62). Thus, it is still an open question whether marine sediment thaumarchaeota per se exhibit variable catabolic lifestyles that include chemo-organotrophic and non-ammonia-based chemolithotrophic catabolism, in addition to the ammonia-dependent chemolithotrophy of the bona fide AOA.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by China NSFC grants 91028011 (H.D.), 41076091 (H.D. and M.G.K.), and 91028001 (N.J.); National Key Basic Research Program of China grants 2013CB955700 (N.J., C.Z., and H.D.) and 2007CB411702 (X.L.); China SOA grant 201105021 (N.J.); and U.S. NSF grants 0541797 and 0948202 (M.G.K.). The lipid work was supported by China NSFC grant 91028005 (C.Z.) and the National “Thousand Talents Program” at the State Key Laboratory of Marine Geology of Tongji University, Shanghai, China.
We thank the reviewers for valuable comments. We also thank the crew and onboard scientists of the 2007 South China Sea Open Cruise by R/V Shiyan 3, SCSIO, CAS, for sediment sampling and Ruipeng Chen, Zheng Wang, Ling Li, Huaiyong Sun, Yingcai Sun, Jun Zuo, Dong Jiang, Jin He, Xinwei Du, Saozheng Li, and Fang Tian for their assistance in the project.
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03204-12.
REFERENCES
- 1. Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252 [DOI] [PubMed] [Google Scholar]
- 2. Pester M, Schleper C, Wagner M. 2011. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr. Opin. Microbiol. 14:300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 4. Stahl DA, de la Torre JR. 2012. Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66:83–101 [DOI] [PubMed] [Google Scholar]
- 5. Hatzenpichler R. 2012. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl. Environ. Microbiol. 78:7501–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. 2009. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 33:855–869 [DOI] [PubMed] [Google Scholar]
- 7. Dang H, Li J, Zhang X, Li T, Tian F, Jin W. 2009. Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J. Appl. Microbiol. 106:1482–1493 [DOI] [PubMed] [Google Scholar]
- 8. Mußmann M, Brito I, Pitcher A, Sinninghe Damsté JS, Hatzenpichler R, Richter A, Nielsen JL, Nielsen PH, Müller A, Daims H, Wagner M, Head IM. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:16771–16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979 [DOI] [PubMed] [Google Scholar]
- 10. Schleper C. 2010. Ammonia oxidation: different niches for bacteria and archaea? ISME J. 4:1092–1094 [DOI] [PubMed] [Google Scholar]
- 11. Berg IA, Kockelkorn D, Buckel W, Fuchs G. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782–1786 [DOI] [PubMed] [Google Scholar]
- 12. Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PS, Chan PP, Gollabgir A, Hemp J, Hügler M, Karr EA, Könneke M, Shin M, Lawton TJ, Lowe T, Martens-Habbena W, Sayavedra-Soto LA, Lang D, Sievert SM, Rosenzweig AC, Manning G, Stahl DA. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:8818–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu AY, Jiao NZ, Zhang CL. 2011. Community structure and function of planktonic Crenarchaeota: changes with depth in the South China Sea. Microb. Ecol. 62:549–563 [DOI] [PubMed] [Google Scholar]
- 14. Hu AY, Jiao NZ, Zhang R, Yang Z. 2011. Niche partitioning of marine group I Crenarchaeota in the euphotic and upper mesopelagic zones of the East China Sea. Appl. Environ. Microbiol. 77:7469–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pitcher A, Wuchter C, Siedenberg K, Schouten S, Sinninghe Damsté JS. 2011. Crenarchaeol tracks winter blooms of ammonia-oxidizing Thaumarchaeota in the coastal North Sea. Limnol. Oceanogr. 56:2308–2318 [Google Scholar]
- 16. Varela MM, van Aken HM, Sintes E, Reinthaler T, Herndl GJ. 2011. Contribution of Crenarchaeota and Bacteria to autotrophy in the North Atlantic interior. Environ. Microbiol. 13:1524–1533 [DOI] [PubMed] [Google Scholar]
- 17. Yakimov MM, La Cono V, Smedile F, Deluca TH, Juárez S, Ciordia S, Fernández M, Albar JP, Ferrer M, Golyshin PN, Giuliano L. 2011. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (central Mediterranean Sea). ISME J. 5:945–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park BJ, Park SJ, Yoon DN, Schouten S, Sinninghe Damsté JS, Rhee SK. 2010. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl. Environ. Microbiol. 76:7575–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Offre P, Nicol GW, Prosser JI. 2011. Community profiling and quantification of putative autotrophic thaumarchaeal communities in environmental samples. Environ. Microbiol. Rep. 3:245–253 [DOI] [PubMed] [Google Scholar]
- 20. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walsh JJ. 1991. Importance of continental margins in the marine biogeochemical cycling of carbon and nitrogen. Nature 350:53–55 [Google Scholar]
- 22. Schouten S, Hopmans EC, Sinninghe Damsté JS. 2013. The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: a review. Org. Geochem. 54:19–63 [Google Scholar]
- 23. Hopmans EC, Weijers JWH, Schefuß E, Herfort L, Sinninghe Damsté JS, Schouten S. 2004. A novel proxy for terrestrial organic matter in sediments based on branched and isoprenoid tetraether lipids. Earth Planet. Sci. Lett. 224:107–116 [Google Scholar]
- 24. Moisander PH, Beinart RA, Voss M, Zehr JP. 2008. Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon. ISME J. 2:954–967 [DOI] [PubMed] [Google Scholar]
- 25. Wu J, Chung SW, Wen LS, Liu KK, Chen YL, Chen H, Karl DM. 2003. Dissolved inorganic phosphorus, dissolved iron, and Trichodesmium in the oligotrophic South China Sea. Global Biogeochem. Cycles 17:1008 [Google Scholar]
- 26. Yin KD, Qian P-Y, Wu MCS, Chen JC, Huang LM, Song XY, Jian WJ. 2001. Shift from P to N limitation of phytoplankton growth across the Pearl River estuarine plume during summer. Mar. Ecol. Prog. Ser. 221:17–28 [Google Scholar]
- 27. Zhang Y, Zhao ZH, Sun J, Jiao NZ. 2011. Diversity and distribution of diazotrophic communities in the South China Sea deep basin with mesoscale cyclonic eddy perturbations. FEMS Microbiol. Ecol. 78:417–427 [DOI] [PubMed] [Google Scholar]
- 28. Dang HY, Luan XW, Chen RP, Zhang XX, Guo LZ, Klotz MG. 2010. Diversity, abundance and distribution of amoA-encoding archaea in deep-sea methane seep sediments of the Okhotsk Sea. FEMS Microbiol. Ecol. 72:370–385 [DOI] [PubMed] [Google Scholar]
- 29. Dang HY, Yang JY, Li J, Luan XW, Zhang YB, Gu GZ, Xue RR, Zong MY, Klotz MG. 2013. Environment-dependent distribution of the sediment nifH-harboring microbiota in the northern South China Sea. Appl. Environ. Microbiol. 79:121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cao H, Hong Y, Li M, Gu JD. 2011. Phylogenetic diversity and ecological pattern of ammonia-oxidizing archaea in the surface sediments of the western Pacific. Microb. Ecol. 62:813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dang HY, Zhang XX, Sun J, Li TG, Zhang ZN, Yang GP. 2008. Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiology 154:2084–2095 [DOI] [PubMed] [Google Scholar]
- 32. Schloss PD, Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altschul S, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 35. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 36. Mincer YJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9:1162–1175 [DOI] [PubMed] [Google Scholar]
- 37. Stephen JR, Chang YJ, Macnaughton SJ, Kowalchuk GA, Leung KT, Flemming CA, White DC. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ge HM, Zhang CL, Dang HY, Zhu C, Jia GD. Distribution of tetraether lipids in surface sediments of the northern South China Sea: implications for TEX86 proxies. Geosci. Front., in press. doi:10.1016/j.gsf.2012.10.002 [Google Scholar]
- 39. Mullins TD, Britschgi TB, Krest RL, Giovannoni SJ. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148–158 [Google Scholar]
- 40. Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wei Y, Wang J, Liu J, Dong L, Li L, Wang H, Wang P, Zhao M, Zhang CL. 2011. Spatial variations in archaeal lipids of surface water and core-top sediments in the South China Sea and their implications for paleoclimate studies. Appl. Environ. Microbiol. 77:7479–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bouskill NJ, Eveillard D, Chien D, Jayakumar A, Ward BB. 2012. Environmental factors determining ammonia-oxidizing organism distribution and diversity in marine environments. Environ. Microbiol. 14:714–729 [DOI] [PubMed] [Google Scholar]
- 43. Mosier AC, Francis CA. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002–3016 [DOI] [PubMed] [Google Scholar]
- 44. Park SJ, Kim JG, Jung MY, Kim SJ, Cha IT, Ghai R, Martín-Cuadrado AB, Rodríguez-Valera F, Rhee SK. 2012. Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus sediminis” AR2, from Svalbard in the Arctic Circle. J. Bacteriol. 194:6948–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, Preston C, de la Torre J, Richardson PM, DeLong EF. 2006. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. U. S. A. 103:18296–18301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcón LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77 doi:10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Konstantinidis KT, DeLong EF. 2008. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2:1052–1065 [DOI] [PubMed] [Google Scholar]
- 48. Xu J, Yin KD, He L, Yuan XC, Ho AYT, Harrison PJ. 2008. Phosphorus limitation in the northern South China Sea during late summer: influence of the Pearl River. Deep Sea Res. I 55:1330–1342 [Google Scholar]
- 49. Ning X, Lin C, Hao Q, Liu C, Le F, Shi J. 2009. Long term changes in the ecosystem in the northern South China Sea during 1976–2004. Biogeosciences 6:2227–2243 [Google Scholar]
- 50. Pitcher A, Hopmans EC, Mosier AC, Park SJ, Rhee SK, Francis CA, Schouten S, Sinninghe Damsté JS. 2011. Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing archaea enriched from marine and estuarine sediments. Appl. Environ. Microbiol. 77:3468–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schouten S, Hopmans EC, Schefuß E, Sinninghe Damsté JS. 2002. Distributional variations in marine crenarchaeotal membrane lipids: a new tool for reconstructing ancient sea water temperatures? Earth Planet. Sci. Lett. 204:265–274 [Google Scholar]
- 52. Lengger SK, Hopmans EC, Reichart GJ, Nierop KGJ, Sinninghe Damsté JS, Schouten S. 2012. Intact polar and core glycerol dibiphytanyl glycerol tetraether lipids in the Arabian Sea oxygen minimum zone. Part II. Selective preservation and degradation in sediments and consequences for the TEX86. Geochim. Cosmochim. Acta 98:244–258 [Google Scholar]
- 53. Schouten S, Hopmans EC, Sinninghe Damsté JS. 2004. The effect of maturity and depositional redox conditions on archaeal tetraether lipid palaeothermometry. Org. Geochem. 35:567–571 [Google Scholar]
- 54. Roussel EG, Sauvadet AL, Chaduteau C, Fouquet Y, Charlou JL, Prieur D, Cambon Bonavita MA. 2009. Archaeal communities associated with shallow to deep subseafloor sediments of the New Caledonia Basin. Environ. Microbiol. 11:2446–2462 [DOI] [PubMed] [Google Scholar]
- 55. Durbin AM, Teske A. 2011. Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 13:3219–3234 [DOI] [PubMed] [Google Scholar]
- 56. Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, Lovejoy C, Tremblay JE, Potvin M, Heinrich F, Estrada M, Riemann L, Bork P, Pedrós-Alió C, Bertilsson S. 2012. Role for urea in nitrification by polar marine Archaea. Proc. Natl. Acad. Sci. U. S. A. 109:17989–17994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu L, Han W, Zhang J, Wu Y, Wang B, Lin X, Zhu J, Cai Z, Jia Z. 2012. Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea. ISME J. 6:1978–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626 doi:10.1371/journal.pone.0016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA. 2012. Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J. Bacteriol. 194:2121–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sintes E, Bergauer K, De Corte D, Yokokawa T, Herndl GJ. 2012. Archaeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Environ. Microbiol. doi:10.1111/j.1462-2920.2012.02801.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lund MB, Smith JM, Francis CA. 2012. Diversity, abundance and expression of nitrite reductase (nirK)-like genes in marine thaumarchaea. ISME J. 6:1966–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Muller F, Brissac T, Le Bris N, Felbeck H, Gros O. 2010. First description of giant Archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ. Microbiol. 12:2371–2383 [DOI] [PubMed] [Google Scholar]
- 63. Teske A, Sørensen KB. 2008. Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J. 2:3–18 [DOI] [PubMed] [Google Scholar]
- 64. Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:e95 doi:10.1371/journal.pbio.0040095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Teira E, van Aken H, Veth C, Herndl GJ. 2006. Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol. Oceanogr. 51:60–69 [Google Scholar]
- 66. Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MM. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. U. S. A. 104:7104–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Coolen MJL, Abbas B, van Bleijswijk J, Hopmans EC, Kuypers MMM, Wakeham SG, Sinninghe Damsté JS. 2007. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ. Microbiol. 9:1001–1016 [DOI] [PubMed] [Google Scholar]
- 68. Beman JM, Popp BN, Francis CA. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429–441 [DOI] [PubMed] [Google Scholar]
- 69. Stewart FJ, Ulloa O, DeLong EF. 2012. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ. Microbiol. 14:23–40 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.