Abstract
Gene transcription (microarrays) and protein levels (proteomics) were compared in cultures of the acidophilic chemolithotroph Acidithiobacillus ferrooxidans grown on elemental sulfur as the electron donor under aerobic and anaerobic conditions, using either molecular oxygen or ferric iron as the electron acceptor, respectively. No evidence supporting the role of either tetrathionate hydrolase or arsenic reductase in mediating the transfer of electrons to ferric iron (as suggested by previous studies) was obtained. In addition, no novel ferric iron reductase was identified. However, data suggested that sulfur was disproportionated under anaerobic conditions, forming hydrogen sulfide via sulfur reductase and sulfate via heterodisulfide reductase and ATP sulfurylase. Supporting physiological evidence for H2S production came from the observation that soluble Cu2+ included in anaerobically incubated cultures was precipitated (seemingly as CuS). Since H2S reduces ferric iron to ferrous in acidic medium, its production under anaerobic conditions indicates that anaerobic iron reduction is mediated, at least in part, by an indirect mechanism. Evidence was obtained for an alternative model implicating the transfer of electrons from S0 to Fe3+ via a respiratory chain that includes a bc1 complex and a cytochrome c. Central carbon pathways were upregulated under aerobic conditions, correlating with higher growth rates, while many Calvin-Benson-Bassham cycle components were upregulated during anaerobic growth, probably as a result of more limited access to carbon dioxide. These results are important for understanding the role of A. ferrooxidans in environmental biogeochemical metal cycling and in industrial bioleaching operations.
INTRODUCTION
The ability of microorganisms to catalyze dissimilatory redox transformations of sulfur and iron has had a major impact on the evolution of planet Earth. Ferrous iron (Fe2+) can act as an electron donor for some aerobic chemolithotrophs, while ferric iron (Fe3+) is used as a terminal electron acceptor by a wide range of obligate and facultative anaerobes, including acidophilic prokaryotes (1). In addition, many acidophiles can obtain energy from the oxidation of elemental sulfur (S0) and inorganic sulfur compounds.
It is recognized that dissimilatory ferric iron reduction has a profound effect on the iron cycle by mediating both the destruction and formation of iron minerals (reviewed in reference 2). Fe3+ reduction has been extensively studied in neutrophiles such as Geobacter and Shewanella spp. which use solid Fe3+ (hydr)oxides as electron acceptors (reviewed in reference 3). In Shewanella oneidensis, outer membrane c-type cytochromes have been suggested to cause the direct reduction of solid Fe3+ (hydr)oxides (4). In addition, electron shuttles (5) and an Fe3+ chelating/solubilizing pathway (6) were suggested for a Shewanella sp., thereby avoiding the need for direct contact with a solid substrate. At a pH of <2.5, Fe3+ is soluble, though some basic ferric iron minerals (jarosites) can form at pHs of <2. Therefore, extremely acidophilic iron-reducing prokaryotes do not have the problem of utilizing a solid-phase electron sink. Dissimilatory Fe3+ reduction is widespread among moderately acidophilic and extremely acidophilic bacteria (reviewed in reference 7). Acidophilic iron reducers are unrelated to their neutrophilic counterparts and display considerable phylogenetic diversity (7). Different strategies to reduce Fe(III) have been described in phylogenetically diverse Fe(III) reducers, suggesting that these mechanisms evolved independently several times (8).
The redox potential (Eh) of the Fe2+/Fe3+ couple is affected by solution pH and associated organic/inorganic ligands. In extremely acidic (pH, ∼2) sulfate-based solutions, the Eh value is ∼+700 mV (9), whereas it is much more electronegative at circumneutral pH (∼+300 mV [10]). In bioenergetic terms, Fe2+ oxidation yields less energy in acidic than in neutral pH environments, whereas Fe3+ is a more energetically favorable electron acceptor at low pH (11).
Acidithiobacillus ferrooxidans is the most widely studied of all acidophiles and is known to use a variety of electron donors (Fe2+, elemental sulfur [S0], reduced inorganic sulfur compounds, hydrogen [H2], and formic acid) and either molecular oxygen, S0, or Fe3+ as electron acceptor (12–14). One strain of A. ferrooxidans has been reported to couple the oxidation of H2 to the reduction of S0 (12), and the type strain genome contains genes predicted to encode hydrogenases and a S0 reductase (15) that have both been biochemically characterized (16–18). Models for substrate oxidation and energy conservation during aerobic growth of A. ferrooxidans on either Fe2+ or inorganic sulfur compounds have been developed on the basis of functional genomic approaches (15, 19–21) as well as molecular genetics and biochemical methods (reviewed in references 3 and 21 to 23). In contrast, relatively little is known about the mechanism of Fe3+ reduction in acidophiles. Based on kinetics and inhibition reaction analysis, Corbett and Ingledew (24) proposed an anaerobic pathway where electrons from S0 oxidation enter the respiratory chain via the bc1 complex and continue through periplasmic transporters involved in the iron-oxidizing system to the final electron acceptor, Fe3+. This model is supported by evidence from inhibition reactions and biochemical studies (14). It was also proposed that the oxidation of S0 by Fe3+ was one of two steps in sulfur metabolism with specific enzymes involved. However, the various results might also be explained because a number of different strains of bacteria were used, complicating the interpretation of these analyses (25–27). Tetrathionate reductase (TetH) (28) and ArsH (29) have been proposed to be candidates for the final electron donor to Fe3+. In addition, Kucera et al. (30) found a significant increase in the abundance of electron transporters, such as rusticyanin and cytochrome c552 (which are known to be involved in Fe2+ oxidation), in cultures of A. ferrooxidans grown anaerobically on Fe3+. These authors suggested that rus operon-encoded proteins were involved in both Fe2+ oxidation and Fe3+ reduction in this acidophile.
Iron- and sulfur-oxidizing acidophiles have widespread use in processing metal ores, a technology generically known as “biomining.” Their key roles are to generate both Fe3+, a powerful chemical oxidant that initiates the oxidative dissolution of sulfidic minerals, and sulfuric acid, which maintains metals released from the minerals in solution, thereby facilitating downstream recovery. Biomining acidophiles, such as A. ferrooxidans, that can reduce as well as oxidize iron may be responsible for net consumption of Fe3+ in anaerobic zones and microsites (e.g., in heap reactors) and, thus, potentially have a negative impact on mineral dissolution. On the other hand, reductive dissolution of Fe3+ minerals has been demonstrated to have potential for bioprocessing of oxidized metal ores, such as nickel laterites (31). Consequently, understanding the nature of Fe3+ reduction by A. ferrooxidans and how it is controlled is of fundamental importance in mineral bioprocessing technologies.
In order to address the lacuna in our knowledge of the mechanism of S0-coupled iron reduction in A. ferrooxidans, a transcriptomic and proteomic study was undertaken. mRNA and protein contents were explored either during aerobic growth with S0 as an electron source and O2 as electron acceptor or under anaerobic conditions with Fe3+ as electron acceptor. Differential RNA and protein levels were related to changes in cellular functions that were used to develop a preliminary model for A. ferrooxidans electron transport during dissimilatory Fe3+ reduction.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and harvesting.
The purity of the A. ferrooxidans culture (ATCC 23270T) was confirmed by streaking onto a variety of overlay media (32). A. ferrooxidans was preadapted to growth on 1% (wt/vol) S0 in a shake flask culture (30°C; initial pH, 2.5) with a basal salts-trace element mix before inoculation into a 2.3-liter bioreactor fitted with pH, temperature, and aeration control (Electrolab, United Kingdom). The basal salts contained (final concentrations) MgSO4 · 7H2O (1 g/liter), (NH4)2SO4 (0.9 g/liter), Na2SO4 · 10H2O (0.3 g/liter), KH2PO4 (0.1 g/liter), KCl (0.1 g/liter), and Ca(NO3)2 · 4H2O (0.03 g/liter) with a trace element solution of (final concentrations) ZnSO4 · 7H2O (20 mg/liter), CuSO4 · 5H2O (2 mg/liter), MnSO4 · 4H2O (2 mg/liter), CoSO4 · 7H2O (2 mg/liter), Cr2(SO4)3 · 15H2O (1 mg/liter), H3BO3 (1.2 mg/liter), Na2MoO4 · 2H2O (1 mg/liter), NiSO4 · 6H2O (2 mg/liter), Na2SeO4 · 10H2O (2 mg/liter), Na2WO4 2H2O (0.2 mg/liter), and NaVO3 (0.2 mg/liter). The reactor contained 100 g S0 (sterilized by Tyndallization) and 2 liters of basal salts-trace element mix. The temperature was maintained at 30°C, the pH was maintained at 1.8 (by automated addition of 1 M H2SO4 or NaOH), and the bioreactor was aerated with sterile air at 1 liter/min and stirred at 150 rpm. Bacterial growth was monitored by using a Thoma counting chamber (Hawksley, United Kingdom). Culture agitation and aeration were stopped when cell numbers increased to ∼109/ml, allowing the S0 particles to collect as a loose sediment. Approximately 1.5 liters of the surface liquor was aseptically withdrawn during exponential growth when nutrients were not depleted, and cells were harvested by centrifugation (10,000 × g for 20 min at 4°C) and washed three times in basal salts solution (pH 2). Part of the obtained cell pellet was frozen (−20°C) ahead of proteomic analysis, while the remainder was suspended in RNAlater (Qiagen, United Kingdom) and stored at −20°C prior to transcriptomic investigations. The bioreactor was topped up with 1.5 liters of sterile basal salts-trace elements, and two further cycles of aerobic growth were carried out under identical conditions.
S0 was also used for anaerobic growth, but the electron acceptor was switched from O2 to Fe3+. Following harvesting of the third aerobic batch culture, the bioreactor was inoculated with A. ferrooxidans grown anaerobically in 25 mM ferric sulfate, 1% (wt/vol) S0, pH 2.0, at 30°C in an anaerobic jar (AnaeroGen system; Oxoid, United Kingdom). Basal salts-trace elements were added to bring the bioreactor culture volume to ∼2 liters, and ferric sulfate was added (from a filter-sterilized 0.5 M solution, pH 1.5) to give a final concentration of ∼20 mM. The culture was deaerated with a stream of oxygen-free N2 (2 liters/min for 30 min), and then the N2 flow was lowered to 1 liter/min. Carbon was provided by intermittently sparging the bioreactor (∼30 min/day) with a 10% CO2–90% N2 gas mix. Temperature and pH were maintained as for aerobic growth. Fe3+ reduction was monitored by measuring Fe2+ concentrations using the ferrozine assay (33), and when >90% of the iron had been reduced, additional ferric sulfate solution (equivalent to 20 mM Fe3+) was added to the bioreactor. Sequential addition of relatively small volumes of ferric sulfate ensured that no Fe3+ precipitates formed, and for each batch culture, the equivalent of ∼100 mM Fe3+ was added. Anaerobic cultures were harvested as described previously, and cell pellets from three separate batch cultures were obtained.
Sulfide generation during A. ferrooxidans anaerobic growth.
Evidence for production of hydrogen sulfide (H2S) by A. ferrooxidans maintained under anaerobic conditions in the presence of S0 was obtained by incubating cells in the presence of Cu2+, which reacts with H2S to form insoluble CuS in acidic liquors (34). Six 25-ml universal bottles, each containing 10 mM ferrous sulfate, 10 mM copper(II) sulfate, 100 mg of S0, and basal salts-trace elements (total volume, 20 ml), were inoculated with 1 ml of an A. ferrooxidans culture (approximately 1 × 108 cells/ml) grown aerobically on S0. Two noninoculated cultures were also prepared. Ferrous iron, rather than Fe3+, was used in this experiment to preclude the reaction 2Fe3+ + H2S → 2Fe2+ + S0 + 2H+. All eight bottles were incubated under anaerobic conditions (AnaeroGen system; Oxoid, United Kingdom) for up to 6 weeks, and concentrations of soluble Cu2+ were measured using ion chromatography (34).
Two-dimensional electrophoresis, image analysis, and protein identification.
Proteomic analysis of total soluble protein and outer membrane-enriched fractions was carried out as described previously (35). This method enriches for outer membrane proteins but is not compatible with inner membrane proteins. Protein samples were extracted from triplicate bacterial samples. First-dimension isoelectric focusing was performed using Immobiline DryStrip immobilized pH gradient gels (18 cm; nonlinear pH range, pH 3 to 10; GE Healthcare), followed by second-dimension SDS-polyacrylamide gel electrophoresis. The Coomassie-stained gels were analyzed with image analysis software (Melanie, version 7.03; Genebio). The criteria used to identify protein spot intensities were defined by Mangold et al. (35). For analysis of the cytoplasmic proteins, all spots detected were taken into account. However, five predominant spots in gels from the anaerobic outer membrane-enriched fraction were excluded to avoid a skewed analysis which would otherwise misleadingly identify many proteins as upregulated under aerobic conditions (see Fig. S2 in the supplemental material). Proteins were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (35). Peptide mass fingerprints were searched against a local database containing the genome sequence of A. ferrooxidans ATCC 23270T using the Mascot search engine, allowing 2 missed cleavages and a peptide tolerance of 50 ppm (36). Database hits were significant, with a score of 47 at a 0.05 significance level.
RNA isolation, transcriptional arrays, and data analysis.
Cells were pelleted from suspensions in RNAlater and washed in 10 mM H2SO4. Total RNA was extracted using a modified acid-phenol extraction method, including an initial step with TRIzol reagent, as described by the manufacturer (Invitrogen). RNA was further purified using a High Pure RNA isolation kit (Roche Applied Science) and treated with DNase I (Roche Applied Science) according to the manufacturer's instructions. The absence of DNA was verified by PCR for each sample. RNA was used for the synthesis of cDNA fluorescently labeled with Cy3 and Cy5 (20, 37). Microarray transcript profiling was carried out as described previously (20, 37). Arrays were scanned for fluorescent signals using a ScanArray 4400A scanner (MDS Analytical Technologies). The processed aerobic (S0/O2) and anaerobic (S0/Fe3+) signal intensities for each spot were used for calculating the aerobic/anaerobic mRNA concentration ratio (37). Each gene expression ratio was calculated from 40 values calculated from 4 biological and 10 technical replicates and normalized by the nonlinear Lowess method using the Acuity (version 4.0) program (Molecular Devices). A 0.7-fold deviation from the 1:1 hybridization (log2) ratio was taken as indicative of differential levels of RNA. Hierarchical cluster analysis (Pearson correlation, average linkage) was performed using the Genesis software suite (38).
qPCRs.
Quantitative real-time PCRs (qPCRs) were performed with an iCycler thermal cycler (Bio-Rad) and a KAPA SYBR FAST qPCR kit (Kapabiosystems). Twenty-microliter PCR mixtures contained 2 μl of a 1:100-diluted cDNA sample, 200 nM each primer, and 1× KAPA SYBR FAST qPCR master mix. The reference dye carboxy-X-rhodamine was included at a final concentration of 5 nM. The cycling protocol was as follows: initial denaturation for 10 min at 95°C, followed by 40 cycles of 30 s each at 95°C, 56°C, and 72°C. Fluorescence was measured after the extension phase at 72°C. Specific amplification was confirmed by a single peak in the melting curve. For each experimental condition, total RNA was extracted from two independent A. ferrooxidans cultures. Each RNA sample was retrotranscribed to evaluate concentration levels relative to the concentration of the constitutive reference gene rpoC (39). The reactions for each target gene were performed in triplicate, and each target gene was simultaneously amplified in the same PCR run. Thus, data sets consist of 6 values per gene per experimental setup generated under standardized PCR cycling conditions. Tenfold dilutions of stationary-phase genomic DNA (range, 10 ng to 1 pg) were used to generate a 5-point standard curve for each gene by using the cycle threshold (CT) value versus the logarithm of each dilution factor. The reaction efficiency (E), which was equal to [10(−1/slope)]−1, for every gene was derived from the slope of the corresponding standard curves. The genes tested by qPCR and the respective primers used in the reactions are listed in Table S1 in the supplemental material.
Bioinformatic analysis.
The sequence and annotation of the complete A. ferrooxidans ATCC 23270T genome were retrieved from Gen-Bank/EMBL/DDBJ (CP001219) (15). The annotated genome was displayed in the Artemis interactive format (www.sanger.ac.uk/Software/Artemis). The bioinformatics programs used to characterize candidate genes and their predicted protein products included blastp, tblastn, blastx, and psiblast (www.ncbi.nlm.nih.gov) and the suite of protein characterization programs available in InterproScan (www.ebi.ac.uk/interpro). Metabolic reconstructions were carried out on the basis of comparisons with the model metabolic pathways obtained from the BIOCYC (www.biocyc.org), KEGG (www.genome.ad.jp/kegg), and ERGO (Integrated Genomics) databases. Comparative genomic analyses were performed using the Microbesonline (www.microbesonline.org) and String (http://string.embl.de/) programs.
Microarray data accession number.
The data for all hybridizations were submitted to the MIAMExpress database under accession number E-MEXP-3361.
RESULTS AND DISCUSSION
With the exception of the electron acceptor and CO2 supply, aerobic (S0/O2) and anaerobic (S0/Fe3+) cultures of A. ferrooxidans ATCC 23270 were grown under similar conditions. Although similar cell yields (∼109/ml) were obtained, cell mass was attained in 3 to 5 days when O2 was the electron acceptor, whereas cell mass was attained in 2 to 3 weeks when Fe3+ was used (data not shown).
Proteomic and transcriptomic analysis of S0/O2 versus S0/Fe3+ growth.
This study compared protein levels and RNA synthesis during aerobic and anaerobic growth of A. ferrooxidans on S0 (Table 1; details are given in Tables S2 and S3 in the supplemental material). A total of 78 upregulated and 13 unique protein spots were identified from the soluble- and outer membrane-enriched aerobic cultures, while 11 upregulated and 6 unique protein spots were identified from the anaerobic cultures (see Fig. S1 and S2 in the supplemental material). Comparative transcriptomic microarrays of A. ferrooxidans grown aerobically and under anaerobic conditions on sulfur identified a total of 42 and 44 upregulated genes, respectively.
Table 1.
Gene product concentration levels (microarrays and qPCR) and protein spot intensities (proteomics) from Acidithiobacillus ferrooxidans grown under aerobic and anaerobic conditionse
| Regulation, use, and protein or gene name | Predicted function | Annotation no.a | Fold change in protein expression | Median gene log2 ratio | qPCRf |
|---|---|---|---|---|---|
| Upregulated under aerobic conditions | |||||
| Energy metabolism | |||||
| HdrB | Heterodisulfide reductase subunit B, homolog | AFE_2586b | 9.4 | 1.0 | |
| HdrB | Heterodisulfide reductase subunit B, homolog | AFE_2586b | 13.5 | ||
| HdrA | Pyridine nucleotide disulfide oxidoreductase | AFE_2553b | 3.3 | ||
| HdrA | Pyridine nucleotide disulfide oxidoreductase | AFE_2553b | 8.1 | ||
| Conserved hypothetical protein (clusters with Hdr) | AFE_2552 | 5.4 | |||
| AtpA | ATP synthase F1, α subunit (EC 3.6.3.14) | AFE_3205 | 2.1c | ||
| atpC | ATP synthase F1, ε subunit | AFE_3202 | 1.0 | ||
| AtpD | ATP synthase F1, β subunit (EC 3.6.3.14) | AFE_3203 | 2.3 | ||
| MotA/TolQ/ExbB proton channel family protein | AFE_0768 | 0.8 | |||
| tetH | Tetrathionate hydrolase | AFE_0029 | 4.3 | ||
| Cyc2 | Outer membrane cytochrome c, Cyc2 | AFE_3153 | 5.6 | 0.9 | |
| cyc1 | Cytochrome c4, Cyc1 | AFE_3152 | 0.7 | ||
| coxA | Cytochrome c oxidase, aa3 type, subunit I | AFE_3149 | 0.9 | ||
| coxC | Cytochrome c oxidase, aa3 type, subunit III | AFE_3148 | 0.9 | ||
| rus | Rusticyanin | AFE_3146 | 1.7 | ||
| Carbon management | |||||
| Fba | Fructose bisphosphate aldolase, class II (EC 4.1.2.13) | AFE_3248b | 2.7 | O2 > Fe3+ | |
| Fructose bisphosphate aldolase, putative | AFE_1802 | 4.9 | |||
| Fba | Fructose-bisphosphate aldolase, class II (EC 4.1.2.13) | AFE_3248b | 3.4 | ||
| Fba | Fructose bisphosphate aldolase, class II (EC 4.1.2.13) | AFE_3248b | 3.0 | ||
| PdhB | Pyruvate dehydrogenase, E1 component, β subunit | AFE_3069 | 2.4 | ||
| PdhB | Pyruvate dehydrogenase, E1 component, α subunit | AFE_1813 | 7.1 | 0.8 | |
| Icd | NADP-dependent isocitrate dehydrogenase (EC 1.1.1.42) | AFE_0424 | 2.2 | ||
| CbbL2 | RubisCO, large subunit 2 (EC 4.1.1.39) | AFE_2155 | 2.9 | O2 > Fe3+ | |
| Pgi | Glucose-6-phosphate isomerase (EC 5.3.1.9) | AFE_2924 | 10.9 | ||
| Rpe | Ribulose phosphate 3-epimerase (EC 5.1.3.1) | AFE_3247 | 2.5 | ||
| Tal | Transaldolase (EC 2.2.1.2) | AFE_0419 | 2.2 | ||
| Tkt-1 | Transketolase (EC 2.2.1.1) | AFE_1843 | 2.1 | ||
| Transketolase pyridine binding domain protein | AFE_1667b | 4.0 | |||
| Transketolase pyridine binding domain protein | AFE_1667b | 7.0 | |||
| GlgB | 1,4-α-Glucan branching enzyme (EC 2.4.1.18) | AFE_2836 | 4.6 | 0.7 | |
| 6-Phosphogluconate dehydrogenase | AFE_2024 | 1.2 | |||
| zwf | Glucose-6-phosphate 1-dehydrogenase | AFE_2025 | 0.9 | ||
| gph-1 | Phosphoglycolate phosphatase | AFE_1823 | 0.7 | ||
| Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | AFE_2053 | 0.9 | |||
| Phosphoglucomutase | AFE_2324 | 1.0 | |||
| Cell wall/membrane/envelope biogenesis | |||||
| OMPP1/FadL/TodX family | AFE_2542b | 1.0 | |||
| OMPP1/FadL/TodX family | AFE_2542b | 2.6 | |||
| Upregulated under anaerobiosis | |||||
| Energy metabolism | |||||
| Pyridine nucleotide disulfide oxidoreductase (DsrE-like) | AFE_2556 | 1.3 | |||
| tusA | Hypothetical protein (similar to SirA) | AFE_2557 | 0.9 | ||
| sreA | Sulfur reductase molybdopterin subunit | AFE_2177 | Fe3+ > O2 | ||
| SreB | Sulfur reductase, iron-sulfur binding subunit | AFE_2178 | Unique | Fe3+ > O2 | |
| sreC | Sulfur reductase, membrane subunit, putative | AFE_2179 | Fe3+ > O2 | ||
| sreD | Sulfur reductase | AFE_2181 | Fe3+ > O2 | ||
| AtpF | ATP synthase F0, subunit b (EC 3.6.3.14) | AFE_3207 | Unique | 1.5 | |
| atpE | ATP synthase C chain | AFE_3208 | 1.4 | ||
| atpB | ATP synthase F0, A subunit | AFE_3209 | 1.0 | ||
| cycA2 | Cytochrome c4, CycA2 | AFE_2727 | 0.9 | ||
| petA2 | Ubiquinol-cytochrome c reductase, iron-sulfur subunit | AFE_2729 | 0.9 | ||
| petB2 | Ubiquinol-cytochrome c reductase, cytochrome b subunit | AFE_2730 | 1.1 | ||
| ubiE | Ubiquinone/menaquinone biosynthesis methyltransferase | AFE_0289 | 0.7 | ||
| Carbon management | |||||
| CbbS1 | RubisCO, small subunit (EC 4.1.1.39) | AFE_1690 | 3.8 | Fe3+ > O2 | |
| CscE | Carboxysome shell peptide | AFE_1683 | 3.1 | ||
| AcnA | Aconitate hydratase, putative | AFE_0423 | 2.0 | ||
| cbbp | Phosphoribulokinase | AFE_0536 | 0.7 | Fe3+ > O2 | |
| cscC | Carboxysome shell peptide | AFE_1685 | 1.6 | ||
| tkt-2 | Transketolase | AFE_3252 | 0.9 | ||
| Cell wall/membrane/envelope biogenesis | |||||
| OMPP1/FadL/TodX family | AFE_2542b | Unique | |||
| OMPP1/FadL/TodX family | AFE_2542b | Unique | |||
| OMPP1/FadL/TodX family | AFE_2542b | 9.6d | |||
| OMPP1/FadL/TodX family | AFE_2542b | 6.8d | |||
| OMPP1/FadL/TodX family | AFE_2542b | 1.7d | |||
| Posttranslational modification/protein turnover/chaperone functions and inorganic ion transport and metabolism | |||||
| surA | Survival protein SurA | AFE_3035 | 1.4 | ||
| Amino acid metabolism and transport | |||||
| Sulfite reductase hemoprotein, β component | AFE_3122 | 0.9 | |||
| cysH | Adenylylsulfate reductase | AFE_3123 | 1.5 |
The annotation number is that of GenBank accession number NC011761.
Identification of multiple spots for a single gene in the proteomics is likely due to posttranslational modifications.
The protein spot contains a mixture or proteins. No conclusions regarding upregulation of the individual proteins can be drawn.
Fold change values are not comparable to the remaining data in the table, as these protein spots were not considered for final analyses. Values are given here to show the gradual increase of the different isoforms of the same protein in gels of outer membrane-enriched fractions for anaerobic conditions.
Details are given in Tables S2 and S3 in the supplemental material.
O2 > Fe3+ denotes that a higher mRNA content was detected by qPCR analysis during aerobic growth than with anaerobic growth; vice versa for Fe3+ > O2.
Some discrepancies between protein levels measured by proteomics and levels of RNA determined by transcriptomics were noted. Such discrepancies have been observed in many instances of transcriptomic/proteomic analyses and have been variously attributed to differences in stability/degradation, posttranscriptional regulation, and posttranslational modification (40–43). In this study, levels of RNA have been validated by qPCR for various genes. A concordance between the results has been observed (Table 1).
Genes and proteins that could potentially be ascribed to S0 energy metabolism, Fe3+ reduction, electron transfer and energy conservation, and chemoautotrophy and carbon metabolism are shown in Table 1. Other genes and proteins that display differential regulation are listed in Table S2 in the supplemental material and are grouped according to Gene Ontology predictions: (i) cell wall, membrane, and envelope biogenesis; (ii) chaperone functions; (iii) translation transcription and cell cycle; (iv) amino acid metabolism; (vi) nucleotide metabolism and transport; and (vii) gene regulation.
Sulfur metabolism.
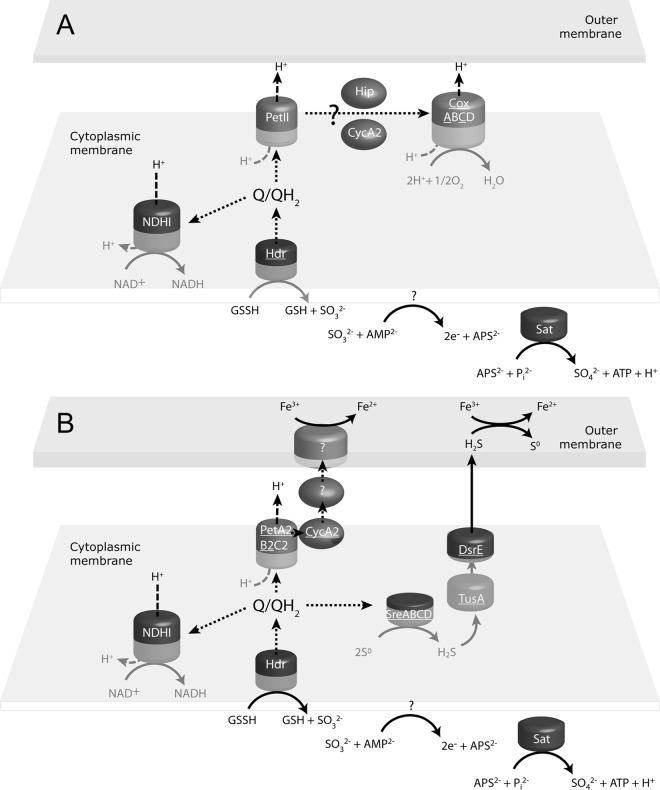
Oxidation of S0 by A. ferrooxidans under aerobic conditions is thought to be mediated by heterodisulfide reductase (Hdr), which is predicted to be an inner membrane-anchored protein with its active site in the cytoplasm (20, 35). S0 is poorly soluble in water, so the likely substrate for Hdr is the sulfane-sulfur compound glutathione persulfide (GSSH), which contains a disulfide bond that has been proposed to be cleaved by Hdr to produce SO32− and glutathione (GSH) (44, 45). There are three predicted copies of the gene for the HdrB catalytic subunit on the A. ferrooxidans genome (20), one of which is found outside the hdr locus (AFE_2586). While products of this gene were detected under both aerobic and anaerobic conditions, suggesting that Hdr mediates S0 oxidation in both situations (Fig. 1), protein levels were greater under aerobic conditions. A hypothetical protein (AFE_2552) and HdrA (AFE_2553) were also found at higher levels in the aerobic culture (see Fig. S3 in the supplemental material). In contrast, both dsrE (AFE_2556) and tusA (AFE_2557) were upregulated during anaerobic growth on S0. Although it has not been experimentally verified in A. ferrooxidans, the products of these genes are predicted to be involved in a sulfur relay system (46–48). All subunits of the sulfur reductase (SreABCD [AFE_2178 to AFE_2181]) were expressed only under anaerobic conditions. The sre operon in Aquifex aeolicus encodes a sulfur-reducing multiprotein complex that has been biochemically confirmed to be a sulfur reductase (49). The product of the Sre complex is H2S, and the upregulation of Sre would explain why both DsrE and TusA were also upregulated (assuming an H2S transfer role rather than a sulfane-sulfur transfer role under these conditions), as it would be important for toxic H2S to be expelled from the bacterial cells. Taken together, these observations suggest that disproportionation of S0 occurs when A. ferrooxidans is grown under anaerobic conditions. Oxidation of S0 to SO32− generates 4 electrons and 6 protons (assuming that oxygen is derived from water, as would necessarily be the case in anaerobic cultures; S0 + 3H2O → SO32− + 4e− + 6H+), and oxidation of SO32− to SO42− generates 2 electrons and 2 protons (SO32− + H2O → SO42− + 2e− + 2H+). The reduction of S0 to H2S consumes 2 electrons and 2 protons (S0 + 2e− + 2H+ → H2S). Therefore, more S0 would need to be reduced than oxidized to maintain the balance of electrons if sulfur disproportionation was the exclusive energy-transducing metabolism occurring in anaerobic cultures. However, Fe3+ respiration (discussed below), where electrons are transferred directly to Fe3+, would modify the stoichiometry of S0 oxidation and reduction. Disproportionation of S0 was initially thought of as surprising, as the process is endergonic under standard conditions, except where sulfide is removed by reaction with metals (50), as is shown here for A. ferrooxidans cultured in the presence of Fe3+.
Fig 1.
Pathway of aerobic sulfur (A) and anaerobic (B) electron transport. The pathways were based on this study and published reports (20, 44). Genes and/or proteins that were identified to be more upregulated in this study are underlined. Electron transfer is designated with short dashes, while proton transport is designated with long dashes. Q/QH2 denotes the quinol pool.
Support for the hypothesis that A. ferrooxidans produces H2S when grown anaerobically on S0 came from anoxic cultures incubated in the presence of Cu2+ and Fe2+. Concentrations of soluble Cu2+ fell by 35 ± 7.7 mg/liter (from 10 mM to ∼9.45 mM, balancing the consumption of approximately 0.5 mM S0 and the production of CuS) under these conditions, while noninoculated controls showed no decrease, and black precipitates (presumed to be CuS) formed in inoculated cultures (Cu2+ + H2S → CuS + 2H+) but not in sterile controls.
The SO32− produced from S0 oxidation may be used for energy production by converting SO32− to adenosine 5′-phosphosulfate (APS), which is then converted to SO42− using ATP sulfurylase (Sat; which was not differentially regulated) to generate SO42− and ATP (Fig. 1) (20). A minor portion of the SO32− may also be reduced to H2S by the sulfite reductase (CysI [AFE_3122]) and assimilated as required for cellular growth (51). Interestingly, cysI and cysH (APS reductase [AFE_3123]) were also upregulated under anaerobic conditions, even though the cellular growth requirement would not be expected to be greater under such conditions.
Ferric iron reduction.
No definitive iron reductase in A. ferrooxidans was identified in this study. TetH (28) and ArsH (29) have been suggested to mediate Fe3+ reduction by A. ferrooxidans. However, in the present study, tetH (AFE_0029; Table 1) is the most highly upregulated gene under oxic conditions, and neither the RNA nor the protein product of arsH was detected above background levels (data not shown), diminishing the likelihood that either TetH or ArsH plays a major role in Fe3+ reduction.
We propose that an important functional coupling of S0 oxidation to Fe3+ reduction occurs via an indirect mechanism involving reduction of Fe3+ by H2S generated by disproportionation of S0 under anaerobic conditions (2Fe3+ + H2S → 2Fe2+ + S0 + 2H+) (Fig. 1B). An alternate hypothesis is based on the observation that the genes encoding the bc1 complex (petA2 [AFE_2729] and petB2 [AFE_2730]) and the cytochrome c (cycA2 [AFE_2727]) were upregulated under anaerobic conditions (Table 1). In this model, electrons could flow from S0 to quinone and onwards to the bc1 complex and then to CycA2, as shown in Fig. 1B. We do not know whether Cyc2 is the final protein of this electron transport chain reducing Fe3+ or whether there are additional members before the final acquisition of electrons by Fe3+, such as rusticyanin, as proposed by Kucera et al. (30). These mechanisms are not mutually exclusive and could potentially operate simultaneously as branched electron pathways or as alternate pathways. In either case, it would be important to understand how electron flow is regulated. An important consideration is that some strains of iron-oxidizing acidithiobacilli can grow by coupling the oxidation of hydrogen to the reduction of Fe3+ (7, 12). In such cases, S0 reduction to H2S would not be expected to occur and an alternative mechanism for iron reduction other than the indirect mechanism must exist.
Electron transfer and energy conservation.
The aerobic electron transfer pathway and energy conservation have been investigated previously (see reference 20 and references therein), and the data in this study support the published model (Fig. 1). A proportion of the electrons generated from the anaerobic disproportionation of S0 could be transported by reverse electron flow to the NADH dehydrogenase. The energy required for this process could come from reverse proton motive force (24, 52).
Energy conservation under aerobic conditions is suggested to be via the F0F1 ATPase (53), and 3 of the 5 F1 subunits (α-atpA [AFE_3205; identified in a mixture; see Table 1 for details], ε-atpC [AFE_3202], and the β subunit of atpD [AFE_3203]) were upregulated. The higher levels of the catalytic F1 portion of the ATPase during aerobic growth suggest enhanced synthesis of ATP, which correlates with the observed higher growth rates under such conditions. In addition, a MotA/TolQ/ExbB proton channel family protein (AFE_0768) was upregulated; this has been shown in other organisms to be involved in electron transfer from NADH to the quinone pool, generating proton motive force (54). In contrast, under anaerobic conditions, the A (atpB [AFE_3209]), C (atpE [AFE_3208]), and B (atpF [AFE_3207]) F0 subunits were upregulated (Table 1). The F0 subunit does not require the F1 catalytic portion to translocate protons, and therefore, a greater proportion of the proton transfer under anaerobic conditions may be used to generate a proton motive force rather than ATP. In addition to energy derived from generating a proton motive force, ATP can be generated from substrate-level phosphorylation (oxidation of SO32− to SO42−) under both aerobic and anaerobic conditions, as described above. Also, the ubiquinone/menaquinone biosynthesis methyltransferase gene (ubiE [AFE_0289]) that is part of the ubiquinone biosynthesis pathway (55) was upregulated under anaerobic conditions.
Chemoautotrophy and central carbon metabolism.
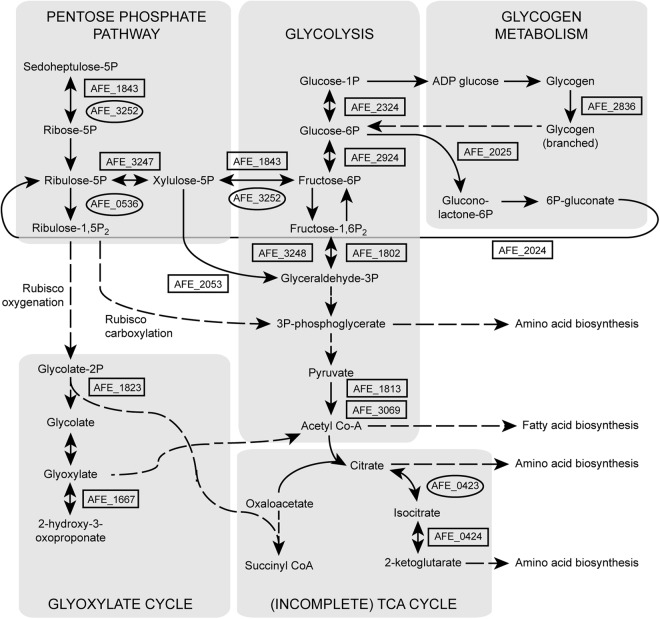
A. ferrooxidans fixes CO2 for growth via the Calvin-Benson-Bassham (CBB) cycle, and its genome contains a total of 5 cbb gene clusters with multiple copies of form I and a copy of form II ribulose-1,5-bisphosphate carboxylase oxygenase (RubisCO) (56). The second RubisCO copy (cbbL2 [AFE_2155]) from the cbb2 operon was upregulated under aerobic conditions (Table 1; see Fig. S3 in the supplemental material), in agreement with earlier predictions (57). Under anaerobic conditions, several genes and proteins linked to CO2 fixation (AFE_0536, AFE_1683, AFE_1685, AFE_1690, and AFE_3252) were upregulated (Table 1), probably due to the CO2 supply being intermittent (56). Differences in carbon fixation in the presence of various levels of CO2 are important for optimizing bioleaching (58).
Many proteins associated with central carbon metabolism were upregulated under aerobic conditions (Fig. 2 and Table 1), reflecting higher growth rates, with relatively few upregulated under anaerobic conditions. The former included the α and β subunits from the pyruvate dehydrogenase multienzyme complex (AFE_1813 and AFE_3069) that connects glycolysis and the tricarboxylic acid (TCA) cycle, a single protein (isocitrate dehydrogenase [AFE_0424]) from the incomplete TCA cycle present in A. ferrooxidans, pentose phosphate pathway proteins (Fig. 2) (including a 6-phosphogluconate dehydrogenase [AFE_2024] and glucose-6-phosphate 1-dehydrogenase [AFE_2025]), and several nonoxidative branch proteins (AFE_1843, AFE_3247, and AFE_0419), possibly to produce reducing power and ribulose-5-phosphate for carbon sequestration. Other proteins identified as being upregulated under aerobic conditions included those with roles in carbon/energy storage and others associated with the glyoxylate cycle (Fig. 2).
Fig 2.
Aerobic (gene numbers in boxes) and anaerobic (gene numbers in ovals) upregulated central carbon metabolism genes and proteins. Solid lines signify direct conversions, while dashed lines signify hidden intermediates (for clarity, not all connections have been included). The gene numbers refer to AFE_1843, tkt-1 transketolase (EC 2.2.1.1); AFE_3252, tkt-2 transketolase; AFE_3247, ribulose phosphate 3-epimerase; AFE_0536, phosphoribulokinase; AFE_2053, phosphoketolase; AFE_1823, phosphoglycolate phosphatase; AFE_1667, transketolase pyridine binding domain protein; AFE_2324, phosphoglucomutase; AFE_2924, glucose-6-phosphate isomerase (EC 5.3.1.9); AFE_1802, fructose-bisphosphate aldolase (class I); AFE_3248, fructose-bisphosphate aldolase (class II) (EC 4.1.2.13); AFE_3069, pyruvate dehydrogenase, E1 component, β subunit (EC 1.2.4.1); AFE_1813, dehydrogenase complex; AFE_2836, 1,4-α-glucan branching enzyme; AFE_2025, glucose-6-phosphate 1-dehydrogenase; AFE_2024, 6-phosphogluconate dehydrogenase family protein; AFE_0423, aconitate hydratase; AFE_0424, isocitrate dehydrogenase, NADP dependent. 5P, 5′-phosphate; 1,5-P2, 1,5-diphosphate; 2P, 2-phosphate; 1P, 1-phosphate; 6P, 6-phosphate; 1,6-P2, 1,6-diphosphate; 3P, 3′-phosphate; Acetyl Co-A, acetyl coenzyme A; 6P-gluconate, 6-phosphogluconate.
Cell wall, membrane, and envelope biogenesis.
The most prominent membrane protein spots on the anaerobic gels were a reproducible train of 5 spots. All the spots were identified as an OMPP1/FadL/TodX family protein (AFE_2542; high levels of the respective gene products were detected under both conditions in the transcriptomics assay) and the major outer membrane protein 40 (AFE_0365; not upregulated between the conditions) (Table 1). The OMPP1/FadL/TodX family protein is suggested to transport hydrophobic compounds from outside the cell to the periplasm (59). The protein spots with acidic isoelectric point (pIs) had higher intensities under anaerobic conditions, while the spots with a more basic pI had higher intensities under aerobic conditions (see Fig. S2 in the supplemental material). OMPP1/FadL/TodX family proteins have been demonstrated to be located in the outer membrane (A. Yarzábal and V. Bonnefoy, unpublished data) and to be upregulated during aerobic S0 metabolism compared to during Fe2+ oxidation (19, 60). The gene encoding survival protein A (surA [AFE_3035]), which is required for correct folding of outer membrane proteins (61), was also upregulated under anaerobic conditions.
Other differentially regulated genes and proteins.
Several oxidative stress and molecular chaperone proteins were upregulated under aerobic growth (Table 1), presumably due to response to production of oxygen free radicals and higher growth rates. Other genes and proteins observed to be differentially regulated during aerobic and anaerobic growth were those involved in translation, transcription, cell cycle, amino acid metabolism, and nucleotide metabolism and transport, which were upregulated under aerobic conditions, and ribosome subunit proteins and different nucleotide metabolism and transport genes/proteins, which were upregulated under anaerobic conditions (see Tables S2 and S3 in the supplemental material).
Conclusions.
The results of this study suggest models for the coupling of S0 oxidation to Fe3+ reduction. It is posited that they provide useful frameworks for the design of future biochemical experiments.
Evidence from transcriptomic and proteomic analysis, supported by some physiological data, suggests that Fe3+ reduction by A. ferrooxidans grown anaerobically using S0 as electron donor is mediated, at least in part, by an indirect mechanism involving H2S. A new model representing the proposed mechanism is shown in Fig. 1. The extent to which indirect iron reduction is complemented by a direct mechanism involving electron flow from S0 to Fe3+ via the bc1 complex, a soluble cytochrome, and possibly other electron carriers is unknown. Since H2S would not be generated as a waste product by A. ferrooxidans when grown on hydrogen (electron donor) and Fe3+ (electron acceptor), an additional mechanism to the indirect pathway in the present study presumably exists. Faster growth rates under aerobic conditions correlated with a number of proteins involved in central carbon pathways being upregulated.
Fe3+ reduction coupled to S0 oxidation results in the generation of Fe2+, which is an energy substrate for many bioleaching microorganisms. This could represent an important mechanism for recycling the Fe3+/Fe2+ couple and might be of considerable significance for heap bioleaching of copper and other minerals where gradients of air and nutrients occur.
Supplementary Material
ACKNOWLEDGMENTS
D.S.H. was supported by Fondecyt 1090451. M.D. thanks the Swedish Research Council for financial support (Vetenskapsrådet contract number 621-2007-3537). Part of this work was financed by the European Union Framework 6 project BioMinE (no. NM2.ct, 2005.500329).
D.B.J., V.B., and M.D. thank the various partners of the BioMinE project for their contributions to the work reported in this paper.
Footnotes
Published ahead of print 25 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03057-12.
REFERENCES
- 1. Johnson DB, Hallberg KB. 2009. Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Adv. Microb. Physiol. 54: 201–255 [DOI] [PubMed] [Google Scholar]
- 2. Liermann LJ, Hausrath EM, Anbar AD, Brantley SL. 2007. Assimilatory and dissimilatory processes of microorganisms affecting metals in the environment. J. Anal. Atom. Spectrom. 22: 867–877 [Google Scholar]
- 3. Bird LJ, Bonnefoy V, Newman DK. 2011. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 19: 330–340 [DOI] [PubMed] [Google Scholar]
- 4. Shi L, Squier TC, Zachara JM, Fredrickson JK. 2007. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65: 12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U. S. A. 105: 3968–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taillefert M, Beckler JS, Carey E, Burns JL, Fennessey CM, DiChristina TJ. 2007. Shewanella putrefaciens produces an Fe(III)-solubilizing organic ligand during anaerobic respiration on insoluble Fe(III) oxides. J. Inorg. Biochem. 101: 1760–1767 [DOI] [PubMed] [Google Scholar]
- 7. Johnson DB, Kanao T, Hedrich S. 2012. Redox transformations of iron at extremely low pH: fundamental and applied aspects. Front. Microbiol. 3: 96 doi:10.3389/fmicb.2012.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lovley DR, Holmes DE, Nevin KP. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49: 219–286 [DOI] [PubMed] [Google Scholar]
- 9. Welham NJ, Malatt KA, Vukcevic S. 2000. The effect of solution speciation on iron-sulphur-arsenic-chloride systems at 298 K. Hydrometallurgy 57: 209–223 [Google Scholar]
- 10. Straub KL, Benz M, Schink B. 2001. Iron metabolism in anoxic environments at near neutral pH. FEMS Microbiol. Ecol. 34: 181–186 [DOI] [PubMed] [Google Scholar]
- 11. Hedrich S, Schlomann M, Johnson DB. 2011. The iron-oxidizing proteobacteria. Microbiology 157: 1551–1564 [DOI] [PubMed] [Google Scholar]
- 12. Ohmura N, Sasaki K, Matsumoto N, Saiki H. 2002. Anaerobic respiration using Fe3+, S0, and H2 in the chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans. J. Bacteriol. 184: 2081–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pronk JT, De Bruyn JC, Bos P, Kuenen JG. 1992. Anaerobic growth of Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 58: 2227–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pronk JT, Liem K, Bos P, Keunen JG. 1991. Energy transduction by anaerobic ferric iron respiration in Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 57: 2063–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valdes J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, Blake R, Eisen JA, Holmes DS. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9: 597 doi:10.1186/1471-2164-9-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fischer J, Quentmeier A, Kostka S, Kraft R, Friedrich CG. 1996. Purification and characterization of the hydrogenase from Thiobacillus ferrooxidans. Arch. Microbiol. 165: 289–296 [DOI] [PubMed] [Google Scholar]
- 17. Schroder O, Bleijlevens B, de Jongh TE, Chen Z, Li T, Fischer J, Forster J, Friedrich CG, Bagley KA, Albracht SP, Lubitz W. 2007. Characterization of a cyanobacterial-like uptake [NiFe] hydrogenase: EPR and FTIR spectroscopic studies of the enzyme from Acidithiobacillus ferrooxidans. J. Biol. Inorg. Chem. 12: 212–233 [DOI] [PubMed] [Google Scholar]
- 18. Ng KY, Sawada R, Inoue S, Kamimura K, Sugio T. 2000. Purification and some properties of sulfur reductase from the iron-oxidizing bacterium Thiobacillus ferrooxidans NASF-1. J. Biosci. Bioeng. 90: 199–203 [DOI] [PubMed] [Google Scholar]
- 19. Chi A, Valenzuela L, Beard S, Mackey AJ, Shabanowitz J, Hunt DF, Jerez CA. 2007. Periplasmic proteins of the extremophile Acidithiobacillus ferrooxidans: a high throughput proteomics analysis. Mol. Cell. Proteomics 6: 2239–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quatrini R, Appia-Ayme C, Denis Y, Jedlicki E, Holmes D, Bonnefoy V. 2009. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10: 394 doi:10.1186/1471-2164-10-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holmes DS, Bonnefoy V. 2007. Genetic and bioinformatic insights into iron and sulfur oxidation mechanisms of bioleaching organisms, p 281–307 In Rawlings DE, Johnson DB. (ed), Biomining. Springer-Verlag, Berlin, Germany [Google Scholar]
- 22. Rohwerder T, Sand W. 2007. Oxidation of inorganic sulfur compounds in acidophilic prokaryotes. Eng. Life Sci. 7: 301–309 [Google Scholar]
- 23. Bonnefoy V, Holmes DS. 2012. Genomic insights into microbial oxidation and iron homeostasis in extremely acidic environments. Environ. Microbiol. 14: 1597–1611 [DOI] [PubMed] [Google Scholar]
- 24. Corbett CM, Ingledew WJ. 1987. Is Fe3+/2+ cycling an intermediate in sulfur oxidation by Fe2+ grown Thiobacillus ferrooxidans? FEMS Microbiol. Lett. 41: 1–6 [Google Scholar]
- 25. Sugio T, Domatsu C, Munukata O, Tano T, Imai K. 1985. Role of a ferric iron-reducing system in sulfur oxidation by Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 49: 1401–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sugio T, Mizunashi W, Magaki K, Tano T. 1987. Purification and some properties of sulfur-ferric iron oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 169: 4916–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sugio T, Hirose T, Ye LZ, Tano T. 1992. Purification and some properties of sulfite-ferric ion oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 174: 4189–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sugio T, Taha TM, Takeuchi F. 2009. Ferrous iron production mediated by tetrathionate hydrolase in tetrathionate-, sulfur-, and iron-grown Acidithiobacillus ferrooxidans ATCC 23270 cells. Biosci. Biotechnol. Biochem. 73: 1381–1386 [DOI] [PubMed] [Google Scholar]
- 29. Mo H, Chen Q, Du J, Tang L, Qin F, Miao B, Wu X, Zeng J. 2011. Ferric reductase activity of the ArsH protein from Acidithiobacillus ferrooxidans. J. Microbiol. Biotechnol. 21: 464–469 [DOI] [PubMed] [Google Scholar]
- 30. Kucera J, Bouchal P, Cerna H, Potesil D, Janiczek O, Zdrahal Z, Mandl M. 2012. Kinetics of anaerobic elemental sulfur oxidation by ferric iron in Acidithiobacillus ferrooxidans and protein identification by comparative 2-DE-MS/MS. Antonie van Leeuwenhoek 101: 561–573 [DOI] [PubMed] [Google Scholar]
- 31. Hallberg KB, Grail BM, du Plessis C, Johnson DB. 2011. Reductive dissolution of ferric iron minerals: a new approach for bioprocessing nickel laterites. Miner. Eng. 24: 620–624 [Google Scholar]
- 32. Johnson DB, Hallberg KB. 2007. Techniques for detecting and identifying acidophilic mineral-oxidizing microorganisms. In Rawlings DE, Johnson DB. (ed), Biomining. Springer-Verlag, Berlin, Germany [Google Scholar]
- 33. Lovley DR, Phillips EJ. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53: 1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ñancucheo I, Johnson DB. 2012. Selective removal of transition metals from acidic mine waters by novel consortia of acidophilic sulfidogenic bacteria. Microb. Biotechnol. 5: 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mangold S, Valdes J, Holmes DS, Dopson M. 2011. Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2: 17 doi:10.3389/fmicb.2011.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- 37. Quatrini R, Appia-Ayme C, Denis Y, Ratouchniak J, Veloso F, Valdes J, Lefimil C, Silver S, Roberto F, Orellana O, Denizot F, Jedlicki E, Holmes D, Bonnefoy V. 2006. Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy 83: 263–272 [Google Scholar]
- 38. Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29: 123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nieto PA, Covarrubias PC, Jedlicki E, Holmes DS, Quatrini R. 2009. Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans. BMC Mol. Biol. 10: 63 doi:10.1186/1471-2199-10-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nie L, Wu G, Culley DE, Scholten JC, Zhang W. 2007. Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit. Rev. Biotechnol. 27: 63–75 [DOI] [PubMed] [Google Scholar]
- 41. Scherl A, Francois P, Bento M, Deshusses JM, Charbonnier Y, Converset V, Huyghe A, Walter N, Hoogland C, Appel RD, Sanchez JC, Zimmermann-Ivol CG, Corthals GL, Hochstrasser DF, Schrenzel J. 2005. Correlation of proteomic and transcriptomic profiles of Staphylococcus aureus during the post-exponential phase of growth. J. Microbiol. Methods 60: 247–257 [DOI] [PubMed] [Google Scholar]
- 42. Nie L, Wu G, Zhang W. 2006. Correlation between mRNA and protein abundance in Desulfovibrio vulgaris: a multiple regression to identify sources of variations. Biochem. Biophys. Res. Commun. 339: 603–610 [DOI] [PubMed] [Google Scholar]
- 43. Greenbaum D, Colangelo C, Williams K, Gerstein M. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4: 117 doi:10.1186/gb-2003-4-9-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rohwerder T, Sand W. 2003. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149: 1699–1709 [DOI] [PubMed] [Google Scholar]
- 45. Chen L, Ren Y, Lin J, Liu X, Pang X. 2012. Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS One 7: e39470 doi:10.1371/journal.pone.0039470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dahl C, Schulte A, Stockdreher Y, Hong C, Grimm F, Sander J, Kim R, Kim SH, Shin DH. 2008. Structural and molecular genetic insight into a widespread sulfur oxidation pathway. J. Mol. Biol. 384: 1287–1300 [DOI] [PubMed] [Google Scholar]
- 47. Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. 2006. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 21: 97–108 [DOI] [PubMed] [Google Scholar]
- 48. Stockdreher Y, Venceslau SS, Josten M, Sahl HG, Pereira IA, Dahl C. 2012. Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: evidence for sulfur transfer from DsrEFH to DsrC. PLoS One 7: e40785 doi:10.1371/journal.pone.0040785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guiral M, Tron P, Aubert C, Gloter A, Iobbi-Nivol C, Giudici-Orticoni MT. 2005. A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus. J. Biol. Chem. 280: 42004–42015 [DOI] [PubMed] [Google Scholar]
- 50. Frederiksen TM, Finster K. 2004. The transformation of inorganic sulfur compounds and the assimilation of organic and inorganic carbon by the sulfur disproportionating bacterium Desulfocapsa sulfoexigens. Antonie van Leeuwenhoek 85: 141–149 [DOI] [PubMed] [Google Scholar]
- 51. Valdes J, Veloso F, Jedlicki E, Holmes D. 2003. Metabolic reconstruction of sulfur assimilation in the extremophile Acidithiobacillus ferrooxidans based on genome analysis. BMC Genomics 4: 51 doi:10.1186/1471-2164-4-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kelly DP. 1999. Thermodynamic aspects of energy conservation by chemolithotrophic sulfur bacteria in relation to the sulfur oxidation pathways. Arch. Microbiol. 171: 219–229 [Google Scholar]
- 53. Ingledew WJ. 1982. Thiobacillus ferrooxidans. The bioenergetics of an acidophilic chemolithotroph. Biochim. Biophys. Acta 683: 89–117 [DOI] [PubMed] [Google Scholar]
- 54. Braun TF, Poulson S, Gully JB, Empey JC, Van Way S, Putnam A, Blair DF. 1999. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J. Bacteriol. 181: 3542–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee PT, Hsu AY, Ha HT, Clarke CF. 1997. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179: 1748–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Esparza M, Bowien B, Jedlicki E, Holmes DS. 2009. Gene organization and CO2-responsive expression of four cbb operons in the biomining bacterium Acidithiobacillus ferrooxidans. Adv. Mater. Res. 71-73: 207–210 [Google Scholar]
- 57. Esparza M, Cardenas JP, Bowien B, Jedlicki E, Holmes DS. 2010. Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans. BMC Microbiol. 10: 229 doi:10.1186/1471-2180-10-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bryan CG, Davis-Belmar CS, van Wyk N, Fraser MK, Dew D, Rautenbach GF, Harrison ST. 2012. The effect of CO2 availability on the growth, iron oxidation and CO2-fixation rates of pure cultures of Leptospirillum ferriphilum and Acidithiobacillus ferrooxidans. Biotechnol. Bioeng. 109: 1693–1703 [DOI] [PubMed] [Google Scholar]
- 59. van den Berg B. 2010. Going forward laterally: transmembrane passage of hydrophobic molecules through protein channel walls. Chembiochem 11: 1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ramirez P, Guiliani N, Valenzuela L, Beard S, Jerez CA. 2004. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds, or metal sulfides. Appl. Environ. Microbiol. 70: 4491–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lazar SW, Kolter R. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178: 1770–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.