Abstract
We describe here a comparative genome analysis of three dairy product isolates of Lactobacillus rhamnosus GG (LGG) and the ATCC 53103 reference strain to the published genome sequence of L. rhamnosus GG. The analysis showed that in two of three isolates, major DNA segments were missing from the genomic islands LGGISL1,2. The deleted DNA segments consist of 34 genes in one isolate and 84 genes in the other and are flanked by identical insertion elements. Among the missing genes are the spaCBA genes, which encode pilin subunits involved in adhesion to mucus and persistence of the strains in the human intestinal tract. Subsequent quantitative PCR analyses of six commercial probiotic products confirmed that two more products contain a heterogeneous population of L. rhamnosus GG variants, including genotypes with or without spaC. These results underline the relevance for quality assurance and control measures targeting genome stability in probiotic strains and justify research assessing the effect of genetic rearrangements in probiotics on the outcome of in vitro and in vivo efficacy studies.
INTRODUCTION
Recently, the concept of “generic probiotics” was introduced, as a practical solution to create access to probiotics for people in the developing world (1). This concept refers to the free use of probiotic bacteria introduced under a novel brand name, after intellectual property rights have expired. In analogy to generic drugs, also patent-expired probiotics are free to be used by others, and health and safety claims from the expired patents can be linked to the generic probiotic strains, provided that the genotype of the original strain is identical to that of the generic strain. This raises questions about the extent of genome stability in probiotic bacterial strains and its impact on probiotic functionality.
Today, only a few examples exist about active components of probiotics, which could be targeted to confirm the presence of the associated functionality (2). Lebeer et al. (3) report on the classification of several genes and molecules that contribute to probiotic and health-promoting actions of Lactobacillus: (i) adaptation factors, including determinants of stress resistance, metabolism in the host, and adherence to the gut mucosa, and (ii) probiotic factors directly mediating health effects including antipathogenic, epithelium barrier-preserving, and immunomodulatory molecules. In another recent study, a collection of lactic acid bacteria isolated from fermented foods has been screened for more than 30 probiotic functionality related genes. The authors defined genes as “probiotic” that are involved in survival in the gastrointestinal tract (resistance to low pH and bile salt), starch metabolism, and folate and riboflavin synthesis (4).
Many of the probiotic strains marketed today originate from intestinal or plant isolates, and the shift from their complex and highly variable niche to the relatively constant and nutrient-rich industrial production environment is likely to be linked with selection for metabolic simplification (5). This simplification can coincide with gene loss that occurs at relatively high frequencies in genomes with a large a number of insertion sequences (IS). These IS elements consist of transposase genes flanked by two inverted repeats, which are mobile through transposition. Accordingly, they play an important role in the occurrence of mutations, the disruption of genes, the overexpression of genes, and chromosomal rearrangements such as deletions, duplications, or inversions (6–8).
In the present study, we carried out a comparative genome analysis to investigate if, and to what extent, the genome of the probiotic Lactobacillus rhamnosus GG (LGG), which contains a relatively high number of 69 IS elements (9, 10), is stable in commercial products. Our findings indicate that major genetic rearrangements occurred in some dairy product isolates, which include the deletion of genes that are hypothesized to play a role in probiotic functionality.
The results of this work emphasize the need for quality control and assurance strategies targeting the presence and maintenance of genes involved in host-microbe interactions underlying probiotic functionality, in particular in instable genome regions or genomic islands with a relatively high occurrence of mobile genetic elements. In addition, these results justify further study of the effect of genetic rearrangements in probiotics on the outcome of in vitro and in vivo efficacy studies.
MATERIALS AND METHODS
Bacterial strain and cultivation conditions.
The strains used in the present study were isolated from products by incubation of dilution series on Rogosa (RO) or MRS agar plates (Tritium Microbiology, Eindhoven, The Netherlands) under aerobic or anaerobic conditions at 37°C as shown in Table 1. Species identities were confirmed by sequencing of 16S-rRNA genes (Baseclear, Leiden, The Netherlands).
Table 1.
Strains used in this study, their origin, and references
| Descriptiona | Sourceb | Species | Strain | Culture collection no. | Novel name | SRA BioSample accession no.e | Source or reference |
|---|---|---|---|---|---|---|---|
| Reference*† | LMG culture collection‡ | L. rhamnosus | GG | LMG 18243, ATCC 53103, TNO 2012.097 | SAMN01831559 | ||
| Product isolate 1* | Liquid dairy formula 1‡ | L. rhamnosus | GG | LMG 25859, TNO 2012.098 | L. rhamnosus yoba 2010 | SAMN01831560 | 1 |
| Product isolate 2A* | Liquid dairy formula 2Ac | L. rhamnosus | GG | TNO 2010.113 | SAMN01831561 | ||
| Product isolate 2B† | Liquid dairy formula 2B‡c | L. rhamnosus | GG | ||||
| Product isolate 3*† | Powder dairy formula 3‡ | L. rhamnosus | GG | LMG 27229, TNO 2012.094 | L. rhamnosus yoba 2012 | SAMN01831562 | This study |
| Product sample 4† | Liquid dairy formula 4 | L. rhamnosus, S. thermophilusd | GG | ||||
| Product sample 5† | Liquid dairy formula 5 | L. rhamnosus | GG | ||||
| Product sample 6† | Liquid dairy formula 6 | L. rhamnosus | GG | ||||
| Product sample 7† | Dried supplement 7 | L. rhamnosus | GG | ||||
| Product sample 8† | Liquid dairy formula 8 | L. caseid | DN-114001 |
*, used for comparative genome sequence; †, qPCR was conducted on the entire product.
‡, qPCR was conducted on product isolates.
Same product, but different batches; 2A was produced 1 year before 2B.
Negative control samples.
The entire study results have been deposited at the NCBI BioSample database, Sequence Read Archive, project number SRP017797.
DNA extraction.
DNA was isolated from bacterial colonies by the use of the AGOWA mag Mini DNA isolation kit (LGC Genomics, Berlin, Germany). When DNA was directly isolated from probiotic dairy products, the isolation was preceded by the addition of 45 ml of 2% sodium citrate (pH 7) to a 5-ml sample, followed by 30 min of incubation at 30°C and centrifugation for 15 min at 3,000 rpm.
qPCR.
Quantitative PCR (qPCR) was adapted from a protocol reported previously (11). Primer-probe combinations for qPCR were specifically designed for the spaC gene (LGG_00444) and a single-copy gene, LGG_00154, by the use of Primer Express Software v2.0 (Applied Biosystems, Bleiswijk. The Netherlands), as listed in Table S1 in the supplemental material. LGG_00444 encodes a pilin subunit for binding to human intestinal mucus. LGG_00154 encodes a hypothetical membrane associated protein, designated map, located outside LGGISL1,2 (locus 168611 to 168105). Moreover, LGG_00154 is located on an amplicon previously selected for specific identification of L. rhamnosus GG (12).
The TaqMan probes contained the minor groove binder (MGB) probe in combination with a nonfluorescent quencher and a reporter. The experiment was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems) with the following settings: 1 step of 2 min at 50°C and 10 min at 95°C, followed by 50 cycles of 15 s at 95°C and 1 min at 60°C. The composition of the qPCR mix included 15 μl of 2× PCR Mastermix (Diagenode, Liège, Belgium), 1.3 μl of primer forward (10 pmol μl−1), 1.3 μl of primer reverse (10 pmol μl−1), 1.3 μl of MGB probe (5 pmol μl−1), 10 μl of ultrapure (type 1) water (Millipore, Amsterdam, The Netherlands), and 1 μl of DNA sample. Dilutions of L. rhamnosus genomic DNA were quantified by optical density measurement at a wavelength of 260 nm (with an extinction coefficient for double-stranded DNA of 0.020 μg ml−1 cm−1) and used as standards for a calibration curve (108 to 1 fg μl−1). The threshold cycle (CT) values were derived from the qPCR and set threshold values. The corresponding amount of DNA is derived from the calibration curve.
Nextera DNA library preparation and MiSeq sequencing.
Nextera DNA library was prepared for analysis on an Illumina MiSeq sequencer according to the Illumina Nextera protocol. Briefly, 50 ng of genomic DNA was tagged and fragmented in the presence of transposomes with adapters, purified, and enriched by a limited-cycle PCR. Cluster generation and sequencing was performed according to the Illumina MiSeq system Quick Reference Guide. Each library was sequenced paired end for 150 cycles on the MiSeq.
TruSeq DNA library preparation and HiSeq next generation sequencing.
For each sample, an indexed sequencing library was prepared of 1 μg of genomic DNA according to the Illumina TruSeq DNA protocol. One microliter of each library was loaded on an Agilent Technologies 2100 Bioanalyzer using a high-sensitivity DNA assay to determine the library concentration and to check for quality. The libraries were clustered on the Illumina cBOT station and sequenced paired end for 101 cycles on the HiSeq 2000 sequencer according to the Illumina cluster and sequencing protocols.
NGS primary data analysis.
The image analysis and base calling was performed on the MiSeq or HiSeq2000 system using the Illumina online basecaller. The resulting data were demultiplexed using NARWHAL software (13) and aligned to the L. rhamnosus GG genome (accession number NC_013198.1) using a Burrows-Wheeler aligner (14). The resulting SAM files were sorted and converted to binary SAM with Samtools (15).
Comparative genome analysis.
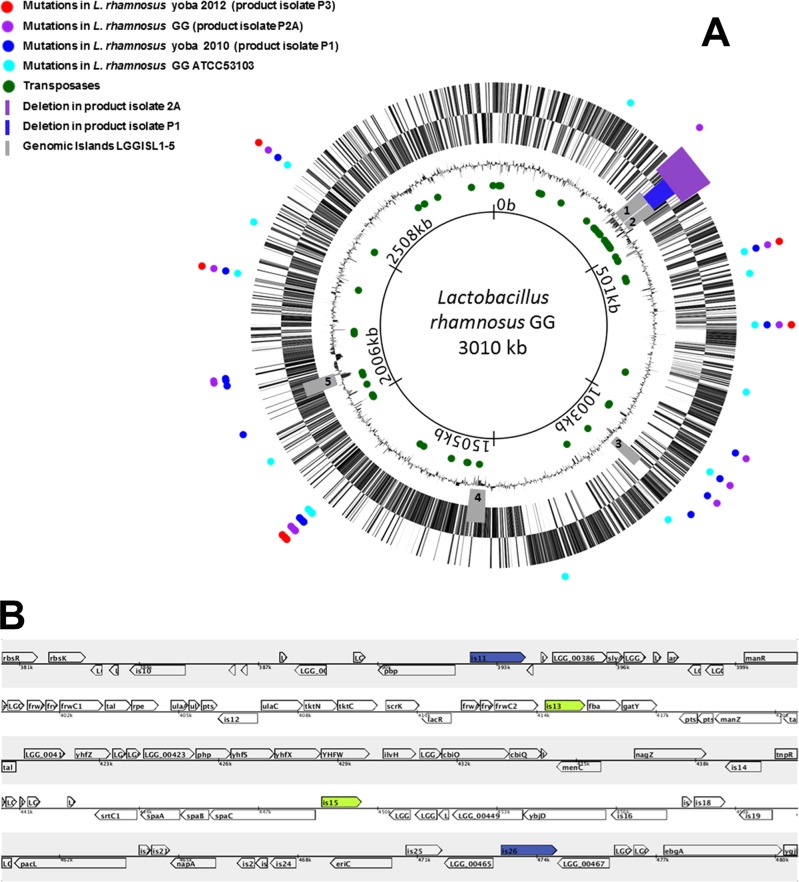
The genome sequence data of the three isolates and reference strain have been deposited at the NCBI BioSample database, Sequence Read Archive, project SRP017797, as indicated in Table 1. Comparative genome analysis was carried out by use of the Breseq tool, version 0.18 (16). The results of the analysis are accessible at http://www.yoba4life.com/yoba-for-life-rd/. The circular and linear genome maps of L. rhamnosus GG in Fig. 1 were created by the use of Microbial Genome Viewer, version 2.0 (17).
Fig 1.
(A) Circular genome map of L. rhamnosus GG (accession number NC_013198.1). Colored circles indicate the loci of the mutations in the ATCC 53103 reference strain and three probiotic product isolates including L. rhamnosus yoba 2010 and 2012. The purple and blue bars indicate the sites of the missing ORFs of isolates P2A and P1, respectively. In gray are the genomic islands LGGISL1 to LGGISL5. Circles indicate from outside to inside: genes for the “+” strand; genes for the “−” strand; the GC percentage; green circles, all IS elements encoding transposases in the genome; genome positions (total length, 3,010,111 bp). (B) Linear map showing the deletions including stretches of 34 genes flanked by two transposases of the IS30 family (is13 and is15 in green) in product isolate 1 and 84 genes flanked by two transposases of the IS5 family (is11 and is26 in blue) in product isolate 2A.
RESULTS
Genomic islands deleted in two propagated strains.
Species identity was confirmed by sequencing of 16S rRNA genes (data not shown). The full genome sequence of three L. rhamnosus GG dairy product isolates and reference strain ATCC 53103, comprising approximately three million base pairs, was determined by massive parallel sequencing. Product isolate 1 was designated L. rhamnosus yoba 2010, and product isolate 3 was designated L. rhamnosus yoba 2012 (Table 1). A comparison between the genomes of the four strains and the published genome of L. rhamnosus GG (9, 10) showed considerable genetic variety (Fig. 1). In two of three product isolates, major parts of the genomic islands LGGISL1,2 were missing, covering stretches of 34 genes and 84 genes, flanked by IS30 and IS5 insertion elements, respectively. The missing genes in both isolates include spaCBA, encoding pilin subunits involved in adhesion of the strains to the intestinal mucus and persistence in the human intestinal tract (10, 18). The DNA fragment of 34 genes missing from product isolate 1 includes two elements at both ends (is13 and is15), which are 100% identical and both encode a 338-amino-acid transposase of the IS30 family (the entire genome contains four copies of this transposase gene). The DNA fragment of 84 genes missing from product isolate 2A also includes two elements at both ends (is11 and is26), which are 100% identical and encode a 471-amino-acid transposase of the IS5 family (the entire genome contains 13 copies of this transposase gene). Strikingly, the LGGISL1,2 region contains a very high density of insertion elements; for example, the deleted DNA sequence of 82 kb in isolate 2A contains 16 insertion elements (is11 to is26), which is almost 10 times more than the average occurrence of these elements in the entire genome. Besides the 2 large deletions, a range of 9 to 23 deletions, duplications, substitutions, and point mutations (silent and nonsilent) were identified in the reference strain and the three product isolates (see Tables S2 to S4 in the supplemental material).
Genetic heterogeneity in commercial products.
Two qPCRs were carried out targeting spaC and map, as a control gene, in order to investigate whether the large deletions, including the spaCBA genes, already had occurred in the L. rhamnosus GG strains present in the commercial products or later during the isolation of the variants by cultivation on selective nutrient agar. In a first series of experiments, DNA was directly isolated from seven commercial products. Six of these products contained L. rhamnosus GG according to the product label specifications, and one product contained a Lactobacillus casei strain (Table 1). It should be noted that two of these products also served as the source for the comparative genome analysis of product isolates 1, 2A, and 3, mentioned before (Table 1). The qPCR results confirmed the presence of the map and spaC genes in all products containing the LGG label. However, in two of these products the detected quantity of spaC genes appeared to be significantly lower than the map control gene. The ratios of spaC to map were 0.38 and 0.31 for products 2B and 4, respectively, while the average ratio in the four other products was 1.11 ± 0.15. This indicates that these two products contain a heterogeneous population of L. rhamnosus GG variants consisting of genotypes with or without spaC.
In a follow-up experiment, a qPCR targeting map and spaC was conducted on 14 and 10 DNA samples extracted from a corresponding number of single colonies obtained from the commercial products 2B and 4, respectively. In order to evaluate whether environmental effects could select for a spaC minus genotype, colonies were selected after cultivation under aerobic and anaerobic conditions on MRS or RO agar. Positive controls included DNA isolates obtained from 12 colonies of cultivated ATCC 53103 strain and 12 colonies of product isolate 3/L. rhamnosus yoba 2012, which, based on the genome sequencing results, had a genome very similar to the reference strain (Fig. 1 and Table 2). As negative controls, one sample without DNA and four samples with DNA extracts from a Streptococcus thermophilus isolate were included.
Table 2.
Results of qPCR on DNA extracted from bacterial isolates from commercial products, control strain ATCC 53103, and Lactobacillus rhamnosus yoba 2012a
| Cultivation condition/sample no. | Product 2B |
Product 4 |
ATCC 53103 |
Product 3/L. rhamnosus yoba 2012 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
map |
spaC |
map |
spaC |
map |
spaC |
map |
spaC w |
|||||||||
| CT | Quantity | CT | Quantity | CT | Quantity | CT | Quantity | CT | Quantity | CT | Quantity | CT | Quantity | CT | Quantity | |
| RA/1 | 17.5 | 3.0E+06 | 33.8 | 3.46E+02 | 18.0 | 2.3E+06 | 35.2 | 1.5E+02 | 17.5 | 3.11E+06 | 18.3 | 3.7E+06 | 17.6 | 2.9E+06 | 18.6 | 3.2E+06 |
| RA/2 | 17.7 | 2.7E+06 | 34.0 | 3.00E+02 | 17.6 | 2.9E+06 | 18.5 | 3.4E+06 | 17.5 | 3.04E+06 | 18.4 | 3.6E+06 | 17.8 | 2.6E+06 | 18.7 | 2.9E+06 |
| RA/3 | 17.5 | 3.0E+06 | 18.4 | 3.46E+06 | 17.5 | 3.0E+06 | 35.9 | 9.6E+01 | 17.5 | 2.96E+06 | 18.3 | 3.8E+06 | 17.7 | 2.7E+06 | 18.7 | 2.9E+06 |
| RA/4 | 17.2 | 3.6E+06 | 32.5 | 7.53E+02 | 16.6 | 5.0E+06 | 17.5 | 6.0E+06 | ND | ND | ND | ND | ND | ND | ND | ND |
| RN/1 | 17.4 | 3.2E+06 | 18.2 | 3.92E+06 | 27.2* | 9.9E+03 | 29.8 | 3.8E+03 | 17.7 | 2.77E+06 | 18.5 | 3.4E+06 | 17.4 | 3.1E+06 | 18.3 | 3.7E+06 |
| RN/2 | 17.3 | 3.4E+06 | 19.4 | 1.91E+06 | 27.2* | 1.0E+04 | 30.5 | 2.4E+03 | 17.4 | 3.14E+06 | 18.3 | 3.9E+06 | 17.8 | 2.6E+06 | 18.6 | 3.1E+06 |
| RN/3 | 17.7 | 2.7E+06 | 32.2 | 8.78E+02 | 17.2 | 3.6E+06 | 35.2 | 1.5E+02 | 17.6 | 2.81E+06 | 18.1 | 4.2E+06 | 17.6 | 2.8E+06 | 19.0 | 2.4E+06 |
| RN/4 | 17.1 | 3.8E+06 | 34.0 | 3.02E+02 | 17.4 | 3.1E+06 | OR | ND | ND | ND | ND | ND | ND | ND | ND | |
| MA/1 | 17.4 | 3.3E+06 | 36.6 | 6.56E+01 | 17.7 | 2.6E+06 | 35.2 | 1.5E+02 | 17.6 | 2.94E+06 | 18.4 | 3.5E+06 | 17.5 | 3.0E+06 | 18.4 | 3.5E+06 |
| MA/2 | 17.6 | 2.8E+06 | 36.0 | 9.14E+01 | 17.7 | 2.8E+06 | OR | 17.5 | 3.00E+06 | 18.4 | 3.5E+06 | 17.7 | 2.8E+06 | 18.6 | 3.2E+06 | |
| MA/3 | 17.6 | 2.9E+06 | OR | 17.7 | 2.6E+06 | OR | 17.2 | 3.68E+06 | 18.1 | 4.2E+06 | 17.7 | 2.6E+06 | 18.6 | 3.1E+06 | ||
| MA/4 | 17.4 | 3.2E+06 | 37.5 | 3.65E+01 | 17.5 | 3.1E+06 | 34.8 | 1.9E+02 | ND | ND | ND | ND | ND | ND | ND | ND |
| MN/1 | 17.3 | 3.4E+06 | 36.6 | 6.38E+01 | ND | ND | ND | ND | 17.4 | 3.27E+06 | 18.5 | 3.3E+06 | 17.3 | 3.4E+06 | 18.5 | 3.3E+06 |
| MN/2 | 29.1* | 3.2E+03 | 32.0 | 1.03E+03 | ND | ND | ND | ND | 17.8 | 2.46E+06 | 18.9 | 2.6E+06 | 17.8 | 2.5E+06 | 18.9 | 2.6E+06 |
| MN/3 | 17.7 | 2.6E+06 | 38.6 | 1.90E+01 | ND | ND | ND | ND | 17.8 | 2.55E+06 | 18.9 | 2.6E+06 | 17.9 | 2.3E+06 | 19.0 | 2.4E+06 |
| MN/4 | OR* | OR | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||
Abbreviations used cultivation conditions in column 1:R, Rogosa agar; M, MRS agar; A, aerobic; N, anaerobic. Other abbreviations: OR, out of range; ND, not determined. Quantities are expressed in fg/μl. *, the CT of the control gene map is much larger than the average value of 17.5; hence, no mathematical comparison of the data is possible (see the text).
In total, 52 qPCRs were run on L. rhamnosus GG strains targeting map and spaC. Each qPCR was performed on DNA isolated from a unique colony. The CT value of the calibration curve of map varied between 18.3 and 38.3, corresponding to 2.3 × 106 fg/μl and 23 fg/μl, respectively, with an R2 value of 0.993. The CT value of the calibration curve of the spaC varied between 19.1 and 34.1, corresponding to 2.3 × 106 fg/μl and 230 fg/μl, respectively, with an R2 value of 0.997. The average CT of 48 qPCRs targeting the map gene was 17.54 ± 0.24, indicative of the highly reproducible DNA extractions and qPCR. The results of 4 of 52 qPCRs were excluded from further data analysis, because the corresponding CT values of the map gene completely deviated from the control samples, suggesting that the amplification of the DNA had not run well, for instance due to poor DNA isolation and/or contamination of the sample. The CT values of the control sample without DNA scored higher than 35, and the four samples with DNA extracts from a Streptococcus thermophilus isolate scored higher than 33 (data not shown).
Table 2 and Fig. 2 show that in all 24 colonies derived from the two positive control strains (i.e., ATCC 53103 and L. rhamnosus yoba 2012, originating from product isolate 3), the spaC and map genes were present in a ratio of ∼1 (mean, 1.14; standard deviation, 0.12). This ratio confirms that spaC was present in all colonies after cultivation under the four described conditions. In contrast, the CT value of spaC required at least 12 cycles more for 11 of the 14 colonies isolated from product 2B and 8 of the 10 colonies isolated from product 4 (Table 2), meaning that the map/spaC ratio in these colonies was >10,000. Since the source of the DNA for each qPCR was a single colony and the ratio map to spaC was ∼1 in all control samples, this high ratio indicates the factual absence of spaC in 11 of 14 colonies and 8 of 10 colonies isolated from product 2B and 4, respectively. Figure 2A and B illustrate the 2log(spaC:map) ratio of the conducted qPCR for all samples derived from products 2B, 3, and 4 and the reference strain.
Fig 2.
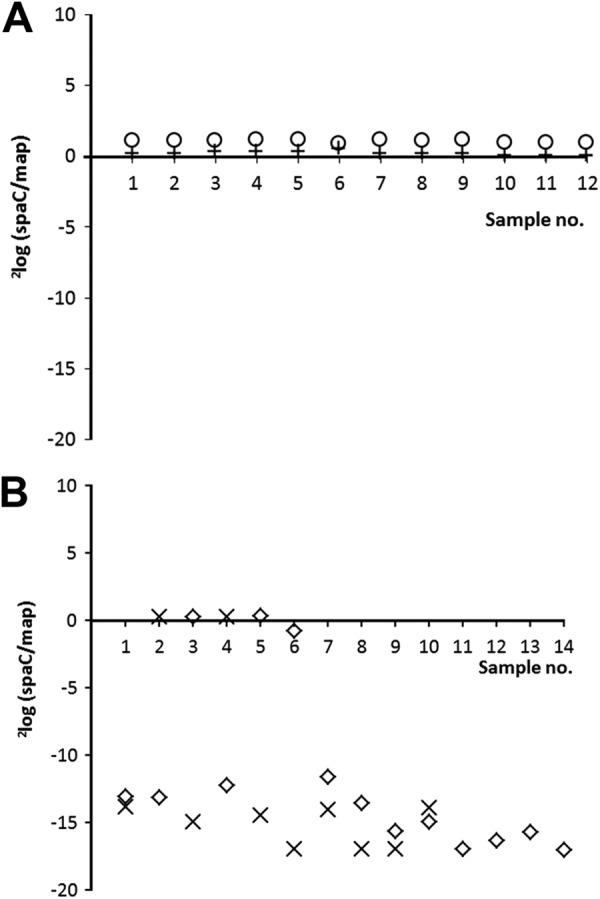
(A) qPCR derived 2log(spaC:map) ratio on DNA extracted from several isolates of control strain ATCC 53103 (+) and L. rhamnosus yoba 2012 (○). (B) qPCR-derived 2log(spaC:map) ratio on DNA extracted from several bacterial isolates of commercial product 2B (♢) and product 4 (×). The data are extracted from Table 2. Note: when the Ct value of spaC was below the detection limit, indicating that spaC was absent, then the 2log(spaC:map) ratio was arbitrarily set at 17.
As already indicated by the outcome of the comparative genome analysis (Fig. 1) for product 1 and 2A, the results from the qPCR analyses confirmed that the declared probiotic L. rhamnosus GG was not present either as a homogenous culture in products 2B and 4 (Table 1). In these products, the majority of the L. rhamnosus bacteria lack the pilin subunit-encoding spaC gene.
DISCUSSION
We conclude from our work that genetic alterations occurred in a major fraction of the population of probiotic L. rhamnosus GG bacteria present in some of today's commercialized products. A typical example of genetic alterations concerns the deletion of the genomic islands LGGISL1,2, including the spaCBA genes encoding multisubunit pilins with mucus-binding proteins (10, 18). Transmission electron microscopy using uranyl acetate staining confirmed the presence or absence of pili in strains with or without spaC, respectively (data not shown). Phenotypic analysis with L. rhamosus yoba 2010 (product isolate 1) and product isolate 2A lacking these pili showed that these isolates had human mucus-binding abilities of only 1.6% ± 0.3% and 1.4% ± 0.5%, respectively, compared to the L rhamnosus GG reference strain (F. Douillard and W. M. de Vos, unpublished data). We reason that these L. rhamnosus GG variants without spaCBA genes will have a reduced residence time in the gastrointestinal tract and even an altered probiotic functionality in case the claimed probiotic effect is related to adhesion of the strain to the intestinal mucus. Recently, the relevance of these genes for probiotic functionality was further demonstrated by showing that the SpaCBA pili could directly induce anti-inflammatory pathways or indirectly lead to anti-inflammatory signaling by promoting the release of anti-inflammatory factors such as the MSP1 and MSP2 soluble proteins (18).
The deleted regions observed in L. rhamnosus GG product isolates 1 and 2A are completely overlapping and comprise DNA stretches of 34 genes (LGG_00412 to LGG_00445) and 84 genes (LGG_00383 to LGG_00466), respectively. They both appear to overlap with the genomic islands LGGISL1 (LGG_00393 to LGG_00427) and LGGISL2 (LGG_00433 to LGG_00450), previously identified as DNA sequences deviating in codon usage, base composition, and dinucleotide frequency from the rest of the genome, suggesting that they originated from horizontal gene transfer. Typical functions of the genomic island LGGISL1 include the capacity to transport or metabolize sugars, while LGGISL2 island appears to encode a set of genes for multisubunit pili (SpaCBA) and a sortase, required for the assembly of pilus structures (10).
The deleted DNA segments were both flanked by two identical insertion elements, encoding IS30 transposases in one product isolate and IS5 transposases in the other. Although ISs are best known to be involved in acquisition of accessory functions, these elements are also known to be involved in chromosomal deletions (7). Since it was found that both missing stretches of DNA were flanked by two identical copies of ISs, they could be involved in the deletions, possibly through the activity of a composite transposon. In case the result of this genetic alteration leads to increased fitness under the conditions used for cultivation, there will be an enrichment for bacterial genotypes without (part of the) genomic islands, as has been observed in the present study for some of the dairy products.
Cai et al. reported that gene decay in lactobacilli is known to be associated with the transition from dynamic and nutritionally variable environments, such as the human gastrointestinal tract and plants, to the relatively constant and nutrient-rich dairy niche (5). This event has also recently been simulated in a laboratory experiment (19). Hence, propagation of strains isolated from the intestinal gut, as is the case for many probiotics, including L. rhamnosus GG, likely results in metabolic simplification and loss of genes, which could be linked to probiotic functionality. A practical example refers to the industrial processing of probiotics, when strains are grown for several generations in relatively large volumes (20) without selective pressure for maintenance of genes with probiotic functionality.
We reason that from a genome stability point of view, future research should evaluate whether traditionally fermented dairy products, where the lactobacilli have been cultivated for numerous generations under nutrient-rich conditions, could be a better source of selection for large scale production of genetically stable probiotic strains than the human gastrointestinal tract. Typically, this was the approach followed by Elia Metchnikoff more than a 100 years ago (21). As a drawback of this approach, one could reason that some specific probiotic functionalities may only be found in isolates from the intestinal tract, illustrating the need for innovative cultivation methods that keep selective pressure on persistence of genes, which are important for health-enhancing microbe-host interactions. In the context of the present study, one might think of growth conditions that maintain selective pressure on the presence of carbohydrate transporting and metabolizing genes encoded by LGGISL1. In addition, it might be worthwhile to avoid exposure to stress conditions that might activate transposons (7). However, pretreatment of cells under nonmutagenic conditions (42°C) appeared to protect them against subsequent increases in the mutation frequency due to oxidative stress, showing that sequential exposure to certain stresses may also be used to protect industrial strains against elevated mutation rates and in this way potentially enhance the stability of these strains (22).
Although we describe here the occurrence of genetic instability, including mutations and deletions of several genes, it should also be noted that the overall genome similarity between the four L. rhamnosus GG variants was >97%, including deleted regions and >99.9% when excluding deleted regions. Even though a single point mutation might lead to a loss of functionality, when checking the mutations and deletions presented in Tables S2 to S4 in the supplemental material, one may conclude that many of the other probiotic functionalities are still present; these include exopolysaccharide production, tolerance to bile salts, stimulation of the immune system, and also all of the probiotic effects beyond the gut, as reviewed by Lebeer et al. (3). Two specific examples to mention are the L. rhamnosus GG secreted proteins MSP1 and MSP2, which have been reported to enhance survival and growth of intestinal epithelial cells and which are still encoded in all of the four sequenced genomes (23). Interestingly, the corresponding genes for these proteins are supposed to remain genetically stable because the secreted proteins are also involved in essential cell metabolic activities such as cell separation (24).
Our results contribute to the debate about both regulatory and scientific aspects of functional claims linked to probiotic products and required quality control measures. The European Food and Safety Authority (EFSA) has scientifically evaluated numerous article 13.1 health claims. The NDA Panel of the EFSA considered that in most of the cases the information provided was not sufficient to characterize a number of foods or constituents with respect to the claimed effects, including some (but not all) probiotic bacteria (25). The NDA Panel proposed species identification by DNA-DNA hybridization or 16S rRNA gene sequence analysis and/or sequence analysis of other relevant genetic markers. Strain identification is proposed by pulsed-field gel electrophoresis of genomic DNA, randomly amplified polymorphic DNA analysis, or other internationally accepted genetic typing molecular methods (26). We reason that even EFSA-proposed strain-specific identification methods such as DNA fingerprinting or other methods, such as enzyme-linked immunosorbent assays, the use of monoclonal antibodies, or strain-specific PCR (27), are not sufficient to guarantee that strains with a claimed functionality are effectively present in probiotic products. Furthermore, and as a general statement, we think that products containing probiotic strains that have a significantly altered genetic content or functionality should be tested in trials to confirm their efficacy. However, it should be noted that bacteria are living entities that always evolve. The suggestion that products cannot be sold because of the potential occurrence of minor genetic alterations as part of an evolutionary process would make it probably impossible to ever sell a probiotic.
Until the impact of genetic instability in general and the absence of pili in particular on probiotic functionality is well understood, we propose a quality assurance approach involving validation steps in the production and release processes of probiotic products. The validation is aimed to confirm the genetic stability of the overall genome, especially around mobile genetic elements that could induce the simultaneous deletion of several genes. In addition, it remains relevant to confirm that strains, which are marketed today, have the same genetic makeup as the strains used in clinical studies on which their health claims have been based. Hereto, and as a starting point for further research, we propose to check whether the results presented here could provide a complementary explanation for the variation of probiotic properties (based on in vitro assays) observed in 16 L. rhamnosus GG product isolates and suggested to be linked to different processing conditions (28).
Finally, we note that when strains such as L. rhamnosus GG are genericized, as has recently been reported (1), it is important that they possess the characteristics that make them efficacious; otherwise, people relying on them will not benefit.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martien Caspers, Alie de Kat Angelino-Bart, and Marianne van Muijlwijk-Harteveld for their expert assistance in cultivation, DNA extraction, qPCR, and data processing. We acknowledge Jan Dijksterhuis for transmission electron microscopy and Ferenc Olasz and Monika Szabo for discussing the content of the manuscript.
ADDENDUM IN PROOF
During the review of this paper, there have been two more reports of the genetic (in)stability of probiotics. Douillard et al. (Appl. Environ. Microbiol. 79:1923–1933, 2013) found that the genomes of two L. rhamnosus GG strains isolated from two different products are virtually identical. Averina et al. (Russ. J. Genet. 48:1103–1111, 2012) noted the loss of the galA and tet(W) genes from the genome of Bifidobacterium longum subsp. longum B379M during cultivation and maintenance under laboratory conditions. Interestingly, tet(W) is located between two IS30 elements, as is described for one of the deletions observed in the present research paper. The latter results indicate that the genetic instability of probiotics may be a rather widespread phenomenon, which further supports a recommendation that regulating authorities, the food industry, and academia focus on controlling the retention of key probiotic genes.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03566-12.
REFERENCES
- 1.Kort R, Sybesma W. 2012. Probiotics for every body. Trends Biotechnol. 30:613–615 [DOI] [PubMed] [Google Scholar]
- 2.Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. 2012. The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 12:728–734 [DOI] [PubMed] [Google Scholar]
- 3.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72:728–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turpin W, Humblot C, Guyot JP. 2011. Genetic screening of functional properties of lactic acid bacteria in a fermented pearl millet slurry and in the metagenome of fermented starchy foods. Appl. Environ. Microbiol. 77:8722–8734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL. 2009. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol. Evol. 1:239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider D, Lenski RE. 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 155:319–327 [DOI] [PubMed] [Google Scholar]
- 7.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Visser JA, Akkermans AD, Hoekstra RF, de Vos WM. 2004. Insertion-sequence-mediated mutations isolated during adaptation to growth and starvation in Lactococcus lactis. Genetics 168:1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita H, Toh H, Oshima K, Murakami M, Taylor TD, Igimi S, Hattori M. 2009. Complete genome sequence of the probiotic Lactobacillus rhamnosus ATCC 53103. J. Bacteriol. 191:7630–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kort R, Keijser BJ, Caspers MP, Schuren FH, Montijn R. 2008. Transcriptional activity around bacterial cell death reveals molecular biomarkers for cell viability. BMC Genomics 9:590 doi:10.1186/1471-2164-9-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahlroos T, Tynkkynen S. 2009. Quantitative strain-specific detection of Lactobacillus rhamnosus GG in human faecal samples by real-time PCR. J. Appl. Microbiol. 106:506–514 [DOI] [PubMed] [Google Scholar]
- 13.Brouwer RW, van den Hout MC, Grosveld FG, van Ijcken WF. 2012. NARWHAL, a primary analysis pipeline for NGS data. Bioinformatics 28:284–285 [DOI] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247 [DOI] [PubMed] [Google Scholar]
- 17.Kerkhoven R, van Enckevort FH, Boekhorst J, Molenaar D, Siezen RJ. 2004. Visualization for genomics: the Microbial Genome Viewer. Bioinformatics 20:1812–1814 [DOI] [PubMed] [Google Scholar]
- 18.Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, Vos WM, Keersmaecker SC, Vanderleyden J. 2012. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 78:185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachmann H, Starrenburg MJ, Molenaar D, Kleerebezem M, van Hylckama Vlieg JE. 2012. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 22:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller J, Ross R, Fitzgerald G, Stanton C. 2009. Manufacture of probiotic bacteria, p 725–759 Charalampopoulos D, Rastall RA. (ed), Prebiotics and probiotics science and technology. Springer, New York, NY [Google Scholar]
- 21.Metchnikoff E. 1907. The prolongation of life: optimistic studies. Butterworth-Heinemann, London, England [Google Scholar]
- 22.Machielsen R, van Alen-Boerrigter IJ, Koole LA, Bongers RS, Kleerebezem M, Van Hylckama Vlieg JE. 2010. Indigenous and environmental modulation of frequencies of mutation in Lactobacillus plantarum. Appl. Environ. Microbiol. 76:1587–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. 2007. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132:562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan F, Polk DB. 2012. Characterization of a probiotic-derived soluble protein which reveals a mechanism of preventive and treatment effects of probiotics on intestinal inflammatory diseases. Gut Microbes 3:25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA) 2011. General guidance for stakeholders on the evaluation of Article 13.1, 13.5 and 14 health claims. EFSA J. 9:2135 [Google Scholar]
- 26.EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA) 2012. Scientific opinion on the substantiation of health claims related to non-characterised micro-organisms (ID 2936, 2937, 2938, 2941, 2944, 2965, 2968, 2969, 3035, 3047, 3056, 3059, further assessment) pursuant to Article 13(1) of Regulation (EC) No 1924/20061. EFSA J. 10:2854 [Google Scholar]
- 27.Endo A, Aakko J, Salminen S. 2012. Evaluation of strain-specific primers for identification of Lactobacillus rhamnosus GG. FEMS Microbiol. Lett. 337:120–125 [DOI] [PubMed] [Google Scholar]
- 28.Grzeskowiak L, Isolauri E, Salminen S, Gueimonde M. 2011. Manufacturing process influences properties of probiotic bacteria. Br. J. Nutr. 105:887–894 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.