Abstract
Vibrio parahaemolyticus is a seafood-borne pathogenic bacterium that is a major cause of gastroenteritis worldwide. We investigated the genetic and evolutionary relationships of 101 V. parahaemolyticus isolates originating from clinical, human carrier, and various environmental and seafood production sources in Thailand using multilocus sequence analysis. The isolates were recovered from clinical samples (n = 15), healthy human carriers (n = 18), various types of fresh seafood (n = 18), frozen shrimp (n = 16), fresh-farmed shrimp tissue (n = 18), and shrimp farm water (n = 16). Phylogenetic analysis revealed a high degree of genetic diversity within the V. parahaemolyticus population, although isolates recovered from clinical samples and from farmed shrimp and water samples represented distinct clusters. The tight clustering of the clinical isolates suggests that disease-causing isolates are not a random sample of the environmental reservoir, although the source of infection remains unclear. Extensive serotypic diversity occurred among isolates representing the same sequence types and recovered from the same source at the same time. These findings suggest that the O- and K-antigen-encoding loci are subject to exceptionally high rates of recombination. There was also strong evidence of interspecies horizontal gene transfer and intragenic recombination involving the recA locus in a large proportion of isolates. As the majority of the intragenic recombinational exchanges involving recA occurred among clinical and carrier isolates, it is possible that the human intestinal tract serves as a potential reservoir of donor and recipient strains that is promoting horizontal DNA transfer, driving evolutionary change, and leading to the emergence of new, potentially pathogenic strains.
INTRODUCTION
Vibrio parahaemolyticus is a seafood-borne pathogenic bacterium and is the leading cause of traveler's diarrhea and gastroenteritis worldwide due to the consumption of undercooked contaminated seafood, particularly shellfish. The organism possesses a number of virulence factors, including a thermostable direct hemolysin (TDH) (1), a thermostable direct hemolysin-related hemolysin (TRH) (2), and type 3 secretion system 1 (TTSS1) and TTSS2 (3). Strains that possess tdh, trh, and TTSS2-encoding genes are generally pathogenic and responsible for the vast majority of clinical cases. While such strains account for only 1 to 2% of isolates sampled from water and different marine species (4–6), they are known to have a widespread global distribution, having been reported from cases of infection in Japan, India, Thailand, Malaysia, China, Indonesia, the United Kingdom, Italy, and the East and Gulf Coasts of the United States (7–19).
The public health and commercial burdens associated with V. parahaemolyticus contamination are very high in Thailand due to the wide consumption of seafood. Although undercooked seafood has been identified as a source of V. parahaemolyticus infection (20, 21), the relative likelihood of contamination from different sources (e.g., the natural marine environment, aquaculture sources, or the market place) has not been established. Such evidence is essential to guide future intervention strategies and minimize both the risk to the consumer and the cost to the producer. Clinical isolates from Thailand typically correspond to the pandemic serovar O3:K6 (9), which was responsible for the Indian pandemic in 1996 (22–25), as well as serotypic variants of this clone (e.g., O1:KUT, O1:K25, and O3:K46) (26–28). Although a number of molecular approaches have been used to study the epidemiology of V. parahaemolyticus in Thailand (9, 28, 29, 30, 31, 32), these studies have not generated detailed sequence-based molecular evolutionary data.
Multilocus sequence typing (MLST) is an important tool for molecular epidemiology and population genetic studies of bacterial pathogens (33–37). The utilization of housekeeping genes encoding core metabolic enzymes means that the data are unlikely to be impacted by strong positive selection (33). González-Escalona et al. (38) developed a successful MLST scheme for V. parahaemolyticus in a study of 100 strains of global origin. The authors demonstrated that V. parahaemolyticus is genetically diverse and has an epidemic population structure in which successful clones emerge from a highly recombining background. Further MLST analyses have confirmed that V. parahaemolyticus is genetically diverse, undergoes frequent recombination, and has an epidemic population structure (39, 40). Furthermore, there is no evidence of linkage between sequence type and serotype in V. parahaemolyticus, indicating that serotype switching by recombination has been common in the species (26, 38, 41). Overall, these studies have demonstrated that MLST presents a powerful tool with which to determine the role of recombination in the diversification of natural populations of V. parahaemolyticus and for understanding the processes leading to the emergence and spread of clinically relevant strains.
In the present study, we modified and applied the MLST scheme of González-Escalona et al. (38) to a strain collection recovered from different epidemiological sources associated with the seafood industry in Thailand in order to further elucidate the population structure of V. parahaemolyticus and to examine the potential of MLST for identifying sources of seafood contamination. Isolates were obtained from clinical samples, human carriers, fresh seafood, frozen shrimp, fresh-farmed shrimp tissue, and shrimp farm water. The data confirm a highly diverse population, with very limited evidence of concordance between ST and epidemiological sources. This points to the requirement for a far higher genetic resolution (such as is provided by next-generation sequencing), combined with a focus on specific clones, in order to robustly pinpoint the most significant sources of contamination. Our data also confirm high rates of recombination, particularly at recA, and we present a novel approach to clustering the isolates on the basis of amino acid sequences.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
V. parahaemolyticus type strain NCTC 10903 (ATCC 17802T) was obtained from the National Collection of Type Cultures (NCTC), Health Protection Agency (HPA), United Kingdom. One hundred and one V. parahaemolyticus isolates were obtained from six categories of sources in Thailand as follows. Clinical isolates (n = 15) were recovered from gastroenteritis patients, human carrier isolates (n = 18) were recovered from healthy volunteer workers at a seafood-processing plant, fresh seafood isolates (n = 18) were recovered from a seafood market located in central Thailand, frozen-shrimp isolates (n = 16) were recovered from frozen shrimp at a processing factory in central Thailand, shrimp tissue isolates (n = 18) were recovered from fresh shrimp at two shrimp farms (farms 1 and 2) in southern Thailand, and water isolates (n = 16) were recovered from water samples at the same two shrimp farms. The clinical and human carrier isolates were all obtained from different individuals, from stool samples and rectal swabs, respectively. Bacterial isolation and identification procedures were adapted from those described by Kaysner and DePaola (42). Briefly, samples were plated onto thiosulfate-citrate-bile salts-sucrose agar at 37°C for 24 h. Selected colonies were confirmed as V. parahaemolyticus by biochemical characterization and by O and K serotype determination (42). Details of the isolates are provided in Table 1. The isolates were stored at −80°C in 50% (vol/vol) glycerol in tryptic soy broth (TSB) (Oxoid) with 3% (wt/vol) NaCl and subcultured on tryptic soy agar (TSA) (Oxoid) with 3% (wt/vol) NaCl by overnight aerobic incubation at 37°C. For preparation of DNA, a few colonies were inoculated into 10-ml volumes of TSB with 3% (wt/vol) NaCl and grown aerobically overnight at 37°C with shaking at 120 rpm.
Table 1.
Properties of 102 V. parahaemolyticus isolates
| Isolate | Source of isolation | Isolation date | Serotype | tdha | trha | ST | Allelic profile (dnaE, gyrB, recA, dtdS, pntA, pyrC, tnaA)b |
|---|---|---|---|---|---|---|---|
| VP2 | Food poisoning (type strain) | 1950 | O1:K1 | − | − | 1 | 5, 52, 27, 13, 17, 25, 10 |
| VP4 | Frozen shrimp from processing plant A | April 1999 | O3:K20 | − | − | 229 | 109, 136, 25, 121, 83, 107, 83 |
| VP6 | Frozen shrimp from processing plant A | April 1999 | O10:K66 | − | − | 230 | 110, 144, 166, 35, 18, 108, 86 |
| VP8 | Frozen shrimp from processing plant A | April 1999 | O3:KUT | − | − | 231 | 111, 17, 3, 123, 85, 37, 87 |
| VP12 | Frozen shrimp from processing plant A | April 1999 | O1:KUT | − | − | 241 | 112, 143, 25, 120, 26, 109, 81 |
| VP14 | Frozen shrimp from processing plant A | April 1999 | O2:K3 | − | − | 232 | 98, 131, 30, 32, 77, 11, 82 |
| VP16 | Frozen shrimp from processing plant A | April 1999 | O3:K20 | − | − | 233 | 109, 136, 114, 121, 83, 107, 83 |
| VP20 | Frozen shrimp from processing plant A | April 1999 | O1:K25 | − | − | 242 | 113, 145, 61, 70, 28, 11, 26 |
| VP22 | Frozen shrimp from processing plant A | April 1999 | O1:K20 | − | − | 234 | 5, 84, 115, 74, 84, 26, 84 |
| VP24 | Frozen shrimp from processing plant A | April 1999 | O9:K44 | − | − | 235 | 10, 69, 27, 76, 46, 65, 29 |
| VP26 | Frozen shrimp from processing plant A | April 1999 | O7:K19 | − | − | 236 | 114, 100, 61, 122, 66, 54, 85 |
| VP28 | Frozen shrimp from processing plant A | April 1999 | O5:K17 | − | − | 237 | 115, 43, 25, 108, 71, 73, 62 |
| VP30 | Frozen shrimp from processing plant A | April 1999 | O3:K20 | − | − | 233 | 109, 136, 114, 121, 83, 107, 83 |
| VP32 | Frozen shrimp from processing plant A | April 1999 | O2:KUT | − | − | 243 | 12, 146, 117, 124, 28, 10, 54 |
| VP36 | Frozen shrimp from processing plant A | April 1999 | O10:KUT | − | − | 238 | 109, 136, 114, 121, 21, 107, 83 |
| VP38 | Frozen shrimp from processing plant A | April 1999 | O3:K20 | − | − | 239 | 6, 6, 3, 17, 11, 26, 23 |
| VP40 | Frozen shrimp from processing plant A | April 1999 | O11:K40 | − | − | 240 | 31, 147, 62, 84, 4, 45, 88 |
| VP44 | Water from shrimp farm 1, pond A | January 2008 | O9:K23 | − | − | 244 | 19, 74, 61, 68, 86, 11, 26 |
| VP46 | Water from shrimp farm 1, pond A | January 2008 | O1:K38 | − | − | 244 | 19, 74, 61, 68, 86, 11, 26 |
| VP48 | Water from shrimp farm 1, pond B | January 2008 | O9:K24 | − | − | 244 | 19, 74, 61, 68, 86, 11, 26 |
| VP50 | Water from shrimp farm 1, pond B | January 2008 | O7:K52 | − | − | 244 | 19, 74, 61, 68, 86, 11, 26 |
| VP52 | Water from shrimp farm 1, pond B | January 2008 | O1:KUT | − | − | 244 | 19, 74, 61, 68, 86, 11, 26 |
| VP54 | Water from shrimp farm 1, pond B | January 2008 | O1:KUT | − | − | 244 | 19, 74, 61, 68, 86, 11, 26 |
| VP56 | Water from shrimp farm 1, pond B | January 2008 | O2:KUT | − | − | 245 | 49, 148, 25, 125, 60, 110, 89 |
| VP58 | Water from shrimp farm 1, pond B | January 2008 | O7:KUT | − | − | 239 | 6, 6, 3, 17, 11, 26, 23 |
| VP60 | Water from shrimp farm 1, pond B | January 2008 | O2:K3 | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP62 | Water from shrimp farm 1, pond C | January 2008 | O1:KUT | − | − | 247 | 116, 149, 72, 76, 45, 62, 26 |
| VP64 | Water from shrimp farm 1, pond C | January 2008 | OUT:KUT | − | − | 248 | 5, 88, 61, 19, 18, 3, 90 |
| VP66 | Water from shrimp farm 1, pond C | January 2008 | O1:K26 | − | − | 248 | 5, 88, 61, 19, 18, 3, 90 |
| VP72 | Water from shrimp farm 2, pond A | January 2008 | O9:K44 | − | − | 249 | 3, 151, 25, 29, 61, 11, 62 |
| VP74 | Water from shrimp farm 2, pond A | January 2008 | O2:K3 | − | − | 180 | 89, 105, 15, 89, 57, 11, 57 |
| VP76 | Water from shrimp farm 2, pond A | January 2008 | O7:K52 | − | − | 249 | 3, 151, 25, 29, 61,11, 62 |
| VP80 | Water from shrimp farm 2, pond B | January 2008 | O6:K46 | − | − | 250 | 118, 87, 119, 117, 54, 111, 91 |
| VP84 | Shrimp hepatopancreas, farm 2, pond B | August 2007 | O10:K71 | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP86 | Shrimp hepatopancreas, farm 2, pond B | August 2007 | O4:K63 | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP88 | Shrimp hepatopancreas, farm 2, pond B | August 2007 | O9:K23 | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP90 | Shrimp hepatopancreas, farm 2, pond B | August 2007 | O2:K28 | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP94 | Shrimp muscle, farm 2, pond B | August 2007 | O1:KUT | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP96 | Shrimp muscle, farm 2, pond B | August 2007 | O10:K52 | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP98 | Shrimp muscle, farm 2, pond B | August 2007 | O2:KUT | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP100 | Shrimp intestine, farm 1, pond B | August 2007 | O1:KUT | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP102 | Shrimp intestine, farm 1, pond B | August 2007 | O1:K1 | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP104 | Shrimp intestine, farm 2, pond B | August 2007 | O9:K23 | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP106 | Shrimp intestine, farm 2, pond B | August 2007 | O1:KUT | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP108 | Shrimp internal body, farm 1, pond B | August 2007 | O2:K3 | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP110 | Shrimp internal body farm 1, pond B | August 2007 | O1:K56 | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP112 | Shrimp internal body, farm 1, pond B | August 2007 | O9:K44 | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP114 | Shrimp internal body, farm 1, pond B | August 2007 | O7:KUT | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP116 | Shrimp shell, farm 1, pond B | August 2007 | OUT:KUT | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP118 | Shrimp shell, farm 1, pond B | August 2007 | OUT:KUT | − | − | 246 | 33, 87, 24, 5, 10, 5, 1 |
| VP122 | Shrimp shell, farm 2, pond B | August 2007 | OUT:KUT | − | − | 251 | 119, 152, 120, 29, 23, 11, 61 |
| VP126 | Healthy carrier from seafood plant B | November 2003 | O10:KUT | − | − | 252 | 36, 153, 121, 126, 27, 112, 17 |
| VP128 | Healthy carrier from seafood plant B | November 2003 | O9:K23 | − | − | 253 | 35, 154, 25, 50, 73, 35, 23 |
| VP130 | Healthy carrier from seafood plant B | November 2002 | O1:KUT | − | − | 254 | 112, 155, 122, 19, 87, 96, 92 |
| VP132 | Healthy carrier from seafood plant B | August 2003 | O3:K46 | + | − | 3 | 3, 4, 19, 4, 29, 4, 22 |
| VP136 | Healthy carrier from seafood plant B | October 2003 | O1:KUT | + | + | 199 | 22, 28, 17, 13, 8, 19, 14 |
| VP138 | Healthy carrier from seafood plant B | November 2003 | O11:K5 | + | + | 255 | 12, 156, 123, 127, 19, 12, 47 |
| VP140 | Healthy carrier from seafood plant B | August 2003 | O4:KUT | + | − | 62 | 19, 4, 88, 2, 34, 18, 23 |
| VP142 | Healthy carrier from seafood plant B | July 2002 | OUT:KUT | − | − | 256 | 42, 157, 59, 128, 6, 113, 57 |
| VP144 | Healthy carrier from seafood plant B | July 2002 | O4:K63 | + | − | 257 | 120, 158, 89, 129, 26, 114, 93 |
| VP146 | Healthy carrier from seafood plant B | July 2002 | O1:K25 | − | − | 258 | 121, 95, 124, 130, 26, 115, 12 |
| VP148 | Healthy carrier from seafood plant B | August 2003 | O1:K41 | − | − | 256 | 42, 157, 59, 128, 6, 113, 57 |
| VP150 | Healthy carrier from seafood plant B | October 2003 | O1:KUT | + | − | 259 | 122, 25, 125, 131, 21, 11, 66 |
| VP152 | Healthy carrier from seafood plant B | November 2002 | O8:K21 | − | − | 259 | 122, 25, 125, 131, 21, 11, 66 |
| VP154 | Healthy carrier from seafood plant B | October 2003 | O4:KUT | + | − | 68 | 41, 40, 36, 41, 36, 39, 32 |
| VP156 | Healthy carrier from seafood plant B | November 2002 | O1:K1 | − | − | 260 | 3, 82, 126, 69, 30, 7, 23 |
| VP158 | Healthy carrier from seafood plant B | August 2003 | O1:KUT | + | − | 3 | 3, 4, 19, 4, 29, 4, 22 |
| VP160 | Healthy carrier from seafood plant B | November 2003 | O11:K40 | − | − | 261 | 123, 100, 127, 50, 66, 11, 31 |
| VP162 | Healthy carrier from seafood plant B | November 2003 | O1:K12 | + | + | 255 | 12, 156, 123, 127, 19, 12, 47 |
| VP164 | Clinical sample from hospital patient | September 1990 | O1:K58 | + | − | 17 | 13, 10, 19, 27, 28, 27, 21 |
| VP166 | Clinical sample from hospital patient | August 1990 | O1:K1 | + | + | 83 | 5, 52, 27, 13, 17, 25, 40 |
| VP168 | Clinical sample from hospital patient | August 1990 | O3:KUT | − | + | 66 | 42, 25, 3, 40, 35, 38, 31 |
| VP170 | Clinical sample from hospital patient | Febuary 1991 | O3:KUT | + | + | 262 | 105, 156, 123, 127, 19, 12, 47 |
| VP172 | Clinical sample from hospital patient | August 1990 | O1:K1 | + | + | 83 | 5, 52, 27, 13, 17, 25, 40 |
| VP174 | Clinical sample from hospital patient | Febuary 1991 | O3:KUT | + | + | 263 | 50, 55, 48, 52, 23, 53, 47 |
| VP176 | Clinical sample from hospital patient | May 1990 | O1:K1 | + | + | 264 | 5, 52, 27, 132, 17, 25, 40 |
| VP178 | Clinical sample from hospital patient | January 1991 | O1:K69 | + | + | 262 | 105, 156, 123, 127, 19, 12, 47 |
| VP180 | Clinical sample from hospital patient | September 1990 | O8:K22 | + | − | 262 | 105, 156, 123, 127, 19, 12, 47 |
| VP182 | Clinical sample from hospital patient | April 1990 | O1:K69 | − | + | 262 | 105, 156, 123, 127, 19, 12, 47 |
| VP184 | Clinical sample from hospital patient | April 1990 | O4:K11 | + | − | 262 | 105, 156, 123, 127, 19, 12, 47 |
| VP188 | Clinical sample from hospital patient | January 1991 | O1:K69 | + | + | 262 | 105, 156, 123, 127, 19, 12, 47 |
| VP190 | Clinical sample from hospital patient | July 1990 | O4:K10 | + | − | 265 | 11, 48, 107, 48, 26, 48, 26 |
| VP194 | Clinical sample from hospital patient | September 1990 | OUT:KUT | + | − | 83 | 5, 52, 27, 13, 17, 25, 40 |
| VP200 | Clinical sample from hospital patient | September 1990 | O4:K8 | + | − | 189 | 11, 48, 3, 48, 26, 48, 26 |
| VP204 | Fresh oysters from market | March 2003 | O1:K64 | − | − | 267 | 126, 161, 43, 19, 54, 10, 26 |
| VP206 | Fresh oysters from market | March 2003 | O2:K3 | − | − | 268 | 127, 67, 24, 133, 10, 5, 95 |
| VP208 | Fresh oysters from market | March 2003 | OUT:KUT | − | − | 269 | 128, 162, 128, 134, 1, 11, 94 |
| VP210 | Fresh bloody clams from market | December 2003 | O7:KUT | − | − | 270 | 129, 151, 98, 12, 88, 117, 26 |
| VP212 | Fresh bloody clams from market | December 2003 | O10:K19 | − | − | 271 | 3, 134, 81, 76, 26, 11, 26 |
| VP214 | Fresh bloody clams from market | December 2003 | O1:KUT | − | − | 272 | 60, 116, 91, 135, 50, 118, 2 |
| VP216 | Boiled crab meat from market | October 2002 | O2:KUT | − | − | 273 | 130, 58, 113, 69, 89, 119, 23 |
| VP218 | Boiled crab meat from market | October 2002 | O1:KUT | − | − | 274 | 69, 92, 69, 27, 54, 71, 24 |
| VP220 | Boiled crab meat from market | October 2002 | OUT:KUT | − | − | 275 | 31, 163, 129, 19, 36, 120, 96 |
| VP222 | Boiled mussels from market | July 2003 | NA | − | − | 276 | 131, 147, 60, 136, 90, 27, 23 |
| VP224 | Boiled mussels from market | July 2003 | O10:KUT | − | − | 276 | 131, 147, 60, 136, 90, 27, 23 |
| VP226 | Boiled mussels from market | July 2003 | OUT:KUT | − | − | 277 | 3, 29, 98, 67, 26, 121, 33 |
| VP228 | Fresh shrimp from market | June 2003 | O1:KUT | − | − | 278 | 103, 164, 130, 137, 50, 122, 57 |
| VP230 | Fresh shrimp from market | June 2003 | O3:K58 | − | − | 279 | 132, 165, 25, 110, 4, 116, 12 |
| VP232 | Fresh shrimp from market | June 2003 | O5:KUT | − | − | 278 | 103, 164, 130, 137, 50, 122, 57 |
| VP234 | Fresh shrimp from market | June 2003 | O5:KUT | − | − | 114 | 55, 15, 31, 55, 18, 58, 46 |
| VP236 | Fresh shrimp from market | June 2003 | O1:K69 | − | − | 280 | 28, 144, 3, 138, 26, 123, 97 |
| VP238 | Fresh shrimp from market | June 2003 | O10:K52 | − | − | 281 | 133, 67, 4, 79, 43, 63, 23 |
+, present; −, absent.
The numbers in this column represent the allele designation at each locus.
Preparation of chromosomal DNA.
Cells from 1.0 ml of overnight culture were harvested by centrifugation for 1 min at 13,000 × g and washed once in sterile distilled H2O. DNA was prepared with InstaGene Matrix (Bio-Rad) according to the manufacturer's instructions and stored at −20°C.
PCR amplification and DNA sequencing.
Selection of the seven loci analyzed by MLST was based on the previously published MLST scheme for V. parahaemolyticus (38), but modifications were made as described below. For chromosome I, the housekeeping genes used were dnaE (DNA polymerase III, alpha subunit), gyrB (DNA gyrase subunit B), and recA (RecA protein). For chromosome II, the housekeeping genes used were dtdS (threonine 3-dehydrogenase), pntA (transhydrogenase, alpha subunit), pyrC (dihydro-oratase), and tnaA (tryophanase). In the original V. parahaemolyticus MLST scheme (38), a single set of PCR and sequencing primers was used. However, separate PCR and sequencing primers give more accurate sequencing results and are recommended for MLST (33). Thus, in the present study, separate sets of PCR and sequencing primers were used for amplification and sequencing of the seven gene fragments. New PCR primers (located upstream and downstream of the previously published forward and reverse PCR/sequencing primers, respectively) were designed, using Primer Designer version 2 (Scientific and Educational Software). In addition, new shorter sequencing primers were designed, using the same software, based on the previously published primers (38). All primers were designed to a length of 18 nucleotides except gyrB-F2, which contains 17 nucleotides. The nucleotide sequences of the primers used for PCR and DNA sequencing are provided in Table S1 in the supplemental material. In some isolates, gene fragments were unable to be amplified by these primers, in which case additional primers were designed (see Table S2 in the supplemental material).
PCR fragments containing partial segments of dnaE (776 bp), gyrB (758 bp), recA (932 bp), dtdS (572 bp), pntA (676 bp), pyrC (681 bp), and tnaA (600 bp) were amplified from chromosomal DNA using a proofreading Taq polymerase kit (Platinum Pfx DNA Polymerase; Invitrogen) according to the manufacturer's instructions. PCRs were carried out in a GeneAmp PCR System 9700 Thermo Cycler (Applied Biosystems) using 30 cycles of the following amplification parameters: denaturation at 94°C for 45 s, annealing at 59°C for 45 s, and extension at 72°C for 2 min. An initial denaturation step of 94°C for 2 min was used, and a final extension step at 72°C for 10 min. However, in some cases (e.g., for dnaE, gyrB, dtdS, and pyrC), improved results were obtained by varying the annealing temperatures between 55 and 60°C. The expected sizes of the PCR products were confirmed by electrophoresis in a 1% (wt/vol) agarose gel incorporating 0.004% (vol/vol) SybrSafe (Invitrogen). DNA was purified with a Qiaquick PCR purification kit (Qiagen) and finally eluted in 30 to 50 μl sterile distilled H2O and stored at −20°C.
Sequencing reactions were performed in 10-μl reaction mixtures using the BigDye Terminator cycle-sequencing kit version 3.1 according to the manufacturer's instructions. Each reaction mixture consisted of 3.5 μl dilution buffer, 3 μl DNA template (approximately 50 ng/μl DNA), 2 μl of 2-pmol/μl forward or reverse sequencing primer, 1 μl distilled H2O (dH2O), and 0.5 μl of a 1:16 dilution of BigDye Terminator Ready Reaction Mix. The reactions were carried out in a GeneAmp PCR System 9700 Thermo Cycler (Applied Biosystems) using 25 cycles of the following amplification parameters: denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 4 min. The DNA fragments were cleaned up by ethanol precipitation and sequenced using an ABI 3730 capillary sequencer (Genepool Sequencing Unit, University of Edinburgh [http://genepool.bio.ed.ac.uk]).
Nucleotide and population structure analyses.
Sequencing data were checked and edited using Lasergene version 5.0 (DNAStar) sequence analysis software. For each of the seven alleles representing each isolate, forward and reverse primers were aligned and a consensus sequence was generated. Each consensus sequence was submitted to the V. parahaemolyticus MLST website (http://pubmlst.org/vparahaemolyticus) to determine its identity against existing alleles. New alleles that did not match any preexisting alleles within the database were submitted to the database curator, who assigned new allele and serotype designations. Nucleotide sequence analyses were conducted with MEGA version 4.0.2 (43), in conjunction with alignment programs written by T. S. Whittam (Michigan State University). Global optimal eBURST (goeBURST) (44) analysis of the 63 sequence types (STs) in the data set was performed using Phyloviz software (http://www.phyloviz.net). Recombination events were detected in concatenated DNA sequences using the recombination detection program version 3.0 (RDP3) software package (45). The Bayesian Analysis of Population Structure (BAPS) software version 5.3 (46, 47) was used to infer the population structure by clustering the STs into genetically distinct groups. The “clustering with linked loci” module was employed to approximate the number of genetically distinct groups, i.e., the genetic-mixture analysis (47). Following the recommendations in the BAPS manual (http://web.abo.fi/fak/mnf/mate/jc/software/BAPS5manual.pdf), several K values (where K is the estimated maximum number of genetically distinct groups) were used to assess how this might affect the results; a range of K values (from 2 to 20) were used, and in all cases, the results were identical. To test for admixture among the genetic groups identified by BAPS, an admixture analysis was performed (46) with a minimum population size of 5 and the following specifications: the number of iterations used to estimate the admixture coefficient for the individuals was set to 100; the number of reference individuals from each population was 200; the number of iterations used to estimate the admixture coefficient for reference individuals was 20. Only STs having P values of less than 0.05 were considered to have “significant” evidence of admixture. BAPS clusters in respect to phylogenetic inference were illustrated by mapping BAPS clusters onto a neighbor-joining tree of concatenated sequences.
aaST designation.
The amino acid sequence types (aaSTs) of 348 nucleotide STs from the MLST database (http://pubmlst.org/vparahaemolyticus), including the 63 STs of the Thai isolates from the present study, were assigned according to scripts written by David Aanensen. Briefly, for each MLST locus, nucleotide allele sequences were translated in frame, and unique amino acid alleles (aaAlleles) were assigned. For every nucleotide ST, replacement of the nucleotide allele with the aaAllele allowed us to assign unique aaSTs for each strain. Further information about aaST assignment of the 348 nucleotide STs from the V. parahaemolyticus MLST database is provided in Table S3 in the supplemental material.
RESULTS
Nucleotide diversity at each locus.
The nucleotide sequence data for the seven housekeeping gene fragments for the 102 V. parahaemolyticus isolates, including the type strain NCTC 10903, are summarized in Table 2. The number of alleles observed for each locus ranged from 39 (pntA) to 49 (gyrB), and the percentage of polymorphic nucleotide sites varied from 7.3% (tnaA) to 25.8% (recA). The most frequently occurring alleles at each locus were dnaE119 (10), gyrB87 and gyrB152 (10), recA24, recA61 and recA120 (10), dtdS29 (12), pntA23 (11), pyrC11 (26), and tnaA26 (13). The dN/dS ratios (ratios of nonsynonymous to synonymous evolutionary changes) were <1 for dnaE, recA, pntA, pyrC, and tnaA; no nonsynonymous changes were detected at gyrB or dtdS. The mean dN/dS ratio for the two genes on chromosome I (0.038) was slightly higher than that of the three genes on chromosome II (0.024).
Table 2.
Nucleotide and allelic diversity of MLST loci for 102 V. parahaemolyticus isolates
| Chromosome and locus | Fragment size (bp) | No. of alleles | No. (%) of polymorphic nucleotide sites | No. (%) of inferred variable amino acid sites | dN/dS ratio |
|---|---|---|---|---|---|
| I | |||||
| dnaE | 558 | 47 | 44 (7.9) | 4 (2.2) | 0.044 |
| gyrB | 591 | 49 | 47 (8.0) | 1 (0.5) | 0.000 |
| recA | 726 | 42 | 187 (25.8) | 17 (7.0) | 0.033 |
| II | |||||
| dtdS | 456 | 48 | 36 (7.9) | 0 (0.0) | 0.000 |
| pntA | 430 | 39 | 36 (8.4) | 7 (4.9) | 0.024 |
| pyrC | 489 | 44 | 39 (8.0) | 10 (6.1) | 0.026 |
| tnaA | 423 | 40 | 31 (7.3) | 6 (4.3) | 0.021 |
Genotypic diversity.
Details of the isolates, including the source and year of isolation, serotype, presence of the hemolysin-encoding genes tdh and trh, ST, and allelic profile, are presented in Table 1. A total of 63 STs were identified among the 102 isolates, and 53 (84%) of these were novel, since they had not previously been recorded in the MLST database (http://pubmlst.org/vparahaemolyticus). The high proportion of novel STs in our study illustrates the high degree of environmental diversity, even on a fairly local scale, and how poorly the current MLST data set represents this diversity within V. parahaemolyticus. We also note that the recovery of novel STs was nonrandom with respect to the epidemiological source. Sixty-eight isolates were recovered from seafood, frozen shrimp, shrimp tissue, or water, and they represented 40 STs, 39 (98%) of which were novel. Eighteen isolates were recovered from human carriers, and they represented 14 STs, 10 (71%) of which were novel. Finally, 16 isolates (including the type strain) were recovered from human disease cases, and they represented 9 STs, only four (44%) of which were novel.
Four of the clinical STs identified in the present study were also associated with clinical isolates in the MLST database. ST83 represents a common clinical ST in Japan and India; ST189 corresponds to clinical isolates from China, Japan, and India; ST66 represents clinical isolates previously recovered from Mozambique; and ST17 corresponds to clinical isolates from Spain and the United States. Similarly, the four human carrier STs identified in the present study that were recorded in the MLST database had also been recovered from cases of disease. Of these, ST3 represents the pandemic sequence type, while ST62 and ST199 were previously recovered from clinical cases in China and ST68 from Mozambique. Significantly, none of the clinical and carrier STs were associated with environmental isolates. Our data strongly suggest that clinical and human carrier isolates do not represent a random sample of the reservoir of diversity present in the environment. Rather, a limited number of genotypes appear to be adapted to human carriage, and a subset of these isolates have the potential to cause disease.
Clonal relationships of V. parahaemolyticus.
The extent of recombination within natural populations of bacteria can be determined by calculating the degree of linkage between alleles relative to a null of random association using the standardized index of association (ISA) (48, 49). A value of zero is indicative of linkage equilibrium or a freely recombining, nonclonal population, whereas a value of unity represents linkage disequilibrium or nonrandom association of alleles. When ISA was calculated for the entire collection of 102 isolates, a value of 0.5966 (P < 0.05) was obtained, indicating that the alleles are in linkage disequilibrium. This value was very similar to the figure of 0.7626 obtained by González-Escalona et al. (38) for 100 V. parahaemolyticus strains recovered from geographically diverse locations. When the analysis was repeated using one isolate to represent each of the 63 STs, the ISA value was calculated to be 0.1350 (P < 0.05). Although this represented a substantial decrease from 0.5966, it also indicated that the alleles are in linkage disequilibrium.
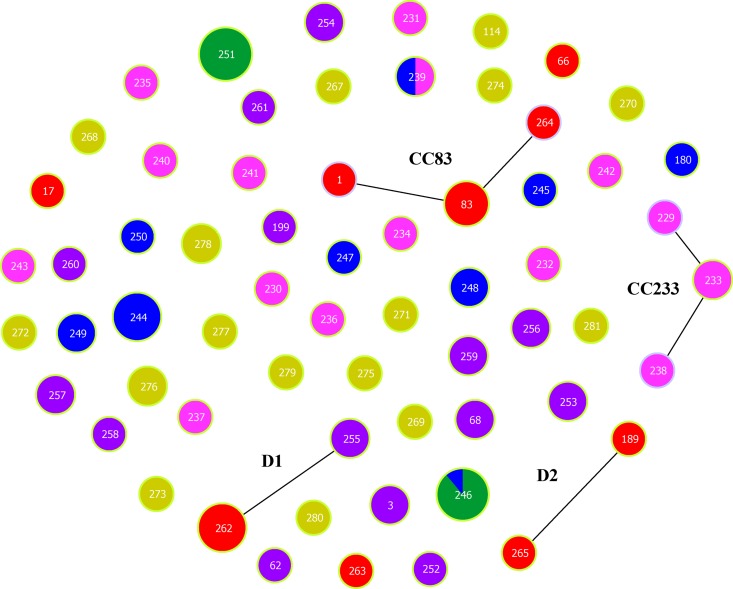
The application of eBURST (50) to our data resolved the 63 STs into two clonal complexes (CC83 and CC233), two doublets (D1 and D2), and 53 singletons (Fig. 1). Clonal complex 83 consists of five clinical isolates and includes the type strain. CC233 comprises four isolates recovered from frozen shrimp. D1 corresponds to six clinical isolates representing ST262 and a single-locus variant, ST255, which includes two human carrier isolates. These two genotypes differ only at the dnaE locus, but inspection of the variant allele sequences reveals that they differ by 5 polymorphisms in 557 sites (0.9%), which suggests an intraspecies recombination event involving the dnaE locus. A far more striking example of recombination is provided by D2. It corresponds to two clinical isolates represented by ST189 and ST265, which differ only at recA. As the corresponding recA alleles differ by 138 polymorphisms in 729 sites (18.9%), this very likely reflects an interspecies recombination event. In support of this, the best BLAST score for recA107 of ST265 corresponded to Vibrio cincinnatiensis, although this sequence was still >10% divergent from the query sequence. Clearly, recA107 of ST265 was likely imported from an as yet unidentified donor species (see Fig. S1 in the supplemental material).
Fig 1.
eBURST analysis of 63 STs of V. parahaemolyticus. The analysis is based on allelic profiles of MLST data and displays clusters of linked and individual unrelated STs. Single-locus variants (SLVs) are illustrated by linkage lines among the nodes. Color coding represents the source of isolation of each ST: red, clinical sample; purple, human carrier; yellow, seafood; green, shrimp tissue; pink, frozen shrimp; dark blue, shrimp farm water. The frequency of each ST is indicated by the size of its node.
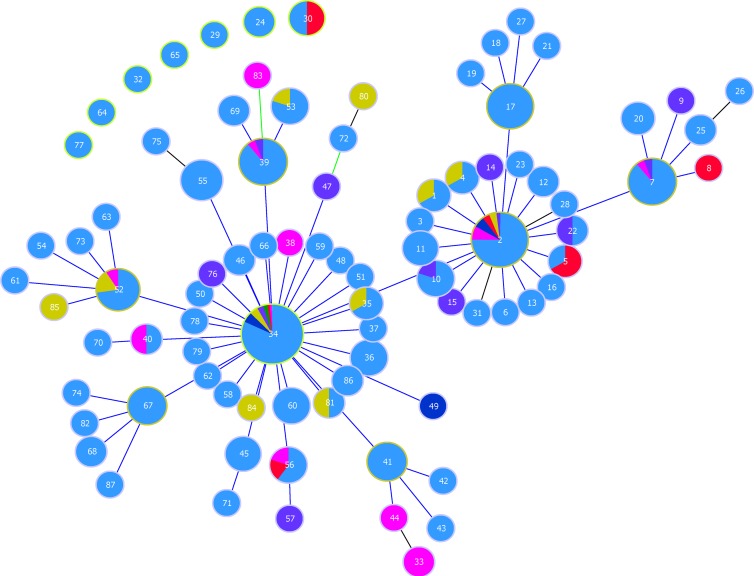
The high degree of allelic diversity in the data meant that eBURST had limited utility for identifying related strains. To counter this, we reassigned allele and ST designations on the basis of amino acid sequences. In total, 87 aaSTs were assigned from the 348 STs in the V. parahaemolyticus MLST database (http://pubmlst.org/vparahaemolyticus), and they were used to perform an eBURST analysis (Fig. 2). Two major predicted ancestors, aaST2 and aaST34, were identified. Each of them was represented by a higher proportion of environmental isolates (aaST2, 76%; aaST34, 62%) than of clinical isolates (aaST2, 24%; aaST34, 38%) regardless of the geographical region. The pandemic ST3 isolates correspond to subgroup founder aaST7, which comprises 18 STs. Unlike aaST2 and aaST34, the majority (92%) of isolates in aaST7 originated from clinical sources, whereas a much lower proportion (8%) of environmental isolates were present. None of the Thai clinical isolates corresponded to ST3, but two ST3 isolates were recovered from human carriers. Although amino acid eBURST analysis was capable of demonstrating clonal relationships within the bacterial population more clearly than nucleotide eBURST, there was no clear evidence that clinical strains were associated with a particular epidemiological source in Thailand.
Fig 2.
Population snapshot of 87 aaSTs of V. parahaemolyticus that were resolved from 348 STs from the V. parahaemolyticus MLST database (http://pubmlst.org/vparahaemolyticus). Two predicted founder groups, aaST2 and aaST34, were identified, and each was surrounded by a ring of subgroup founders and SLVs. Blue represents isolates recovered from the V. parahaemolyticus MLST database, while other colors represent the source of isolation of the Thai isolates in the present study: red, clinical samples; purple, human carrier; yellow, seafood; green, shrimp tissue; pink, frozen shrimp; dark blue, shrimp farm water. The frequency of each aaST is indicated by the size of its node.
Phylogenetic analysis.
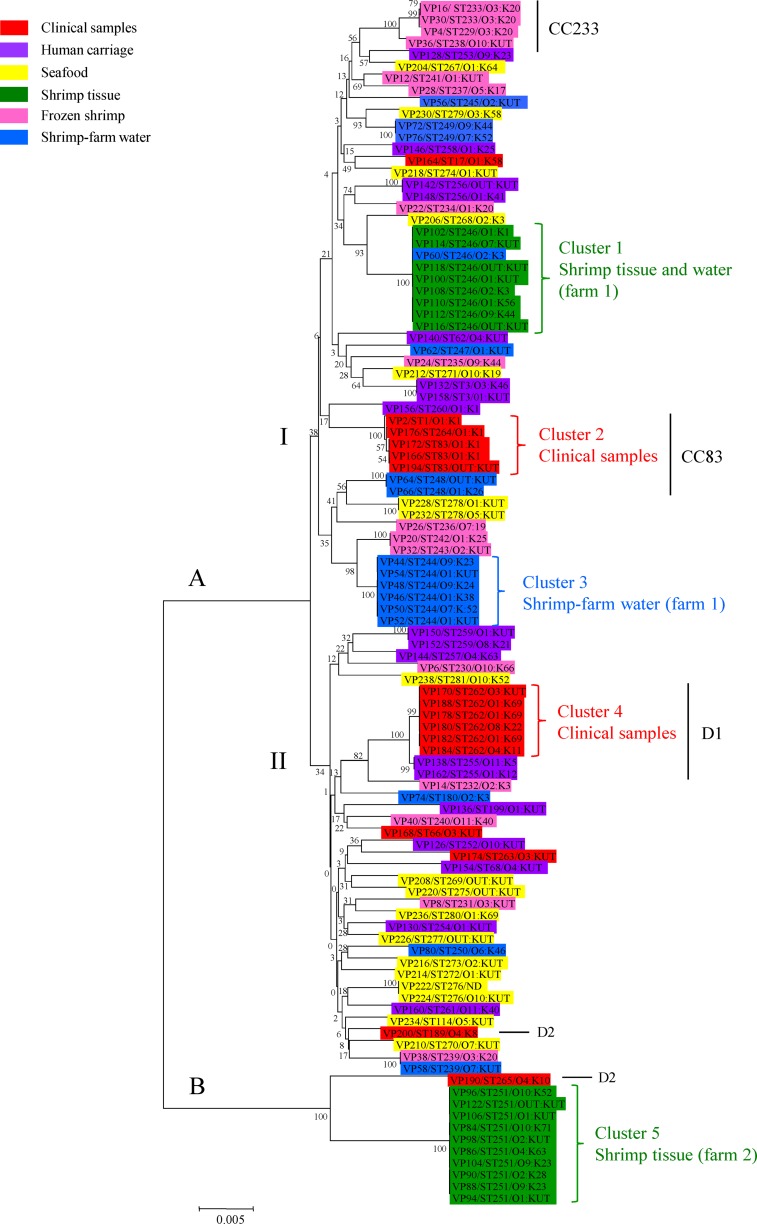
A neighbor-joining tree representing the concatenated sequences of the seven housekeeping gene fragments in 102 isolates is shown in Fig. 3. The phylogenetic tree consists of two major lineages, A and B, which are separated by a relatively large genetic distance and have a high bootstrap value. Lineage A is further subdivided into two clades, I and II, although they are closely related and have low bootstrap values. The isolates recovered from human carriers, seafood, and frozen shrimp were very diverse and were distributed widely throughout the tree. The short internal nodes and low bootstrap scores are consistent with a history of frequent recombination and indicate that the topology of the tree is poorly supported. However, five clear clusters, 1 to 5, representing isolates of the same and closely related STs, were apparent within the tree, and they were strongly supported by high bootstrap values. Each of the clusters represents isolates almost exclusively recovered from a single source. Cluster 1 was represented by eight isolates recovered from shrimp tissue at farm 1 in August 2007 and a single isolate from water at the same farm in January 2008; cluster 2 corresponded to four clinical isolates obtained from a single hospital in Bangkok, Thailand, between May and September 1990, as well as the type strain; cluster 3 corresponded to six isolates recovered from shrimp farm water at farm 1 in January 2008; cluster 4 corresponded to six clinical isolates originating from the same hospital as those present in cluster 2 between April 1990 and February 1991; and cluster 5 corresponded to 10 isolates recovered from shrimp tissue at farm 2 in August 2007. The contemporaneous recovery of clinical isolates representing distinct clusters, 2 and 4, from patients in the same hospital in Bangkok in 1990–1991 clearly indicates that two distinct disease-causing clones were circulating at this time. We also note a close association between these clinical clusters and three isolates from human carriers (VP156, cluster 2, and VP138 and VP162, cluster 4). Different hemolysin gene profiles were also observed among clinical isolates within each of the clusters 2 and 4 (Table 1). In cluster 2, isolate VP194 possesses only tdh whereas isolates VP166, VP172, and VP176 possess both tdh and trh; in cluster 4, VP170, VP178, and VP188 contain both tdh and trh whereas VP180 and VP184 contain only tdh and VP182 contains only trh. With the exception of VP194 (OUT:KUT), all isolates in cluster 2 represent serotype O1:K1; in contrast, isolates in cluster 4 represent multiple serotypes (O1:K69, O4:K11, O8:K22, and O3:KUT).
Fig 3.
Neighbor-joining tree of 102 concatenated sequences of V. parahaemolyticus from multiple sources in Thailand. The numbers at the nodes represent bootstrap values based on 500 replications.
Clusters 1 and 5 represent predominantly shrimp tissue isolates and correspond to isolates recovered from two different farms, 1 and 2, respectively. The existence of these clusters points to very limited diversity within the farms at any given point in time. In support of this, cluster 3 represents isolates recovered from two separate ponds (A and B) at farm 1 in January 2008. However, this cluster is distinct from cluster 1, which represents isolates recovered from shrimp tissue at the same farm 5 months earlier, in August 2007. These observations suggest that the clusters may represent temporal effects resulting from cycles of rapid clonal expansion and replacement within a single farm.
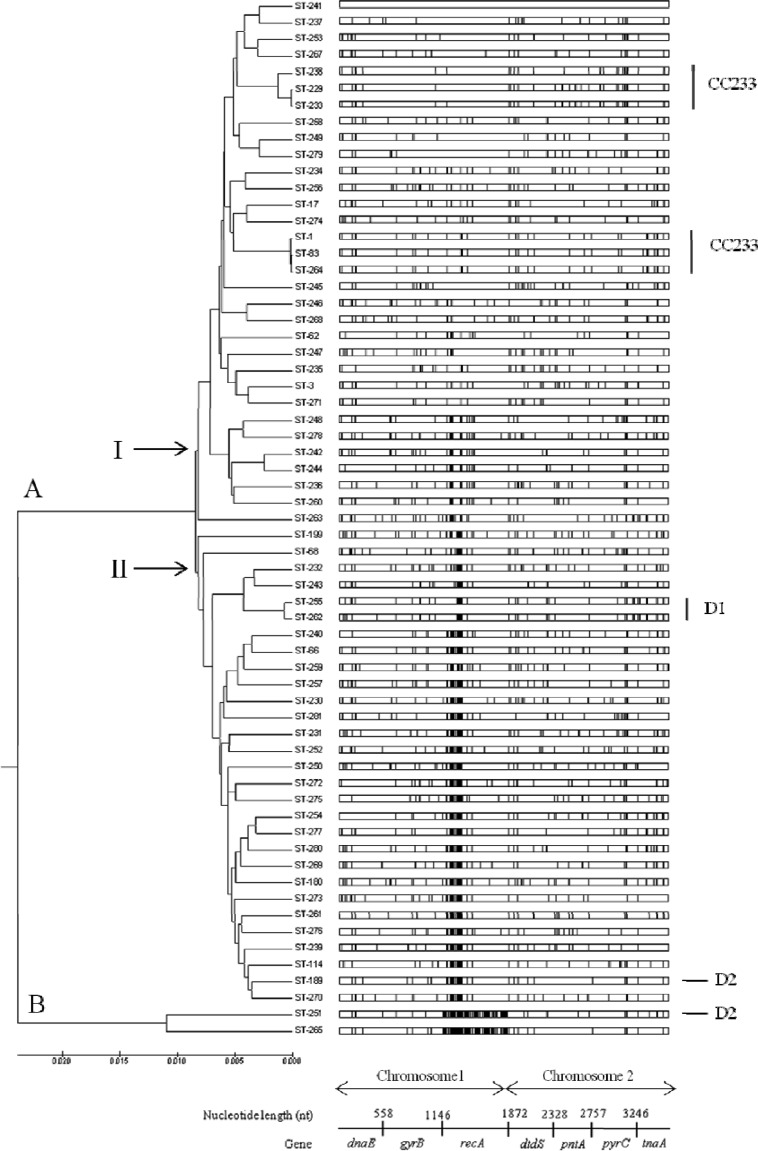
The distribution of polymorphic nucleotide sites among concatenated sequences of 63 STs clearly showed that isolates of ST251 and ST265 (lineage B) have highly divergent recA alleles (Fig. 4). As discussed above, these recA alleles have most probably been acquired by horizontal DNA transfer. When recA is removed from the concatenated sequences, a neighbor-joining tree is recovered, which lacks well-defined clusters (see Fig. S2 in the supplemental material). A high level of divergence at the recA locus is also apparent within other V. parahaemolyticus STs in the MLST database (http://pubmlst.org/vparahaemolyticus). However, it is also clear that intragenic recombination has occurred within recA involving STs from both clades I and II (Fig. 4). Detailed patterns of intragenic recombination within recA from our data are further illustrated in Fig. S3 in the supplemental material, and it is possible that recombinational exchanges occur more frequently within the human host (i.e., within the intestinal tract) than in the environment.
Fig 4.
Distribution of polymorphic nucleotide sites among concatenated sequences of 63 STs. The vertical lines represent polymorphic nucleotide sites with respect to the top sequence, ST241. The demarcation and nucleotide lengths of the seven genes are indicated along the bottom scale.
Bayesian clustering analysis (BAPS) was employed to identify genetically distinct subpopulations. ST251 and ST265 (Fig. 4) were removed from the clustering analysis to offset the effects of recombination, resulting in two distinct genetic groups (see Fig. S4 in the supplemental material). BAPS admixture analysis (see Fig. S5 in the supplemental material) revealed six hybrid STs, shown in blue in Fig. S4 in the supplemental material. The six hybrid STs predicted by BAPS analysis and described above were further examined using RDP3. The predicted parents of the isolates representing these six STs are shown in Table 3. Recombination events within the MLST data set were examined in further detail using RDP3 (45). Thirteen unique recombination events and 175 recombination signals were detected by the RDP, GENECONV, and MaxChi programs within the RDP3 package. The recombination rate per mutation (σ/θ) within the 63 STs was 12.922, indicating that the observed diversity was driven predominantly by recombination. A graphic representation of the concatenated sequences of the 63 STs representing recombination breakpoints shows that a majority of recombination events occur at the position of recA (between base positions 1147 and 1872) (see Fig. S6 in the supplemental material).
Table 3.
Strain information for six hybrid STs predicted by BAPS and RDP
| Recombinant predicted by BAPS and RDP |
Donor |
Recipient |
|||
|---|---|---|---|---|---|
| ST | Source | ST | Source | ST | Source |
| 262 | Clinical sample | 68 | Human carrier | 279 | Seafood |
| 263 | Clinical sample | 235 | Frozen shrimp | 239 | Water |
| 255 | Human carrier | 68 | Human carrier | 278 | Seafood |
| 242 | Frozen shrimp | 248 | Water | 232 | Frozen shrimp |
| 244 | Water | 248 | Water | Unknown | NAa |
| 68 | Human carrier | —b | NA | — | NA |
NA, not applicable.
—, none identified.
DISCUSSION
Here, we show that V. parahaemolyticus isolates are highly diverse, even when recovered from a single country. High rates of recombination have previously been demonstrated in V. parahaemolyticus (38, 39), as well as in other Vibrio species (51–53). Our analyses are in agreement with those of González-Escalona et al. (38), who suggested that V. parahaemolyticus has an epidemic population structure in which new, better-adapted clone complexes emerge and spread from a highly recombining genetic background. This hypothesis is supported by the observed decrease in the value of ISA from 0.5966 to 0.1350 when only 63 STs were considered compared to the entire strain collection. Similar findings have been described for Vibrio cholerae (54) and Staphylococcus epidermidis (55). We were unable to identify the most likely epidemiological source of the clinical or human carrier isolates, and our results suggest that virulent, and potentially virulent, strains of V. parahaemolyticus are not routinely associated with specific sources of seafood production.
Five isolates from human carriers represented STs (ST3, ST62, ST68, and ST199) that were identical to those of isolates recovered from clinical sources of worldwide distribution. These findings provide some evidence of a genetic link between human carriage and clinical isolates and demonstrate that symptomless carriers could become sources of infection by transmitting potentially pathogenic bacteria to seafood products within the processing factory or directly to uninfected individuals. However, 10 novel STs were associated with human carrier isolates but not with clinical and environmental isolates (Fig. 3). Thus, the human intestinal tract also serves as a potential reservoir of unique V. parahaemolyticus isolates that are not commonly seen among clinical or environmental isolates.
Substantial serotypic diversity was observed among isolates within clusters 1 (ST246), 3 (ST244), 4 (ST262), and 5 (ST251), but not among isolates of cluster 2 (ST1, ST83, and ST264) (Fig. 3). All of the isolates in clinical cluster 2, with the exception of isolate VP194 (OUT:KUT), as well as the closely related carrier isolate VP156, were of serotype O1:K1. In contrast, clusters 1, 3, 4, and 5 comprised isolates of seven, five, four, and eight combinations of O and K serotypes, respectively. These results clearly demonstrate a remarkably high degree of serotypic diversity among V. parahaemolyticus isolates and confirm previous findings that multiple serotypes of V. parahaemolyticus occur within a single ST, or closely related STs (26, 27, 38, 40, 41). It has previously been demonstrated that the serotype O4:K68 strain has most likely evolved from the pandemic O3:K6 strain by replacement of both the O and K antigens as a consequence of recombination events involving the entire O- and K-antigen-encoding gene clusters (56, 57). Unlike other Vibrio species, including V. cholerae and Vibrio vulnificus, which have a single region encoding both O and K antigens, the O-antigen-encoding region of pandemic O3:K6 V. parahaemolyticus is not present in the same location as the K-antigen gene cluster (58). Therefore, V. parahaemolyticus is likely to have greater potential for a larger number of O- and K-antigen combinations than other Vibrio species as a consequence of genetic recombination of the O- and K-antigen-encoding genes. There was also evidence of horizontal gene transfer influencing the distribution of the hemolysin genes (tdh and trh) among isolates of the same, or closely related, STs. Clinical isolates within cluster 2 (ST1, ST83, and ST264) possessed two different hemolysin gene profiles (tdh positive trh positive and tdh positive trh negative) whereas those within cluster 4 (ST262) possessed three different hemolysin gene profiles (tdh positive trh positive, tdh positive trh negative, and tdh negative trh positive).
Bayesian analysis was unable to discriminate between isolates from different epidemiological sources in Thailand. However, the analysis did confirm that the population structure of Thai V. parahaemolyticus isolates is strongly influenced by recA. Clearly, recA has a major influence on the apparent phylogenetic relationships and population structure of V. parahaemolyticus based on the current MLST scheme. In the present study, we identified two highly divergent recA alleles, recA107 and recA120, that have been acquired by horizontal DNA transfer by isolates representing ST265 and ST251, respectively. Nucleotide Blast analysis of the recA107 (ST265; clinical isolate) and recA120 (ST251; environmental isolates) alleles were best matched to the recA sequences of V. cincinnatiensis (83% similarity) and Vibrio halioticoli (83%), respectively. V. cincinnatiensis has been identified as a human-pathogenic bacterium (59), whereas V. halioticoli is commonly found in the guts of abalones (60). It is clear, therefore, that certain V. parahaemolyticus strains have acquired highly divergent recA alleles by horizontal gene transfer from other Vibrio species. Furthermore, the evidence suggests that this has occurred on at least two occasions among the clinical and environmental strains investigated in the present study alone. High recA diversity within Vibrio species has also been reported in a number of previous MLST studies (26, 38, 61). Highly divergent recA alleles and evidence for frequent recombination at this locus were also observed in previous MLST studies of V. parahaemolyticus isolates from the southeastern China coast (40) and the Chinese mainland (41), respectively. Based on our current analysis of Thai V. parahaemolyticus isolates, together with the findings of previous studies, recA is clearly not an ideal molecular marker for evolutionary analyses of V. parahaemolyticus and other Vibrio species.
In addition to potential assortative recombination events involving the entire recA gene from other species, intragenic recombination has also played a significant role in V. parahaemolyticus evolution, since recA itself has a complex mosaic structure (see Fig. S3 in the supplemental material). Notably, intragenic recombination affecting recA has also been reported in V. cholerae and Vibrio mimicus (51, 61), suggesting that the gene may represent a hot spot of recombination in Vibrio species. The mosaic recA alleles of V. parahaemolyticus (see Fig. S3 in the supplemental material) were present predominantly in clinical or human carrier isolates, suggesting that recombinational exchange has occurred more frequently in the human intestinal tract than in the environment. These findings imply that carriage of different strains of V. parahaemolyticus within the human intestinal tract is acting as a driving force in the evolution of the pathogen and in the emergence of new strains. In support of this hypothesis, a previous study of carriage rates among healthy workers at two seafood-processing plants in Thailand (62) demonstrated that although the carriage rate was relatively low (varying from 0.6 to 2.6%) overall, a relatively high percentage (31.1%) of these individuals carried strains of multiple serotypes (from 2 to 5 serotypes) at the same time. Furthermore, a small number of individuals were culture positive at different time points over the course of the 12-month sampling period, suggesting that strains were resident within the intestinal tract for periods of weeks or months. The identification of multiple serotypes of V. parahaemolyticus within the same individuals at the same time point provides evidence that environmental conditions do exist within the human intestinal tract, albeit for a limited period of time, that allow the colocalization of donor and recipient strains and for DNA exchange to occur. Transcriptional-profiling studies of V. cholerae isolates recovered from patient specimens during early and late stages of infection also suggested that the human upper intestine may be a particularly suitable niche for the horizontal transfer of genetic material that is important in the pathogenicity of the pathogen (63). Furthermore, it has also been demonstrated that inflammation within the mammalian gut leads to increased densities of pathogenic Salmonella and commensal Escherichia coli strains (64). It was proposed that such pathogen-induced blooms drive conjugative horizontal gene transfer between different Enterobacteriaceae and are responsible for the evolution of both pathogens and commensals. Thus, the human intestinal tract may represent an ecological niche that stimulates the occurrence of horizontal gene transfer both among different strains of V. parahaemolyticus and between V. parahaemolyticus and other commensal species (65–68). What remains unknown is the frequency of such evolutionary events, although it is likely to be relatively low. Clearly, further information about the frequency of simultaneous human carriage of V. parahaemolyticus strains, the residence time of V. parahaemolyticus in the human gut, and rates of horizontal gene transfer are required to assess the role of carriage within the human intestinal tract in the evolution of this important pathogen. Equally important are further studies to investigate how frequently newly evolved strains are then returned to the environment, where they may reinfect human hosts.
In conclusion, MLST analyses of Thai V. parahaemolyticus isolates from different sources indicated that clinical strains are unrelated to those recovered from seafood and other environmental sources. Furthermore, the majority of STs represented by human carrier isolates are novel and are not associated with clinical or environmental isolates. However, a small number of STs associated with human carrier isolates were genetically related to clinical strains both from Thailand and from worldwide sources. Thus, a limited number of carrier isolates represent pathogenic phenotypes and may be positively selected for within the human population. Very high levels of serotypic diversity were observed among isolates representing the same ST and recovered from a single source at the same time. Extensive recombination was also observed to be affecting the recA locus, particularly within clinical and carrier isolates. Recombinational exchange clearly plays an important role in the evolution of V. parahaemolyticus, but the human intestinal tract provides an environment that appears to be driving the emergence of new, potentially pathogenic strains.
Supplementary Material
Footnotes
Published ahead of print 1 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03067-12.
REFERENCES
- 1.Sakurai J, Matsuzaki A, Takeda Y, Miwatani T. 1974. Existence of two distinct hemolysins in Vibrio parahaemolyticus. Infect. Immun. 9:777–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honda T, Ni YX, Miwatani T. 1988. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56:961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749 [DOI] [PubMed] [Google Scholar]
- 4.Miyamoto Y, Kato T, Obara Y, Akiyama S, Takizawa K, Yamai S. 1969. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J. Bacteriol. 100:1147–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joseph SW, Colwell RR, Kaper JB. 1982. Vibrio parahaemolyticus and related halophilic vibrios. Crit. Rev. Microbiol. 10:77–124 [DOI] [PubMed] [Google Scholar]
- 6.Su Y, Liu C. 2007. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24:549–558 [DOI] [PubMed] [Google Scholar]
- 7.Bilung LM, Radu S, Bahaman AR, Rahim RA, Napis S, Ling MW, Tanil GB, Nishibuchi M. 2005. Detection of Vibrio parahaemolyticus in cockle (Anadara granosa) by PCR. FEMS Microbiol. Lett. 252:85–88 [DOI] [PubMed] [Google Scholar]
- 8.Caburlotto G, Gennari M, Ghidini V, Tafi M, Lleo MM. 2009. Presence of T3SS2 and other virulence-related genes in tdh-negative Vibrio parahaemolyticus environmental strains isolated from marine samples in the area of the Venetian Lagoon, Italy. FEMS Microbiol. Ecol. 70:506–514 [DOI] [PubMed] [Google Scholar]
- 9.Vuddhakul V, Chowdhury A, Laohaprertthisan V, Pungrasamee P, Patararungrong N, Thianmontri P, Ishibashi M, Matsumoto C, Nishibuchi M. 2000. Isolation of a pandemic O3:K6 clone of a Vibrio parahaemolyticus strain from environmental and clinical sources in Thailand. Appl. Environ. Microbiol. 66:2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagley S, Koofhethile K, Wing JB, Rangdale R. 2008. Comparison of Vibrio parahaemolyticus isolated from seafoods and cases of gastrointestinal disease in the UK. Int. J. Environ. Health Res. 18:283–293 [DOI] [PubMed] [Google Scholar]
- 11.Chao G, Jiao X, Zhou X, Yang Z, Huang J, Zhou L, Qian X. 2009. Distribution, prevalence, molecular typing, and virulence of Vibrio parahaemolyticus isolated from different sources in coastal province Jiangsu, China. Food Control 20:907–912 [Google Scholar]
- 12.Deepanjali A, Kumar HS, Karunasagar I. 2005. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 71:3575–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePaola A, Kaysner CA, Bowers J, Cook DW, Ray B, Wiles K, Depaola A, Cook D, Kaysner C, Puhr N. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66:4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara-Kudo Y, Sugiyama K, Nishibuchi M, Chowdhury A, Yatsuyanagi J, Ohtomo Y, Saito A, Nagano H, Nishina T, Nakagawa H, Konuma H, Miyahara M, Kumagai S. 2003. Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan. Appl. Environ. Microbiol. 69:3883–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keneko T, Colwell R. 1978. The annual cycle of Vibrio parahaemolyticus in Chesapeake Bay. Microb. Ecol. 155:135–155 [DOI] [PubMed] [Google Scholar]
- 16.Parveen S, Hettiarachchi KA, Bowers JC, Jones JL, Tamplin ML, Mckay R, Beatty W, Brohawn K, Dasilva LV, Depaola A. 2008. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int. J. Food Microbiol. 128:354–361 [DOI] [PubMed] [Google Scholar]
- 17.Di Pinto A, Ciccarese G, De Corato R, Novello L, Terio V. 2008. Detection of pathogenic Vibrio parahaemolyticus in southern Italian shellfish. Food Control 19:1037–1041 [Google Scholar]
- 18.Marlina, Radu S, Kqueen CY, Napis S, Zakaria Z. 2007. Detection of tdh and trh genes in Vibrio parahaemolyticus isolated from Corbicula Moltkiana prime in West Sumatera, Indonesia. Southeast Asian J. Trop. Med. Public Health 38:349–355 [PubMed] [Google Scholar]
- 19.Raghunath P, Acharya S, Bhanumathi A, Karunasagar I. 2008. Detection and molecular characterization of Vibrio parahaemolyticus isolated from seafood harvested along the southwest coast of India. Food Microbiol. 25:824–830 [DOI] [PubMed] [Google Scholar]
- 20.Barker WH, Jr, Gangarosa EJ. 1974. Food poisoning due to Vibrio parahaemolyticus. Annu. Rev. Med. 25:75–81 [DOI] [PubMed] [Google Scholar]
- 21.Fujino T, Okuno Y, Nakada D, Aoyama A, Fukai K, Mukai T, Ueho T. 1953. On the bacteriological examination of shirasu food poisoning. Med. J. Osaka Univ. 4:299–304 [Google Scholar]
- 22.Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, Wong HC, Depaola A, Kim YB, Albert MJ, Nishibuchi M. 2000. Pandemic spread of an O3: K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasu H, Iida T, Sugahara T, Yamaichi Y, Park KS, Yokoyama K, Makino K, Shinagawa H, Honda T. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay AK, Garg S, Bhattacharya SK, Nair GB, Nishibuchi M. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury NR, Stine OC, Morris JG, Nair GB. 2004. Assessment of evolution of pandemic Vibrio parahaemolyticus by multilocus sequence typing. J. Clin. Microbiol. 42:1280–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chowdhury NR, Chakraborty S, Ramamurthy T. 2000. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg. Infect. Dis. 6:631–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serichantalergs O, Bhuiyan NA, Nair GB, Chivaratanond O, Srijan A, Bodhidatta L, Anuras S, Mason CJ. 2007. The dominance of pandemic serovars of Vibrio parahaemolyticus in expatriates and sporadic cases of diarrhoea in Thailand, and a new emergent serovar (O3:K46) with pandemic traits. J. Med. Microbiol. 56:608–613 [DOI] [PubMed] [Google Scholar]
- 29.Bhoopong P, Palittapongarnpim P, Pomwised R, Kiatkittipong A, Kamruzzaman M, Nakaguchi Y, Nishibuchi M, Ishibashi M, Vuddhakul V. 2007. Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J. Clin. Microbiol. 45:1544–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laohaprertthisan V, Chowdhury A, Kongmuang U, Kalnauwakul S, Ishibashi M, Matsumoto C, Nishibuchi M. 2003. Prevalence and serodiversity of the pandemic clone among the clinical strains of Vibrio parahaemolyticus isolated in southern Thailand. Epidemiol. Infect. 130:395–406 [PMC free article] [PubMed] [Google Scholar]
- 31.Wootipoom N, Bhoopong P, Pomwised R, Nishibuchi M, Ishibashi M, Vuddhakul V. 2007. A decrease in the proportion of infections by pandemic Vibrio parahaemolyticus in Hat Yai. J. Med. Microbiol. 56:1630–1638 [DOI] [PubMed] [Google Scholar]
- 32.Suthienkul O, Iida T, Park KS, Ishibashi M, Supavej S, Yamamoto K, Honda T. 1996. Restriction fragment length polymorphism of the tdh and trh genes in clinical Vibrio parahaemolyticus strains. J. Clin. Microbiol. 34:1293–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiden MCJ. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561–588 [DOI] [PubMed] [Google Scholar]
- 34.Urwin R, Maiden MCJ. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479–487 [DOI] [PubMed] [Google Scholar]
- 35.Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper JE, Feil EJ. 2004. Multilocus sequence typing—what is resolved? Trends Microbiol. 12:373–377 [DOI] [PubMed] [Google Scholar]
- 37.Turner KME, Feil EJ. 2007. The secret life of the multilocus sequence type. Int. J. Antimicrob. Agents 29:129–135 [DOI] [PubMed] [Google Scholar]
- 38.González-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus L, DePaola A. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan Y, Cui Y, Han H, Xiao X, Wong H, Tan Y, Guo Z, Liu X, Yang R, Zhou D. 2011. Extended MLST-based population genetics and phylogeny of Vibrio parahaemolyticus with high levels of recombination. Int. J. Food Microbiol. 145:106–112 [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, Hu W, Wu B, Zhang P, Chen J, Wang S, Fang W. 2011. Vibrio parahaemolyticus isolates from southeastern Chinese coast are genetically diverse with circulation of clonal complex 3 strains since 2002. Foodborne Pathog. Dis. 8:1169–1176 [DOI] [PubMed] [Google Scholar]
- 41.Chao G, Wang F, Zhou X, Jiao X, Huang J, Pan Z, Zhou L, Qian X. 2011. Origin of Vibrio parahaemolyticus O3:K6 pandemic clone. Int. J. Food Microbiol. 145:459–463 [DOI] [PubMed] [Google Scholar]
- 42.Kaysner CA, DePaola A. 2004. Vibrio. Bacteriological analytical manual. U.S. Food and Drug Administration, Silver Spring, MD. http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/ucm070830.htm
- 43.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 44.Francisco AP, Bugalho M, Ramirez M, Carriço J. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin DP, Williamson C, Posada D. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262 [DOI] [PubMed] [Google Scholar]
- 46.Corander J, Marttinen P. 2006. Bayesian identification of admixture events using multilocus molecular markers. Mol. Ecol. 15:2833–2843 [DOI] [PubMed] [Google Scholar]
- 47.Corander J, Tang J. 2007. Bayesian analysis of population structure based on linked molecular information. Math. Biosci. 205:19–31 [DOI] [PubMed] [Google Scholar]
- 48.Smith JM, Smith NH, O'Rourke M, Spratt BG. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haubold B, Hudson RR. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847–848 [DOI] [PubMed] [Google Scholar]
- 50.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byun R, Elbourne LD, Lan R, Reeves PR. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keymer DP, Boehm AB. 2011. Recombination shapes the structure of an environmental Vibrio cholerae population. Appl. Environ. Microbiol. 77:537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, Munn CB, Swings J. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garg P, Aydanian A, Smith D, Glenn JM, Nair GB, Stine OC. 2003. Molecular epidemiology of O139 Vibrio cholerae: mutation, lateral gene transfer, and founder flush. Emerg. Infect. Dis. 9:810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miragaia M, Thomas JC, Couto I, Enright MC, De Lencastre H. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189:2540–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okura M, Osawa R, Tokunaga A, Morita M, Arakawa E, Watanabe H. 2008. Genetic analyses of the putative O and K antigen gene clusters of pandemic Vibrio parahaemolyticus. Microbiol. Immunol. 52:251–264 [DOI] [PubMed] [Google Scholar]
- 57.Chen Y, Stine OC, Badger JH, Gil AI, Nair GB, Nishibuchi M, Fouts DE. 2011. Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics 12:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Dai J, Morris JG, Johnson JA. 2010. Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol. 10:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brayton PR, Bode RB, Colwell RR, Macdonell MT, Hall HL, Grimes DJ. 1986. Vibrio cincinnatiensis sp. nov., a new human pathogen. Microbiology 23:104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sawabe T, Ohtsuka M, Nakano K. 1998. Vibrio halioticoli sp. nov., a non-motile alginolytic marine bacterium isolated from the gut of the abalone Haliotis discus hannai. Int. J. Syst. Bacteriol. 48:573–580 [DOI] [PubMed] [Google Scholar]
- 61.Thompson CC, Thompson FL, Vicente ACP. 2008. Identification of Vibrio cholerae and Vibrio mimicus by multilocus sequence analysis (MLSA). Int. J. Syst. Evol. Microbiol. 58:617–621 [DOI] [PubMed] [Google Scholar]
- 62.Athajariya T. 2004. The study of serotypes and virulence genes of Vibrio parahaemolyticus in healthy carriers at frozen seafood plants. M.S. thesis. Faculty of Public Health, Mahidol University, Bangkok, Thailand [Google Scholar]
- 63.Larocque RC, Harris JB, Dziejman M, Li X, Khan AI, Faruque ASG, Shah M, Nair GB, Ryan ET, Qadri F, John J, Calderwood SB, Faruque SM, Mekalanos JJ. 2005. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect. Immun. 73:4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt K, Walker AW, Ackermann M, Dobrindt U, Thomson NR, Hardt W-D. 2012. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. U. S. A. 109:1269–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruwandeepika HAD, Defoirdt T, Bhowmick PP, Shekar M, Bossier P, Karunasagar I. 2010. Presence of typical and atypical virulence genes in vibrio isolates belonging to the Harveyi clade. J. Appl. Microbiol. 109:888–899 [DOI] [PubMed] [Google Scholar]
- 66.Wang D, Wang H, Zhou Y, Zhang Q, Zhang F, Du P, Wang S, Chen C, Kan B. 2011. Genome sequencing reveals unique mutations in characteristic metabolic pathways and the transfer of virulence genes between V. mimicus and V. cholerae. PLoS One 6:e21299 doi:10.1371/journal.pone.0021299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okada N, Iida T, Park K, Goto N, Yasunaga T, Hiyoshi H, Matsuda S, Kodama T, Honda T. 2009. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77:904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haley BJ, Grim CJ, Hasan N, Choi SY, Chun J, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Huq A, Colwell RR. 2010. Comparative genomic analysis reveals evidence of two novel Vibrio species closely related to V. cholerae. BMC Microbiol. 10:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.