Abstract
The possibility that metatranscriptomic analysis could distinguish between direct interspecies electron transfer (DIET) and H2 interspecies transfer (HIT) in anaerobic communities was investigated by comparing gene transcript abundance in cocultures in which Geobacter sulfurreducens was the electron-accepting partner for either Geobacter metallireducens, which performs DIET, or Pelobacter carbinolicus, which relies on HIT. Transcript abundance for G. sulfurreducens uptake hydrogenase genes was 7-fold lower in cocultures with G. metallireducens than in cocultures with P. carbinolicus, consistent with DIET and HIT, respectively, in the two cocultures. Transcript abundance for the pilus-associated cytochrome OmcS, which is essential for DIET but not for HIT, was 240-fold higher in the cocultures with G. metallireducens than in cocultures with P. carbinolicus. The pilin gene pilA was moderately expressed despite a mutation that might be expected to repress pilA expression. Lower transcript abundance for G. sulfurreducens genes associated with acetate metabolism in the cocultures with P. carbinolicus was consistent with the repression of these genes by H2 during HIT. Genes for the biogenesis of pili and flagella and several c-type cytochrome genes were among the most highly expressed in G. metallireducens. Mutant strains that lacked the ability to produce pili, flagella, or the outer surface c-type cytochrome encoded by Gmet_2896 were not able to form cocultures with G. sulfurreducens. These results demonstrate that there are unique gene expression patterns that distinguish DIET from HIT and suggest that metatranscriptomics may be a promising route to investigate interspecies electron transfer pathways in more-complex environments.
INTRODUCTION
Interspecies electron transfer is essential to the proper functioning of a diversity of anaerobic microbial communities, but there is little information on the actual mechanisms by which microorganisms cooperate to exchange electrons in most anaerobic environments. The best-studied mechanism for interspecies electron exchange is H2 interspecies transfer (HIT) in which the electron-donating partner reduces protons to produce H2 and the electron-accepting partner oxidizes H2 with the reduction of an electron acceptor (1–4). Alternatively, in formate interspecies transfer (FIT), formate can serve as the electron carrier rather than H2 (3–5). Organic compounds with quinone moieties (6, 7) and sulfur compounds (8–10) can also serve as interspecies electron shuttles in laboratory cultures, but their environmental significance is not known.
Direct interspecies electron transfer (DIET) is an alternative strategy whereby electron exchange does not require soluble molecules. For example, adaptive evolution of a coculture of Geobacter metallireducens and Geobacter sulfurreducens, grown under conditions in which the two species had to exchange electrons in order to utilize ethanol as an electron donor with fumarate as an electron acceptor, led to a coculture in which electrons were transferred directly through electrically conductive cell aggregates rather than via HIT or FIT (11). Gene deletion and biochemical studies demonstrated that important components of DIET in the coculture were the conductive pili of G. sulfurreducens (12–15) and OmcS (16, 17), a multiheme c-type cytochrome aligned along the pili (11). The possibility of HIT or FIT could be ruled out by the finding that the coculture had a poor capacity for H2 or formate metabolism and the rapid formation of cocultures initiated with mutants of G. sulfurreducens unable to utilize H2 and formate (11, 18).
DIET also appeared to be the mechanism for electron exchange in natural methanogenic communities that formed aggregates in an anaerobic wastewater digester converting brewery wastes to methane (19). The aggregates had electrical conductivity comparable to that of Geobacter cocultures, and the temperature-dependent response of the conductivity suggested that long-range electron transport in the aggregates was like metallic electron transport (19), similar to the metal-like conductivity of G. sulfurreducens pili (12–15). Geobacter species were abundant within the aggregates and oxidized ethanol and fatty acids to drive methanogenesis. Conductive materials such as magnetite and granular activated carbon can facilitate DIET (7, 20–22).
DIET has the potential to be more efficient than HIT or FIT (23). However, not all microorganisms can make biological electrical connections with other species. This was apparent in a study in which Pelobacter carbinolicus was grown in culture with G. sulfurreducens (11, 18). Unlike G. metallireducens, which could transfer electrons to a strain of G. sulfurreducens unable to use H2 and formate just as effectively as to wild-type G. sulfurreducens (11, 18), P. carbinolicus required a G. sulfurreducens partner that could use at least one of these electron transfer molecules (18). The inability of P. carbinolicus to function via DIET is consistent with its poor ability to transfer electrons directly to other external electron acceptors such as electrodes (24) or Fe(III) oxides (25).
The diversity of mechanisms by which species might exchange electrons leads to the question of how electrons are actually transferred in anaerobic environments and the development of strategies to elucidate which mechanisms predominate. Metatranscriptomic analysis offers the means to query the physiological status of microorganisms in natural environments (26–28). Therefore, in a proof-of-concept study, we compared the gene transcript abundance in cocultures exchanging electrons via HIT with that in cocultures cooperating via DIET to determine whether metatranscriptomic data can provide information diagnostic of these two different modes of interspecies electron transfer.
MATERIALS AND METHODS
Cultures.
Transcriptomic analyses were performed on previously described anaerobic cocultures in which G. sulfurreducens was the electron-accepting partner and either G. metallireducens (11) or P. carbinolicus (18) was the electron-donating partner. Both cocultures were grown in strictly anaerobic, bicarbonate-buffered medium as previously described (11, 18) with ethanol (20 mM) as the electron donor and fumarate (40 mM) as the electron acceptor. Metabolism of ethanol and reduction of fumarate were monitored over time as previously described (18, 19, 29). Cultures were harvested for transcriptomic or protein analysis at mid-log phase.
Mutant strains of G. metallireducens deficient in pilA, fliC (30), or the putative outer surface c-type cytochrome gene Gmet_2896 (31) were obtained from our laboratory culture collection maintained in pure cultures as previously described (30, 31) but adapted to ethanol (20 mM) instead of acetate as the sole electron donor. To determine whether these strains could grow in coculture with G. sulfurreducens, 0.5-ml cultures of each partner at mid-log phase were added to 10 ml anoxic medium containing 20 mM ethanol and 40 mM fumarate and substrate consumption was monitored in parallel with that of a coculture established with the wild-type G. metallireducens strain.
Western blotting.
For Western blotting, whole-cell lysate (5 μg) proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and either visualized by Coomassie staining with the SeeBlue Plus prestained protein marker (Invitrogen) or transferred to polyvinylidene difluoride immunoblotting membrane (Bio-Rad) for Western analysis, blocked in 3% bovine serum albumin in Tris-buffered saline with Tween for 1 h at room temperature, probed with PilA-specific antiserum diluted 1:1,000, washed twice in phosphate-buffered saline–Tween (10 mM sodium phosphate, 0.15 M NaCl, 0.05% Tween 20, pH 7.5), and incubated at 4°C overnight with a horseradish peroxidase (HRP)-coupled anti-rabbit secondary antibody diluted 1:20,000 (Dako). Bands were then visualized with Ready Pour HRP–3,3′,5,5′-tetramethylbenzidine (Roche) by exposure to X-ray film.
Total mRNA extraction.
For extraction of mRNA from G. metallireducens-G. sulfurreducens cocultures, the overlying medium of cultures grown in 50 ml was removed anaerobically with a syringe and needle. The aggregates left at the bottom of the bottle were mixed with RNAlater (Ambion) as previously described (27). The stopper was removed from the culture bottle, the aggregates were removed, and RNA was extracted with TRIzol (Sigma) as previously described (18). For the P. carbinolicus-G. sulfurreducens cocultures, RNAlater (10% of the culture volume) was directly injected into the culture medium and cells were harvested at 4°C with centrifugation at 6,000 × g for 20 min as previously described (18) and then extracted with TRIzol.
RNA was purified with the MinElute PCR purification kit (Qiagen) prior to recombinant DNase I (Ambion) digestion in accordance with the manufacturers' protocols, followed by an additional treatment with the MinElute PCR purification kit. The absence of genomic DNA contamination was verified by 16S rRNA gene analysis as described previously (27). Aliquots of total RNA were preserved for real-time quantitative PCR (RT-qPCR), and the remainder was used for mRNA isolation. mRNA was isolated with the MICROBExpress kit (Ambion) by following the manufacturer's protocol. Aliquots of triplicate mRNA extracts were analyzed with the Experion RNA HiSens kit (Bio-Rad). The electropherogram result showed highly reproducible mRNA extracts (see Fig. S1 in the supplemental material). Equal concentrations of mRNA extracts from the triplicate samples were then pooled to make composite samples (32–34) for each type of coculture for sequencing.
Illumina sequencing and data analysis.
Illumina sequencing libraries were prepared with the TruSeq RNA Sample Preparation kit by following the manufacturer's protocol (Illumina). Briefly, 100 ng of the total mRNA was chemically fragmented and converted into single-stranded cDNA by random-hexamer priming. Next, the second strand was generated to create double-stranded cDNA. The overhangs resulting from fragmentation were converted to blunt ends by using an end repair mixture, and then the 3′ ends were adenylated. Adenylated products were ligated with individual adapters containing unique hexameric indices/barcodes and enriched with a final 10-cycle PCR. The two enriched and purified samples containing unique barcodes and representing the two coculture types that were obtained were mixed in equimolar concentrations and used for hybridization in a HiSeq2000 flow cell for paired-end sequencing. Illumina sequence reads (100 bases) were functionally assigned by mapping against the published genomes of G. sulfurreducens strain PCA (35) and G. metallireducens (36) with ArrayStar (DNASTAR), BOWTIE tools (37), and DeSeq (38). DeSeq is a free R software package (http://www.r-project.org/) that uses a method based on the negative binomial distribution with variance and mean linked by local regression. DeSeq allows significant-difference calculation in RNA-seq experiments both with and without biological replicates (38). Reads belonging to 16S/23S rRNA, reads that matched more than one segment of a genome, and reads with more than two mismatches were discarded. The remaining mRNA reads were reanalyzed and normalized with the RPKM (reads assigned per kilobase of target per million mapped reads) method (39, 40) by using ArrayStar. Expression levels were considered significant only when the log2 RPKM value was ≥8. This was equivalent to the log2 RPKM value of the housekeeping gene recA (GSU0145) in the present experiment, which was previously used as the internal standard during RT-qPCR (18). The n-fold change in the expression level was calculated by comparing the normalized reads from G. sulfurreducens-G.metallireducens cocultures with those of P. carbinolicus-G. sulfurreducens cocultures. The n-fold changes were computed only for those genes that had an RPKM value of ≥8 in one of the samples, and only ≥2-fold changes in expression (up or down) were considered significant (see Fig. S2 in the supplemental material). In addition to this, significant n-fold changes were also calculated by using DeSeq (see Table S1a in the supplemental material).
RT-qPCR.
To verify the differential expression detected by sequencing, RT-qPCR was performed for some genes that were highly upregulated or downregulated in G. metallireducens-G. sulfurreducens versus P. carbinolicus-G. sulfurreducens cocultures. For the RT-qPCR, about 300 ng of total RNA was converted into WTA (whole-transcriptome amplification) cDNA libraries and enriched by WTA PCR with reagents and protocols supplied with or recommended by the manufacturer (Sigma). Briefly, 300 ng of total RNA was mixed with 2.5 ml WTA library synthesis buffer and 2.5 ml WTA library stabilization solution and the total volume was adjusted to 24 μl with nuclease-free water; the mixture was heated at 70°C for 5 min and immediately cooled. One microliter of library synthesis enzyme was added, and WTA cDNA libraries were synthesized by using the following thermocycler program: 24°C for 15 min, 42°C for 2 h, and 95°C for 5 min. Aliquots (5 μl) were WTA PCR amplified with JumpStart Taq DNA polymerase (Sigma), WTA amplification master mix, and deoxynucleoside triphosphate mix in accordance with the manufacturer's protocol, except that the number of cycles was reduced to 15. The enriched product was purified with the QIAquick PCR purification kit (Qiagen) and used as the template in a qPCR experiment (18, 41). RT-PCR was carried out with the ABI prism 7900 (Applied Biosystems). Primers targeting the genes pilA, pilR, omcS, omcT, omcZ, and recA were designed (Table 1) from the G. sulfurreducens genome sequence (35), and representative products from all of these primer sets were verified by sequencing. Reactions were performed in triplicate for each gene tested in a total volume of 20 μl as described previously (18). A t test was used to calculate significant differences in gene expression levels at a significance level of P = 0.05.
Table 1.
Oligonucleotides used in this study
| Primer | Sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|
| pilAF | ATCGGTATTCTCGCTGCAAT | 117 |
| pilAR | AATGCGGACTCAAGAGCAGT | |
| pilRF | TTTCCGGGAGGATCTCTTTT | 143 |
| pilRR | TTATGATGCGGTCGCTGTAG | |
| omcSF | TGGTTGGCGAAGGCATAGG | 138 |
| omcSR | CCATCAAGAACAGCGGTTCC | |
| omcTF | GGCTTCTGCGGTACTGATGT | 145 |
| omcTR | CCAGCAGATGAACAACGCTA | |
| omcZF | AAGGTTGCTGACCTTGTTGG | 158 |
| omcZR | CCACCTATCAGCCCACTGTT | |
| recAF | CACCGGCATAATCTCCAAGT | 150 |
| recAR | ATCTTGCGGATATCGAGACG |
Nucleotide sequence accession number.
The sequence reads determined in this study have been submitted to the EMBL databases under accession no. ERP001972.
RESULTS AND DISCUSSION
In order to gain insight into physiological differences during growth by DIET versus growth by HIT, the gene transcript abundances in G. sulfurreducens were compared in two coculture systems that differ in their modes of interspecies electron exchange. In cocultures supplied with ethanol as the electron donor, G. sulfurreducens can reduce the electron acceptor, fumarate, with electrons obtained either from G. metallireducens via DIET (11) or from P. carbinolicus via HIT (18). In both types of cocultures, G. sulfurreducens has the option to use acetate derived from ethanol metabolism as an additional electron donor but continued metabolism of ethanol is not possible without removal of the electrons released from the oxidation of ethanol to acetate and carbon dioxide.
Unique Illumina sequence reads of G. sulfurreducens transcripts in the cocultures ranged from 1.8 × 106 to 2.9 × 106, yielding 47- to 76-fold coverage of the G. sulfurreducens genome. Of the 3,466 predicted protein-coding genes of G. sulfurreducens (35), 910 had significant expression levels (RPKM ≥ 8) in at least one of the cocultures (see Fig. S2a and Table S1b in the supplemental material) and 640 genes had ≥2-fold higher or lower transcript abundance in one coculture set than in the other (see Fig. S2b). In this study, the focus was on genes predicted to encode proteins localized at the outer cell surface and/or involved in the metabolism of electron donors.
Evaluation of electron transfer via H2 or formate.
G. sulfurreducens has only one hydrogenase that functions as an uptake hydrogenase, membrane-bound Hyb (42). Transcripts of all five of the genes encoding Hyb subunits (GSU0782 to GSU0786) were significantly (P = 0.05) downregulated (>7-fold) in G. sulfurreducens growing in cocultures with G. metallireducens compared to those in P. carbinolicus-G. sulfurreducens cocultures (Fig. 1; see Table S1c in the supplemental material). Consistent with the lower transcript abundance of Hyb subunit genes in G. metallireducens-G. sulfurreducens cocultures was the lower expression (>3-fold down) of the genes for Hyp (GSU0305 to GSU0309 and GSU0374), which are expected to catalyze Ni-Fe cofactor acquisition by hydrogenases and are necessary for hydrogenase expression (43). Other gene sets such as hya (GSU0120 to GSU0123), hox (GSU2717 to GSU2722), mvh (GSU2416 to GSU2423), hdr (GSU0085 to GSU0092), and ehr (GSU0739 to GSU0745), which by annotation might be expected to function in H2 metabolism, had low expression levels in both types of cocultures (Fig. 1; see Table S1c in the supplemental material), consistent with previous studies indicating that they are not involved in H2 uptake (42, 44).
Fig 1.

Heat map comparison of expression levels of genes associated with hydrogenase (black letter) and formate dehydrogenase (green letter) in cocultures of G. metallireducens-G. sulfurreducens (GS/GM) and G. sulfurreducens/P. carbinolicus (GS/PC). The n-fold change shown at the right represents the gene transcript upregulation or downregulation in G. sulfurreducens in the GS/GM coculture compared to the transcript level in the GS/PC coculture. The n-fold change is presented only for G. sulfurreducens genes with significant expression (log2 RPKM ≥ 8) in one of the cocultures.
The finding that genes associated with H2 uptake were more highly expressed in G. sulfurreducens growing with P. carbinolicus than in G. sulfurreducens growing with G. metallireducens is consistent with the conclusions from other experimental results that P. carbinolicus provides electrons from ethanol oxidation to G. sulfurreducens with H2 serving as the electron carrier whereas G. metallireducens transfers electrons to G. sulfurreducens through direct electrical connections (11, 18). Thus, the results presented here demonstrated that the transcriptomic approach could reveal expected differences in physiology under different syntrophic conditions and suggest that low expression of uptake hydrogenase genes in potential electron-accepting partners might help rule out this mode of interspecies electron transfer in environmental studies.
When P. carbinolicus is grown in coculture with a strain of G. sulfurreducens that cannot consume H2, formate can serve as an alternative interspecies electron carrier (18). However, in the cocultures investigated here, which had been transferred over 400 times prior to transcript analysis, transcript abundance for all four formate dehydrogenase genes (GSU0777 to GSU0780) was low (Fig. 1; see Table S1c in the supplemental material). This result suggests that there was long-term selection for H2 as the preferred interspecies electron carrier.
Transcript abundance for formate dehydrogenase genes was also very low in G. sulfurreducens grown with G. metallireducens (Fig. 1; see Table S1c in the supplemental material). Formate was previously ruled out as an important electron carrier in G. metallireducens-G. sulfurreducens cocultures because cocultures initiated with a strain of G. sulfurreducens that could not utilize H2 or formate still grew well (18). The low transcript abundance of the formate dehydrogenase genes further indicates that formate was unlikely to be an important interspecies electron carrier in G. metallireducens-G. sulfurreducens cocultures, consistent with electron transfer via DIET.
Expression of G. sulfurreducens genes previously implicated in DIET.
One of the greatest (P = 0.01) increases in gene transcript abundance (>308-fold up) in G. sulfurreducens cocultures with G. metallireducens versus cocultures with P. carbinolicus was for the multiheme c-type cytochrome OmcS (GSU2504) (Fig. 2; see Table S1d in the supplemental material). Gene deletion studies demonstrated that OmcS is required for DIET in G. metallireducens-G. sulfurreducens cocultures (11). OmcS is associated with the conductive pili (13, 14) of G. sulfurreducens, but the distance between OmcS molecules precludes electron hopping/tunneling between them (16, 45). Rather, OmcS is thought to promote electrical contacts between pili and extracellular electron acceptors or donors, and in DIET, OmcS may provide a connection for electron transfer to the pili, which then transport electrons to the cell (12, 46).
Fig 2.
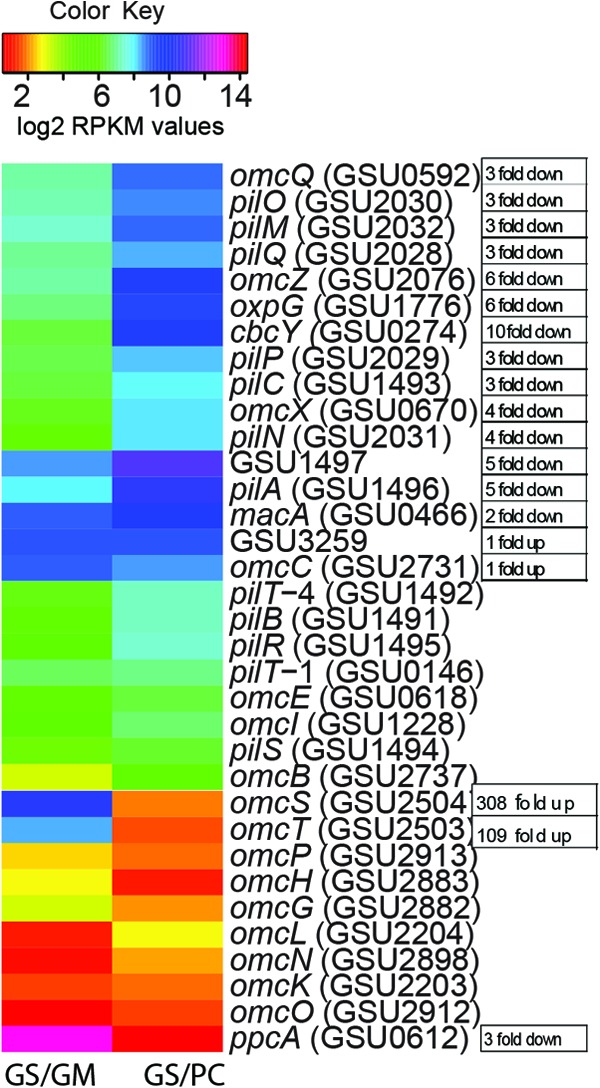
Heat map comparison of expression levels of genes associated with DIET in cocultures. For details, see the legend to Fig. 1.
The high abundance of OmcS in G. metallireducens-G. sulfurreducens cocultures was previously attributed to point mutations that accumulated in adapted cocultures in the gene for PilR, an RpoN-dependent enhancer-binding protein that regulates the expression of a variety of genes in G. sulfurreducens (11, 47). There was a low abundance of PilR gene transcripts (4-fold down) in G. metallireducens-G. sulfurreducens cocultures (see Table S1d in the supplemental material), and this was confirmed by RT-qPCR (Fig. 3a).
Fig 3.
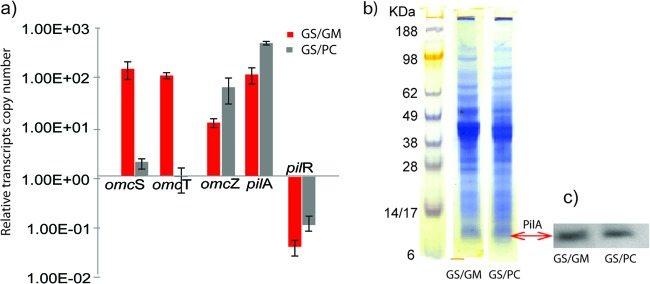
(a) RT-qPCR assays of selected genes involved in DIET. The y axis shows normalized gene expression values based on the housekeeping gene recA. Standard deviations were calculated from triplicate independent samples. (b) Coomassie-stained 12% SDS-PAGE of equal amounts of protein from cocultures of G. sulfurreducens with G. metallireducens (GS/GM) and P. carbinolicus (GS/PC). (c) Western blot probed for G. sulfurreducens PilA protein.
In a previous study, deletion of pilR resulted in increased expression of omcS but also decreased expression of the gene for PilA (GSU1496), the structural pilin protein (47). However, pili with OmcS were abundant in the cocultures (11), suggesting that regulation of pilA expression is complex. Consistent with this observation, the number of normalized reads (RPKM ≥ 8) for transcripts of G. sulfurreducens pilA and a gene downstream of pilA (for pilin domain 2 protein; GSU1497) indicated that although transcript abundance was 5-fold lower in G. metallireducens-G. sulfurreducens cocultures (Fig. 2; see Table S1d in the supplemental material), pilA and GSU1497 were moderately expressed genes. RT-qPCR confirmed a slightly lower expression of pilA in G. metallireducens-G. sulfurreducens cocultures (Fig. 3a), and Western blot analysis suggested that there was little difference between the quantities of G. sulfurreducens PilA protein in the two cocultures (Fig. 3b and c).
The expression of many genes for outer surface redox-active proteins was lower in G. metallireducens-G. sulfurreducens cocultures (see Table S1d in the supplemental material), suggesting that they do not play an important role in DIET. For example, transcript abundance of omcZ was reduced (>6-fold) and lower expression of omcZ was confirmed with qRT-PCR (Fig. 3a). The only other gene considered to encode an outer surface redox-active protein with significantly (P = 0.01) higher transcript abundance (109-fold) in G. metallireducens-G. sulfurreducens cocultures than in P. carbinolicus-G. sulfurreducens cocultures was omcT (GSU2503; Fig. 2 and 3a). This gene is in an operon with omcS (48). The two genes are cotranscribed, but omcS expression levels are generally higher because omcS is also transcribed individually (17, 49). Like OmcS, OmcT is predicted to be a multiheme outer surface c-type cytochrome but was 40-fold less abundant during growth on Fe(III) oxide than OmcS was (50).
OmcB (GSU2737), another outer membrane multiheme c-type cytochrome that is required for optimal Fe(III) reduction in G. sulfurreducens (51, 52), had lower expression in G. metallireducens-G. sulfurreducens cocultures, consistent with changes in expression levels expected as the result of a mutation in pilR (47). Another example of changes in gene expression that might be attributed to a pilR mutation (47) was significantly (P = 0.01) higher expression of genes for the hypothetical proteins GSU0967 and GSU2490 in G. metallireducens-G. sulfurreducens cocultures (Fig. 2; see Table S1b in the supplemental material).
The periplasmic cytochrome PpcA (GSU0612) is one of the most abundant G. sulfurreducens cytochromes (53). Transcript abundance for ppcA (RPKM > 13) and its homolog ppcD (RPKM > 10) was high in G. sulfurreducens growing with either G. metallireducens or P. carbinolicus (Fig. 2; see Table S1e in the supplemental material). Transcript abundance for ppcA was somewhat lower (3-fold) in G. metallireducens-G. sulfurreducens cocultures (Fig. 2), suggesting a lack of selection for increased expression during DIET. Only PpcA and PpcD can accept and donate protons along with electrons (54). The expression level of the remaining three homologs of ppcA in G. sulfurreducens (ppcB, ppcC, and ppcE) was low in both types of cocultures (see Table S1e).
The gene for the putative periplasmic c-type cytochrome PccH (GSU3274) had higher transcript abundance in G. sulfurreducens directly accepting electrons from electrodes and was essential for this process (55). Therefore, it might be expected that PccH would be important in cells accepting electrons via DIET. However, no reads for pccH were detected in either of the cocultures, suggesting that it does not play an important role in DIET. GSU1018, which encodes a protein of unknown function predicted to be in the periplasm, had significantly (P = 0.01) higher (6-fold) transcript abundance in G. metallireducens-G. sulfurreducens cocultures (see Table S1b in the supplemental material). Its function in DIET warrants further investigation.
Expression of G. sulfurreducens genes related to central metabolism.
The accumulation of acetate in P. carbinolicus-G. sulfurreducens cocultures (18), but not in G. metallireducens-G. sulfurreducens cocultures, may be further evidence for the differences in mechanisms of interspecies electron transfer in the two systems. The availability of H2 in P. carbinolicus-G. sulfurreducens cocultures could potentially repress the expression of genes for acetate metabolism through the regulator HgtR (18, 56). This possibility was further evaluated by using gene transcript data.
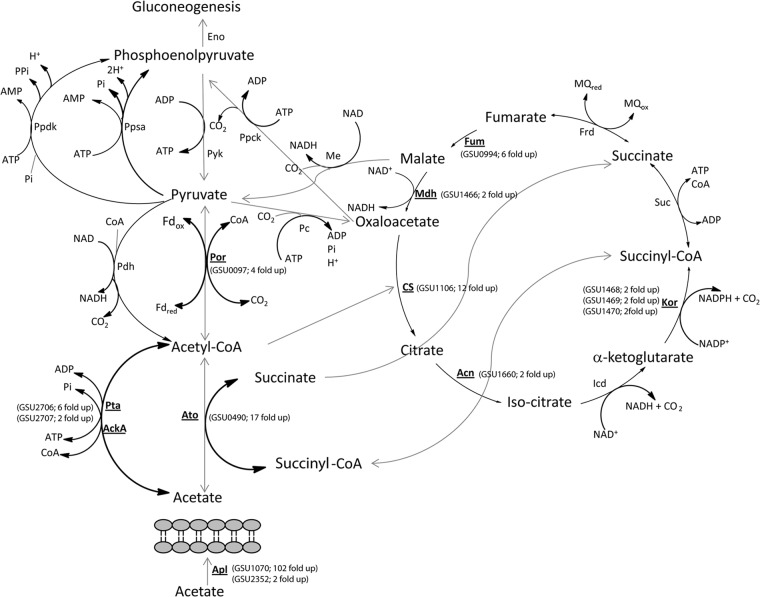
Gene expression patterns suggested that G. sulfurreducens in coculture with G. metallireducens was physiologically poised to oxidize acetate as an electron donor, whereas G. sulfurreducens in coculture with P. carbinolicus was not (Fig. 4 and 5; see Table S1c in the supplemental material). For example, transcripts for the G. sulfurreducens genes aplB (GSU1070, >102-fold up) and aplC (GSU2352, 2-fold up) had high expression levels in cocultures with G. metallireducens. These genes, along with aplA (GSU1068), are associated with acetate uptake, and the expression of at least two of these three genes is necessary for effective acetate uptake (57, 58).
Fig 4.

Heat map comparison of expression levels of genes associated with central metabolism in cocultures. For details, see the legend to Fig. 1.
Fig 5.
Central metabolism in G. sulfurreducens showing metabolic pathways when acetate is available. Genes that are upregulated (≥2-fold) in G. metallireducens-G. sulfurreducens cocultures compared to those in P. carbinolicus-G. sulfurreducens are in bold and underlined. Double-headed arrows represent reversible reactions.
G. sulfurreducens genes, such as those for succinyl:acetate coenzyme A (CoA)-transferase (ato-1; GSU0490, 17-fold up), acetate kinase (ackA; GSU2707, 2-fold up), and phosphotransacetylase (pta; GSU2706, 6-fold up), that activate acetate for oxidation and gluconeogenesis (59) had higher transcript abundance in the G. metallireducens-G. sulfurreducens cocultures. Trichloroacetic acid (TCA) cycle gene transcripts were also in significantly (P = 0.01) higher abundance, including transcripts for citrate synthase (GSU1106; 12-fold up), required for entry of acetyl-CoA into the TCA cycle (35), and many others (Fig. 5), consistent with oxidation of acetate. Furthermore, genes in the nuo-1 cluster (GSU0338 to GSU0351) encoding one of the two NADH dehydrogenase complexes were more (P = 0.01) highly expressed (>2-fold up) in G. metallireducens-G. sulfurreducens cocultures (see Table S1c in the supplemental material), consistent with efficient electron transfer from NADH produced by the TCA cycle to menaquinone, which delivers electrons to fumarate, the terminal electron acceptor. Thus, gene expression patterns suggest that acetate is poorly metabolized in P. carbinolicus-G. sulfurreducens cocultures because of the reduced expression of acetate metabolism genes in G. sulfurreducens, which can most likely be attributed to higher H2 availability.
G. metallireducens components important for DIET.
G. metallireducens has not been observed to grow syntrophically by HIT, eliminating the possibility of comparing G. metallireducens gene expression patterns during growth by DIET versus other syntrophic growth modes. However, the transcriptomic analysis of the G. metallireducens-G. sulfurreducens cocultures yielded 1.8 × 105 reads that could be assigned to G. metallireducens, providing 9-fold coverage of the G. metallireducens genome. Of the 3,518 protein-coding genes of G. metallireducens (36), the expression of 2,890 genes was detected in G. metallireducens-G. sulfurreducens cocultures but only 194 genes had significant expression levels (RPKM ≥ 8; see Table S1f in the supplemental material).
The relative expression levels of some genes compared to others in the genome provided insight into G. metallireducens proteins of potential importance in DIET. For example, the genes encoding PilA (Gmet_1399) and an associated pilin domain 2 protein (Gmet_1400) were highly expressed (RPKM > 9) in the G. metallireducens-G. sulfurreducens cocultures (see Table S1f in the supplemental material). In order to evaluate further the potential role of G. metallireducens pili in DIET, the previously described PilA-deficient mutant of G. metallireducens (30) was used as the inoculum for cocultures with G. sulfurreducens. However, repeated attempts to initiate cocultures in this manner failed (see Fig. S3 in the supplemental material). This finding is consistent with the model in which electrons derived from ethanol oxidation in G. metallireducens are transported along pili in the initial step of extracellular electron transport to G. sulfurreducens (12).
It has been proposed (12) that just as pilus-associated OmcS is essential for DIET with G. sulfurreducens, there may be one or more G. metallireducens c-type cytochromes necessary for DIET. The protein encoded by Gmet_2896, homologous to OmcE of G. sulfurreducens, is localized on the outer surface of G. metallireducens and is highly expressed during growth on Fe(III) oxide but not during growth on Fe(III) citrate, analogous to OmcS (31). Gmet_2896 was highly expressed (RPKM > 8) in cocultures with G. sulfurreducens (see Table S1f in the supplemental material), and attempts to establish cocultures with a strain of G. metallireducens from which Gmet_2896 was deleted were unsuccessful. These results suggested that the Gmet_2896 cytochrome plays a key role in DIET. Further investigation to determine whether it is associated with the pili of G. metallireducens is warranted. Genes for several other c-type cytochromes were among the most highly expressed G. metallireducens genes (see Table S1f), but gene deletion and other physiological studies are required in order to determine whether any of these cytochromes directly contribute to DIET. For example, the PpcA homolog, Gmet_2902, had high transcript abundance (see Table S1f) but its predicted periplasmic location (31) suggests that it does not directly aid in making extracellular electrical contacts.
Other G. metallireducens genes that were highly expressed (RPKM > 8; see Table S1f) included genes for flagellar components such as FliC (Gmet_0442), FliS (Gmet_0445), FliD (Gmet_0444), and FliW (Gmet_0441). Genes for FliC and other flagellar components were among the most highly upregulated genes in G. metallireducens growing on Fe(III) oxide versus cells grown on Fe(III) citrate (31), and deletion of fliC inhibited growth on Fe(III) oxide but not growth on Fe(III) citrate (30). The fliC mutant was unable to form cocultures with G. sulfurreducens even after long-term incubations (see Fig. S2), suggesting that the presence of flagella is important for aggregate formation. One possibility is that the flagella aid in establishing cell-to-cell contact (60).
Implications.
These results demonstrate that differences in gene transcript abundance can aid in discriminating whether syntrophic partners are exchanging electrons via DIET or HIT. Thus, this study represents the foundation for the transcriptional analysis of more complex environments to determine the potential environmental relevance of DIET. Furthermore, the results provide further insight into the mechanisms of DIET in G. metallireducens-G. sulfurreducens cocultures.
The gene expression patterns provide multiple lines of evidence consistent with electron transfer via DIET in G. metallireducens-G. sulfurreducens cocultures and via HIT in P. carbinolicus-G. sulfurreducens cocultures. For example, the results support the proposed importance of electron transfer to G. sulfurreducens via OmcS and conductive pili and reveal for the first time the importance of pili, flagella, and at least one outer surface c-type cytochrome of G. metallireducens in DIET. The low abundance of transcripts for uptake hydrogenase and formate dehydrogenase subunits provide strong evidence that H2 and formate do not serve as electron carriers between G. metallireducens and G. sulfurreducens. Transcript abundances for many genes suggested that G. sulfurreducens is poised for acetate metabolism when in coculture with G. metallireducens, further indicating that acetate is released from G. metallireducens and serves as an additional electron donor for G. sulfurreducens in the cocultures.
The clear differences in gene expression patterns in G. sulfurreducens growing via DIET versus HIT suggest that it should be possible to determine whether other electron-accepting organisms in syntrophic associations are participating in DIET and demonstrate that insights into the mechanisms of electron exchange may be obtained from such analyses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mollie Murnane and Devesh Shrestha for lab assistance and Muktak Aklujkar for critical reading of the manuscript.
This research was supported by Office of Science (BER), U.S. Department of Energy, award DESC0004485.
Footnotes
Published ahead of print 1 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03837-12.
REFERENCES
- 1.McInerney MJ, Sieber JR, Gunsalus RP. 2009. Syntrophy in anaerobic global carbon cycles. Curr. Opin. Biotechnol. 20:623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. 1967. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch. Mikrobiol. 59:20–31 [DOI] [PubMed] [Google Scholar]
- 3.Stams AJM, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7:568–577 [DOI] [PubMed] [Google Scholar]
- 4.Stams AJM, de Bok FAM, Plugge CM, van Eekert MHA, Dolfing J, Schraa G. 2006. Exocellular electron transfer in anaerobic microbial communities. Environ. Microbiol. 8:371–382 [DOI] [PubMed] [Google Scholar]
- 5.Thiele JH, Zeikus JG. 1988. Control of interspecies electron flow during anaerobic digestion—significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl. Environ. Microbiol. 54:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovley DR, Fraga JL, Coates JD, Blunt-Harris EL. 1999. Humics as an electron donor for anaerobic respiration. Environ. Microbiol. 1:89–98 [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Rotaru AE, Shrestha PM, Malvankar NS, Nevin KP, Lovley DR. 2012. Promoting direct interspecies electron transfer with activated carbon. Energy Environ. Sci. 5:8982–8989 [Google Scholar]
- 8.Kaden J, Galushko AS, Schink B. 2002. Cysteine-mediated electron transfer in syntrophic acetate oxidation by cocultures of Geobacter sulfurreducens and Wolinella succinogenes. Arch. Microbiol. 178:53–58 [DOI] [PubMed] [Google Scholar]
- 9.Straub KL, Schink B. 2003. Evaluation of electron-shuttling compounds in microbial ferric iron reduction. FEMS Microbiol. Lett. 220:229–233 [DOI] [PubMed] [Google Scholar]
- 10.Straub KL, Schink B. 2004. Ferrihydrite-dependent growth of Sulfurospirillum deleyianum through electron transfer via sulfur cycling. Appl. Environ. Microbiol. 70:5744–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS, Lovley DR. 2010. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330:1413–1415 [DOI] [PubMed] [Google Scholar]
- 12.Lovley DR. 2011. Live wires: direct extracellular electron exchange for bioenergy and the bioremediation of energy-related contamination. Energy Environ. Sci. 4:4896–4906 [Google Scholar]
- 13.Malvankar NS, Vargas M, Nevin KP, Franks AE, Leang C, Kim BC, Inoue K, Mester T, Covalla SF, Johnson JP, Rotello VM, Tuominen MT, Lovley DR. 2011. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 6:573–579 [DOI] [PubMed] [Google Scholar]
- 14.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101 [DOI] [PubMed] [Google Scholar]
- 15.Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 72:7345–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leang C, Qian X, Mester T, Lovley DR. 2010. Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl. Environ. Microbiol. 76:4080–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta T, Coppi MV, Childers SE, Lovley DR. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:8634–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotaru A-E, Shrestha PM, Liu F, Ueki T, Summers ZM, Lovley DR. 2012. Interspecies electron transfer via hydrogen and formate rather than direct electrical connections in cocultures of Pelobacter carbinolicus and Geobacter sulfurreducens. Appl. Environ. Microbiol. 78:7645–7651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita M, Malvankar NS, Franks AE, Summers ZM, Giloteaux L, Rotaru AE, Rotaru C, Lovley DR. 2011. Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. mBio 2(4):e00159–00111 doi:10.1128/mBio.00159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato S, Hashimoto K, Watanabe K. 2012. Microbial interspecies electron transfer via electric currents through conductive minerals. Proc. Natl. Acad. Sci. U. S. A. 109:10042–10046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato S, Hashimoto K, Watanabe K. 2012. Methanogenesis facilitated by electric syntrophy via (semi)conductive iron-oxide minerals. Environ. Microbiol. 14:1646–1654 [DOI] [PubMed] [Google Scholar]
- 22.Kato S, Nakamura R, Kai F, Watanabe K, Hashimoto K. 2010. Respiratory interactions of soil bacteria with (semi)conductive iron-oxide minerals. Environ. Microbiol. 12:3114–3123 [DOI] [PubMed] [Google Scholar]
- 23.Lovley DR. 2011. Reach out and touch someone: potential impact of DIET (direct interspecies energy transfer) on anaerobic biogeochemistry, bioremediation, and bioenergy. Rev. Environ. Sci. Biotechnol. 10:101–105 [Google Scholar]
- 24.Richter H, Lanthier M, Nevin KP, Lovley DR. 2007. Lack of electricity production by Pelobacter carbinolicus indicates that the capacity for Fe(III) oxide reduction does not necessarily confer electron transfer ability to fuel cell anodes. Appl. Environ. Microbiol. 73:5347–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haveman SA, DiDonato RJ, Jr, Villanueva L, Shelobolina ES, Postier BL, Xu B, Liu A, Lovley DR. 2008. Genome-wide gene expression patterns and growth requirements suggest that Pelobacter carbinolicus reduces Fe(III) indirectly via sulfide production. Appl. Environ. Microbiol. 74:4277–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konopka A, Wilkins M. 2012. Application of meta-transcriptomics and proteomics to analysis of in situ physiological state. Front. Microbiol. 3:184 doi:10.3389/fmicb.2012.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shrestha PM, Kube M, Reinhardt R, Liesack W. 2009. Transcriptional activity of paddy soil bacterial communities. Environ. Microbiol. 11:960–970 [DOI] [PubMed] [Google Scholar]
- 28.Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT, Morales R, Allen AE, Armbrust EV. 2012. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl. Acad. Sci. U. S. A. 109:E317–E325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nevin KP, Richter H, Covalla SF, Johnson JP, Woodard TL, Orloff AL, Jia H, Zhang M, Lovley DR. 2008. Power output and columbic efficiencies from biofilms of Geobacter sulfurreducens comparable to mixed community microbial fuel cells. Environ. Microbiol. 10:2505–2514 [DOI] [PubMed] [Google Scholar]
- 30.Tremblay P-L, Aklujkar M, Leang C, Nevin KP, Lovley DR. 2011. A genetic system for Geobacter metallireducens: role of the flagellin and pilin in the reduction of Fe(III) oxide. Environ. Microbiol. Rep. 4:82–88 [DOI] [PubMed] [Google Scholar]
- 31.Smith JA, Lovley DR, Tremblay P-L. 2013. Outer cell surface components essential for Fe(III) oxide reduction in Geobacter metallireducens. Appl. Environ. Microbiol. 79:901–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrestha PM, Kammann C, Lenhart K, Dam B, Liesack W. 2012. Linking activity, composition and seasonal dynamics of atmospheric methane oxidizers in a meadow soil. ISME J. 6:1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jager M, Ott CE, Grunhagen J, Hecht J, Schell H, Mundlos S, Duda GN, Robinson PN, Lienau J. 2011. Composite transcriptome assembly of RNA-seq data in a sheep model for delayed bone healing. BMC Genomics 12:158 doi:10.1186/1471-2164-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu C, Li Y, Holmes A, Szafranski K, Faulkes CG, Coen CW, Buffenstein R, Platzer M, de Magalhaes JP, Church GM. 2011. RNA sequencing reveals differential expression of mitochondrial and oxidation reduction genes in the long-lived naked mole-rat compared to mice. PLoS One 6:e26729 doi:10.1371/journal.pone.0026729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Methé BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967–1969 [DOI] [PubMed] [Google Scholar]
- 36.Aklujkar M, Krushkal J, DiBartolo G, Lapidus A, Land ML, Lovley DR. 2009. The genome sequence of Geobacter metallireducens: features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 9:109 doi:10.1186/1471-2180-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Gen. Biol. 10:R25 doi:10.1186-Gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Gen. Biol. 11:R106 doi:10.1186-Gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 40.Klevebring D, Bjursell M, Emanuelsson O, Lundeberg J. 2010. In-depth transcriptome analysis reveals novel TARs and prevalent antisense transcription in human cell lines. PLoS One 5:e9762 doi:10.1371/journal.pone.0009762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlins SA, Mehra R, Rhodes DR, Shah RB, Rubin MA, Bruening E, Makarov V, Chinnaiyan AM. 2006. Whole transcriptome amplification for gene expression profiling and development of molecular archives. Neoplasia 8:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coppi MV, O'Neil RA, Lovley DR. 2004. Identification of an uptake hydrogenase required for hydrogen-dependent reduction of Fe(III) and other electron acceptors by Geobacter sulfurreducens. J. Bacteriol. 186:3022–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sargent F, Ballantine SP, Rugman PA, Palmer T, Boxer DH. 1998. Reassignment of the gene encoding the Escherichia coli hydrogenase 2 small subunit-identification of a soluble precursor of the small subunit in a hypB mutant. Eur. J. Biochem. 255:746–754 [DOI] [PubMed] [Google Scholar]
- 44.Tremblay PL, Lovley DR. 2012. Role of the NiFe hydrogenase Hya in oxidative stress defense in Geobacter sulfurreducens. J. Bacteriol. 194:2248–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malvankar NS, Tuominen MT, Lovley DR. 2012. Lack of cytochrome involvement in long-range electron transport through conductive biofilms and nanowires of Geobacter sulfurreducens. Energy Environ. Sci. 5:8651–8659 [Google Scholar]
- 46.Lovley DR. 2012. Electromicrobiology. Annu. Rev. Microbiol. 66:391–409 [DOI] [PubMed] [Google Scholar]
- 47.Juárez K, Kim BC, Nevin K, Olvera L, Reguera G, Lovley DR, Methé BA. 2009. PilR, a transcriptional regulator for pilin and other genes required for Fe(III) reduction in Geobacter sulfurreducens. J. Mol. Microbiol. Biotechnol. 16:146–158 [DOI] [PubMed] [Google Scholar]
- 48.Mahadevan R, Yan B, Postier B, Nevin KP, Woodard TL, O'Neil R, Coppi MV, Methé BA, Krushkal J. 2008. Characterizing regulation of metabolism in Geobacter sulfurreducens through genome-wide expression data and sequence analysis. OMICS 12:33–59 [DOI] [PubMed] [Google Scholar]
- 49.Holmes DE, Chaudhuri SK, Nevin KP, Mehta T, Methé BA, Liu A, Ward JE, Woodard TL, Webster J, Lovley DR. 2006. Microarray and genetic analysis of electron transfer to electrodes in Geobacter sulfurreducens. Environ. Microbiol. 8:1805–1815 [DOI] [PubMed] [Google Scholar]
- 50.Ding YH, Hixson KK, Aklujkar MA, Lipton MS, Smith RD, Lovley DR, Mester T. 2008. Proteome of Geobacter sulfurreducens grown with Fe(III) oxide or Fe(III) citrate as the electron acceptor. Biochim. Biophys. Acta 1784:1935–1941 [DOI] [PubMed] [Google Scholar]
- 51.Leang C, Coppi MV, Lovley DR. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leang C, Lovley DR. 2005. Regulation of two highly similar genes, omcB and omcC, in a 10 kb chromosomal duplication in Geobacter sulfurreducens. Microbiology 151:1761–1767 [DOI] [PubMed] [Google Scholar]
- 53.Lloyd JR, Leang C, Myerson ALH, Coppi MV, Cuifo S, Methé B, Sandler SJ, Lovley DR. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgado L, Bruix M, Pessanha M, Londer YY, Salgueiro CA. 2010. Thermodynamic characterization of a triheme cytochrome family from Geobacter sulfurreducens reveals mechanistic and functional diversity. Biophys. J. 99:293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strycharz SM, Glaven RH, Coppi MV, Gannon SM, Perpetua LA, Liu A, Nevin KP, Lovley DR. 2011. Gene expression and deletion analysis of mechanisms for electron transfer from electrodes to Geobacter sulfurreducens. Bioelectrochemistry 80:142–150 [DOI] [PubMed] [Google Scholar]
- 56.Ueki T, Lovley DR. 2010. Genome-wide gene regulation of biosynthesis and energy generation by a novel transcriptional repressor in Geobacter species. Nucleic Acids Res. 38:810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Risso C, Methé BA, Elifantz H, Holmes DE, Lovley DR. 2008. Highly conserved genes in Geobacter species with expression patterns indicative of acetate limitation. Microbiology 154:2589–2599 [DOI] [PubMed] [Google Scholar]
- 58.Mahadevan R, Palsson BO, Lovley DR. 2011. In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modelling. Nat. Rev. Microbiol. 9:39–50 [DOI] [PubMed] [Google Scholar]
- 59.Segura D, Mahadevan R, Juárez K, Lovley DR. 2008. Computational and experimental analysis of redundancy in the central metabolism of Geobacter sulfurreducens. PLoS Comput. Biol. 4:e36 doi:10.1371/journal.pcbi.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato S, Watanabe K. 2010. Ecological and evolutionary interactions in syntrophic methanogenic consortia. Microbes Environ. 25:145–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.