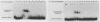Abstract
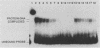
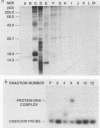
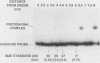
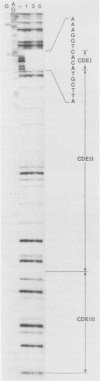
A centromere-specific DNA-binding protein has been purified to homogeneity by a combination of conventional and sequence-affinity chromatography from the yeast Saccharomyces cerevisiae. This protein (designated CBP-I) has an apparent molecular weight of 16,000. It binds specifically to the CDEI (centromere DNA element I) region of yeast centromere DNA, as shown by the electrophoretic mobility retardation assay and DNase I protection analysis, but does not bind specifically to other regions of yeast centromere DNA such as CDEII and CDEIII. The relative binding affinity of purified CBP-I to five different point mutations of CDEI correlates directly with the previously determined ability of each point mutation to convey centromere function in a mitotic chromosome segregation assay (J. H. Hegemann, J. H. Shero, G. Cottarel, P. Philippsen, and P. Hieter, Mol. Cell. Biol. 8:2523-2535, 1988). This supports the authenticity of CBP-I as a functional component of the yeast kinetochore.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol. 1987 Jan;7(1):403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. V., Schenk E. A., Olmsted J. B. Human anticentromere antibodies: distribution, characterization of antigens, and effect on microtubule organization. Cell. 1983 Nov;35(1):331–339. doi: 10.1016/0092-8674(83)90236-2. [DOI] [PubMed] [Google Scholar]
- Cumberledge S., Carbon J. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics. 1987 Oct;117(2):203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91(3-4):313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Sullivan K. F., Machlin P. S., Cooke C. A., Kaiser D. A., Pollard T. D., Rothfield N. F., Cleveland D. W. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987 Apr;104(4):817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet A., Fitzgerald-Hayes M. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):68–75. doi: 10.1128/mcb.7.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldner H. H., Lakomek H. J., Bautz F. A. Human anti-centromere sera recognise a 19.5 kD non-histone chromosomal protein from HeLa cells. Clin Exp Immunol. 1984 Oct;58(1):13–20. [PMC free article] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hegemann J. H., Pridmore R. D., Schneider R., Philippsen P. Mutations in the right boundary of Saccharomyces cerevisiae centromere 6 lead to nonfunctional or partially functional centromeres. Mol Gen Genet. 1986 Nov;205(2):305–311. doi: 10.1007/BF00430443. [DOI] [PubMed] [Google Scholar]
- Hegemann J. H., Shero J. H., Cottarel G., Philippsen P., Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jun;8(6):2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Inoué S. Cell division and the mitotic spindle. J Cell Biol. 1981 Dec;91(3 Pt 2):131s–147s. doi: 10.1083/jcb.91.3.131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. M., Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974 Apr;118(1):8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer L., del Mazo J., Avila J. Identification of centromere proteins in different mammalian cells. Eur J Cell Biol. 1988 Apr;46(1):196–199. [PubMed] [Google Scholar]
- Mann C., Davis R. W. Structure and sequence of the centromeric DNA of chromosome 4 in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):241–245. doi: 10.1128/mcb.6.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K., Mecke D. Isolation and characterization of nuclei and nuclear membranes from Saccharomyces cerevisiae protoplasts. FEBS Lett. 1980 Dec 15;122(1):95–99. doi: 10.1016/0014-5793(80)80410-8. [DOI] [PubMed] [Google Scholar]
- McGrew J., Diehl B., Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh N. J., James I. E., Maddison P. J. Differential isotype recognition of two centromere associated polypeptides by immunoblotting in connective tissue disease. Clin Exp Immunol. 1988 Jun;72(3):457–464. [PMC free article] [PubMed] [Google Scholar]
- McNeilage L. J., Whittingham S., McHugh N., Barnett A. J. A highly conserved 72,000 dalton centromeric antigen reactive with autoantibodies from patients with progressive systemic sclerosis. J Immunol. 1986 Oct 15;137(8):2541–2547. [PubMed] [Google Scholar]
- Moroi Y., Peebles C., Fritzler M. J., Steigerwald J., Tan E. M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Dec;7(12):4522–4534. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D. K., O'Day K., Wener M. H., Andrews B. S., Margolis R. L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987 Apr;104(4):805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri L., Landonio L., Stotz A., Philippsen P. Role of conserved sequence elements in yeast centromere DNA. EMBO J. 1985 Jul;4(7):1867–1874. doi: 10.1002/j.1460-2075.1985.tb03862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri L., Philippsen P. Centromeric DNA from chromosome VI in Saccharomyces cerevisiae strains. EMBO J. 1982;1(12):1605–1611. doi: 10.1002/j.1460-2075.1982.tb01362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld P. J., Kelly T. J. Purification of nuclear factor I by DNA recognition site affinity chromatography. J Biol Chem. 1986 Jan 25;261(3):1398–1408. [PubMed] [Google Scholar]
- Valdivia M. M., Brinkley B. R. Fractionation and initial characterization of the kinetochore from mammalian metaphase chromosomes. J Cell Biol. 1985 Sep;101(3):1124–1134. doi: 10.1083/jcb.101.3.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]