Abstract
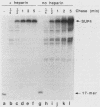
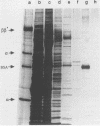
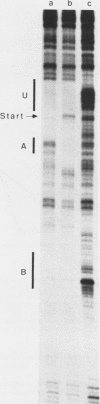
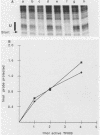
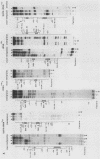
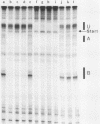
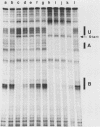
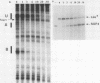
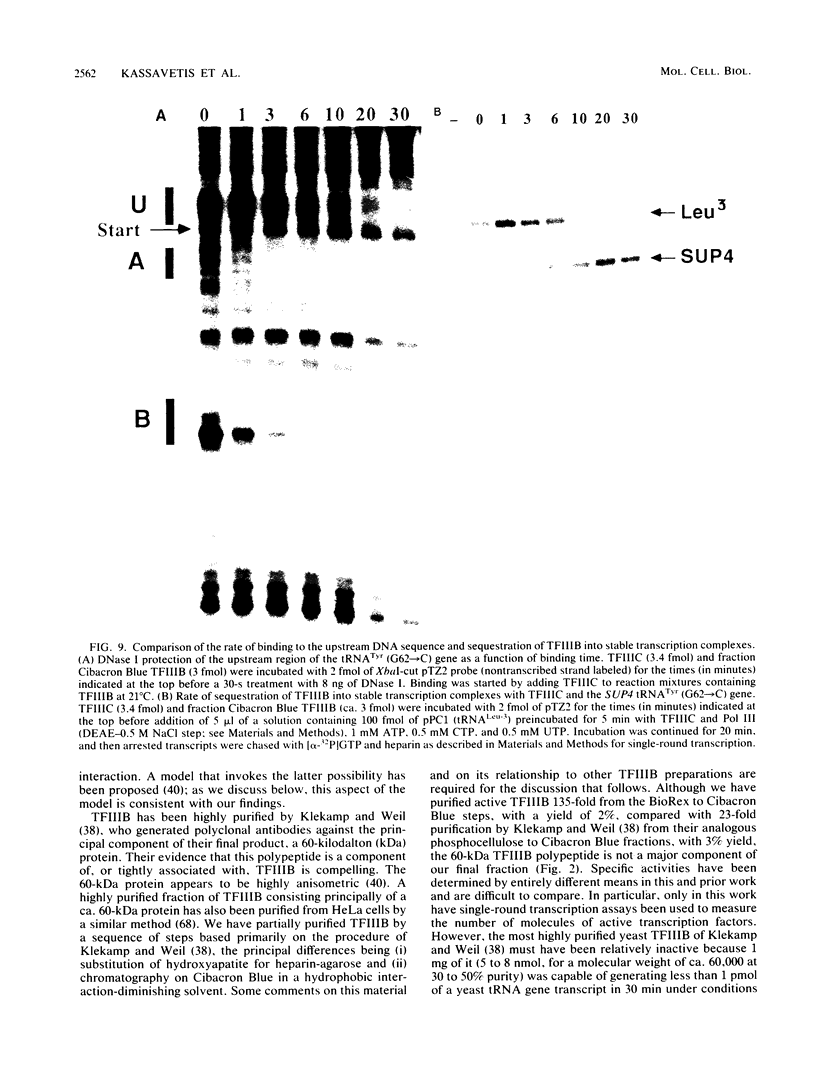
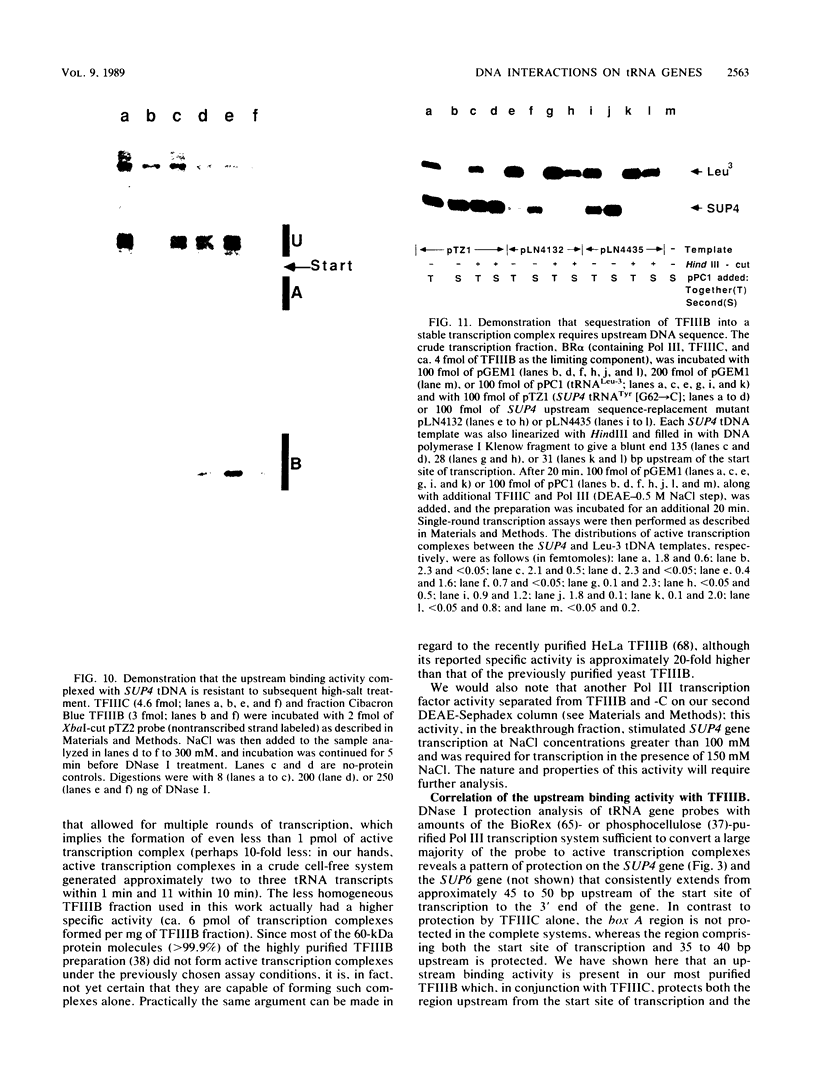
Transcription complexes that assemble on tRNA genes in a crude Saccharomyces cerevisiae cell extract extend over the entire transcription unit and approximately 40 base pairs of contiguous 5'-flanking DNA. We show here that the interaction with 5'-flanking DNA is due to a protein that copurifies with transcription factor TFIIIB through several steps of purification and shares characteristic properties that are normally ascribed to TFIIIB: dependence on prior binding of TFIIIC and great stability once the TFIIIC-TFIIIB-DNA complex is formed. SUP4 gene (tRNATyr) DNA that was cut within the 5'-flanking sequence (either 31 or 28 base pairs upstream of the transcriptional start site) was no longer able to stably incorporate TFIIIB into a transcription complex. The TFIIIB-dependent 5'-flanking DNA protein interaction was predominantly not sequence specific. The extension of the transcription complex into this DNA segment does suggest two possible explanations for highly diverse effects of flanking-sequence substitutions on tRNA gene transcription: either (i) proteins that are capable of binding to these upstream DNA segments are also potentially capable of stimulating or interfering with the incorporation of TFIIIB into transcription complexes or (ii) 5'-flanking sequence influences the rate of assembly of TFIIIB into stable transcription complexes.
Full text
PDF







Images in this article
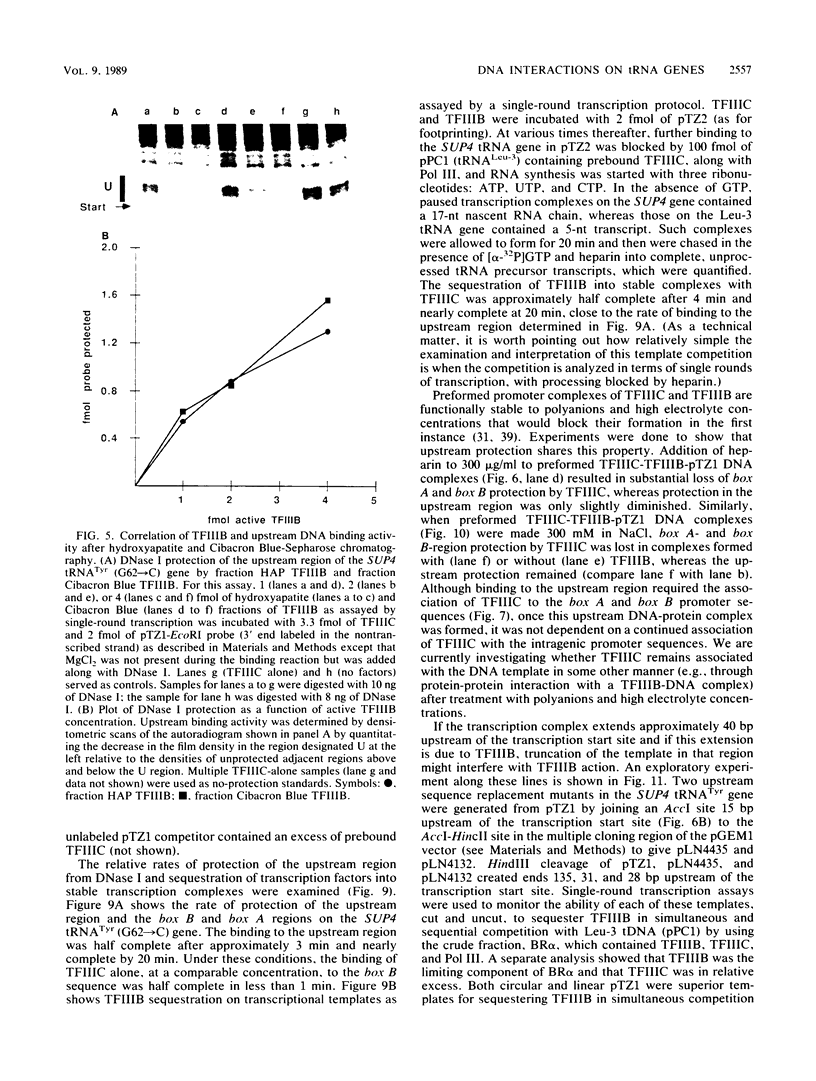
Selected References
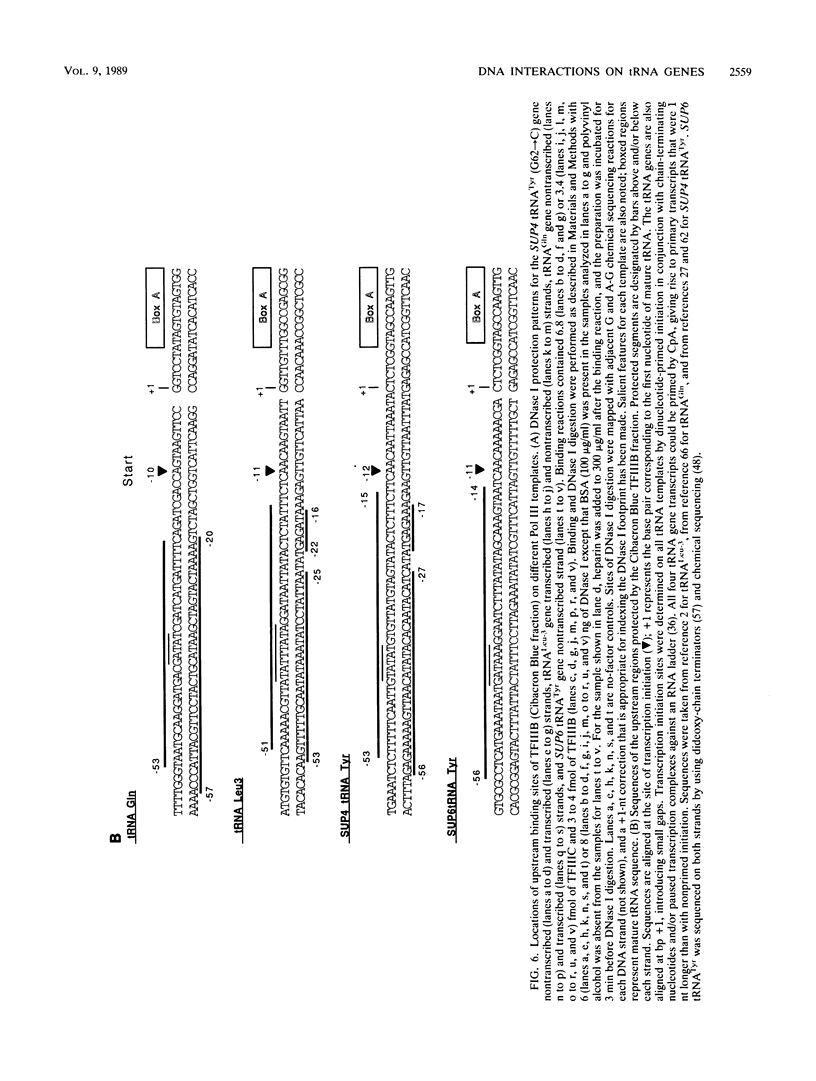
These references are in PubMed. This may not be the complete list of references from this article.
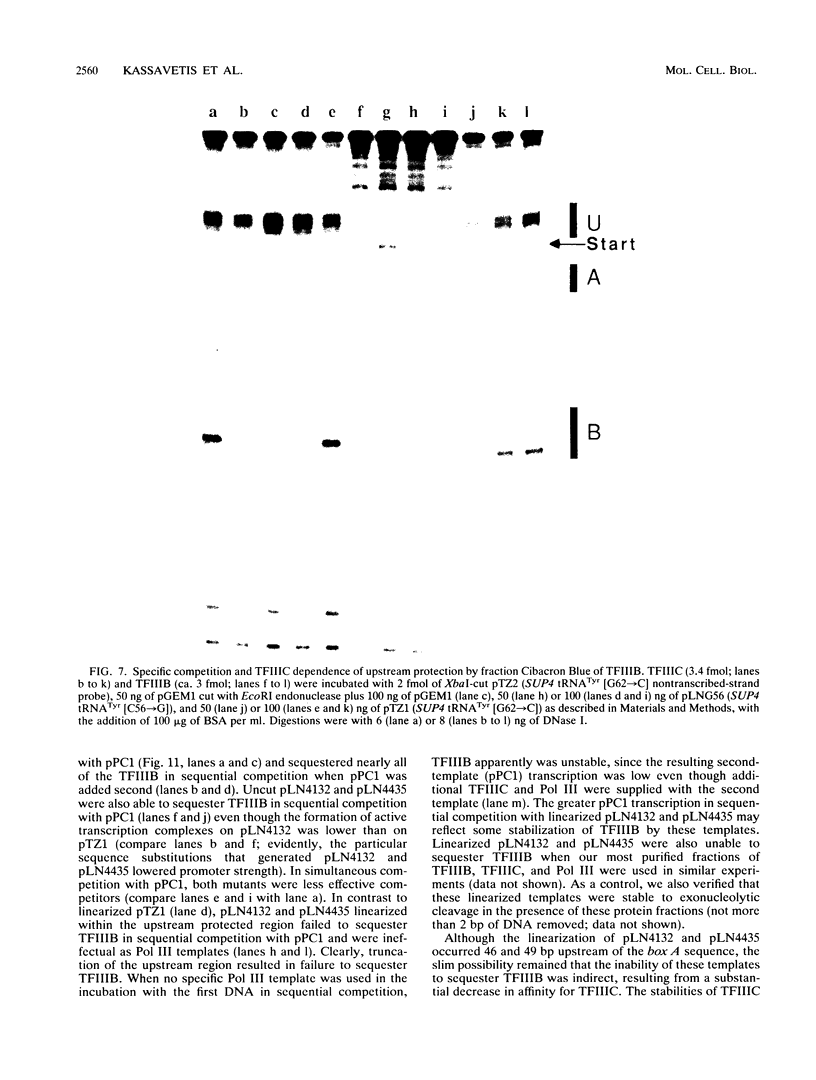
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
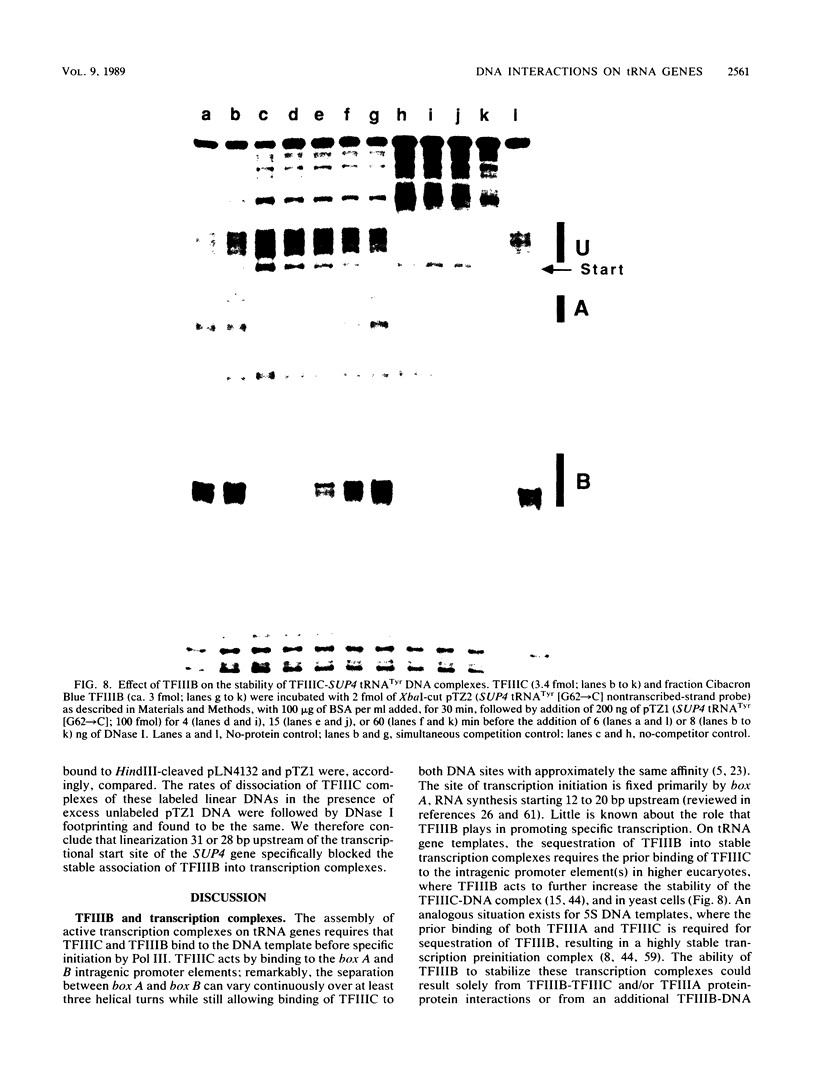
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Arnold G. J., Gross H. J. Unrelated leader sequences can efficiently promote human tRNA gene transcription. Gene. 1987;51(2-3):237–246. doi: 10.1016/0378-1119(87)90312-x. [DOI] [PubMed] [Google Scholar]
- Arnold G. J., Schmutzler C., Gross H. J. Functional dissection of 5' and 3' extragenic control regions of human tRNA(Val) genes reveals two different regulatory effects. DNA. 1988 Mar;7(2):87–97. doi: 10.1089/dna.1988.7.87. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Camier S., Sentenac A., Hall B. D. Gene size differentially affects the binding of yeast transcription factor tau to two intragenic regions. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8768–8772. doi: 10.1073/pnas.84.24.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Gabrielsen O., Hall B. D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986 Apr 25;261(12):5275–5282. [PubMed] [Google Scholar]
- Bark C., Weller P., Zabielski J., Janson L., Pettersson U. A distant enhancer element is required for polymerase III transcription of a U6 RNA gene. Nature. 1987 Jul 23;328(6128):356–359. doi: 10.1038/328356a0. [DOI] [PubMed] [Google Scholar]
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Boulanger P. A., Yoshinaga S. K., Berk A. J. DNA-binding properties and characterization of human transcription factor TFIIIC2. J Biol Chem. 1987 Nov 5;262(31):15098–15105. [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Böhme H. J., Kopperschläger G., Schulz J., Hofmann E. Affinity chromatography of phosphofructokinase using Cibacron blue F3G-A. J Chromatogr. 1972 Jun 28;69(1):209–214. doi: 10.1016/s0021-9673(00)83103-9. [DOI] [PubMed] [Google Scholar]
- Carbon P., Murgo S., Ebel J. P., Krol A., Tebb G., Mattaj L. W. A common octamer motif binding protein is involved in the transcription of U6 snRNA by RNA polymerase III and U2 snRNA by RNA polymerase II. Cell. 1987 Oct 9;51(1):71–79. doi: 10.1016/0092-8674(87)90011-0. [DOI] [PubMed] [Google Scholar]
- Carey M. F., Gerrard S. P., Cozzarelli N. R. Analysis of RNA polymerase III transcription complexes by gel filtration. J Biol Chem. 1986 Mar 25;261(9):4309–4317. [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Cortese R. Transcription by RNA polymerase III. Curr Top Dev Biol. 1983;18:59–88. doi: 10.1016/s0070-2153(08)60579-7. [DOI] [PubMed] [Google Scholar]
- Das G., Henning D., Wright D., Reddy R. Upstream regulatory elements are necessary and sufficient for transcription of a U6 RNA gene by RNA polymerase III. EMBO J. 1988 Feb;7(2):503–512. doi: 10.1002/j.1460-2075.1988.tb02838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N., Berk A. J. Separation of TFIIIC into two functional components by sequence specific DNA affinity chromatography. Nucleic Acids Res. 1987 Dec 10;15(23):9895–9907. doi: 10.1093/nar/15.23.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Dugaiczyk A., Boyer H. W., Goodman H. M. Ligation of EcoRI endonuclease-generated DNA fragments into linear and circular structures. J Mol Biol. 1975 Jul 25;96(1):171–184. doi: 10.1016/0022-2836(75)90189-8. [DOI] [PubMed] [Google Scholar]
- Elliott T., Geiduschek E. P. Defining a bacteriophage T4 late promoter: absence of a "-35" region. Cell. 1984 Jan;36(1):211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Coppo A., Fruscoloni P., Benedetti P., Di Segni G., Tocchini-Valentini G. P. Comparative mutational analysis of wild-type and stretched tRNA3(Leu) gene promoters. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8763–8767. doi: 10.1073/pnas.84.24.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts S. J., Weil P. A., Chalkley R. Novobiocin inhibits interactions required for yeast TFIIIB sequestration during stable transcription complex formation in vitro. Nucleic Acids Res. 1987 Feb 25;15(4):1493–1506. doi: 10.1093/nar/15.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Olson M. V., Hall B. D. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C. I., Holland M. J. Purification of yeast RNA polymerases using heparin agarose affinity chromatography. Transcriptional properties of the purified enzymes on defined templates. J Biol Chem. 1983 Mar 10;258(5):3230–3241. [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Evans C. F., Engelke D. R. Comparison of tRNA gene transcription complexes formed in vitro and in nuclei. Mol Cell Biol. 1987 Sep;7(9):3212–3220. doi: 10.1128/mcb.7.9.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn D., Wingender E., Seifart K. H. Transcription complexes for various class III genes differ in parameters of formation and stability towards salt. J Mol Biol. 1987 Jan 20;193(2):303–313. doi: 10.1016/0022-2836(87)90221-x. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983 Apr 21;302(5910):681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Elliott T., Rabussay D. P., Geiduschek E. P. Initiation of transcription at phage T4 late promoters with purified RNA polymerase. Cell. 1983 Jul;33(3):887–897. doi: 10.1016/0092-8674(83)90031-4. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. Bacteriophage T4 late promoters: mapping 5' ends of T4 gene 23 mRNAs. EMBO J. 1982;1(1):107–114. doi: 10.1002/j.1460-2075.1982.tb01132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Zentner P. G., Geiduschek E. P. Transcription at bacteriophage T4 variant late promoters. An application of a newly devised promoter-mapping method involving RNA chain retraction. J Biol Chem. 1986 Oct 25;261(30):14256–14265. [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Partial purification and characterization of the Saccharomyces cerevisiae transcription factor TFIIIB. J Biol Chem. 1986 Feb 25;261(6):2819–2827. [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Properties of yeast class III gene transcription factor TFIIIB. Implications regarding mechanism of action. J Biol Chem. 1987 Jun 5;262(16):7878–7883. [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Specific transcription of homologous class III genes in yeast-soluble cell-free extracts. J Biol Chem. 1982 Jul 25;257(14):8432–8441. [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Yeast class III gene transcription factors and homologous RNA polymerase III form ternary transcription complexes stable to disruption by N-lauroyl-sarcosine (sarcosyl). Arch Biochem Biophys. 1986 May 1;246(2):783–800. doi: 10.1016/0003-9861(86)90335-8. [DOI] [PubMed] [Google Scholar]
- Krol A., Carbon P., Ebel J. P., Appel B. Xenopus tropicalis U6 snRNA genes transcribed by Pol III contain the upstream promoter elements used by Pol II dependent U snRNA genes. Nucleic Acids Res. 1987 Mar 25;15(6):2463–2478. doi: 10.1093/nar/15.6.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R., Dingermann T. Identification of a protein factor binding to the 5'-flanking region of a tRNA gene and being involved in modulation of tRNA gene transcription in vivo in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14B):6737–6752. doi: 10.1093/nar/16.14.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoccia E., Baldi I. M., Gandini-Attardi D., Ciafrè S., Tocchini-Valentini G. P. Site selection by the tRNA splicing endonuclease of Xenopus laevis. Cell. 1988 Nov 18;55(4):731–738. doi: 10.1016/0092-8674(88)90231-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Murphy S., Di Liegro C., Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987 Oct 9;51(1):81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- Ottonello S., Rivier D. H., Doolittle G. M., Young L. S., Sprague K. U. The properties of a new polymerase III transcription factor reveal that transcription complexes can assemble by more than one pathway. EMBO J. 1987 Jul;6(7):1921–1927. doi: 10.1002/j.1460-2075.1987.tb02452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond G. J., Johnson J. D. The role of non-coding DNA sequences in transcription and processing of a yeast tRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5969–5988. doi: 10.1093/nar/11.17.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., Henning D., Das G., Harless M., Wright D. The capped U6 small nuclear RNA is transcribed by RNA polymerase III. J Biol Chem. 1987 Jan 5;262(1):75–81. [PubMed] [Google Scholar]
- Ruet A., Camier S., Smagowicz W., Sentenac A., Fromageot P. Isolation of a class C transcription factor which forms a stable complex with tRNA genes. EMBO J. 1984 Feb;3(2):343–350. doi: 10.1002/j.1460-2075.1984.tb01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sandmeyer S. B., Bilanchone V. W., Clark D. J., Morcos P., Carle G. F., Brodeur G. M. Sigma elements are position-specific for many different yeast tRNA genes. Nucleic Acids Res. 1988 Feb 25;16(4):1499–1515. doi: 10.1093/nar/16.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Söll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983 Feb 25;258(4):2447–2453. [PubMed] [Google Scholar]
- Segall J. Assembly of a yeast 5 S RNA gene transcription complex. J Biol Chem. 1986 Sep 5;261(25):11578–11584. [PubMed] [Google Scholar]
- Selker E. U., Morzycka-Wroblewska E., Stevens J. N., Metzenberg R. L. An upstream signal is required for in vitro transcription of Neurospora 5S RNA genes. Mol Gen Genet. 1986 Oct;205(1):189–192. doi: 10.1007/BF02428052. [DOI] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Caspers P., Geiduschek E. P. Effects of temperature and single-stranded DNA on the interaction of an RNA polymerase III transcription factor with a tRNA gene. Cell. 1985 Feb;40(2):311–317. doi: 10.1016/0092-8674(85)90145-x. [DOI] [PubMed] [Google Scholar]
- Taylor M. J., Segall J. Characterization of factors and DNA sequences required for accurate transcription of the Saccharomyces cerevisiae 5 S RNA gene. J Biol Chem. 1985 Apr 10;260(7):4531–4540. [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Delta sequences and double symmetry in a yeast chromosomal replicator region. J Mol Biol. 1982 Apr 5;156(2):293–307. doi: 10.1016/0022-2836(82)90330-8. [DOI] [PubMed] [Google Scholar]
- Tyler B. M. Transcription of Neurospora crassa 5 S rRNA genes requires a TATA box and three internal elements. J Mol Biol. 1987 Aug 20;196(4):801–811. doi: 10.1016/0022-2836(87)90406-2. [DOI] [PubMed] [Google Scholar]
- Waldschmidt R., Jahn D., Seifart K. H. Purification of transcription factor IIIB from HeLa cells. J Biol Chem. 1988 Sep 15;263(26):13350–13356. [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- Wu G. J., Railey J. F., Cannon R. E. Defining the functional domains in the control region of the adenovirus type 2 specific VARNA1 gene. J Mol Biol. 1987 Apr 5;194(3):423–442. doi: 10.1016/0022-2836(87)90672-3. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S. K., Boulanger P. A., Berk A. J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]