Abstract
Human beta-defensins (hBDs) are crucial peptides for the innate immune response and are thus prime candidates as therapeutic agents directed against infective diseases. Based on the properties of wild-type hBD1 and hBD3 and of previously synthesized analogs (1C, 3I, and 3N), we have designed a new analog, 3NI, and investigated its potential as an antimicrobial drug. Specifically, we evaluated the antimicrobial activities of 3NI versus those of hBD1, hBD3, 1C, 3I, and 3N. Our results show that 3NI exerted greater antibacterial activity against Pseudomonas aeruginosa, Escherichia coli, and Enterococcus faecalis than did hBD1 and hBD3, even with elevated salt concentrations. Moreover, its antiviral activity against herpes simplex virus 1 was greater than that of hBD1 and similar to that of hBD3. Subsequently, we investigated the cytotoxic effects of all peptides in three human epithelial carcinoma cell lines: A549 from lung, CaCo-2 from colon, and Capan-1 from pancreas. None of the analogs significantly reduced cell viability versus wild-type hBD1 and hBD3. They did not induce genotoxicity or cause an increase in the number of apoptotic cells. Using confocal microscopy, we also investigated the localization of the peptides during their incubation with epithelial cells and found that they were distributed on the cell surface, from which they were internalized. Finally, we show that hBD1 and hBD3 are characterized by high resistance to serum degradation. In conclusion, the new analog 3NI seems to be a promising anti-infective agent, particularly given its high salt resistance—a feature that is relevant in diseases such as cystic fibrosis.
INTRODUCTION
Innate immunity, which is the first line of defense against pathogens, is attracting interest consequent to increasing antibiotic resistance. Short antimicrobial peptides, including defensins, are key components of the innate immune system (1). Human beta-defensins (hBDs) are cationic cysteine-rich molecules with a three-dimensional structure stabilized by three disulfide bridges (2–4). They exert a broad spectrum of antimicrobial activity against Gram-positive and Gram-negative bacteria, fungi, and enveloped viruses (5–7). Although the role played by hBDs in antimicrobial reactions has been well established (6, 7), the intracellular molecular mechanisms have not been fully elucidated. hBD analogs that differ in structural characteristics from the wild-type peptides appear to preserve their antimicrobial activities (8–10). This finding prompted a race to design hBD analogs potentially endowed with therapeutic properties (9, 11–21).
In this scenario, we focused on hBD1 and hBD3. hBD1 is constitutively expressed and not upregulated during infection or inflammation, and its antibacterial activity is impaired at high NaCl concentrations (22–24); hBD3 is upregulated during infection and inflammation and is less sensitive to high salt concentrations (25–28). In a previous study, we synthesized hBD1 and hBD3 analogs (1C, 3I, and 3N) that maintained antimicrobial activities at salt concentrations approaching those found in the lung fluid of patients with cystic fibrosis (9). We also found that the charged C-terminal domain of hBD3 (RRKK) and the internal domain of hBD1 (PIFTKIQGT) are crucial for the activities of defensins and that deletion of six residues at the hBD3 N terminus did not reduce the activity (9).
Based on these findings, we designed a new chimeric molecule, 3NI, that might have enhanced antimicrobial effects and salt resistance. To evaluate the therapeutic potential of such molecules, we investigated the effects of wild-type hBD1 and hBD3, of analogs 1C, 3I, and 3N, and of the new analog (3NI) on cell viability, apoptosis, and DNA status in three human epithelial cell lines, i.e., lung carcinoma (A549), colon carcinoma (CaCo-2), and pancreas adenocarcinoma (Capan-1), exposed to Pseudomonas aeruginosa, Escherichia coli, Enterococcus faecalis, and herpes simplex virus 1 (HSV-1), which are the most frequent causes of infections. Lastly, using confocal microscopy, we evaluated the interactions of these peptides with cells to determine their cellular fates.
MATERIALS AND METHODS
Synthesis and NBD labeling of peptides.
Peptides were synthesized using the standard solid-phase 9-fluorenylmethoxy carbonyl (Fmoc) method (9). Resin-bound peptides were labeled as reported elsewhere (29). Briefly, 30 to 70 mg of resin-bound peptide (10 to 25 μmol) was treated with piperidine in dimethylformamide (DMF) to remove the Fmoc-protecting group of the peptide's N-terminal amino acid. The resin-bound peptide was reacted with 4-chloro-7-nitrobenz-2-oxa-1,3-diazole (NBD-Cl) in DMF (3 to 4 eq). After 24 h, peptides were washed with methylene chloride, cleaved from the resin, and purified as reported for the unlabeled peptides (9). This procedure generated a major product with an NBD moiety attached to the peptide's N-terminal amino group. Its identity was confirmed by liquid chromatography-mass spectrometry.
Antibacterial activity assay.
CFU assays of the antibacterial activities of hBDs against P. aeruginosa (ATCC 27853; American Type Culture Collection, Manassas, VA), E. coli (ATCC 25922), and E. faecalis (ATCC 29212) were performed. The strains were grown under aerobic conditions in tryptic soy broth (Difco Laboratories, Detroit, MI) at 37°C and were incubated with hBDs for 2 h at 37°C. We used two concentrations of peptides (2.5 μM and 12.5 μM). For the salt dependence assay, 0, 50, 100, and 200 mM concentrations of NaCl were included in the incubation buffer, as described previously (9). Each assay was performed in triplicate. The MIC of the new molecule 3NI was determined with a modified version of the broth microdilution assay of the Clinical and Laboratory Standards Institute using a final inoculum concentration of 105 CFU/ml. The peptide concentrations were 100.0, 50.0, 25.0, 12.5, 6.25, 3.12, and 1.56 μM.
Antiviral activity assay.
Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich, MO) supplemented with 10% fetal bovine serum (FBS). HSV-1 carrying a lacZ gene driven by the cytomegalovirus IE-1 promoter to express beta-galactosidase was propagated as described elsewhere (9). Experiments were conducted in parallel with no-peptide controls. The inhibitory effects of wild-type hBD1 and hBD3 and their analogs on HSV-1 infectivity were assessed by incubating cell monolayers with increasing peptide concentrations (1.5, 10.0, 20.0, 50.0, 100.0, and 250.0 μM) and with the viral inoculum for 45 min at 37°C. Experiments were performed in triplicate, and the percentages of inhibition were calculated with respect to the no-peptide control experiments.
Cell culture.
A549, CaCo-2, and Capan-1 cells were from the Bank of Human and Animal Continuous Cell Lines (CEINGE-Biotecnologie Avanzate, Naples, Italy). A549 and CaCo-2 cells were grown in 90% DMEM (Sigma-Aldrich) supplemented with 10% FBS (Lonza, Basel, Switzerland) and 1% l-glutamine (Sigma-Aldrich). Capan-1 cells were grown in 80% RPMI 1640 medium (Sigma-Aldrich) supplemented with 20% FBS (Lonza, Basel, Switzerland) and 1% l-glutamine (Sigma-Aldrich). The cells were grown as a monolayer in a flask at 37°C in 5% CO2.
MTT test: viability assay.
A549, CaCo-2, and Capan-1 cells were plated in triplicate at 4 × 104 cells/ml in a 96-well plate for 24 h. Cell viability was evaluated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma-Aldrich) assay. The next day, cells were incubated with and without 2.5, 12.5, or 25.0 μM wild-type hBD1, or hBD3, analog 1C, 3N, or 3NI for 0, 4, 8, 12, 16, 20, 24, 48, and 72 h, or analog 3I for 0, 4, 8, 24, 48, and 72 h. Then, all wells were treated with 20 μl 1:10-diluted MTT stock solution (5 mg/ml), mixed, and incubated at 37°C in a humidified incubator (5% CO2) for 4 h; 200 μl dimethyl sulfoxide was added to each well to dissolve the purple formazan, and absorbance was measured at 550 nm. The viability index of untreated cells was compared to that of the negative control (cells plus medium). Experiments were performed in triplicate. Cells incubated with medium alone represented 100% viability.
Annexin V assay: apoptosis assay.
A549, CaCo-2, and Capan-1 cells were plated at 1 × 105 cells/ml in a 24-well plate and left to attach for 24 h. The next day, cells were exposed to 2.5, 12.5, or 25.0 μM wild-type hBD1 or hBD3 or analog 1C, 3I, 3N, or 3NI for 24, 48, and 72 h. Harvested washed cells were resuspended at 1 × 105 cells/ml. Cells were centrifuged (5 min at 1,300 rpm), resuspended in 100 μl annexin V binding buffer (1×; Becton, Dickinson), 5 μl annexin V-fluorescein isothiocyanate (FITC) (Becton, Dickinson), and 5 μl propidium iodide (PI), and stained for 15 min at room temperature. Four hundred microliters of annexin V binding buffer was added before acquisition. Cells not stained with PI and annexin V-FITC were deemed to be live cells; PI-negative annexin V-FITC-positive cells were deemed to be in early apoptosis. Experiments were performed in triplicate.
Comet assay: single-cell gel electrophoresis.
DNA breakage was evaluated using a comet assay kit (Trevigen, MD). Three independently reproduced experiments were performed for each cell line. To determine DNA strand breakage, A549, CaCo-2, and Capan-1 cells were plated at 1 × 105 cells/ml in a 24-well plate and left to attach for 24 h. The next day, cells were exposed to two concentrations (2.5 and 12.5 μM) of wild-type hBD1 or hBD3 or analog 1C, 3I, 3N, or 3NI for 4 and 24 h. Comet slides were stained with diluted SYBR green and examined under an automated robotic epifluorescence microscope with an excitation filter (510 to 550 nm). One hundred cells were analyzed per slide. Experiments were performed in triplicate. As positive controls, we treated A549, CaCo-2, and Capan-1 cells with 250 μM H2O2 for 30 min and 60 μM etoposide for 1 h. Untreated cells served as negative controls.
Confocal microscopy.
A549 and CaCo-2 cells were seeded at 25% confluence in 6-well chambers (Ibidi GmbH, Martinsried, Germany). They were incubated with 15 μM concentrations of each NBD-labeled peptide and 1 μM FM4-64FX for 10 min at 37°C in 5% CO2 (“pulse”); defensin-containing medium was then replaced with fresh growth medium (“chase”), and cells were incubated at 37°C in 5% CO2 for 0 min and 2 h. After this chase, cells were fixed with 3.7% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), permeabilized with PBS–0.1% Triton X-100 for 10 min at room temperature, and blocked with 1% bovine serum albumin (BSA) in PBS for 30 min. Cells were mildly prefixed with 1.8% PFA in PBS (2 min at room temperature); cells stained with FM4-64FX and incubated with NBD-hBDs were observed after 10 min and 2 h with an LSM 510 Meta confocal microscope (Carl Zeiss, Jena, Germany), equipped with a Plan Apo 63× objective (1.4 numerical aperture), at the following settings: green channel to detect NBD, excitation with a 488-nm argon laser, 505- to 550-nm emission band-pass filter, and red channel to detect FM4-64FX, excitation with a 543-nm helium/neon laser, 560- to 700-nm emission band-pass filter (using the meta monochromator).
Serum stability.
The serum stability of defensins was assessed at 0, 0.5, 1, 2, 4, 6, 8, 24, and 48 h as reported (30). Supernatants were injected into a reverse-phase high-performance liquid chromatography (HPLC) column (Applied Biosystems, CA). Components were separated using a Jupiter C18 microanalytical column (5-μm particle size, 300-Å pore size, 2 mm by 250 mm) (Phenomenex, Aschaffenburg, Germany) with a linear gradient of 20 to 60% acetonitrile in acidified water (0.1% trifluoroacetic acid [TFA]) over 30 min. To examine peptide cleavage in serum, HPLC fractions were analyzed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry (Voyager DE-STR; Applied Biosystems). Briefly, a sinapinic acid (Sigma-Aldrich) matrix solution of 10 mg/ml in acetonitrile (ACN)-H2O with 0.1% TFA (70:30) was placed on a MALDI target (1 μl), mixed with equal amounts of sample, placed on top of the seed layer, and allowed to dry. The MALDI-time of flight mass spectrometer was operated in the linear mode and calibrated externally using a protein mixture of insulin, myoglobin, and cytochrome c.
RESULTS
Design and synthesis of the novel peptide 3NI.
Based on our previous results (9) and to maximize antimicrobial activity, we designed and synthesized a novel chimeric molecule, 3NI, which is an hBD3 analog without the N-terminal hBD3 domain and with the internal domain of hBD1 (Fig. 1). The 3NI sequence is KYYCRVRGGRCAVLSCPIFTKIQGTCSTRGRKCCRRKK, with a net charge of +12. Figure 1 shows schematically the hBD1 and hBD3 sequences and their domains in the chimeric peptide.
Fig 1.

Sequence alignments of wild-type hBD1 and hBD3, the analogs 1C, 3I, and 3N, and the novel analog 3NI. The segments of sequences derived from hBD1 are shown in red, and those derived from hBD3 are shown in blue. The sequence of 3NI is KYYCRVRGGRCAVLSCPIFTKIQGTCSTRGRKCCRRKK, with a net charge of +12.
Antibacterial and antiviral activities of the novel 3NI analog and the wild-type hBDs.
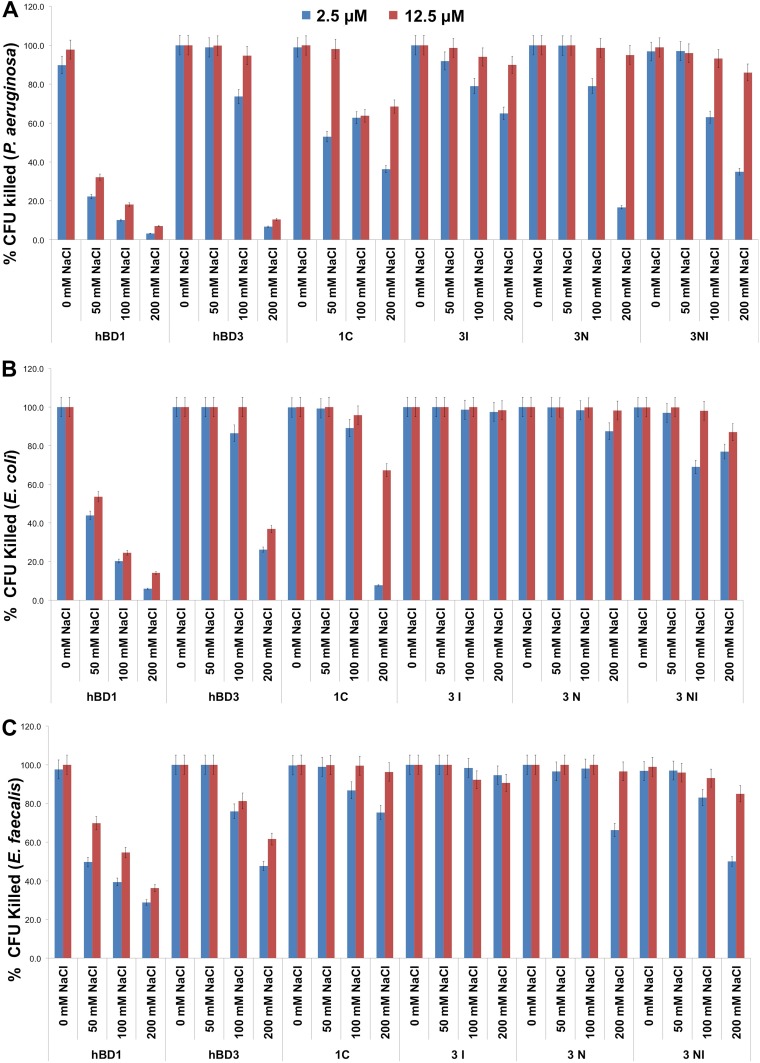
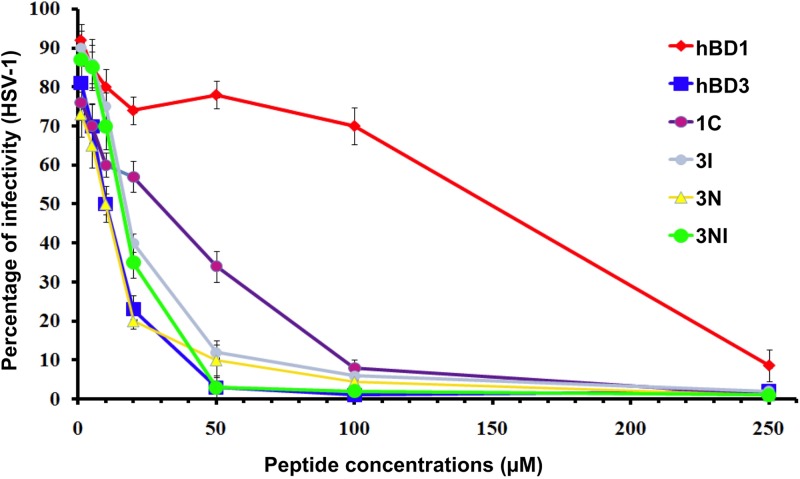
To evaluate the antibacterial activity of the new analog 3NI versus those of peptides hBD1, hBD3, 1C, 3I, and 3N (9), we tested two concentrations of each peptide (2.5 and 12.5 μM) and four concentrations of NaCl (0, 50, 100, and 200 mM) against P. aeruginosa, E. coli, and E. faecalis. At 2.5 μM, 3NI exerted good antibacterial activity against P. aeruginosa up to a concentration of 100 mM NaCl; at 12.5 μM, it was active up to 200 mM NaCl (Fig. 2A). Interestingly, the antibacterial activity of 3NI against E. coli started at concentrations as low as 2.5 μM in the presence of up to 200 mM NaCl (Fig. 2B). It was similarly active against E. faecalis and P. aeruginosa, although it was more effective than hBD3 at 2.5 μM in the presence of the two highest salt concentrations (Fig. 2C). Its MICs ranged between 12.5 and 25.0 μM for the three microorganisms. All peptides exerted strong antibacterial effects at 12.5 μM against P. aeruginosa, E. coli, and E. faecalis. To evaluate anti-HSV-1 activity, we tested all peptides at 1.5, 20.0, 50.0, 100.0, and 250.0 μM and found that analog 3NI and hBD3 (both at 50 μM) inhibited HSV-1 infectivity (Fig. 3). Wild-type hBD1 and the previously synthesized analogs were much less effective.
Fig 2.
The novel 3NI analog has enhanced antibacterial activity, particularly against P. aeruginosa, in comparison to the wild-type peptides. The antibacterial activities of wild-type hBD1 and hBD3, the analogs 1C, 3I, and 3N, and the novel analog 3NI were tested at two concentrations (2.5 and 12.5 μM) against P. aeruginosa (A), E. coli (B), and E. faecalis (C) with 0, 50, 100, and 200 mM NaCl. Error bars show the standard deviations (SDs) from three independent experiments.
Fig 3.
The novel 3NI analog greatly reduces HSV-1 infectivity. The antiviral activities of wild-type hBD1 and hBD3 and the analogs 1C, 3I, 3N, and 3NI were tested against HSV-1 at a concentration of 20.0 μM. Error bars show the SDs from three independent experiments.
Cytotoxicities of wild-type hBD1 and hBD3, analogs 1C, 3N, and 3I, and the novel 3NI analog in A549, CaCo-2, and Capan-1 cells.
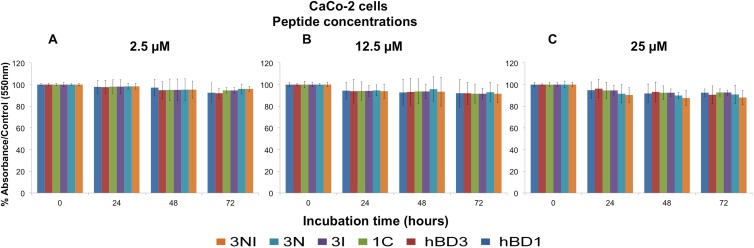
The MTT test was performed on A549, CaCo-2, and Capan-1 cells to evaluate the cytotoxic effects of wild-type hBD1 and hBD3 and the analogs 1C, 3N, 3I, and 3NI at concentrations of 2.5, 12.5, and 25.0 μM. Exposure times were 4, 8, 12, 16, 20, 24, 48, and 72 h, except for 3I, for which the exposure times were 4, 8, 24, 48, and 72 h. Because similar results were obtained in the three cell lines and no significant differences were observed at the first five time points, here we report the findings obtained with CaCo-2 cells at 24, 48, and 72 h. Figure 4 shows the percent viability of CaCo-2 cells exposed to the peptides at concentrations of 2.5, 12.5, and 25.0 μM (Fig. 4A to C, respectively). With all peptides at all concentrations used, cell viability was only slightly reduced after 24 h (2 to 10%), whereas it was reduced by about 15% after 48 h and 72 h. However, with all peptides under all conditions tested, the reductions in cell viability never exceeded 15%. These experiments show that wild-type hBDs and their analogs do not induce relevant cytotoxic effects in vitro.
Fig 4.
Wild-type hBD1 and hBD3, the 1C, 3I, and 3N analogs, and the novel 3NI analog do not affect cell viability. The peptides were used for the MTT test at three concentrations, 2.5 μM (A), 12.5 μM (B), and 25.0 μM (C), with CaCo-2 cells. The data are expressed as the means ± standard errors of three independent experiments. Similar results were obtained with A549 and Capan-1 cells.
We next evaluated whether the wild-type peptides and the analogs 1C, 3I, 3N, and 3NI damaged DNA in the A549, CaCo-2, and Capan-1 cell lines. Comet assays with the three cell lines after 4 h and 24 h of exposure to the peptides at concentrations of 2.5 and 12.5 μM showed lack of DNA damage in all three cell lines (data not shown). We then carried out flow-cytometric analyses of A549 (Table 1), CaCo-2 (Table 2), and Capan-1 (Table 3) cells to evaluate the apoptotic effects of wild-type hBD1 and hBD3 and their analogs at concentrations of 2.5, 12.5, and 25.0 μM; the exposure times were 24, 48, and 72 h. The numbers of apoptotic cells did not differ between the peptide-treated and untreated cell lines, although the difference increased in Capan-1 cells treated with 25.0 μM 3NI for 72 h.
Table 1.
Effects of wild-type peptides (hBD1 and hBD3) and analog peptides (1C, 3I, 3N, and 3NI) on A549 cell apoptosisa
| Peptide | % of apoptotic cells |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 24-h incubation |
48-h incubation |
72-h incubation |
|||||||
| 2.5 μM | 12.5 μM | 25.0 μM | 2.5 μM | 12.5 μM | 25.0 μM | 2.5 μM | 12.5 μM | 25.0 μM | |
| Negative control | 1.2 | 1.2 | 1.2 | 1.4 | 1.4 | 1.4 | 2.3 | 2.3 | 2.3 |
| hBD1 | 2 | 2.2 | 2.7 | 1.4 | 2.2 | 2.1 | 0.8 | 1 | 0.9 |
| hBD3 | 3.6 | 2.9 | 4.5 | 1.5 | 2.2 | 3.6 | 0.8 | 2 | 3.7 |
| 1C | 2.2 | 1.7 | 2.3 | 3.2 | 2.5 | 2.9 | 1.6 | 1.6 | 1.2 |
| 3I | 1.8 | 1.8 | 2.0 | 2.8 | 2.9 | 3.0 | 1.8 | 2 | 2.5 |
| 3N | 2.2 | 1.6 | 3.4 | 4.2 | 3.3 | 4.8 | 1.3 | 2.1 | 1.5 |
| 3NI | 1.9 | 1.8 | 1.1 | 4.7 | 5.7 | 14.3 | 2 | 2.2 | 2.7 |
Cells were incubated for 24, 48, or 72 h with all peptides at concentrations of 2.5, 12.5, and 25.0 μM. Data are expressed as a percentage of apoptotic cells after the subtraction of negative control values (untreated cells). No synthetic peptide significantly increased apoptosis versus the negative control. Experiments were performed in triplicate.
Table 2.
Effects of wild-type peptides (hBD1 and hBD3) and analog peptides (1C, 3I, 3N, and 3NI) on CaCo-2 cell apoptosisa
| Peptide | % of apoptotic cells |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 24-h incubation |
48-h incubation |
72-h incubation |
|||||||
| 2.5 μM | 12.5 μM | 25.0 μM | 2.5 μM | 12.5 μM | 25.0 μM | 2.5 μM | 12.5 μM | 25.0 μM | |
| Negative control | 4.2 | 4.2 | 4.2 | 6.6 | 6.6 | 6.6 | 6.3 | 6.3 | 6.3 |
| hBD1 | 7.1 | 10.3 | 9.6 | 6.1 | 6.2 | 7 | 8.2 | 7.5 | 4.1 |
| hBD3 | 12.6 | 12.8 | 20.8 | 8.7 | 6 | 34.1 | 9.2 | 7.6 | 9.8 |
| 1C | 9 | 12.3 | 11.7 | 7.8 | 13.6 | 20.6 | 5.2 | 10.9 | 8 |
| 3I | 6.1 | 6.8 | 7.0 | 5.8 | 6.2 | 7.1 | 5.3 | 6.1 | 7.2 |
| 3N | 8.2 | 12.3 | 12.9 | 10.8 | 10.3 | 10.1 | 6 | 5.9 | 9.1 |
| 3NI | 6.6 | 9.2 | 14.3 | 7.5 | 6.2 | 14.2 | 7.3 | 6.3 | 6.2 |
Cells were incubated for 24, 48, or 72 h with all peptides at concentrations of: 2.5, 12.5, and 25.0 μM. Data are expressed as a percentage of apoptotic cells after the subtraction of negative control values (untreated cells). No synthetic peptide significantly increased apoptosis versus the negative control. Experiments were performed in triplicate.
Table 3.
Effects of wild-type peptides (hBD1 and hBD3) and analog peptides (1C, 3I, 3N, and3NI) on Capan-1 cell apoptosisa
| Peptide | % of apoptotic cells |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 24-h incubation |
48-h incubation |
72-h incubation |
|||||||
| 2.5 μM | 12.5 μM | 25.0 μM | 2.5 μM | 12.5 μM | 25.0 μM | 2.5 μM | 12.5 μM | 25.0 μM | |
| Negative control | 15.2 | 15.2 | 15.2 | 10.2 | 10.2 | 10.2 | 19.6 | 19.6 | 19.6 |
| hBD1 | 14.4 | 12.6 | 12.9 | 17.8 | 11.3 | 10.7 | 15.1 | 8.1 | 17.2 |
| hBD3 | 14 | 8.3 | 9.2 | 10.9 | 14.8 | 18.7 | 10.8 | 15.9 | 26.2 |
| 1C | 9.2 | 12.3 | 14 | 16.1 | 16.2 | 16.2 | 21 | 18 | 22 |
| 3I | 7.0 | 7.3 | 8.1 | 9.2 | 10.3 | 11.4 | 15.2 | 16.8 | 17.6 |
| 3N | 8.4 | 7.4 | 16.7 | 14.7 | 12.5 | 11.2 | 12.8 | 25.2 | 22.3 |
| 3NI | 10.7 | 15.5 | 13.1 | 15.4 | 16.9 | 23.4 | 25.6 | 24.5 | 31.8 |
Cells were incubated for 24, 48, and 72 h with all peptides at concentrations of 2.5, 12.5, and 25.0 μM. Data are expressed as a percentage of apoptotic cells after the subtraction of negative control values (untreated cells). No synthetic peptide significantly increased apoptosis versus the negative control. Experiments were performed in triplicate.
Wild-type hBD1 and hBD3, analogs 1C, 3N, and 3I, and the novel 3NI analog actively enter cells after localization on the plasma membrane.
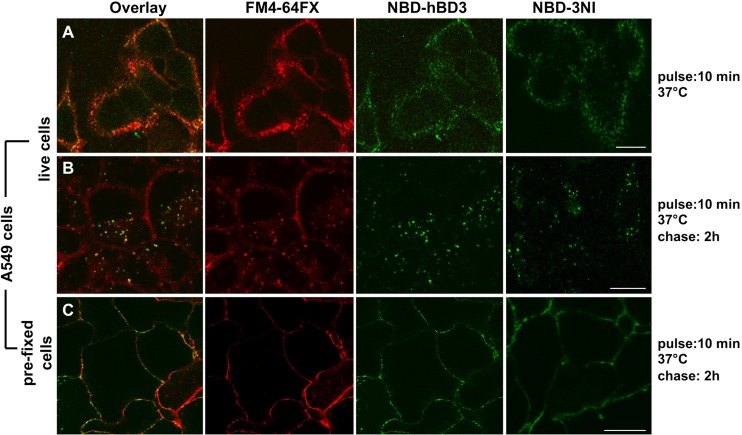
To determine whether hBDs and their analogs interact with eukaryotic cells, we investigated the localization of NBD-labeled wild-type hBD1 and hBD3 and analogs 1C, 3I, 3N, and 3NI in A549 and CaCo-2 cells. As all of the NBD-labeled peptides displayed comparable behavior in time course experiments in the two cell lines, we report the results obtained with NBD-hBD3 and NBD-3NI in A549 cells. In these experiments, we incubated live cells with the lipophilic probe FM4-64FX and the NBD-labeled peptides for 10 min (pulse), and then we added fresh medium at 37°C for 0 min (Fig. 5A) and for 2 h (chase) (Fig. 5B) to monitor the distribution of NBD-hBD3 and NBD-3NI on the plasma membrane and within cells, respectively. We found that the NBD-labeled peptides colocalized with FM4-64FX at both times. These data indicate that peptides bind to the plasma membrane and suggest that the peptides enter the cells via an endocytosis-mediated mechanism. To distinguish between active and passive mechanisms of internalization, we carried out experiments with mild prefixation of cells (see Materials and Methods). Within 10 min, NBD-hBD3, NBD-3NI, and FM4-64X localized on the plasma membrane (data not shown), where they remained for at least 2 h (Fig. 5C).
Fig 5.
Wild-type hBD3 and the new analog 3NI, as examples of all of the peptides studied, interact with and enter cells after localization on the plasma membrane. (A and B) A549 cells were treated with NBD-hBD3 or NBD-3NI and a plasma membrane/endocytic marker (FM4-64FX) for 10 min (A) and then were incubated with fresh medium for 2 h (B). (C) A549 cells were subjected to mild prefixation to block active mechanisms of internalization and were incubated with NBD-hBD3 or NBD-3NI and FM4-64FX for 2 h. Similar results were obtained with the other peptides. Scale bars, 10 μm. Images represent single 1-μm-thick optical slices. hBD3 and 3NI are green, and FM4-64FX is red; colocalizing regions appear orange-yellow in the overlay image. Three independent experiments were performed, and images were acquired from at least 100 cells per experiment.
Serum stability.
The experiments conducted to evaluate the serum stability of hBD1 and hBD3 showed that hBD1 was quite stable (80% intact even after 48 h of incubation), whereas hBD3 was susceptible to serum degradation. In fact, only 50% of the peptide was intact after 4 h and up to 24 h of incubation, and 20% was intact at 48 h. hBD3 peptide degradation started at the C terminus (-RKK), as detected by mass spectrometric analysis, which suggests that degradation is mediated by a carboxypeptidase.
DISCUSSION
Because of their antimicrobial and chemotactic activities, hBD1 and hBD3 may be exploited in the quest for new therapeutic molecules to fight infections (4, 31). The main differences between hBD1 and hBD3 lie in the N and C termini and in the internal domain between the third and fourth Cys residues (3, 8). The positively charged C-terminal region in hBD3 is packed against the internal domain, which contains two positive residues, whereas the internal domain of hBD1 contains one positive residue (8, 9). The internal domain of hBD1 and the C-terminal domain of hBD3 are fundamental for antimicrobial and chemotactic activities, and the disordered N-terminal domain of hBD3 is not necessary for this activity (9, 20). Therefore, we designed and synthesized a new molecule (3NI) that lacks the N-terminal domain of hBD3 and contains the internal domain of hBD1 between the third and fourth cysteine residues. Given the evidence that the antimicrobial properties of hBDs depend largely on the overall charge, we created the 3NI peptide considering that (i) the C-terminal domain (8 amino acids) of hBD3 has more positive charges (net charge, +6) than the C-terminal domain (5 amino acids) of hBD1 (net charge, +2), (ii) hBD1 has one lysine residue, whereas hBD3 has two glutamine and two lysine residues at the C terminus, and (iii) the charged residues in the internal domain between the third and fourth cysteine residues differ between hBD1 (net charge, +1) and hBD3 (net charge, +2). The resulting peptide, like the previously designed analogs (1C, 3I, and 3N), has a higher net positive charge than hBD1 (3NI, +12; 1C, +8; 3I, +12; 3N, +11). Given these structural and charge features, 3NI would theoretically possess greater antimicrobial power under both “physiological” and high-salt concentrations.
Our finding that 3NI exerts relevant antibacterial activity, also in the presence of high salt concentrations, against P. aeruginosa, E. coli, and E. faecalis confirms that the C terminus of hBD3 and the internal domain of hBD1 are crucial for antibacterial activity (8, 9). The antimicrobial activity of the new peptide was higher than those of wild-type hBD1 and hBD3 and comparable to those of analogs 1C, 3N, and 3I. Interestingly, 3NI maintained enhanced killing activity under high-salt conditions (up to 200 mM NaCl). Bacterial infections, particularly chronic P. aeruginosa lung infections, are the major cause of morbidity and mortality associated with cystic and noncystic fibrosis. In addition, P. aeruginosa is responsible for bacterial biofilm formation, which causes up to 80% of infections that are very difficult to treat and eradicate (13). Thus, 3NI is particularly promising for the treatment of recurrent infections in cystic fibrosis patients. The anti-HSV-1 activity of 3NI resembled that of hBD3. In fact, at 50 μM, it reduced infectivity by 99%, in accordance with HIV studies (12, 25, 31).
Given the antimicrobial efficacy of wild-type defensins and their analogs, we investigated whether these peptides exerted cytotoxic effects on the human epithelial cell lines A549, CaCo-2, and Capan-1, which normally express beta-defensins. Exposure of the cells to the wild-type and chimeric peptides for up to 48 h did not affect cell viability or apoptosis, and it was not genotoxic. A small decrease in cell viability (about 20%) and a small increase in apoptosis (2 to 31%) occurred after 72 h. However, these effects were greatest at 25 μM, which is twice as high as the concentration at which these peptides exert antibacterial effects. Interestingly, in terms of apoptosis, CaCo-2 cells were less sensitive to the peptides than were the other two cell lines, whereas Capan-1 cells were more sensitive to the peptides. It is noteworthy that the Capan-1 cell line lends itself to investigations of human pancreatic processes because its three-dimensional structure resembles that of human pancreatic ducts (32). In particular, the sensitivity of Capan-1 cells to the peptides may be due to their morphological and structural features. In fact, Capan-1 cells are organized in empty spheroids whose three-dimensional structure resembles that of human pancreatic duct cells; hence, they represent a good model for studies of Cl− and HCO3− ion exchange processes typical of the human pancreas (32). Taken together, our findings suggest that different epithelial districts respond differently to hBDs.
Confocal microscopy confirmed that these peptides are not toxic to the cell lines tested, at least not with up to 2 h of treatment. In fact, they localize on the cell surface and are internalized by the cells by an active mechanism(s). The toxicity-free internalization of defensins indicates that internalization may mediate the antiviral effect of these molecules. Further endocytosis-based assays are required to identify the pathways governing peptide internalization.
It is noteworthy that hBD1 and hBD3 are characterized by high resistance to serum degradation (80% and 50% intact peptide after 48 h, respectively). Moreover, we found that degradation starts at the C terminus and ends at the first cysteine residue, suggesting that proteolysis is probably prevented by the presence of a disulfide bond. In conclusion, our results show that the tested analogs, including the new 3NI analog, are not toxic and might have therapeutic potential due to their intrinsic structural features. Further studies are required to better understand the roles of defensins and their analogs in primary cells and animal models.
ACKNOWLEDGMENTS
This study was supported by the Regione Campania (DGRC no. 1901 del 22/12/2009), the MIUR (grant no. PS35-126/IND), the IRCCS-SDN Foundation, and the Ministero della Salute.
We thank Jean Ann Gilder (Scientific Communication srl, Naples, Italy) for revision and editing of the text, Vittorio Lucignano (CEINGE-Biotecnologie Avanzate, Naples, Italy) for assistance in designing and assembling the display items of this study, and Daniela Sarnataro for discussions concerning Fig. 5 and interpretation of the corresponding experiments.
Footnotes
Published ahead of print 28 January 2013
REFERENCES
- 1. Wiesner J, Vilcinskas A. 2010. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–464 [DOI] [PubMed] [Google Scholar]
- 2. de la Fuente-Núñez C, Korolik V, Bains M, Nguyen U, Breidenstein EB, Horsman S, Lewenza S, Burrows L, Hancock RE. 2012. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56:2696–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klüver E, Adermann K, Schulz A. 2006. Synthesis and structure—activity relationship of beta-defensins, multi-functional peptides of the immune system. J. Pept. Sci. 12:243–257 [DOI] [PubMed] [Google Scholar]
- 4. Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. 2006. Human beta-defensins. Cell. Mol. Life Sci. 63:1294–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710–720 [DOI] [PubMed] [Google Scholar]
- 6. Lehrer RI. 2004. Primate defensins. Nat. Rev. Microbiol. 2:727–738 [DOI] [PubMed] [Google Scholar]
- 7. Sahl HG, Pag U, Bonness S, Wagner S, Antcheva N, Tossi A. 2005. Mammalian defensins: structures and mechanism of antibiotic activity. J. Leukoc. Biol. 77:466–475 [DOI] [PubMed] [Google Scholar]
- 8. Krishnakumari V, Singh S, Nagaraj R. 2006. Antibacterial activities of synthetic peptides corresponding to the carboxy-terminal region of human beta-defensins 1–3. Peptides 27:2607–2613 [DOI] [PubMed] [Google Scholar]
- 9. Scudiero O, Galdiero S, Cantisani M, Di Noto R, Vitiello M, Galdiero M, Naclerio G, Cassiman JJ, Pedone C, Castaldo G, Salvatore F. 2010. Novel synthetic, salt-resistant analogs of human beta-defensins 1 and 3 endowed with enhanced antimicrobial activity. Antimicrob. Agents Chemother. 54:2312–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Buchner J, Schaller M, Stange EF, Wehkamp J. 2011. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 469:419–423 [DOI] [PubMed] [Google Scholar]
- 11. Chandrababu KB, Ho B, Yang D. 2009. Structure, dynamics, and activity of an all-cysteine mutated human β defensin-3 peptide analogue. Biochemistry 48:6052–6061 [DOI] [PubMed] [Google Scholar]
- 12. Cole AM, Lehrer RI. 2003. Minidefensins: antimicrobial peptides with activity against HIV-1. Curr. Pharm. Des. 9:1463–1473 [DOI] [PubMed] [Google Scholar]
- 13. Dartois V, Sanchez-Quesada J, Cabezas E, Chi E, Dubbelde C, Dunn C, Granja J, Gritzen C, Weinberger D, Ghadiri MR, Parr TR., Jr 2005. Systemic antibacterial activity of novel synthetic cyclic peptides. Antimicrob. Agents Chemother. 49:3302–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoover DM, Wu Z, Tucker K, Lu W, Lubkowski J. 2003. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob. Agents Chemother. 47:2804–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jung S, Mysliwy J, Spudy B, Lorenzen I, Reiss K, Gelhaus C, Podschun R, Leippe M, Grötzinger J. 2011. Human beta-defensin 2 and beta-defensin 3 chimeric peptides reveal the structural basis of the pathogen specificity of their parent molecules. Antimicrob. Agents Chemother. 55:954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klüver E, Schulz-Maronde S, Scheid S, Meyer B, Forssmann WG, Adermann K. 2005. Structure-activity relation of human beta-defensin 3: influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry 44:9804–9816 [DOI] [PubMed] [Google Scholar]
- 17. Krishnakumari V, Rangaraj N, Nagaraj R. 2009. Antifungal activity of human beta defensins HBD-1 to HBD-3 and their C-terminal analogs Phd1-3. Antimicrob. Agents Chemother. 53:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu S, Zhou L, Chen L, Dastidar SG, Verma C, Li J, Tan D, Beuerman R. 2009. Effect of structural parameters of peptides on dimer formation and highly oxidized side products in the oxidation of thiols of linear analogs of human beta-defensin 3 by DMSO. J. Pept. Sci. 15:95–106 [DOI] [PubMed] [Google Scholar]
- 19. Taylor K, McCullough B, Clarke DJ, Langley RJ, Pechenick T, Hill A, Campopiano DJ, Barran PE, Dorin JR, Govan JR. 2007. Covalent dimer species of beta-defensin Defr1 display potent antimicrobial activity against multidrug-resistant bacterial pathogens. Antimicrob. Agents Chemother. 51:1719–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taylor K, Clarke DJ, McCullough B, Chin W, Seo E, Yang D, Oppenheim J, Uhrin D, Govan JR, Campopiano DJ, MacMillan D, Barran P, Dorin JR. 2008. Analysis and separation of residues important for the chemoattractant and antimicrobial activities of beta-defensin 3. J. Biol. Chem. 283:6631–6639 [DOI] [PubMed] [Google Scholar]
- 21. Wu Z, Hoover DM, Yang D, Boulègue C, Santamaria F, Oppenheim JJ, Lubkowski J, Lu W. 2003. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc. Natl. Acad. Sci. U. S. A. 100:8880–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. 1997. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553–560 [DOI] [PubMed] [Google Scholar]
- 23. Hoover DM, Chertov O, Lubkowski J. 2001. The structure of human beta-defensin-1: new insights into structural properties of beta-defensins. J. Biol. Chem. 276:39021–39026 [DOI] [PubMed] [Google Scholar]
- 24. Lehrer RI. 2011. Peptide gets in shape for self-defence. Nature 469:309–310 [DOI] [PubMed] [Google Scholar]
- 25. Conejo-Garcia JR, Jaumann F, Schulz S, Krause A, Rodríguez-Jiménez J, Forssmann U, Adermann K, Klüver E, Vogelmeier C, Becker D, Hedrich R, Forssmann WG, Bals R. 2001. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Cell Tissue Res. 306:257–264 [DOI] [PubMed] [Google Scholar]
- 26. Harder J, Bartels J, Christophers E, Schroder JM. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707–5713 [DOI] [PubMed] [Google Scholar]
- 27. Maisetta G, Batoni G, Esin S, Florio W, Bottai D, Favilli F, Campa M. 2006. In vitro bactericidal activity of human beta-defensin 3 against multidrug-resistant nosocomial strains. Antimicrob. Agents Chemother. 50:806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sass V, Schneider T, Wilmes M, Körner C, Tossi A, Novikova N, Shamova O, Sahl HG. 2010. Human β-defensin 3 inhibits cell wall biosynthesis in staphylococci. Infect. Immun. 78:2793–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rapaport D, Shai Y. 1991. Interaction of fluorescently labelled pardaxin and its analogues with lipid bilayers. J. Biol. Chem. 266:23769–23775 [PubMed] [Google Scholar]
- 30. Antcheva N, Morgera F, Creatti L, Vaccari L, Pag U, Pacor S, Shai Y, Sahl HG, Tossi A. 2009. Artificial beta-defensin based on a minimal defensin template. Biochem. J. 421:435–447 [DOI] [PubMed] [Google Scholar]
- 31. Weinberg A, Quiñones-Mateu ME, Lederman MM. 2006. Role of human beta-defensins in HIV infection. Adv. Dent. Res. 19:42–48 [DOI] [PubMed] [Google Scholar]
- 32. Fanjul M, Hollande E. 1993. Morphogenesis of “duct-like” structures in three-dimensional cultures of human cancerous pancreatic duct cells (Capan-1). In Vitro Cell Dev. Biol. Anim. 29A:574–584 [DOI] [PubMed] [Google Scholar]