Abstract
Although the genus Enterovirus contains many important human pathogens, there is no licensed drug for either the treatment or the prophylaxis of enterovirus infections. We report that fluoxetine (Prozac)—a selective serotonin reuptake inhibitor—inhibits the replication of human enterovirus B (HEV-B) and HEV-D but does not affect the replication of HEV-A and HEV-C or human rhinovirus A or B. We show that fluoxetine interferes with viral RNA replication, and we identified viral protein 2C as the target of this compound.
TEXT
The genus Enterovirus (family Picornaviridae) includes many medically and socioeconomically important human pathogens (e.g., poliovirus, coxsackievirus, echovirus, rhinovirus, and enterovirus 71 [EV71]). These can cause a wide range of diseases, including acute flaccid paralysis, aseptic meningitis, respiratory infections, viral myocarditis, fulminant pancreatitis, hand-foot-and-mouth disease, and conjunctivitis. Though often self-limiting, these infections can result in severe complications that are fatal in some cases. No approved antiviral therapeutics currently exist, and treatment remains limited to supportive care.
Enteroviruses are small, nonenveloped viruses. The positive-sense, single-stranded RNA genome of around 7.6 kb encodes a single large polyprotein that is autocatalytically processed into seven nonstructural (2A to 2C and 3A to 3D) and four structural (VP1 to VP4) proteins.
To identify new inhibitors of enteroviruses, we screened the 281-compound-containing NIH Clinical Collection 2 library (National Institutes of Health) by using a Renilla luciferase-expressing coxsackie B3 virus (CVB3-Rluc) (1). BGM cells were incubated with the different molecules (10 μM) and infected with CVB3-Rluc. Luciferase activity was analyzed with the Renilla-Glo Luciferase Assay System (Promega) at 6 h postinfection (p.i.), a time point at which replication is maximal. This assay allows the rapid and cost-efficient antiviral screening of compound libraries and allows the detection of inhibitors of viral attachment/entry, translation, or replication but not assembly/egress. In parallel, potential cytotoxic effects of the drugs were assessed with an 3-(4,5dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium-based assay (CellTiter 96 AQueous One Solution Cell Proliferation Assay; Promega). Compounds were selected for further study if they reduced luciferase activity by more than 10-fold, had little or no cytotoxicity, and displayed reproducible activity when purchased from another vendor. Using these criteria, we identified fluoxetine HCl, a selective serotonin reuptake inhibitor (brand name, Prozac) that is used for the treatment of major depression and anxiety disorders (MedlinePlus drug information, http://www.nlm.nih.gov/medlineplus/druginfo/meds/a689006.html), as an inhibitor of in vitro CVB3 replication. Fluoxetine HCl was recently also identified as inhibitor of CVB3 in an independent screening (2).
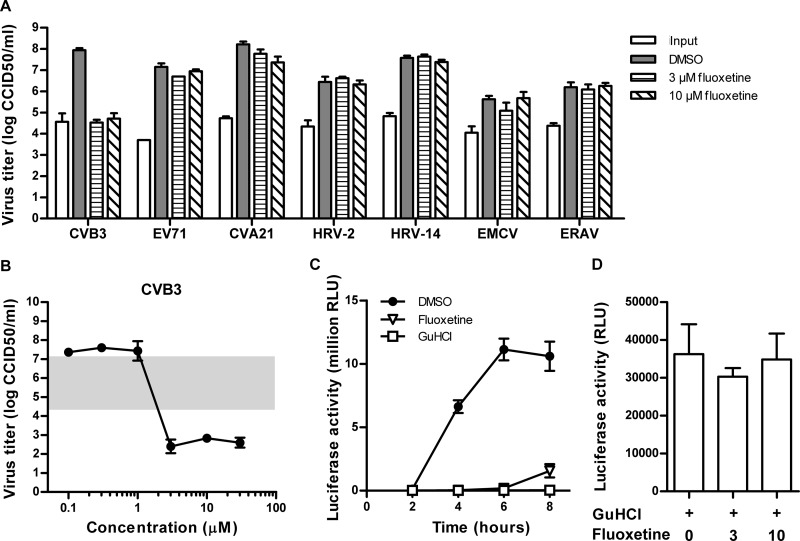
To evaluate the activity of fluoxetine in more detail, we performed a cytopathic-effect (CPE)-based multicycle antiviral assay (3). The compound inhibited the CPE of CVB3 on Vero cells with a 50% effective concentration (EC50) of 3.36 ± 0.47 μM (50% cytotoxic concentration [CC50], 28 μM) (Table 1). Antiviral activity was confirmed on CVB3-infected HeLa R19 and BGM cells (data not shown). CVB3 is a member of the species HEV-B. Fluoxetine proved also active against the other HEV-Bs tested, echovirus 1 (E-1), E-9, and E-11, as well as against members of the species HEV-D, EV68, and EV70. However, no activity against members of other enterovirus species, i.e., HEV-A and HEV-C and human rhinovirus A (HRV-A) or HRV-B, was detected. To confirm this antiviral activity further, the effect of fluoxetine in a single cycle of viral growth was assessed. BGM or HeLa R19 cells were infected at a multiplicity of infection (MOI) of 4 and then incubated with or without the compound for 8 h at 37°C. Viruses were then collected by freeze-thawing, and titers were determined by endpoint titration (4). In the presence of 3 or 10 μM fluoxetine, the titers of CVB3 were reduced by more than 3 log10 units (Fig. 1A). Consistent with the CPE-based antiviral assay, no inhibitory effect on members of the HEV-A or HRV species was observed. Fluoxetine proved also inactive against more distantly related picornaviruses, i.e., encephalomyocarditis virus (EMCV; genus Cardiovirus) and equine rhinitis A virus (ERAV; genus Aphthovirus). We then tested the effect of fluoxetine on CVB3 replication in a single-cycle assay over a range of concentrations. An EC50 of 1.23 ± 0.38 μM was calculated (Fig. 1B), which is comparable to the effect observed in the multicycle assay.
Table 1.
Antiviral and cytotoxic activitiesa of fluoxetine against various enteroviruses
| Virus | Species | Cell line | EC50 (μM) | CC50 (μM) |
|---|---|---|---|---|
| EV71 | HEV-A | RD | >40 | >40 |
| CVA16 | HEV-A | HeLa Rh | >40 | >40 |
| CVB3 | HEV-B | Vero | 3.36 ± 0.47 | 28 |
| E-1 | HEV-B | BGM | 3.93 ± 0.35 | 28 |
| E-9 | HEV-B | BGM | 4.58 ± 0.09 | 28 |
| E-11 | HEV-B | BGM | 5.76 ± 0.22 | 28 |
| PV1 | HEV-C | BGM | >28 | 28 |
| PV2 | HEV-C | BGM | >28 | 28 |
| PV3 | HEV-C | BGM | >28 | 28 |
| EV68 | HEV-D | HeLa R19 | 1.35 ± 0.08 | 29 |
| EV70 | HEV-D | HeLa R19 | 0.81 ± 0.06 | 29 |
| HRV-2 | HRV-A | HeLa Rh | >40 | >40 |
| HRV-14 | HRV-B | HeLa Rh | >40 | >40 |
Data are mean values ± standard deviations of at least three independent experiments.
Fig 1.
Effects of fluoxetine on selected picornaviruses in a single-cycle assay. (A) Single-cycle assay of the effects of 10 μM fluoxetine on the indicated viruses. Briefly, cells were infected with the indicated viruses at an MOI of 4 CCID50 per cell and treated with fluoxetine or mock treated. Viruses were harvested at 8 h p.i., and virus titers were determined by endpoint titration. Assays were performed in triplicate, and means and standard deviations were calculated. (B) Single-cycle assay of the effect of fluoxetine on CVB3 over a range of concentrations. The shaded area shows the increase in the virus titer after 8 h, where the lower edge and upper edge indicate the titers of the untreated controls at 0 and 8 h p.i., respectively. Assays were performed in triplicate, and means and standard deviations were calculated. (C) BGM cells were transfected with in vitro-transcribed CVB3-Luc replicon RNA and treated with the respective compounds. In this replicon, the coding region of the structural proteins has been replaced with that of firefly luciferase. The well-characterized enterovirus inhibitor GuHCl was used at 2 mM as a positive control. At the indicated times posttransfection, firefly luciferase activity was determined (displayed in relative light units [RLU]). Assays were performed in triplicate, and means and standard deviations were calculated. (D) Fluoxetine does not affect firefly luciferase activity. BGM cells were transfected with CVB3-Luc replicon RNA and treated with the replication inhibitor GuHCl and 3 or 10 μM fluoxetine or 0.1% DMSO. Firefly luciferase activity was analyzed 8 h after transfection.
To gain insight into the mode of action of fluoxetine, we tested its effect on the replication of CVB3 RNA with a firefly luciferase-expressing CVB3 replicon (5). In this replicon, firefly luciferase is encoded in the modified polyprotein in place of the structural proteins, and thus, luciferase levels correspond to the amount of genomic RNA that is synthesized. Fluoxetine significantly reduced luciferase activity compared to that of the control (Fig. 1C), indicating that it inhibits RNA replication. In fact, at a concentration of 3 μM, the antiviral activity of fluoxetine was comparable to that of the well-established enterovirus RNA replication inhibitor guanidine hydrochloride (GuHCl). No change in luciferase counts was observed in fluoxetine-treated controls in which RNA replication had been inhibited by the addition of GuHCl, confirming that the observed reduction of firefly luciferase activity was not due to an effect of the compound on translation or luciferase enzymatic activity (Fig. 1D).
To test whether the observed inhibition of RNA synthesis is a result of inhibition of polyprotein processing, we labeled CVB3-infected cells with [35S]methionine and incubated them with fluoxetine between 4.5 and 5 h p.i., a time when replication is in the logarithmic phase and host protein synthesis shutoff has occurred. No change in viral polyprotein processing was observed (data not shown). We therefore conclude that fluoxetine inhibits viral RNA replication but that this inhibition is not due to an effect on polyprotein processing.
When comparing the chemical structure of fluoxetine with that of known inhibitors of enterovirus RNA replication, we noted a certain degree of similarity to TBZE-029 {1-(2,6-difluorophenyl)-6-(trifluoromethyl)-1,3-dihydro-[1,3]thiazolo[3,4-a]benzimidazole}, a viral protein 2C-targeting molecule (Fig. 2A). Saliently, TBZE-029 inhibits enteroviruses belonging to the species HEV-B and HEV-D, but not those belonging to the species HEV-A and HEV-C (3; A. M. De Palma, unpublished data). To study whether mutations in 2C that confer resistance to TBZE-029 confer cross-resistance to fluoxetine, BGM cells were transfected with in vitro-transcribed full-length genomic RNA of (i) wild-type (WT) CVB3 or the (ii) 2C-A224V,I227V,A229V or (iii) 3A-H57Y mutant virus (as a control) (6, 7). Following transfection, cells were treated with 3 μM fluoxetine or mock treated. At 8 h p.i., cells were lysed and virus titers were determined by endpoint dilution assay. In the presence of fluoxetine, the 2C mutant virus replicated to titers similar to those of the control samples, whereas WT CVB3 titers were markedly decreased (>4 log10 units) in the presence of fluoxetine (Fig. 2C). Also, the replication of the 3A-H57Y mutant virus was efficiently inhibited. Thus, amino acid substitutions in protein 2C confer resistance to the compound.
Fig 2.
Fluoxetine inhibits CVB3 by acting on viral protein 2C. (A) Structural formulae of fluoxetine and TBZE-029. (B) Schematic of the organization of the genome of CVB3 (top) and of protein 2C (bottom). The locations of conserved ATPase motifs A, B, and C; active-site residue K135; and the residues substituted in the TBZE-029-resistant virus (A224V, I227V, and A229V) are indicated. Amino acids are numbered according to their positions in protein 2C. (C) BGM cells were transfected with in vitro-transcribed full-length genomic RNA of WT CVB3 or the 2C_A224V,I227V,A229V or 3A_H57Y mutant (mut) virus and treated with 3 μM fluoxetine or 0.1% DMSO. At 8 h p.i., cells were lysed and the virus titer was determined by endpoint titration. Titers are displayed as log CCID50 per milliliter, and means and standard deviations were calculated from three replicates. (D) Analysis of the effect of fluoxetine on the ATPase activity of protein 2C with Pi ColorLock Gold (Innova Biosiences) in a reaction mixture containing 3 μM recombinant protein, 1 mM ATP, 10 mM HEPES (pH 7.5), 150 mM NaCl, and 10 mM MgCl2. Where indicated, 25 μM fluoxetine was added. Phosphate release was measured through the determination of absorption at 635 nm at the indicated time points. The background of spontaneous phosphate release was subtracted. (E) Assay performed essentially as described for panel D, except that the concentration of fluoxetine was varied as indicated. Absorption was measured after 30 min.
The precise function of protein 2C in the viral life cycle is unknown. Protein 2C has been shown to bind to membranes and viral RNA (8–12) and to interact with several viral and host proteins (13–17) and has been implicated in RNA replication, viral rearrangement of intracellular membranes, and genome encapsidation (12, 14, 18–20). Protein 2C is highly conserved among enteroviruses. In particular, it contains three highly conserved motifs typical of AAA+ translocases, i.e., proteins that use the energy of ATP hydrolysis to translocate along other molecules (Fig. 2B) (21). Congruously, ATPase activity has been confirmed for the 2C proteins of several enteroviruses in vitro (6, 22–27). The substitutions that confer resistance to fluoxetine are located close to motif C, raising the possibility that this compound affects ATPase activity. To test this, we analyzed the effect of fluoxetine on the ATPase activity of a CVB3 2C recombinant protein from which the 36 N-terminal amino acids were removed (2C-Del36). The remaining part of the protein contains the ATPase domain. The sequence was cloned downstream of a cleavable thioredoxin coding sequence. The protein was purified under nondenaturing conditions in accordance with a previously described protocol (28). The mutated CVB3 protein 2C-Del36 (K135A) was included as an ATPase-negative control, as residue K135 is located in motif A and is crucial for ATPase activity (24). At 25 μM, nearly 10 times the physiological concentration needed for full inhibition of virus replication, phosphate release was reduced to only 50% of that of the dimethyl sulfoxide (DMSO)-treated control (Fig. 2D). Indeed, at 3 μM, the concentration needed for full inhibition of virus replication in a single-cycle assay, ATPase activity was reduced by only 15% (Fig. 2E). Even at the highest concentration tested, 500 μM, ATPase activity was not reduced by more than 50%. Thus, while some effect of fluoxetine on ATPase activity could be observed, it appears unlikely that, as for TBZE-029 (6), this is the major mechanism of action of this compound.
The effect of various 2C-targeting compounds on 2C's ATPase activity remains controversial. For example, GuHCl was reported to inhibit the ATPase activity of protein 2C of poliovirus type 1 in one study (25), whereas it failed to elicit an effect in another (26). For other 2C-targeting inhibitors, i.e., compounds to which resistance was provided by mutations in the ATPase domain, no effect on ATPase activity was reported (6, 23, 26). The precise molecular mechanism by which these 2C-targeting compounds, including fluoxetine, inhibit viral replication therefore remains to be established.
The amino acids in protein 2C whose substitution confers resistance to fluoxetine, i.e., A224, I227, and A229, are located in a short stretch of amino acids 224AGSINA229 located immediately C terminal of ATPase motif C. The AGSINA motif is conserved in HEV-B and HEV-D but is not present in other enteroviruses. The finding that the substitution of some of these residues confers resistance to fluoxetine suggests that they play a role in the sensitivity of these viruses to this inhibitor. To test whether the AGSINA motif sensitizes enteroviruses to TBZE-029 and fluoxetine, we introduced this motif into the poliovirus replicon. Unfortunately, this mutant construct failed to replicate. Further work, in particular, the elucidation of the structure of this protein, is required to resolve this question.
While this report was in preparation, Zuo et al. reported the inhibitory activity of fluoxetine on the RNA replication of CVB1, CVB2, and CVB3, all serotypes of the species HEV-B (27). A broader analysis of the activity against related species was not undertaken in that study. One of the authors of that paper speculated that other serotonin reuptake inhibitors might also inhibit enterovirus replication (California NanoSystems Institute webpage, http://www1.cnsi.ucla.edu/news/item?item_id=2073996). Our finding that fluoxetine selectively inhibits HEV-B and HEV-D shows that key differences between closely related enteroviruses may exist and emphasize the importance of analyzing representatives of different species belonging to this genus. On the basis of the idea that fluoxetine appears to act on a viral protein and not on a cellular host factor, we deem it unlikely that other selective serotonin reuptake inhibitors would exhibit an antiviral effect, unless they are similar in structure to fluoxetine. In fact, we did not observe any inhibitory activity of three other selective serotonin reuptake inhibitors, i.e., citalopram (EC50, >160 μM; CC50, >160 μM), paroxetine (EC50, >22.18 μM; CC50, 22.18 ± 0.41 μM), and sertraline (EC50, >17.12 μM; CC50, 17.12 ± 1.17 μM), on CVB3, which is further evidence that the antiviral effect of fluoxetine relates to its specific pharmacophore interacting with 2C and that the antiviral effect is unrelated to serotonin uptake.
Plasma fluoxetine levels in patients receiving the standard dose of 20 mg/day have been reported to be in the mid-to-high nanomolar range (29) and are thus lower than the EC50s for inhibition of enterovirus replication determined in this study. Additionally, fluoxetine, like other selective serotonin reuptake inhibitors, has been associated with side effects that might impede its use as an antiviral drug for the treatment of enterovirus infections. Thus, rather than a treatment option, in its current form, fluoxetine provides a new scaffold for the design of compounds acting on viral protein 2C. Fluoxetine showed an intriguing specificity for enterovirus species B and D, a characteristic shared with the—chemically somewhat similar—antiviral TBZE-029. Intriguingly, the TBZE-029- and fluoxetine-resistant 2C-A224V,I227V,A229V mutant virus exhibits cross-resistance to 2-(alpha-hydroxybenzyl)-benzimidazole, 1-(4-fluorophenyl)-2-[(4-imino-1,4-dihydropyridin-1-yl)methyl]benzimidazole hydrochloride, and GuHCl (6), 2C-targeting compounds that do not possess this sensitivity profile of fluoxetine and TBZE-029. Thus, while drug development is generally directed toward compounds targeting a broad range of species, we would argue that elucidation of the molecular determinant of this specificity will provide information invaluable for the rational design of—at will—narrow- or broad-range inhibitors.
ACKNOWLEDGMENTS
We acknowledge Stijn Delmotte and Caroline Collard for their excellent assistance in the acquisition of part of the antiviral data.
This work was supported by research grants from the SILVER Large Scale Collaborative Project (grant agreement 260644) of the European Union 7th Framework, the European Virus Archive (EVA) project (European FP7 Capacities Project 228292), the Convenant K. U. Leuven-Radboud University Nijmegen framework, GOA/10/014 (for work performed at K. U. Leuven), and FWO Krediet aan Navorsers no. 1.5.206.11 (A. M. de Palma). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
We have no conflicts of interest.
Footnotes
Published ahead of print 18 January 2013
REFERENCES
- 1. Lanke KHW, van der Schaar HM, Belov GA, Feng Q, Duijsings D, Jackson CL, Ehrenfeld E, van Kuppeveld FJM. 2009. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J. Virol. 83:11940–11949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zuo J, Quinn KK, Kye S, Cooper P, Damoiseaux R, Krogstad P. 2012. Fluoxetine is a potent inhibitor of coxsackievirus replication. Antimicrob. Agents Chemother. 56:4838–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Palma AM, Heggermont W, Leyssen P, Pürstinger G, Wimmer E, Clercq ED, Rao A, Monforte AM, Chimirri A, Neyts J. 2007. Anti-enterovirus activity and structure-activity relationship of a series of 2,6-dihalophenyl-substituted 1H,3H-thiazolo[3,4-a]benzimidazoles. Biochem. Biophys. Res. Commun. 353:628–632 [DOI] [PubMed] [Google Scholar]
- 4. Reed L, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 27:493–497 [Google Scholar]
- 5. Wessels E, Duijsings D, Notebaart RA, Melchers WJG, van Kuppeveld FJM. 2005. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 79:5163–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Palma AM, Heggermont W, Lanke K, Coutard B, Bergmann M, Monforte AM, Canard B, De Clercq E, Chimirri A, Pürstinger G, Rohayem J, van Kuppeveld F, Neyts J. 2008. The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif C in the nonstructural protein 2C. J. Virol. 82:4720–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsu N-Y, Ilnytska O, Belov G, Santiana M, Chen Y-H, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJM, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banerjee R, Dasgupta A. 2001. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 82:2621–2627 [DOI] [PubMed] [Google Scholar]
- 9. Echeverri A, Banerjee R, Dasgupta A. 1998. Amino-terminal region of poliovirus 2C protein is sufficient for membrane binding. Virus Res. 54:217–223 [DOI] [PubMed] [Google Scholar]
- 10. Echeverri AC, Dasgupta A. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540–553 [DOI] [PubMed] [Google Scholar]
- 11. Rodríguez PL, Carrasco L. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105–10112 [DOI] [PubMed] [Google Scholar]
- 12. Teterina NL, Gorbalenya AE, Egger D, Bienz K, Ehrenfeld E. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banerjee R, Weidman MK, Echeverri A, Kundu P, Dasgupta A. 2004. Regulation of poliovirus 3C protease by the 2C polypeptide. J. Virol. 78:9243–9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y, Wang C, Mueller S, Paul AV, Wimmer E, Jiang P. 2010. Direct interaction between two viral proteins, the nonstructural protein 2C and the capsid protein VP3, is required for enterovirus morphogenesis. PLoS Pathog. 6:e1001066 doi:10.1371/journal.ppat.1001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang W-F, Yang S-Y, Wu B-W, Jheng J-R, Chen Y-L, Shih C-H, Lin K-H, Lai H-C, Tang P, Horng J-T. 2007. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J. Biol. Chem. 282:5888–5898 [DOI] [PubMed] [Google Scholar]
- 16. Teterina NL, Gorbalenya AE, Egger D, Bienz K, Rinaudo MS, Ehrenfeld E. 2006. Testing the modularity of the N-terminal amphipathic helix conserved in picornavirus 2C proteins and hepatitis C NS5A protein. Virology 344:453–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yin J, Liu Y, Wimmer E, Paul AV. 2007. Complete protein linkage map between the P2 and P3 non-structural proteins of poliovirus. J. Gen. Virol. 88:2259–2267 [DOI] [PubMed] [Google Scholar]
- 18. Cho MW, Teterina N, Egger D, Bienz K, Ehrenfeld E. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129–145 [DOI] [PubMed] [Google Scholar]
- 19. Vance LM, Moscufo N, Chow M, Heinz BA. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang C, Jiang P, Sand C, Paul AV, Wimmer E. 2012. Alanine scanning of poliovirus 2CATPase reveals new genetic evidence that capsid protein/2CATPase interactions are essential for morphogenesis. J. Virol. 86:9964–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gorbalenya AE, Koonin EV, Wolf YI. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145–148 [DOI] [PubMed] [Google Scholar]
- 22. Adams P, Kandiah E, Effantin G, Steven AC, Ehrenfeld E. 2009. Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J. Biol. Chem. 284:22012–22021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klein M, Hadaschik D, Zimmermann H, Eggers HJ, Nelsen-Salz B. 2000. The picornavirus replication inhibitors HBB and guanidine in the echovirus-9 system: the significance of viral protein 2C. J. Gen. Virol. 81:895–901 [DOI] [PubMed] [Google Scholar]
- 24. Mirzayan C, Wimmer E. 1994. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology 199:176–187 [DOI] [PubMed] [Google Scholar]
- 25. Pfister T, Wimmer E. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992–7001 [DOI] [PubMed] [Google Scholar]
- 26. Rodríguez PL, Carrasco L. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105–8110 [PubMed] [Google Scholar]
- 27. Sweeney TR, Cisnetto V, Bose D, Bailey M, Wilson JR, Zhang X, Belsham GJ, Curry S. 2010. Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism. J. Biol. Chem. 285:24347–24359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lantez V, Dalle K, Charrel R, Baronti C, Canard B, Coutard B. 2011. Comparative production analysis of three phlebovirus nucleoproteins under denaturing or non-denaturing conditions for crystallographic studies. PLoS Negl Trop. Dis. 5:e936 doi:10.1371/journal.pntd.0000936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. LLerena A, Dorado P, Berecz R, González A, Jesús Norberto M, De la Rubia A, Cáceres M. 2003. Determination of fluoxetine and norfluoxetine in human plasma by high-performance liquid chromatography with ultraviolet detection in psychiatric patients. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 783:25–31 [DOI] [PubMed] [Google Scholar]