Abstract
Cryptosporidiosis, a diarrheal disease usually caused by Cryptosporidium parvum or Cryptosporidium hominis in humans, can result in fulminant diarrhea and death in AIDS patients and chronic infection and stunting in children. Nitazoxanide, the current standard of care, has limited efficacy in children and is no more effective than placebo in patients with advanced AIDS. Unfortunately, the lack of financial incentives and the technical difficulties associated with working with Cryptosporidium parasites have crippled efforts to develop effective treatments. In order to address these obstacles, we developed and validated (Z′ score = 0.21 to 0.47) a cell-based high-throughput assay and screened a library of drug repurposing candidates (the NIH Clinical Collections), with the hopes of identifying safe, FDA-approved drugs to treat cryptosporidiosis. Our screen yielded 21 compounds with confirmed activity against C. parvum growth at concentrations of <10 μM, many of which had well-defined mechanisms of action, making them useful tools to study basic biology in addition to being potential therapeutics. Additional work, including structure-activity relationship studies, identified the human 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) reductase inhibitor itavastatin as a potent inhibitor of C. parvum growth (50% inhibitory concentration [IC50] = 0.62 μM). Bioinformatic analysis of the Cryptosporidium genomes indicated that the parasites lack all known enzymes required for the synthesis of isoprenoid precursors. Additionally, itavastatin-induced growth inhibition of C. parvum was partially reversed by the addition of exogenous isopentenyl pyrophosphate, suggesting that itavastatin reduces Cryptosporidium growth via on-target inhibition of host HMG-CoA reductase and that the parasite is dependent on the host cell for synthesis of isoprenoid precursors.
INTRODUCTION
Cryptosporidium parvum and Cryptosporidium hominis, the most common etiologic agents of human cryptosporidiosis (1), are gastrointestinal parasites of the phylum Apicomplexa. While infection in immunocompetent hosts typically results in self-limited disease (2), cryptosporidiosis has been identified as a major cause of recurrent diarrhea in children under 5 years old which, as a whole, is estimated to cause 30 to 50% of global deaths in that demographic (3, 4). Moreover, susceptibility to cryptosporidial infection correlates with nutritional status (5), and infection early in life has been associated with persistent developmental deficits (6). The parasite is also an important opportunistic pathogen in AIDS patients, resulting in poor outcomes, including fulminant diarrhea, extraintestinal manifestations, and death. Despite its significant disease burden, the treatment options for cryptosporidiosis are extremely limited. Nitazoxanide (NTZ), the current standard of care, exhibits only moderate clinical efficacy in immunocompetent adults and children, and it is not effective in patients with advanced AIDS (7).
Two major obstacles have impeded the development of improved anticryptosporidials. First, the prohibitive cost of de novo drug development, estimated to be between $500 million and $2 billion per compound successfully brought to market (8), poses a barrier to drug development for pathogens like Cryptosporidium that disproportionately affect residents of poor countries (9). The discovery of new indications for existing medications, known as drug repurposing (or repositioning), provides an attractive alternative (10) to de novo drug development. Second, technical limitations have significantly hindered advancements in understanding Cryptosporidium biology. The lack of a widely accepted method to continuously culture Cryptosporidium species in the laboratory makes genetic manipulation impossible and limits the utility of target-based approaches for drug development. Cell-based high-throughput screening (HTS) facilitates the parallel interrogation of many host and pathogen processes, resulting in the identification of valuable drug leads as well as chemical probes that enable insight into the biology of genetically intractable microbes. Given the relatively small number of chemically diverse drugs available for repurposing, cell-based approaches are also well suited for this strategy.
To address both the financial and technical barriers of drug development for cryptosporidiosis, while simultaneously generating insights into the biology of these intracellular pathogens, we developed a cell-based HTS and used it to screen the NIH Clinical Collections (NCC) for novel C. parvum growth inhibitors. Because these libraries are comprised of FDA-approved drugs and drug-like molecules which often have well-described mechanisms of action, we reasoned that this screen could provide valuable drug leads for the treatment of cryptosporidiosis and chemical probes to study Cryptosporidium biology. Here, we describe the development and validation of the first cell-based HTS for C. parvum, identify lead repurposing candidates for the treatment of human cryptosporidiosis, and, using one of these leads, identify the host mevalonic acid (MVA) pathway and its intermediate isopentenyl pyrophosphate (IPP) as critical to parasite replication. Our screen identified the widely prescribed statin class of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors as potent inhibitors of C. parvum growth. Additional in vitro and bioinformatics data indicated that, unlike other apicomplexans, Cryptosporidium parasites are dependent on host cells for synthesis of isoprenoid precursors, providing a potential mechanism of action for these drugs. This work illustrates the dual utility of a drug repurposing campaign to identify therapeutic leads as well as tools to elucidate microbe and host biology.
MATERIALS AND METHODS
Cell culture and cryptosporidium infection.
Human ileocecal adenocarcinoma (HCT-8) cells were obtained from ATCC and maintained in T-75 tissue culture flasks with RPMI 1640 medium with HEPES, sodium pyruvate (1 mM), and l-glutamine (ATCC) supplemented with 10% horse serum (ATCC) and 120 U/ml penicillin and 120 μg/ml streptomycin. Cells were plated into 384-well, tissue culture-treated, black-walled, clear-bottom microwell plates (BD Falcon) at a density of 8,850 cells/well and allowed to grow to confluence. They were then inoculated with 5.5 × 103 primed C. parvum oocysts (Bunchgrass Farms, Deary, ID) suspended in inoculation medium (RPMI 1640 as described above without horse serum). Oocysts were primed for excystation by following a previously described protocol (11). Briefly, oocysts were treated for 10 min with 10 mM HCl at 37°C, centrifuged, and treated with a 2 mM solution of sodium taurocholate (Sigma-Aldrich) in phosphate-buffered saline (PBS) with Ca2+ and Mg2+. The suspension was incubated for 10 min at 16°C and then diluted in inoculation medium and added to each well. Infected cells were incubated at 37°C for 3 h, at which point an equal volume of growth medium containing 20% horse serum (total serum concentration of 10%) and each experimental compound (see below) was added. The infected cells were then incubated for 48 h.
Experimental compounds. (i) HTS.
The NIH Clinical Collection (NCC) libraries, provided through the National Institutes of Health Molecular Libraries Roadmap Initiative (obtained through Evotec, San Francisco, CA), were received as 10 mM compounds in dimethyl sulfoxide (DMSO). The compounds were further diluted by a factor of 1:1.4 in 100% DMSO, arrayed into the center 308 wells of V-bottom polypropylene 384-well source plates (Whatman), and stored at −80°C until use. A 384 solid pin Multi-Blot replicator (V&P Scientific) was used to transfer approximately 120 nl of the compound from the source plate to the assay plate for a final concentration of approximately 10 μM. Controls on each compound source plate included wells containing DMSO (vehicle) only and wells containing nitazoxanide at a concentration that resulted in delivery of the 90% inhibitory concentration (IC90) of nitazoxanide to ensure consistent loading across the pin transfer tool.
(ii) Secondary assays.
Compounds were recovered from the NCC source plates (confirmatory testing) or repurchased (additional follow-up) (Table 1), diluted in 100% DMSO and further diluted in growth medium with 20% serum (total DMSO concentration never exceeded 0.5%), and immediately added to infected monolayers. Cerivastatin, simvastatin, fluvastatin, mevastatin, lovastatin, and pravastatin were purchased from Sigma-Aldrich, and itavastatin was purchased from Sequoia Research Products. Compound purity was assessed using liquid chromatography-mass spectrometry (LC-MS). LC-MS mass spectra were obtained on an Agilent 6120 single quadrupole mass spectrometer with an Agilent 1200 high-pressure liquid chromatography (HPLC) system with a Zorbax Eclipse XDB-C8 reversed-phase column (4.6 by 50 mm, 3.5 μm). Samples were eluted using a linear gradient from 95:5 to 5:95 water–0.1% formic acid-acetonitrile/0.1% formic acid at a flow of 1 ml/min over a 6-min period.
Table 1.
Confirmed screening hits
| Compound name | PubChem Identifier | Classification | Supplier (catalog no.) | Puritya | IC50 in μM (95% CI)b | TC50 (μM)c | TId |
|---|---|---|---|---|---|---|---|
| Chloroxine | 2722 | Topical antibacterial | Sigma (D64600) | >85% | 0.26 (0.22–0.29) | 35 | 135 |
| Hexachlorophene | 3598 | Topical antibacterial | Sigma (H4625) | ND | 1.4 (1.1–1.7) | 90 | 64 |
| Digoxin | 30322 | Channel inhibitor | Sigma (D6003) | >85% | 0.44 (0.3–0.60) | < 5 | <11 |
| Glipizide | 3478 | Channel inhibitor | Sigma (G117) | >85% | 5.8 (5.2–6.4) | >100 | >17 |
| Oligomycin C | 93857 | Channel inhibitor | Sequoia (SRP01150o) | ND | 6.7 (4.5–10.1) | >100 | 15 |
| Indomethacin | 3715 | Cyclooxygenase inhibitor | Sigma (I8280) | >85% | 5.4 (4.9–5.9) | >100 | >19 |
| Homoharringtonine | 285033 | Experimental | Sequoia (SRP02125h) | >85% | 0.86 (0.50–1.5) | ND | ND |
| Mevastatin | 64715 | HMG-CoA reductase inhibitor | Sigma (A1882) | >85% | 5.1 (3.7–7.0) | >100 | >20 |
| Docetaxel | 148124 | Microtubule agent | Sigma (1885) | >85% | 0.13 (0.079–0.20) | >100 | >769 |
| Floxuridine | 3363 | Pyrimidine analog | Sigma (F0503) | ND | 0.0063 (0.0020–0.020) | >100 | >15,873 |
| 6-Azauridine | 5901 | Pyrimidine analog | Sigma (A1882) | >85% | 3.2 (2.9–3.6) | >100 | >31 |
| Carmofur | 2577 | Pyrimidine analog | Sigma (C1494) | >85% | 4.4 (3.5–5.6) | >100 | >23 |
| 5-Fluorouracil | 3385 | Pyrimidine analog | Sigma (F6627) | >85% | 4.9 (3.7–6.3) | >100 | >20 |
| Doxycycline | 54671203 | Tetracycline analog | Sigma (D9891) | >85% | 6.3 (5.7–6.9) | 50 | 8 |
| Minocycline hydrochloride | 54685925 | Tetracycline analog | Sigma (M9511) | >85% | 6.5 (5.0–8.5) | >100 | >15 |
| Tegaserod maleate | 6918369 | 5HT4 receptor agonist | Sequoia (SRP01187t) | >85% | 4.8 (3.9–5.8) | 24 | 5 |
ND, not determined.
IC50 indicates 50% inhibitory concentration.
TC50 indicates 50% toxic concentration.
TI, therapeutic index. TI calculated as TC50 divided by IC50.
(iii) Immunofluorescence assay.
Following incubation, monolayers were washed three times with PBS containing 111 mM d-galactose with an EL406 automated plate washer (Biotek, Winooski, VT) fixed with 4% paraformaldehyde in PBS for 15 min at room temperature and then permeabilized with 0.25% Triton X in PBS for 10 min at 37°C. The monolayers were then washed three times in PBS with 0.1% Tween 20 and blocked with 4% bovine serum albumin (BSA) in PBS for 2 h. Biotinylated Vicia villosa lectin (VVL; Vector Laboratories) and streptavidin-conjugated Alexa Flour 568 (Invitrogen) were diluted in 1% BSA-PBS with 0.1% Tween 20 (optimal concentrations were determined empirically for each application), added to the wells, and allowed to incubate for 1 h. Fifteen microliters of 0.29 mM Hoechst 33258 (Anaspec) in water was added, and the plates were incubated for an additional 15 min.
The monolayers were then washed 5 times and imaged utilizing a Nikon Eclipse Ti2000 epifluorescence microscope with motorized stage. Images were captured with an EXi blue fluorescent microscopy camera (QImaging, Surrey, British Columbia, Canada). NIS-Elements Advanced Research software (Nikon USA, Melville, NY) was used to program the instrument to automatically focus on the center of each of the middle 308 wells of the plate and then acquire a three-by-three 20× field image around the focal point (corresponding to approximately 13% of the surface area of the well). The images were exported as .tif files and imported into ImageJ (National Institutes of Health) where the batch process function was used to execute a macro to segment and enumerate the Hoechst-stained nuclei (appendix A) and streptavidin-Alexa Flour 568-labeled parasites (appendix B).
(iv) Data handling and statistical analysis.
The ImageJ output was imported into Microsoft Excel for data organization and analysis. Additional statistical analysis and construction of graphs were conducted in GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA).
(v) HTS.
The robustness of the HTS assay was determined by calculating Z′ scores as described previously (12) using equation 1.
| (1) |
The percent inhibition was calculated for each experimental compound (equation 2), and compounds that exhibited ≥80% in one of two biological replicates were recovered from source plates for confirmation.
| (2) |
(vi) Confirmatory IC50 curves.
Compounds recovered from screening plates were diluted and assayed at fixed doses of 0.12, 0.37, 1.1, 3.3, and 10 μM (each concentration, n = 14) for the generation of IC50 curves. Repurchased compounds were diluted to 0.03×, 0.11×, 0.33×, 0.8×, 0.9×, 1.0×, 1.1×, 3×, 9×, and 30× (where possible) the confirmatory IC50 in order to generate final IC50 curves. In the case of final IC50 curves, three wells were left uninfected but treated with each of the corresponding concentrations of the compound to assess for background staining. In experiments utilizing isopentenyl pyrophosphate (IPP), IPP was obtained as the dry trisammonium salt (Isoprenoids, LC, Tampa, FL), reconstituted in inoculation medium at 8.12 mM, and then further diluted into assay plates to the indicated concentration. All curves were generated using the log[inhibitor] versus response − variable slope equation in GraphPad Prism (equation 3), with the bottom constraint set equal to 0.
| (3) |
Cell toxicity assays. (i) Toxicity in confluent monolayers.
HCT-8 cells were grown to confluence in 384-well plates as described above. Growth medium was then replaced with growth medium spiked with 5 μM, 10 μM, 20 μM, 50 μM, or 100 μM (replicates per concentration, n = 6) of the experimental compound, and plates were incubated for 48 h. Following incubation, the outer 76 wells of the plate were treated with trypsin (blank wells), and the medium was replaced with 20 μl of fresh growth medium. The CellTiter AQueous assay (Promega) was then used according to the manufacturer's instructions in order to assess cell viability. Briefly, 5 μl of a solution of the tetrazolium compound MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)2-(4-sulfophenyl)-2H-tetrazolium] and the electron coupling agent phenazine methosulfate was added to each well and plates were incubated for 70 min, at which point absorbance was read at 490 nm with a PowerWave microtiter plate reader (Biotek). The average absorbance of all blank wells was subtracted from the absorbance of experimental wells, and the 50% toxic concentration (TC50) doses were calculated in GraphPad Prism (equation 3), with the bottom constraint set equal to 0.
(ii) Bioinformatic analysis.
Evidence for the presence or absence of Cryptosporidium metabolic pathways was collected using BLAST on 1 November 2012; all alignments are based on the amino acid sequence using default parameters (13). A number of representative proteins were chosen from each pathway using KEGG (14) and ApiCyc annotation (15). Plasmodium falciparum sequences were used for methylerythritol phosphate pathway analysis and Homo sapiens sequences were used for all other pathways. A full list of search candidates and relevant E values is provided in Table S3 in the supplemental material.
RESULTS
A sensitive and specific screening assay for C. parvum growth.
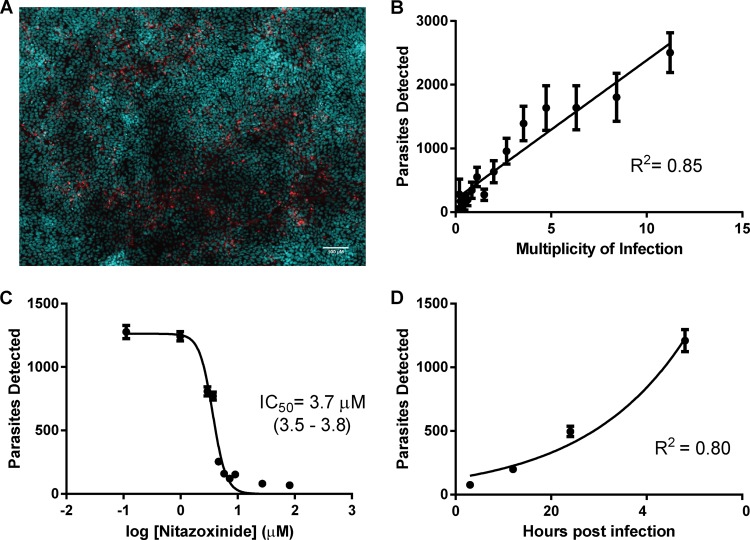
We adapted existing in vitro models of Cryptosporidium infection and growth (11, 16) to develop a robust, sensitive, and specific cell-based assay in a 384-well microtiter plate format. Briefly, we induced excystation of C. parvum oocysts and inoculated them onto a confluent monolayer of ileocecal adenocarcinoma (HCT-8) cells and then incubated them for 48 h. After staining of host cell nuclei and parasites, large (three-by-three tiled) low-magnification images were acquired using automated epifluorescence microscopy (Fig. 1A; host cells, cyan; parasites, red), and parasites and host cell nuclei were enumerated with ImageJ utilizing macros developed in-house (see Fig. S1 in the supplemental material and the appendix). This workflow provided a straightforward means to detect anticryptosporidial activity and potential host cell toxicity of experimental compounds.
Fig 1.
A sensitive and specific assay to detect C. parvum growth in intestinal epithelial cells. (A) Representative merged image of host cell nuclei stained with Hoechst (cyan) and parasites labeled with biotinylated VVL and streptavidin-Alexa Fluor 568 secondary (red). The image is a three-by-three tiled composite of low-magnification fields that corresponds to the middle ≈13% of the plate well surface area. The scale bar represents 100 μm. (B) Primed oocysts were inoculated onto confluent HCT-8 cell monolayers at various multiplicities of infection, incubated for 48 h, and then fixed, stained, and imaged. Each data point is the mean of 16 technical replicates. The solid line represents a linear regression (R2 = 0.85), demonstrating good correlation between the number of parasites detected and the multiplicity of infection (MOI). (C) Primed oocysts were inoculated onto confluent monolayers and then treated with various concentrations of nitazoxanide at 3 h following infection. The half-maximal inhibitory concentration (IC50) for nitazoxanide was determined to be 3.7 μM (95% CI = 3.5 to 3.8 μM) (n = 12 for log [nitazoxanide] = 1.9 and n = 24 for all other data points). (D) Primed oocysts were inoculated onto confluent monolayers and then fixed at 3 (n = 33), 12 (n = 21), 24 (n = 22), and 48 (n = 21) h, stained, imaged, and analyzed. Panels B to D are representative of at least two biological replicates, and error bars represent standard errors of the means.
The number of C. parvum oocysts inoculated onto a confluent monolayer correlated strongly with the number of parasites detected following a 48-h incubation period (R2 = 0.85) (Fig. 1B). The half-maximal inhibitory concentration (IC50) of nitazoxanide (3.7 μM; 95% confidence interval [CI] = 3.5 to 3.8) (Fig. 1C) determined by this assay was in agreement with previously published findings utilizing quantitative reverse transcription-PCR (qRT-PCR) (17) and microscopy (18) to detect C. parvum growth in vitro. Moreover, C. parvum growth kinetics observed in our assay matched those previously described for in vitro culture systems (19). Despite low infection efficiency (the number of intracellular parasites detected at 3 h is approximately one log less than in the original inoculum), we observed exponential parasitic growth in infected wells that were fixed and stained at 3, 12, 24, and 48 h following infection (R2 = 0.80) with a calculated doubling time between 15 and 17 h (Fig. 1D). These data illustrate the sensitivity and specificity of this miniaturized, semiautomated assay to measure C. parvum growth in vitro.
Assay is sufficiently robust for high-throughput screening applications.
We sought to determine our ability to specifically identify infected wells treated with a known C. parvum inhibitor from a larger group of infected, untreated wells in order to evaluate the utility of the assay in a higher-throughput context. The assay readily distinguished infected wells that were randomly selected and treated with an inhibitory concentration of nitazoxanide (41 μM) (n = 17) 3 h following infection from those that were treated with the DMSO vehicle only (n = 269) (Fig. 2A). We then employed the screening window coefficient known as the Z score to assess the robustness of the assay as an HTS. A Z score of ≥0.5 is considered ideal for high-throughput applications; however, successful screens that employed assays with Z scores as low as 0.2 have been reported. The Z′ score, a permutation of the Z score in which only control data are considered, assesses the quality of the assay itself (12). We calculated the Z′ score for our assay for three biological replicates (performed on different days) and found it to be between 0.21 and 0.47 in the abstract (Fig. 2B). These data, combined with a workflow capable of testing >1,700 compounds per week in duplicate, suggest that our assay is well suited to conduct high-throughput screening.
Fig 2.
Assay is sufficiently robust for high-throughput applications. (A) Primed oocysts were inoculated onto confluent monolayers (n = 286) and 96 were left uninfected (gray triangles). Seventeen randomly located infected wells were treated with an inhibitory concentration of nitazoxanide (41 μM; open diamonds), while the remaining 269 wells were treated with DMSO vehicle only (filled circles). Dashed lines represent the separation band (see reference 12), the upper bound of which is equal to μDMSO-treated wells − 3σDMSO-treated wells and the lower bound of which is equal to μuninfected wells + 3σuninfected wells. (B) Confluent monolayers were inoculated with primed oocysts (n = 154 per plate; filled circles) or left uninfected (n = 152 for plate 1 and n = 154 for plates 2 and 3; open triangles) and incubated for 48 h, at which point cells were processed as described in Materials and Methods, and Z′ scores (equation 1, Materials and Methods) were calculated for each biological replicate. The signal band is shown for each plate. Vertical dotted lines separate the data from each of three experiments which were performed on separate days.
Screening of a compound library.
After validating the utility of our assay as an HTS, we conducted a screen of the NIH Clinical Collections libraries, a collection of 727 FDA-approved drugs or drug-like compounds with a history of use in human clinical trials. Each compound was screened at approximately 10 μM in two separate biological replicates (performed on separate days) with good agreement between screening replicates. Figure 3 shows screening results as a scatter plot (Fig. 3A) and as a frequency distribution of percent inhibition by each compound (Fig. 3B) for screening replicate number one. Compound names, simplified molecular input line entry specification (SMILES) strings, PubChem identification numbers, supplier information, compound purity (when available, provided by supplier), and percent inhibition for both screening replicates are presented in Table S1 in the supplemental material for each of the compounds in the library.
Fig 3.
Screening results. The results from screening the 727-compound NIH Clinical Collection libraries are shown for the first screening replicate as a scatter plot (A) and a frequency distribution (B). The solid horizontal lines in panel A represent the hit limit (80% inhibition) for each plate screened. The data are divided into their corresponding assay plates by the dashed vertical lines.
Compounds that exhibited ≥80% inhibition in at least one replicate were selected for follow-up testing. Twenty-four compounds met this definition, and these were recovered from the original library source plates and assessed for purity with LC-MS, and IC50 curves were generated by testing each drug in our growth assay at 5 different concentrations (0.12, 0.37, 1.1, 3.3, and 10 μM; n = 14 wells for each dose). Nineteen compounds had confirmatory IC50s of <10 μM, 2 compounds were inhibitory at all doses tested, and 3 compounds did not show any inhibition (see Table S2 in the supplemental material). Overall, the assay exhibited a 3.3% hit rate with a true positive rate of 87.5% when a hit limit of 80% inhibition was applied.
Secondary assays. (i) Hit prioritization and confirmation.
The ideal drug candidate to repurpose for the treatment of cryptosporidiosis would be safe enough for administration in a pediatric population, exhibit potent and selective toxicity against Cryptosporidium parasites, be formulated for oral administration, be available at low cost, and have FDA approval. We used this product profile as a framework to inform further prioritization of candidate compounds based on our preliminary data as well as information compiled from the Drugbank database (20), ChEMBL database (21), additional literature sources, and clinical experience. Additionally, careful review of our preliminary screening data revealed that we had identified potent inhibitors of C. parvum with well-characterized mechanisms of action that may prove useful as chemical probes to help elucidate basic parasite and host cell biology. Based on these criteria, we identified 16 compounds for additional follow-up which were repurchased, determined to be ≥85% pure by LC-MS (with the exception of tetracycline [purity, <50%] and hexachlorophene, oligomycin C, and floxuridine, which could not be tested by LC-MS), and used to generate final IC50 curves (see Fig. S2 in the supplemental material). Candidate compounds that did not demonstrate host cell toxicity as evidenced by a significant decrease in nuclear counts were further assessed for potential toxicity using a commercially available assay of cellular dehydrogenase activity (CellTiter AQueous; Promega). These findings are summarized in Table 1.
(ii) Additional follow-up.
We further prioritized the compounds in Table 1 with respect to our product profile. Our screen identified representative chemical scaffolds from three chemical families that we considered to have therapeutic potential against cryptosporidiosis. Floxuridine, a pyrimidine analog nucleoside, was the most potent candidate compound (IC50 = 0.0063 μM [95% CI = 0.0020 to 0.020]), while carmofur and 5-fluorouracil (analogs of floxuridine), as well as 6-azauridine (another pyrimidine analog nucleoside), exhibited potency in the single micromolar range (Table 1; see also Fig. S2 in the supplemental material), suggesting that pyrimidine analogs may be a rich source of anticryptosporidial compounds. A previous study demonstrated the ability of C. parvum thymidine kinase to activate pyrimidine analogs including floxuridine. Additionally, trifluoromethyl thymidine (TFT; another fluoropyrimidine) exhibited efficacy in a mouse model of acute C. parvum infection (22). Furthermore, widespread use of nucleoside analogs in the prevention of mother-to-child transmission of HIV (23) suggests that certain formulations of the chemical family are safe for use in pediatric populations. The screen also identified two tetracycline analogs (doxycycline and minocycline) (Table 1; see also Fig. S2 in the supplemental material), a family of drugs with demonstrated activity against other apicomplexans (24–26). Since tetracyclines are believed to act directly on the apicoplast, which Cryptosporidium has lost (27, 28), our findings raise the possibility that these drugs may also have non-apicoplast-dependent mechanisms of action against other Apicomplexa.
Finally, our results revealed that mevastatin, the prototypical HMG-CoA reductase inhibitor (statin), has single micromolar efficacy in vitro against C. parvum. The statins fit well into our product profile, and numerous clinically utilized chemical analogs with excellent safety profiles already exist. Consequently, while the pyrimidine analogs and tetracyclines may have potential as therapeutics, we chose to focus on the chemical scaffold represented by mevastatin.
(iii) HMG-CoA reductase inhibitors.
Confirmatory testing with mevastatin demonstrated inhibition of C. parvum growth in vitro, with an IC50 of 5.1 μM (95% CI = 3.7 to 7.0), a TC50 of >100 μM, and a therapeutic index (TI) of >20. Additionally, a number of other statins in the NCC library came close to, but failed to meet, the hit limit of 80% inhibition (see Table S1 in the supplemental material). These data, combined with the outstanding safety profile of many of the clinically utilized statins in the general population (29), prompted us to conduct structure-activity relationship (SAR) studies utilizing additional statin drugs. Analog compounds were purchased, determined to have ≥85% purity by LC-MS, and used to determine the IC50 against C. parvum and the TC50 on confluent HCT-8 monolayers (Fig. 4). Cerivastatin (IC50 = 0.27 μM [95% CI = 0.21 to 0.36], TC50 > 100, TI > 250) was the most potent statin derivative tested. However, the drug was withdrawn from the market in August 2001 (30), making it a poor repurposing candidate. As a result, we selected itavastatin (also known as pitavastatin) (IC50 = 0.62 μM [95% CI = 0.45 to 0.86], TC50 >100, TI > 167) for additional follow-up studies.
Fig 4.
Lead optimization and structure-activity relationships of HMG-CoA reductase inhibitors. Mevastatin and six additional statin analogs were purchased and used to determine structure-activity relationships. The compounds were repurchased, assessed for purity, and used to generate refined IC50 curves. Each data point is the average of 11 technical replicates (except for the highest concentrations of pravastatin, fluvastatin, and lovastatin, where n = 22). Each IC50 curve is representative of at least two biological replicates. Error bars represent standard errors of the means.
Bioinformatic analysis suggests that itavastatin reduces isopentenyl pyrophosphate pools.
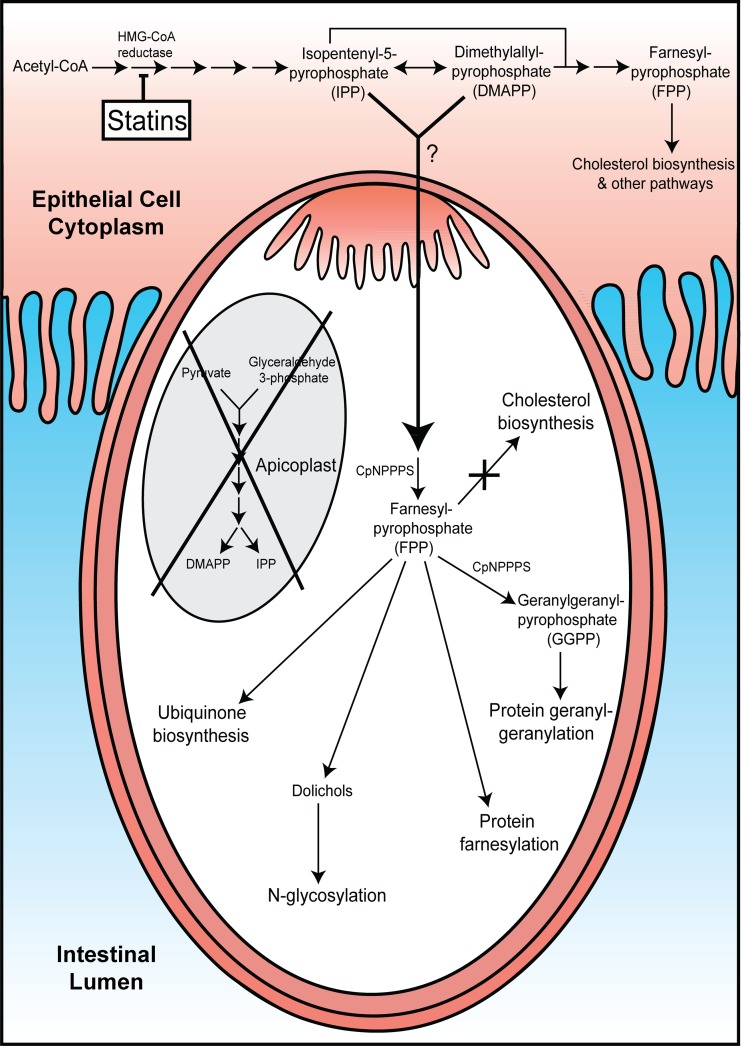
HMG-CoA reductase is the rate-limiting step in the mevalonic acid (MVA) pathway that produces isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP). These five carbon molecules provide the building blocks for isoprenoid synthesis by a family of enzymes known as prenyltransferases (31). Isoprenoids are essential precursors for many basic biological functions, including N-glycosylation, protein farnesylation and geranylgeranylation, and the biosynthesis of cholesterol and ubiquinone (32, 33). The MVA pathway is carried out in the cytosol of mammalian cells and archaebacteria, while IPP and DMAPP synthesis occurs via the methylerythritol phosphate (MEP) pathway in pathogenic bacteria, plant chloroplasts (34), and the apicoplast of Plasmodium parasites and Toxoplasma gondii (Fig. 5) (35, 36). Bioinformatic analysis suggests that the apicomplexa Toxoplasma gondii, Plasmodium falciparum, and Eimeria tenella all encode MEP (but not MVA) machinery (37), and recent evidence suggests that isoprenoid precursor synthesis is a major function of the apicoplast in P. falciparum (38). Interestingly, C. parvum and C. hominis do not encode any of the proteins known to be associated with the MEP or MVA pathways, but they do encode orthologs of prenyltransferases, implying that they utilize IPP but do not synthesize it by any previously described pathway (see Table S3 in the supplemental material) (37, 39). Moreover, additional bioinformatic analysis conducted by our group revealed evidence that supports the existence of prenyltransferases and downstream enzymes associated with the protein prenylation, geranylgeranylation, N-glycosylation, and ubiquinone biosynthesis pathways in C. parvum and C. hominis (see Table S3), underscoring the parasite's requirement for isoprenoids.
Fig 5.
Model for C. parvum growth inhibition by statins. The host cell is represented by the red shaded area (top of figure), while the intracellular parasite (white area) is depicted as separated from the host cell and intestinal lumen (blue area) by the parasitophorous vacuole that is continuous with the parasite's feeder organelle (darker red). The gray oval represents the apicoplast that is present in other apicomplexan parasites but absent from Cryptosporidium parasites (as indicated by “x”). The mevalonic acid (MVA) pathway, including HMG-CoA reductase, is depicted in the host cell. This pathway converts acetyl-CoA to the isoprenoid precursor isopentenyl-5-pyrophosphate (IPP), which can be converted to dimethylallyl pyrophosphate (DMAPP) by isopentenyl pyrophosphate isomerase (IPPI; double-headed arrow). IPP and DMAPP provide substrates for additional prenyltransferases to synthesize larger isoprenoids which are utilized in various cellular processes. These critical isoprenoid precursors are synthesized by the nonmevalonate (MEP) pathway in non-Cryptosporidium apicomplexans through the generation of deoxyxyluose 5-phosphate (DOXP) from pyruvate and glyceraldehyde 3-phosphate. Our bioinformatic analysis (see Table S3 in the supplemental material) revealed strong evidence for the existence of additional prenyltransferases, including CpNPPS enzymes involved in ubiquinone biosynthesis, N-glycosylation, and protein farnesylation and geranylgeranylation. However, there was no evidence of endogenous synthesis of isoprenoid precursors by C. parvum or C. hominis.
These observations, combined with the absence of a detectable apicoplast (the site of isoprenoid synthesis in other apicomplexan parasites) in C. parvum (27, 28) and the fact that itavastatin is a potent inhibitor of growth in vitro, led us to hypothesize that C. parvum exclusively utilizes host-derived IPP that is synthesized via the MVA pathway. Therefore, we reasoned that the addition of exogenous IPP to the growth medium might relieve the inhibition of C. parvum growth by itavastatin (a specific inhibitor of the MVA pathway). Consistent with this notion, the addition of 50 μM IPP to infected monolayers treated with itavastatin increased the IC50 to greater than 5-fold (IC50 itavastatin + IPP of 2.3 μM [95% CI = 0.7 to 7.5] versus IC50 itavastatin only of 0.44 μM [95% CI = 0.31 to 0.62]) (Fig. 6A). Moreover, the ability of IPP to rescue C. parvum growth was specific to itavastatin, as the IC50 did not change in infected monolayers treated with nitazoxanide and up to 200 μM IPP (IC50 NTZ + IPP of 3.2 μM [95% CI = 2.6 to 3.9] versus IC50 NTZ only of 2.9 μM [95% CI = 2.4 to 3.5]) (Fig. 6B). These data suggest that itavastatin inhibits Cryptosporidium growth, at least in part, through inhibition of human HMG-CoA reductase rather than off-target effects of the drug.
Fig 6.
Exogenous isopentenyl pyrophosphate (IPP) partly reverses C. parvum growth inhibition by itavastatin. (A) Infected monolayers were treated with various concentrations of itavastatin and then supplied growth medium with or without 50 μM IPP. The IC50 for itavastatin in wells that received IPP was >5-fold the IC50 in wells with no IPP. (B) Infected monolayers were treated with various concentrations of nitazoxanide and then supplied growth medium with or without 50 μM IPP. The IC50 for nitazoxanide was not affected by the addition of IPP.
DISCUSSION
By miniaturizing, optimizing, and applying automated liquid handling techniques to existing methods of growth and detection of C. parvum (11, 16), we were able to conduct the first cell-based HTS for C. parvum inhibitors. We screened the NIH Clinical Collections libraries in order to pilot the HTS capabilities of our assay, and through a combination of screening, hit prioritization, and secondary assays, we identified itavastatin as a promising lead candidate. The compound's well-defined mechanism of action as an inhibitor of HMG-CoA reductase makes it a useful tool to begin to model isoprenoid utilization in C. parvum, while its demonstrated safety record in patient populations and favorable pharmacokinetic/pharmacodynamic (PK/PD) profile make it an excellent candidate for clinical trials in humans.
We employed the NIH Clinical Collections drug libraries to validate the utility of our cell-based platform as a high-throughput screen. Although our Z' prime score (0.27 to 0.41) was lower than that reported for many target-based screens, we were able to identify a feasible number (n = 24) of compounds for follow-up, the vast majority (87.5%) of which were confirmed to have activity. We chose a hit limit of ≥80% inhibition in a single replicate, which could be easily changed as the size of the screening library varies in order to maintain a practical number of candidate compounds for follow-up testing. In addition to its utility as an HTS, our assay provides a platform upon which additional testing can be carried out.
Many of the active compounds we identified have well-defined mechanisms of action and may serve as valuable tools to help elucidate basic biology in the genetically intractable cryptosporidia. For example, the confirmed efficacy of tegaserod, a 5-hydroxytryptamine receptor 4 (5-HT4) agonist, suggests that host cell serotonin-dependent signaling may be important in the C. parvum life cycle. Additionally, the identification of known channel inhibitors (i.e., digoxin and glipizide) may provide novel tools to examine the effects of ion flux on various life cycle stages of the parasite. While these findings require additional follow-up, they suggest that screening campaigns utilizing libraries that are biased toward drug-like compounds with known mechanisms of action can rapidly generate mechanistic hypotheses while also yielding quality leads for drug repurposing.
The identification of itavastatin as a potent inhibitor of C. parvum growth is an excellent example of the dual utility of a drug repurposing campaign. Six of the seven HMG-CoA reductase inhibitors that we tested exhibited an IC50 of <10 μM against C. parvum. Additionally, the ability of exogenous IPP to reduce the effects of itavastatin suggests that the effect of statins on C. parvum growth is via specific inhibition of human HMG-CoA reductase rather than an off-target effect of the drug and that IPP may be an essential metabolite in the parasite's life cycle. The importance of isoprenoid metabolism is well established for other apicomplexa. Suprapharmacologic doses of HMG-CoA reductase inhibitors have been shown to inhibit Babesia divergens (40), Plasmodium falciparum (40, 41), and Toxoplasma gondii (42) growth in vitro, while P. falciparum growth can be inhibited by blocking IPP synthesis with fosmidomycin or curing parasites of their apicoplasts. Growth can then be rescued with the addition of exogenous IPP (38), proving that it is essential, regardless of its source.
Our bioinformatics analysis of the C. parvum and C. hominis genomes revealed evidence for the existence of multiple pathways in the Cryptosporidium genomes that utilize isoprenoids derived from IPP and DMAPP. However, like other groups (37), we did not find any evidence to suggest that either parasite is capable of IPP or DMAPP synthesis, leading us to propose that the parasite is completely dependent on host synthesis of these precursors (see Table S3 in the supplemental material). Our model is supported by the fact that pharmacologic inhibition of the MVA pathway with statins leads to inhibition of C. parvum growth, which can be relieved by the addition of exogenous IPP. Following entry into the parasitophorous vacuole, isoprenoid precursors most likely provide substrates for the C. parvum enzyme nonspecific polyprenyl pyrophosphate synthase (CpNPPPS), which is capable of synthesizing various polyprenyl pyrophosphates (39) that are utilized by Cryptosporidium enzymes in the ubiquinone biosynthesis, N-glycosylation, and protein farnesylation and geranylgeranylation pathways. Notably, we did not find any evidence for de novo sterol (cholesterol or ergosterol) synthesis from isoprenoid precursors by C. parvum or C. hominis, which is consistent with recent in vitro work demonstrating that Plasmodium and Cryptosporidium parasites are dependent on salvage from host cells to meet their cholesterol requirements (43, 44). IPP's ability to enter host cells and/or the parasitophorous vacuole may be limited by its highly charged nature, which could help explain the compound's inability to fully rescue parasite growth. Finally, we cannot discount the possibility that the effect of itavastatin on C. parvum growth is not exclusively the result of HMG-CoA reductase inhibition, as statins appear to have numerous HMG-CoA reductase-independent effects in vitro and in vivo (45).
Fifteen of the 21 confirmed compounds identified by our screen are currently FDA approved for use in patients, but only five of these compounds, floxuridine (22), clofazimine (46), indomethacin (47), minocycline (48, 49), and doxycycline (50), are described in the literature as having activity against Cryptosporidium species in vitro or in vivo (animal models or human patients), suggesting that the list is rich in new repurposing candidates. While we chose to pursue the HMG-CoA reductase inhibitor itavastatin, we have made the screening results of all 727 compounds available as supplemental data, which we hope will inform additional studies aimed at repurposing drugs for cryptosporidiosis and other apicomplexan infections as well as elucidate apicomplexan biology and novel host-microbe interactions.
The demonstrated importance of isoprenoid metabolism to the apicomplexan life cycle combined with in vitro potency against C. parvum and the fact that HMG-CoA reductase inhibition is extremely well tolerated in humans makes the statins excellent repurposing candidates for the treatment of cryptosporidiosis. Of the statins we tested, itavastatin was the most attractive repurposing candidate. The compound exhibits a favorable pharmacokinetic profile including a peak plasma concentration (Cmax) (51) that exceeds its submicromolar IC50 against C. parvum. Moreover, the drug is not subjected to CYP3A4 metabolism and only minimally metabolized by CYP2C9, resulting in extensive enterohepatic recirculation of the active compound (52). This may lead to a relative concentration of the compound in the lumens of the intestine and biliary tract, the most common sites of Cryptosporidium infection. Additionally, minimal CYP induction may reduce the risk of drug-drug interactions (52), leading to an improved safety profile. Thus, in addition to providing new insight into Cryptosporidium isoprenoid metabolism, the drug's unique combination of in vitro potency against C. parvum and its excellent safety profile provides evidence to support the initiation of human trials to assess the efficacy of itavastatin for the treatment of cryptosporidiosis in patients.
ACKNOWLEDGMENTS
Parts of this project were funded by The Campbell Foundation, a pilot project under grants NIAID R21 AI101381-01, NIH COBRE P20 RR021905, NIH U54AI057168, and NIH U54 AI057159. K.B. is supported by grant T32 AI0055402-06A1 and A.S. is supported by grant NIAID R01 AI072021.
We thank Gary Ward, Jose Teixeira, and Peter Miller for their insight. K.B. and C.D.H. are listed as inventors on U.S. and international patent applications covering methods and screening hits described in the manuscript.
APPENDIX
The ImageJ macro for nucleus separation and enumeration is as follows.
run(“8-bit”);
run(“Subtract Background …,” “rolling = 50”);
run(“Unsharp Mask …,” “radius = 2 mask = 0.70”);
//run(“Threshold …”);
setAutoThreshold(“Huang dark”);
run(“Convert to Mask”);
run(“Watershed”);
run(“Set Measurements …,” “area mean min centroid center perimeter integrated median kurtosis area_fraction stack limit display redirect = None decimal = 2”);
run(“Analyze Particles …,” “size = 159.5125-Infinity circularity = 0.00–1.00 show = Nothing display clear summarize add”);
The ImageJ macro for parasite separation and enumeration is as follows.
run(“Subtract Background …,” “rolling = 3”);
setAutoThreshold(“Huang dark”);
//run(“Threshold …”);
setThreshold(1000, 16383);
run(“Convert to Mask”);
run(“Fill Holes”);
run(“Watershed”);
run(“Analyze Particles …,” “size = 16.48282906–185.4936891 circularity = 0.00–1.00 show = Nothing display clear summarize add”);
Footnotes
Published ahead of print 4 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02460-12.
REFERENCES
- 1. Xiao L, Fayer R, Ryan U, Upton SJ. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen X-M, Keithly JS, Paya CV, LaRusso NF. 2002. Cryptosporidiosis. N. Engl. J. Med. 346:1723–1731 [DOI] [PubMed] [Google Scholar]
- 3. Ochoa TJ, Salazar-Lindo E, Cleary TG. 2004. Management of children with infection-associated persistent diarrhea. Semin. Pediatr. Infect. Dis. 15:229–236 [DOI] [PubMed] [Google Scholar]
- 4. Snelling WJ, Xiao L, Ortega-Pierres G, Lowery CJ, Moore JE, Rao JR, Smyth S, Millar BC, Rooney PJ, Matsuda M, Kenny F, Xu J, Dooley JS. 2007. Cryptosporidiosis in developing countries. J. Infect. Dev. Ctries. 1:242–256 [PubMed] [Google Scholar]
- 5. Macfarlane DE, Horner-Bryce J. 1987. Cryptosporidiosis in well-nourished and malnourished children. Acta Paediatr. Scand. 76:474–477 [DOI] [PubMed] [Google Scholar]
- 6. Guerrant DI, Moore SR, Lima AA, Patrick PD, Schorling JB, Guerrant RL. 1999. Association of early childhood diarrhea and cryptosporidiosis with impaired physical fitness and cognitive function four-seven years later in a poor urban community in northeast Brazil. Am. J. Trop. Med. Hyg. 61:707–713 [DOI] [PubMed] [Google Scholar]
- 7. Abubakar I, Aliyu SH, Arumugam C, Hunter PR, Usman NK. 2007. Prevention and treatment of cryptosporidiosis in immunocompromised patients. Cochrane Database Syst. Rev. 24:CD004932 doi:10.1002/14651858.CD004932.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adams CP, Brantner VV. 2006. Estimating the cost of new drug development: is it really $802 million? Health Aff. (Millwood) 25:420–428 [DOI] [PubMed] [Google Scholar]
- 9. Reich MR. 2000. The global drug gap. Science 287:1979–1981 [DOI] [PubMed] [Google Scholar]
- 10. Ashburn TT, Thor KB. 2004. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3:673–683 [DOI] [PubMed] [Google Scholar]
- 11. Gut J, Nelson RG. 1999. Cryptosporidium parvum: synchronized excystation in vitro and evaluation of sporozoite infectivity with a new lectin-based assay. J. Eukaryot. Microbiol. 46:56S–57S [PubMed] [Google Scholar]
- 12. Zhang JH. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67–73 [DOI] [PubMed] [Google Scholar]
- 13. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27:29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer ET, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Srinivasamoorthy G, Stoeckert CJ, Jr, Thibodeau R, Treatman C, Wang H. 2010. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 38:D415–D419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharling L, Liu X, Gollapalli DR, Maurya SK, Hedstrom L, Striepen B. 2010. A screening pipeline for antiparasitic agents targeting Cryptosporidium inosine monophosphate dehydrogenase. PLoS Negl. Trop. Dis. 4:e794 doi:10.1371/journal.pntd.0000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cai X, Woods KM, Upton SJ, Zhu G. 2005. Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro. Antimicrob. Agents Chemother. 49:4437–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gargala G, Delaunay A, Li X, Brasseur P, Favennec L, Ballet JJ. 2000. Efficacy of nitazoxanide, tizoxanide and tizoxanide glucuronide against Cryptosporidium parvum development in sporozoite-infected HCT-8 enterocytic cells. J. Antimicrob. Chemother. 46:57–60 [DOI] [PubMed] [Google Scholar]
- 19. Arrowood MJ. 2002. In vitro cultivation of Cryptosporidium Species. Clin. Microbiol. Rev. 15:390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. 2011. DrugBank 3.0: a comprehensive resource for “omics” research on drugs. Nucleic Acids Res. 39:D1035–D1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. 2012. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40:D1100–D1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun XE, Sharling L, Muthalagi M, Mudeppa DG, Pankiewicz KW, Felczak K, Rathod PK, Mead J, Striepen B, Hedstrom L. 2010. Prodrug activation by Cryptosporidium thymidine kinase. J. Biol. Chem. 285:15916–15922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Connor EM, Sperling RS, Gelber R, Kiselev P, Scott G, O'Sullivan MJ, VanDyke R, Bey M, Shearer W, Jacobson RL, Jimenez E, O'Neill E, Bazin B, Delfraissy J-F, Culnane M, Coombs R, Elkins M, Moye J, Stratton P, Balsley J. 1994. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. N. Engl. J. Med. 331:1173–1180 [DOI] [PubMed] [Google Scholar]
- 24. Andersen SL, Oloo AJ, Gordon DM, Ragama OB, Aleman GM, Berman JD, Tang DB, Dunne MW, Shanks GD. 1998. Successful double-blinded, randomized, placebo-controlled field trial of azithromycin and doxycycline as prophylaxis for malaria in western Kenya. Clin. Infect. Dis. 26:146–150 [DOI] [PubMed] [Google Scholar]
- 25. Chang HR, Comte R, Pechère JC. 1990. In vitro and in vivo effects of doxycycline on Toxoplasma gondii. Antimicrob. Agents Chemother. 34:775–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pradines B, Rogier C, Fusai T, Mosnier J, Daries W, Barret E, Parzy D. 2001. In vitro activities of antibiotics against Plasmodium falciparum are inhibited by iron. Antimicrob. Agents Chemother. 45:1746–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445 [DOI] [PubMed] [Google Scholar]
- 28. Zhu G, Marchewka MJ, Keithly JS. 2000. Cryptosporidium parvum appears to lack a plastid genome. Microbiology 146:315–321 [DOI] [PubMed] [Google Scholar]
- 29. Tonelli M, Lloyd A, Clement F, Conly J, Husereau D, Hemmelgarn B, Klarenbach S, McAlister FA, Wiebe N, Manns B. 2011. Efficacy of statins for primary prevention in people at low cardiovascular risk: a meta-analysis. CMAJ 183:E1189–E1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davidson MH. 2002. Controversy surrounding the safety of cerivastatin. Expert Opin. Drug Saf. 1:207–212 [DOI] [PubMed] [Google Scholar]
- 31. Liang PH, Ko TP, Wang AH. 2002. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269:3339–3354 [DOI] [PubMed] [Google Scholar]
- 32. Odom AR. 2011. Five questions about non-mevalonate isoprenoid biosynthesis. PLoS Pathog. 7:e1002323 doi:10.1371/journal.ppat.1002323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thurnher M, Nussbaumer O, Gruenbacher G. 2012. Novel aspects of mevalonate pathway inhibitors as antitumor agents. Clin. Cancer Res. 18:3524–3531 [DOI] [PubMed] [Google Scholar]
- 34. Kuzuyama T. 2002. Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci. Biotechnol. Biochem. 66:1619–1627 [DOI] [PubMed] [Google Scholar]
- 35. Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. 1999. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285:1573–1576 [DOI] [PubMed] [Google Scholar]
- 36. Nair SC, Brooks CF, Goodman CD, Strurm A, McFadden GI, Sundriyal S, Anglin JL, Song Y, Moreno SNJ, Striepen B. 2011. Apicoplast isoprenoid precursor synthesis and the molecular basis of fosmidomycin resistance in Toxoplasma gondii. J. Exp. Med. 208:1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clastre M, Goubard A, Prel A, Mincheva Z, Viaud-Massuart M-C, Bout D, Rideau M, Velge-Roussel F, Laurent F. 2007. The methylerythritol phosphate pathway for isoprenoid biosynthesis in coccidia: presence and sensitivity to fosmidomycin. Exp. Parasitol. 116:375–384 [DOI] [PubMed] [Google Scholar]
- 38. Yeh E, DeRisi JL. 2011. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol. 9:e1001138 doi:10.1371/journal.pbio.1001138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Artz JD, Dunford JE, Arrowood MJ, Dong A, Chruszcz M, Kavanagh KL, Minor W, Russell RG, Ebetino FH, Oppermann U, Hui R. 2008. Targeting a uniquely nonspecific prenyl synthase with bisphosphonates to combat cryptosporidiosis. Chem. Biol. 15:1296–1306 [DOI] [PubMed] [Google Scholar]
- 40. Grellier P, Valentin A, Millerioux V, Schrevel J, Rigomier D. 1994. 3-Hydroxy-3-methylglutaryl coenzyme A reductase inhibitors lovastatin and simvastatin inhibit in vitro development of Plasmodium falciparum and Babesia divergens in human erythrocytes. Antimicrob. Agents Chemother. 38:1144–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pradines B, Torrentino-Madamet M, Fontaine A, Henry M, Baret E, Mosnier J, Briolant S, Fusai T, Rogier C. 2007. Atorvastatin is 10-fold more active in vitro than other statins against Plasmodium falciparum. Antimicrob. Agents Chemother. 51:2654–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cortez E, Stumbo AC, Oliveira M, Barbosa HS, Carvalho L. 2009. Statins inhibit Toxoplasma gondii multiplication in macrophages in vitro. Int. J. Antimicrob. Agents. 33:185–186 [DOI] [PubMed] [Google Scholar]
- 43. Ehrenman K, Wanyiri JW, Bhat N, Ward HD, Coppens I. 2013. Cryptosporidium parvum scavenges LDL-derived cholesterol and micellar cholesterol internalized into enterocytes. Cell. Microbiol. [Epub ahead of print.] doi:10.1111/cmi.12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Labaied M, Jayabalasingham B, Bano N, Cha S-J, Sandoval J, Guan G, Coppens I. 2011. Plasmodium salvages cholesterol internalized by LDL and synthesized de novo in the liver. Cell. Microbiol. 13:569–586 [DOI] [PubMed] [Google Scholar]
- 45. Bolego C, Poli A, Cignarella A, Catapano AL, Paoletti R. 2002. Novel statins: pharmacological and clinical results. Cardiovasc. Drugs Ther. 16:251–257 [DOI] [PubMed] [Google Scholar]
- 46. Samson VE, Brown WR. 1996. Pneumatosis cystoides intestinalis in AIDS-associated cryptosporidiosis. More than an incidental finding? J. Clin. Gastroenterol. 22:311–312 [DOI] [PubMed] [Google Scholar]
- 47. Hornok S, Szell Z, Shibalova TA, Varga I. 1999. Study on the course of Cryptosporidium baileyi infection in chickens treated with interleukin-1 or indomethacin. Acta Vet. Hung. 47:207–216 [DOI] [PubMed] [Google Scholar]
- 48. Giacometti A, Cirioni O, Del Prete MS, Barchiesi F, Fineo A, Scalise G. 2001. Activity of buforin II alone and in combination with azithromycin and minocycline against Cryptosporidium parvum in cell culture. J. Antimicrob. Chemother. 47:97–99 [DOI] [PubMed] [Google Scholar]
- 49. Giacometti A, Cirioni O, Scalise G. 1996. In-vitro activity of macrolides alone and in combination with artemisin, atovaquone, dapsone, minocycline or pyrimethamine against Cryptosporidium parvum. J. Antimicrob. Chemother. 38:399–408 [DOI] [PubMed] [Google Scholar]
- 50. Armson A, Meloni BP, Reynoldson JA, Thompson RCA. 1999. Assessment of drugs against Cryptosporidium parvum using a simple in vitro screening method. FEMS Microbiol. Lett. 178:227–233 [DOI] [PubMed] [Google Scholar]
- 51. Lv H, Sun J-G, Wang G-J, Zhu X-Y, Zhang Y, Gu S-H, Liang Y, Sun J. 2007. Determination of pitavastatin in human plasma via HPLC-ESI-MS/MS and subsequent application to a clinical study in healthy Chinese volunteers. Clin. Chim. Acta 386:25–30 [DOI] [PubMed] [Google Scholar]
- 52. Saito Y. 2011. Pitavastatin: an overview. Atheroscler. Suppl. 12:271–276 [DOI] [PubMed] [Google Scholar]