Abstract
Six multiresistant, NDM-1-producing Klebsiella pneumoniae strains were recovered from an outbreak that affected six neonatal patients in a Colombian hospital. Molecular analysis showed that all of the isolates harbored the blaNDM-1, qnrA, and intI1 genes and were clonally related. Multilocus sequence typing showed that the isolates belonged to a new sequence type (ST1043) that was different from the sequence types that had previously been reported. This is the first report of NDM-1-producing isolates in South America.
TEXT
The emergence of carbapenem-resistant Gram-negative bacteria is a worldwide clinical problem that is associated principally with the production of carbapenemases (1). New Delhi metallo-β-lactamase 1 (NDM-1) is a carbapenemase that confers resistance to all β-lactam antibiotics, except aztreonam (2). NDM-1 was first identified in Klebsiella pneumoniae and Escherichia coli recovered from a Swedish patient who was transferred from India and Pakistan in 2008 (2, 3). However, NDM-1-producing Enterobacteriaceae and Acinetobacter spp. have been reported in Europe, Asia, Oceania, North America (4), and recently Central America (5). Here we report a nosocomial outbreak of NDM-1-producing K. pneumoniae in six patients who were admitted to the neonatal unit of a general hospital in Bogotá, Colombia. This is the first report of NDM-1-producing K. pneumoniae isolation in South America.
Table 1 shows the demographic and clinical characteristics of the six patients who were involved in the outbreak of NDM-1-producing K. pneumoniae. In August 2011, a preterm neonate who was 3 weeks of age developed symptoms of late neonatal sepsis. A carbapenem-resistant K. pneumoniae isolate was recovered from the patient's blood. The patient was treated with meropenem and rifampin and had an adequate clinical response and negative blood cultures on day 4 of treatment. The patient had a history of necrotizing enterocolitis IIB that required a total colectomy and ileostomy. There is no documented clinical history of the mother traveling outside the country, and there are no documented contacts of her or her daughter with individuals or health care workers from countries that have previously reported NDM-1-producing bacteria (case 1).
Table 1.
Clinical features of the patients in this study and MICs for their NDM-1-producing K. pneumoniae isolates
| Case | Sex | Pregnancy duration (wk) | Birth wt (g) | Delivery modea | Comorbidity(ies) | Empirical antimicrobial therapyb | Infectionc | Age (days) | MICb (μg/ml) |
Definitive antimicrobial therapyb | Clinical outcome | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | ETP | ATMe | CTX | AMK | CIP | TET | TGCf | CSTf | |||||||||||
| 1 | F | 32 | 1,600 | C | Placenta abruptio, severe perinatal asphyxia, enterocolitis, multidrug-resistant Acinetobacter colonization | TZP, GEN | BSI | 23.0 | 8 | >8 | >4 | 0.32 | >32 | 32 | ≤1 | ≤4 | 0.25 | 0.25 | MEM + RIF | Improvement |
| 2 | M | 32 | 1,660 | C | Chorioamnionitis | AMP + AMK, TZP | NEC | 9.0 | 8 | >8 | 4 | 0.32 | >32 | 32 | ≤1 | ≤4 | 0.25 | 0.25 | IPM + CIP | Improvement |
| 3 | M | 42 | 3,000 | C | Umbilical cord prolapse, meconium aspiration syndrome, severe perinatal asphyxia, hypoxic-ischemic encephalopathy | TZP | BSI | 90.0 | 4 | >8 | >4 | 0.32 | >32 | 32 | ≤1 | ≤4 | 0.25 | 0.25 | IPM + CIP | Improvement |
| 4 | M | 29 | 1,000 | C | Toxemic mother, severe perinatal asphyxia, pneumonia in utero | AMP + AMK, TZP | BSI | 10.0 | >8 | >8 | >4 | 0.32 | >32 | 32 | ≤1 | ≤4 | 0.25 | 0.25 | IPM + CIP | Death |
| 5 | F | 38 | 2,760 | V | Pneumonia in utero with spontaneous pneumothorax, closed thoracostomy (3 days) | AMP + AMK | BSI | 1.5 | 4 | 8 | 4 | 0.32 | >32 | 32 | ≤1 | ≤4 | 0.25 | 0.25 | IPM + CIP | Improvement |
| 6 | F | 30 | 1,150 | C | Chorioamnionitis, pneumonia in utero | AMP + AMK | Fulminant NEC | 13.0 | 8 | >8 | 4 | 0.32 | >32 | 32 | ≤1 | ≤4 | 0.25 | 0.25 | NTd | Death |
C, cesarean; V, vaginal.
TZP, piperacillin-tazobactam; GEN, gentamicin; AMP, ampicillin; AMK, amikacin; RIF, rifampin; CIP, ciprofloxacin; IPM, imipenem; MEM, meropenem; ETP, ertapenem; ATM, aztreonam; CTX, cefotaxime; TET, tetracycline; TGC, tigecycline; CST, colistin.
BSI, bloodstream infection; NEC, necrotizing enterocolitis.
NT, no therapy. The patient died.
Data were obtained by Etest.
Data were interpreted in accordance with 2012 EUCAST breakpoints.
In December 2011, three new cases were identified. One case was identified in a preterm newborn with enterocolitis IIB who was initially treated with piperacillin-tazobactam. Subsequently, a K. pneumoniae isolate was recovered from the patient's blood and the patient was treated with imipenem and ciprofloxacin because the patient's creatinine clearance was less than 25 mg/dl. The patient had an adequate clinical response and was discharged at the end of treatment (case 2). The next case occurred in a patient with sequela of hypoxic-ischemic encephalopathy. An isolate of K. pneumoniae was isolated from the patient's blood. The patient was treated with imipenem and ciprofloxacin and had an adequate clinical response. This patient was in the neonatal unit at the same time as the first patient and had previously been treated with piperacillin-tazobactam and gentamicin on two occasions for aspiration pneumonia. Additionally, carbapenem-resistant K. pneumoniae and E. coli isolates that did not have clinical repercussions had been recovered from the patient's urine the previous month (case 3). The last of the December 2011 cases was that of an extreme preterm neonate with severe perinatal asphyxia with myocardial and renal involvement, who on day 10 of hospitalization developed sepsis. K. pneumoniae was isolated from the patient's blood, catheter tip, and feces. On day 3 of treatment with imipenem and ciprofloxacin, the patient improved. However, the patient later developed a superior vena cava thrombosis and died (case 4).
In January 2012, there were two new cases of K. pneumoniae infection. One case was in a term infant with respiratory distress syndrome from pneumonia in utero. This patient started to deteriorate at 36 h of life and exhibited increased respiratory distress, right apical opacity, spontaneous pneumothorax, leukopenia, and thrombocytopenia. K. pneumoniae was recovered from the patient's blood. The patient was treated with imipenem (25-mg/kg doses) and ciprofloxacin (15 mg/kg). On day 3 of treatment, the blood cultures were negative for K. pneumoniae and the patient had a favorable clinical outcome (case 5). The last case was that of a very preterm infant who was born early because of maternal chorioamnionitis. On day 13 of hospitalization, the infant exhibited fulminant enterocolitis that evolved very rapidly and the patient died. K. pneumoniae was isolated from the patient's blood and peritoneal fluid (case 6).
The isolates were identified and antimicrobial susceptibility was determined with the automated MicroScan WalkAway plus system (Dade Behring). The MICs of meropenem, cefotaxime, ceftazidime, piperacillin-tazobactam, amikacin, gentamicin, ciprofloxacin, and tetracycline were confirmed by the agar dilution method. The MICs of imipenem, ertapenem, and aztreonam were confirmed by the Etest method (6). All isolates were confirmed as K. pneumoniae by amplification of the khe gene (7). Production of carbapenemases was determined with a modified Hodge test (6). Molecular characterization of the blood isolates obtained included detection of the blaCTX-M, blaTEM, blaSHV, blaOXA-48, blaGES, blaVIM, blaKPC, blaNDM, qnrA, qnrB, qnrS, aac(6′)-Ib-cr, sul1, and intl1 genes and the genes coding for six plasmid AmpC enzymes (ACC, FOX, MOX, EBC, DHA, and CIT) (8, 9). The NDM types of the isolates were determined by sequencing of the amplification product. The genetic relatedness of the isolates was determined by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) (10).
All six isolates were resistant to all β-lactams except aztreonam. Additionally, the isolates were resistant to gentamicin, amikacin, and trimethoprim-sulfamethoxazole but were susceptible to ciprofloxacin, tetracycline (6), tigecycline, and colistin (breakpoints available at http://www.eucast.org/clinical_breakpoints/, version 2.0) (Table 1). The blaSHV, blaNDM-1, qnrA, and intl1 genes were detected in all of the isolates. The remaining resistance genes that were evaluated were not detected. PFGE analysis demonstrated a single pulsotype, which suggests that the six isolates were clonally related. The alleles found for the gapA, infB, mdh, pgi, phoE, rpoB, and tonB genes used for MLST were 2, 38, 2, 20, 7, 4, and 89, respectively. This allelic combination corresponds to ST1043 (http://www.pasteur.fr/mlst). To date, there are no reports of NDM-positive or -negative isolates that belong to this sequence type (ST). The plasmid profile analysis of the clinical isolates showed the presence of two plasmids of ∼23 and ∼6 kbp (11). Transconjugants were obtained from the clinical isolates at 30 and 37°C with E. coli strain CAG12177 as the recipient and selected with tetracycline and ceftazidime (12–14). The transconjugants were resistant to all β-lactams (except aztreonam) but susceptible to ciprofloxacin (MIC, 0.125 μg/ml). In the NDM-positive E. coli transconjugants, only the ∼23-kbp plasmid was detected and the qnrA, blaSHV, and intI1 genes were not found. Replicon typing classified this plasmid within the IncA/C incompatibility group (15).
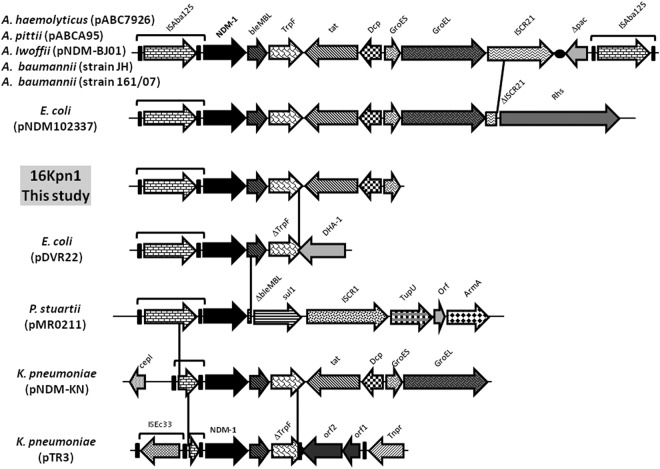
The genetic environment of blaNDM-1 was determined with previously reported primers (16). Additionally, specific primers (TAATGCGGTGCTCAGCTTCG, TTTGACATCGCGCGCAGC, GGCGATGACAGCATCATCCG, CGTGGCACAGCATGATCG, AACACCATGATCGGCTGCAC, and ATCTTGGAGCGCGCGTCTT) were designed for bleMBL, trpF, tat, groES, and groEL gene amplification. Sequencing of the PCR product revealed the presence of the same genetic elements surrounding the blaNDM-1 gene found in several Acinetobacter spp. (17) and E. coli isolate 102337 (Fig. 1).
Fig 1.
Comparative map of the genetic elements surrounding the blaNDM-1 gene found in the 16Kpn1 isolate and Acinetobacter pittii strain ABCA95 plasmid pABCA95, Acinetobacter lwoffii strain WJ10621 plasmid pNDM-BJ01, Acinetobacter haemolyticus strain ABC7926 plasmid pABC7926, A. baumannii strain 161/07, A. baumannii strain JH, E. coli strain N10-2337 plasmid pNDM102337, E. coli strain DVR22, Providencia stuartii plasmid pMR0211, K. pneumoniae strain Kp7 plasmid pNDM-KN, and K. pneumoniae plasmid pTR3 (GenBank accession numbers JQ739157.1, JQ001791.1, JQ080305.1, HQ857107.1, JN872329.1, JF714412.1, JF922606.1, JN687470.1, JN157804.1, and JQ349086.2, respectively).
To break the chain of transmission of this organism, the staff of the unit (including the cleaning crew) received information regarding the outbreak and the importance of hand hygiene. Also, strict contact isolation precautions were implemented for the affected patients. All patients who were infected with K. pneumoniae were clustered, and additional supervised disinfection sessions were performed with sodium dichloroisocyanurate (1,000 ppm). Furthermore, the entry of new patients was restricted. These measures controlled the spread of the identified clone. Epidemiological surveillance was intensified, and no new cases of carbapenem-resistant K. pneumoniae were found in the neonatal unit during the 3 months following the last isolation.
This is the first report of an outbreak of NDM-1-producing K. pneumoniae in Colombia and South America. Additionally, it is the first report of an outbreak in a neonatal unit of isolates that produce NDM-1 and that had a clinical response to imipenem and ciprofloxacin treatment. The patients were hospitalized in the neonatal unit from birth, and contact with people from countries that have reported the presence of NDM-1-producing bacteria was not documented. It is therefore likely that these isolates represent a novel ST and that autochthonous clones are locally acquiring plasmids carrying the gene for NDM-1, as has been reported in Europe (18). This hypothesis should be investigated further, including the possibility of horizontal transmission of plasmids from other bacteria that have been reported as NDM-1 carriers, such as Acinetobacter baumannii and E. coli (16, 19–21).
The clonality of the isolates suggested that the bacteria were transmitted between patients who were concurrently hospitalized in the same room. This route was also strongly implied by the fact that K. pneumoniae was not recovered from samples taken from hospital surfaces, eliminating the environment as a source of infection. In hospitalized adult patients, carbapenem-resistant K. pneumoniae isolates were identified; however, these isolates were resistant to aztreonam and molecular characterization indicated that they were KPC-3 positive, NDM negative, and genetically unrelated to NDM-1-producing K. pneumoniae, as has been previously reported by our group (22).
Our findings are consistent with other reports that have demonstrated the ease of nosocomial spread of resistant members of the family Enterobacteriaceae that cause infections and that manifest limited treatment options and high mortality rates. For this reason, it is important to reinforce infection control measures, to avoid cross-dissemination of multidrug-resistant microorganisms, to strengthen epidemiological surveillance systems that provide feedback to those involved in the management of patients, and to implement appropriate intervention measures.
ACKNOWLEDGMENTS
This work was financially supported by the Research Division of the Universidad El Bosque and Hospital El Tunal ESE.
We thank the neonatal unit nurses of Hospital El Tunal and Diana Cruz and Leidy Rojas for their technical assistance. We thank Harold Stokes for valuable comments and technical review of the manuscript. We thank the platform Genotyping of Pathogens and Public Health (Institut Pasteur, Paris, France) for coding MLST alleles and profiles and making them available at www.pasteur.fr/mlst.
Footnotes
Published ahead of print 28 January 2013
REFERENCES
- 1. Sidjabat H, Nimmo GR, Walsh TR, Binotto E, Htin A, Hayashi Y, Li J, Nation RL, George N, Paterson DL. 2011. Carbapenem resistance in Klebsiella pneumoniae due to the New Delhi metallo-beta-lactamase. Clin. Infect. Dis. 52:481–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595 [DOI] [PubMed] [Google Scholar]
- 3. Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poirel L, Hombrouck-Alet C, Freneaux C, Bernabeu S, Nordmann P. 2010. Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect. Dis. 10:832. [DOI] [PubMed] [Google Scholar]
- 5. Pasteran F, Albornoz E, Faccone D, Gomez S, Valenzuela C, Morales M, Estrada P, Valenzuela L, Matheu J, Guerriero L, Arbizu E, Calderon Y, Ramon-Pardo P, Corso A. 2012. Emergence of NDM-1-producing Klebsiella pneumoniae in Guatemala. J. Antimicrob. Chemother. 67:1795–1797 [DOI] [PubMed] [Google Scholar]
- 6. CLSI 2012. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement, vol 32 CLSI document M100-S22 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Yin-Ching C, Jer-Horng S, Ching-Nan L, Ming-Chung C. 2002. Cloning of a gene encoding a unique haemolysin from Klebsiella pneumoniae and its potential use as a species-specific gene probe. Microb. Pathog. 33:1–6 [DOI] [PubMed] [Google Scholar]
- 8. Monstein HJ, Ostholm-Balkhed A, Nilsson MV, Nilsson M, Dornbusch K, Nilsson LE. 2007. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 115:1400–1408 [DOI] [PubMed] [Google Scholar]
- 9. Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. 2012. Rapid detection of carbapenemase genes by multiplex real-time PCR. J. Antimicrob. Chemother. 67:906–909 [DOI] [PubMed] [Google Scholar]
- 10. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Canton R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schlesinger J, Navon-Venezia S, Chmelnitsky I, Hammer-Munz O, Leavitt A, Gold HS, Schwaber MJ, Carmeli Y. 2005. Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrob. Agents Chemother. 49:1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nichols BP, Shafiq O, Meiners V. 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 16. Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55:5403–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y, Wu C, Zhang Q, Qi J, Liu H, He T, Ma L, Lai J, Shen Z, Liu Y, Shen J. 2012. Identification of New Delhi metallo-beta-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One 7:e37152 doi:10.1371/journal.pone.0037152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nordmann P, Couard JP, Sansot D, Poirel L. 2012. Emergence of an autochthonous and community-acquired NDM-1-producing Klebsiella pneumoniae in Europe. Clin. Infect. Dis. 54:150–151 [DOI] [PubMed] [Google Scholar]
- 19. Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66:2002–2005 [DOI] [PubMed] [Google Scholar]
- 20. Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. 2010. Emergence of metallo-beta-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. 2012. Dissemination of New Delhi metallo-beta-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 18:E362–E365 [DOI] [PubMed] [Google Scholar]
- 22. Saavedra SY, Alvarez CA, Cuervo SI, Olarte N, Escobar JA, Leal AL, Castillo Londoño JS. 2010. Dissemination of a clone KPC-3-producing Klebsiella pneumoniae in hospitals, Bogota, Colombia, abstr 3244. 20th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland [Google Scholar]