Abstract
Since the discovery of streptomycin's bactericidal activity against Mycobacterium tuberculosis, aminoglycosides have been utilized to treat tuberculosis (TB). Today, the aminoglycosides kanamycin and amikacin are used to treat multidrug-resistant (MDR) TB, and resistance to any of the second-line injectable antibiotics, including kanamycin, amikacin, or capreomycin, is a defining characteristic of extensively drug-resistant (XDR) TB. Resistance to kanamycin and streptomycin is thought to be due to the acquisition of unlinked chromosomal mutations. However, we identified eight independent mutations in the 5′ untranslated region of the transcriptional activator whiB7 that confer low-level resistance to both aminoglycosides. The mutations lead to 23- to 145-fold increases in whiB7 transcripts and subsequent increased expression of both eis (Rv2416c) and tap (Rv1258c). Increased expression of eis confers kanamycin resistance in these mutants, while increased expression of tap, which encodes an efflux pump, is a previously uncharacterized mechanism of low-level streptomycin resistance. Additionally, high-level resistance to streptomycin arose at a much higher frequency in whiB7 mutants than in a wild-type (WT) strain. Although whiB7 is typically associated with intrinsic antibiotic resistance in M. tuberculosis, these data suggest that mutations in an uncharacterized regulatory region of whiB7 contribute to cross-resistance against clinically used second-line antibiotics. As drug resistance continues to develop and spread, understanding the mechanisms and molecular basis of antibiotic resistance is critical for the development of rapid molecular tests to diagnose drug-resistant TB strains and ultimately for designing regimens to treat drug-resistant cases of TB.
INTRODUCTION
Drug-resistant strains of Mycobacterium tuberculosis present a great challenge to global tuberculosis (TB) control efforts (1). Multidrug-resistant (MDR) strains of M. tuberculosis, defined as resistant to rifampin (RIF) and isoniazid (INH), are difficult to diagnose and are complicated and expensive to treat. Furthermore, fewer treatment options are available for patients who contract or develop extensively drug-resistant (XDR) TB, which is MDR and has gained additional resistance to a fluoroquinolone and at least one of the second-line injectable antibiotics kanamycin (KAN), amikacin (AMK), or capreomycin (CAP) (1). In 2008, the World Health Organization estimated that 440,000 cases of MDR TB occurred and XDR TB was reported in 58 countries worldwide (1). As the number of drug-resistant cases increases each year, the need to supplement the current time-consuming, growth-based resistance detection methods with more rapid molecular drug susceptibility tests becomes more pressing. However, a better understanding of the molecular mechanisms that confer drug resistance is required for the development of these methods. The combination of new technologies, such as next-generation sequencing, with improved molecular genetic systems in M. tuberculosis creates a robust method to identify mutations in strains with unknown mechanisms of resistance and confirm their role in drug resistance. These strategies are especially important for understanding resistance mechanisms to the less commonly studied second-line antibiotics that are needed to treat patients with drug-resistant TB. The aminoglycoside KAN is central in treating MDR TB, and resistance to KAN (Kanr) is one of the defining characteristics of XDR TB strains (1, 2), yet our understanding of KAN resistance in M. tuberculosis remains incomplete. KAN inhibits protein synthesis by binding to the 16S rRNA in the 30S ribosomal subunit, and resistance is observed at two phenotypic levels: high and low (3, 4). High-level resistance (MIC of ≥80 μg/ml) occurs as a result of mutations in the 1,400-bp region of the 16S rRNA (rrs) where KAN binds to the ribosome (5–8). These rrs mutations (such as the A1401G or G1484T mutation) are also associated with cross-resistance to other ribosome-binding antibiotics, including the structurally similar aminoglycoside AMK and the cyclic peptide CAP (6, 9). In contrast, low-level KAN-resistant (5 μg/ml < MIC < 80 μg/ml) strains generally exhibit resistance to KAN only (3, 5–9). In a recent study, we found that up to 80% of strains displaying low-level KAN resistance harbored mutations in the promoter region of the aminoglycoside acetyltransferase gene eis (Rv2416c) (3). These mutations enhance eis expression, which leads to increased acetylation and inactivation of KAN. However, the mechanism of KAN resistance in the remaining isolates was unexplained. Therefore, other mechanisms for low-level resistance to KAN must exist. In this study, we used whole-genome sequencing and molecular genetics to identify mutations in the 5′ untranslated region (UTR) of whiB7 (Rv3197A), a transcriptional activator of eis (10), that confer low-level resistance in M. tuberculosis by enhancing eis expression. Further analysis also revealed that these mutations confer cross-resistance to streptomycin (STR) due to overexpression of tap (Rv1258c), which encodes an efflux pump.
MATERIALS AND METHODS
Bacterial strains, media, and DNA manipulation.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. M. tuberculosis strains were cultured in Middlebrook 7H9 broth (7H9) supplemented with 10% (vol/vol) albumin-dextrose-catalase (ADC) enrichment (Difco Laboratories) and 0.05% (vol/vol) Tween 80 (Sigma-Aldrich) or on Middlebrook 7H10 plates (7H10) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC) enrichment (Difco-Laboratories) at 37°C. The media were supplemented with hygromycin (HYG) (75 μg/ml), kanamycin (KAN) (5 μg/ml or 25 μg/ml), or streptomycin (STR) (2 μg/ml) when needed for M. tuberculosis. For Escherichia coli strains, Luria-Bertani medium was supplemented with HYG (200 μg/ml) or KAN (50 μg/ml) when needed. Antibiotics were purchased from Sigma. Standard protocols or the manufacturer's instructions were used for all DNA manipulation (New England BioLabs [NEB], Invitrogen, or Finnzymes). All oligonucleotide primers (see Table S2 in the supplemental material) were synthesized at the Biotechnology Core Facility, National Center for Preparedness, Detection, and Control of Infectious Diseases, CDC. Clinical isolates were obtained from the culture collection at the Mycobacteriology Laboratory Branch, CDC. Human subject information linked to the clinical isolates used in this study is protected by the protocol approved by the CDC institutional review board.
MIC determination.
The susceptibilities to KAN and STR were determined according to guidelines and definitions stated by the Clinical and Laboratory Standards Institute (11), using 7H10 agar containing KAN (Sigma) at concentrations of 1, 2, 3, 4, 5, 10, 15, 20, 25, 30, 35, 40, 50, and 80 μg/ml and STR, isoniazid (INH), capreomycin (CAP), rifampin (RIF), and ofloxacin (OFX) at 0.25, 0.5, 1, 2, 4, 8, 10, and 16 μg/ml. The MIC was defined as the lowest concentration of drug resulting in inhibition of growth (>99%) of the initial inoculum after 4 weeks of incubation at 37°C.
Whole-genome sequencing.
High-molecular-weight DNA was extracted as previously described (12). DNA libraries were prepared following manufacturer's recommended protocols and were sequenced on the Illumina GAII platform as single-end 36-bp reads. Sequence reads were aligned to the previously published reference genome sequence of CDC1551 using Maq (13) and allowing 3 mismatches in the first 24 bp (maq map -n3) and ignoring the reads that map to multiple locations, thus ignoring the variations in the direct repeat proline-proline-glutamic acid (PPE)/polymorphic G-rich sequence (PGRS) family genes. Single nucleotide polymorphisms (SNPs) were called and filtered, keeping only the ones that had a minimum depth of 20 reads, a minimum consensus quality of 20, and were not within 10 bp of other SNPs (maq.pl SNPfilter -d20 -q20 -W10). Insertions/deletions (indels) were called using the VAAL package by the Broad Institute using the default parameters of the package (14). The folds coverage of the genomes were 58× and 95× for CDC1551 (parental strain) and K301, respectively.
Generating spontaneous Kanr mutants.
Spontaneous Kanr mutants were generated from the pansusceptible strains H37Rv and CDC1551 as described previously (6). Each strain was grown to a high cell density 3(∼1 × 108 CFU/ml), and portions of the cell suspension were plated on 7H10 plates supplemented with 5 μg/ml of KAN. Serial 10-fold dilutions were plated on 7H10 agar containing no antibiotics to determine the number of viable cells in the suspension. Isolated colonies were inoculated into 7H9 broth containing 5 μg/ml of KAN. Mutants isolated from CDC1551 were given strain names with the K300 numbering, and mutants from H37Rv are referred to with the K200 numbering (Table 1; see Fig. 2). For each isolate, genomic DNA was harvested and the rrs, eis, and whiB7 loci were analyzed by PCR and sequencing. Each 25-μl PCR mixture contained 12.5 μl HotStarTaq master mix (Qiagen), 1.5 μl of the forward and reverse 5 μM primers (see Table S2 in the supplemental material), 8.5 μl distilled water (dH2O), and 1 μl of genomic DNA. The amplification parameters included an initial denaturation step at 95°C for 15 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 30 s, with a final elongation step at 72°C for 10 min. PCR products were analyzed on 1.5% agarose-Tris-EDTA gels and stained with ethidium bromide. PCR products were sequenced with a forward and reverse primer at each locus. Sequencing reactions (20 μl) were completed with ABI's BigDye Terminator v.3.1 kit according to the manufacturer's instructions. The sequencing reactions were analyzed using the ABI 3130xl genetic analyzer with standard run conditions for electrophoresis and data collection. Sequence data were analyzed for the presence or absence of mutations by comparison with published sequences of H37Rv using the SeqMan alignment application of DNASTAR Lasergene 8.0. To determine mutation frequency, CDC1551 and K301 were grown to late log phase, diluted into 7H9 broth in 12 independent cultures, and plated on 7H10 agar containing no drug, 40 μg/ml of KAN, 2 μg/ml STR, or 10 μg/ml STR. Mutation frequency was calculated by dividing the number of colonies on selective media by the total number of cells plated.
Table 1.
Summary of spontaneous mutants and derivatives used in this study
| Strain(s)a | whiB7 genotype | MIC (μg/ml)b |
|
|---|---|---|---|
| KAN | STR | ||
| WT | |||
| CDC1551 | WT | 2 | 0.5 |
| H37Rv | WT | 2 | 0.5 |
| Spontaneous mutants | |||
| K301, K203, K302, K304, K305, K307 | ΔC+133 | 20 | 8 |
| K303 | ΔC+86 | 20 | 8 |
| K306 | ΔG+179 | 20 | 8 |
| K308 | ΔG+128 | 20 | 8 |
| K309 | ΔC+124 | 20 | 8 |
| K205 | +C+133 | 20 | 10 |
| K206 | G+297A | 10 | 8 |
| K209 | A+237G | 10 | 8 |
| CL-27c | +C+133 | 20 | 10 |
| K204 | WT, eis C−14T | 25 | 0.5 |
| Allelic exchange derivatives | |||
| K301-WT | WT | 2 | 0.5 |
| K205-WT | WT | 4 | 2 |
| K209-WT | WT | 2 | 0.5 |
| Deletion strains | |||
| CDC1551ΔwhiB7 | NAd | 2 | 0.25 |
| CDC1551Δtap | WT | 2 | 0.1 |
| CDC1551ΔRv1257c | WT | 2 | 0.5 |
| K301Δtap | ΔC+133 | 10 | 0.1 |
| K301ΔRv1257c | ΔC+133 | 10 | 8 |
| H37RvΔeis | WT | 2 | 0.5 |
K200 strains are derivatives of H37Rv, and K300 strains are derivatives of CDC1551.
All strains had AMK MICs of <4 μg/ml.
Clinical isolate.
NA, not applicable.
Fig 2.
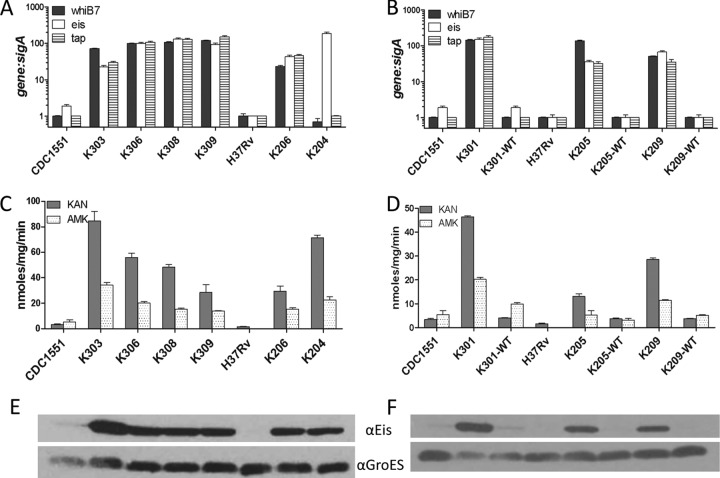
Mutations confer increased expression of whiB7, eis, and tap. The ratio of gene transcripts to sigA (A and B) was determined by qRT-PCR and normalized to H37Rv. Error bars represent the standard errors of the mean (SEM) from at least 3 experiments. (A) whiB7 mutants and parental strains. The mutation harbored in each strain is listed in Table 1. (B) Gene expression from one representative strain of each type of whiB7 mutation and the corresponding allelic exchange strain reverted to the wild-type sequence: deletion strain K301 (ΔC+133), insertion strain K205 (+C+133), and transversion strain K209 (A+237G). Parental strains CDC1551 and H37Rv are included for comparison. (C and D) Acetyltransferase activity due to Eis was measured from crude cell lysates and is expressed in nmol/mg/min. (C) Mutants and parental strains; (D) whiB7 mutants and representative allelic exchange strain. (E and F) Immunoblot analysis of cell lysates from mutants and parental strains (E) and representative allelic exchange isolates (F). Lysates were probed with either anti-Eis (αEis) or anti-GroES (αGroES) serum.
Construction of whiB7 revertants and deletion strains.
The specialized transducing phage phAZ70 was constructed using methods previously described (15) and harbors the wild-type whiB7 gene and promoter and downstream gene Rv3197 separated by a Hygr cassette (see Table S1 in the supplemental material). M. tuberculosis K301, K205, and K209 bacteria were infected with phAZ70 at a multiplicity of infection (MOI) of 10:1 (phage to bacteria) and selected on 7H10 containing HYG. The whiB7 promoter region was amplified and sequenced in each Hygr transductant to confirm the reversion of the whiB7 promoter point mutations to wild-type sequence. whiB7 deletion strains (with bases −40 to 266 removed) were constructed as previously described (3, 15). M. tuberculosis strains H37Rv, K204, and CDC1551 were transduced with phΔwhiB7 at an MOI of 10:1 (phage to bacteria) and selected on 7H10 containing HYG. The marked deletion of whiB7 was confirmed by PCR using the primers listed in Table S2 in the supplemental material. The tap (Rv1258c) (bases 90 to 1230 removed) and Rv1257c deletion (bases 134 to 1188 removed) strains were similarly constructed and confirmed. All plasmids and phage used in this study are listed in Table S1.
Immunoblot analysis.
Immunoblots were performed as described previously (3). Briefly, either 10 or 20 μg of whole-cell lysate was separated on 12% Bis-Tris SDS-PAGE gels (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membranes by electrophoresis at 30 V for 1 h. Membranes were incubated with primary antibody specific to Eis (1:2,500) or GroES (1:20) for 1 h in 1 mM phosphate-buffered saline (PBS) with Tween 20 containing 0.25% skim milk, washed three times with PBS-Tween 20, and then probed for 1 h with anti-IgG secondary antibody (1:10,000) conjugated to horseradish peroxidase (HRP). The reactive bands were visualized using ECL detection reagents (Amersham). Anti-GroES antibodies were obtained from Colorado State University TB Vaccine Testing and Research Materials Contract.
Cell lysate collection and acetyltransferase assays.
M. tuberculosis strains were grown to mid-log phase, washed, harvested, suspended in 50 mM Tris HCl (pH 8.0), transferred to tubes containing glass beads (Biospec Products, Inc.), and lysed using a MiniBead beater (BioCold Scientific). Acetyltransferase assays were carried out as described previously (3). Reaction mixtures (1 ml) contained 0.1 mM acetyl coenzyme A (acetyl-CoA) (Sigma), 1 mM KAN or AMK, 1 mM 5,5′-dithio-bis(2-nitrobenzoate) (DTNB) (Sigma), 50 mM Tris-HCl (pH 8.0), and 50 μg cell lysate. To quantify acetylation, the degree of colorimetric change was subtracted from that of a control mixture that lacked antibiotic substrate. One unit of enzyme activity is defined as the amount of enzyme that catalyzes the formation of 1 nmol 2-nitro-5-benzoate (TNB) (14,151 M−1 cm−1) per min at room temperature at A412.
qRT-PCR.
M. tuberculosis strains were grown to mid-log phase (optical density at 600 nm [OD600] of 0.4 to 0.8) in Middlebrook 7H9 medium, bacteria were harvested and lysed using a FastPrep 120 (Bio 101 Savant), and RNA was purified using an RNeasy kit (Qiagen). One microgram of DNase-treated RNA from each M. tuberculosis strain was used to generate cDNA using the Promega reverse transcriptase system. Quantification of transcripts from 2 μl cDNA was performed by quantitative real-time PCR (qRT-PCR) using Probes master mix (Roche) in a LightCycler 480 detection system. Reactions to detect specific transcripts used 0.4 μM each primer and 0.1 μM probe (Roche) using the probe and primer pairs listed in Table S2 in the supplemental material. The relative amounts of each PCR product were calculated from standard curves obtained from PCR with the same primers and probes and serially diluted K204 or K301 cDNA and normalized to H37Rv cDNA.
RESULTS
Identification of novel mutations in KAN-resistant M. tuberculosis strains.
In a previous study, we isolated 20 spontaneous KAN-resistant (Kanr) mutants derived from the pansusceptible M. tuberculosis strains H37Rv and CDC1551 (6). A low concentration of KAN (5 μg/ml) was used to provide selection, and mutants with both high- and low-level KAN resistance phenotypes were recovered. The frequencies of recovery of the mutants were consistent with single-step mutational events and measured 2.9 × 10−7 for H37Rv mutants (denoted as K200 mutants) (Table 1) and 3.8 × 10−6 for CDC1551 mutants (denoted as K300 mutants) (Table 1). Each spontaneous mutant was analyzed by amplification and sequencing of the rrs and eis loci. Five of the 20 mutants (25%) harbored rrs mutations (A1401G or G1484T), and 2 of 20 (10%) had an eis promoter mutation. The remaining 13 of 20 (65%) spontaneous mutants were resistant to KAN at a low level (MICs of 10 to 30 μg/ml) and susceptible to AMK (MIC of ≤4 μg/ml) but did not harbor any mutations at either locus (Table 1). To identify possible mutations responsible for conferring KAN resistance, the spontaneous Kanr mutant K301 and the parental wild-type strain CDC1551 were analyzed by whole-genome sequencing. By using the previously annotated CDC1551 genome for comparison, we identified 17 single nucleotide polymorphisms (SNPs) present only in K301 compared to the parental strain CDC1551, and the majority of the mutations mapped to the highly repetitive PE-PGRS and hypothetical genes. (The complete SNP analysis is presented in Table S3 in the supplemental material.) In addition, a C deletion was identified in the region upstream of the gene encoding the transcriptional activator, WhiB7 (Fig. 1). In relation to the recently identified transcription start site of whiB7, this mutation maps within the 5′ untranslated region (UTR) of the whiB7 transcript at the position 133 bp downstream of the putative transcription start site (16). Subsequently, amplification and sequencing of the whiB7 5′ UTR and open reading frame in the remaining 13 low-level Kanr spontaneous mutants identified eight distinct whiB7 mutations (Fig. 1 and Table 1). The most commonly identified mutations occurred in a string of six cytosine residues 133 bp downstream of the whiB7 transcription start site and included either a deletion (6 mutants) or an insertion (1 mutant) of a cytosine (Fig. 1 and Table 1). Six other mutants harbored either cytosine or guanine deletions or transversions scattered throughout this region (Fig. 1 and Table 1).
Fig 1.

5′ untranslated region of whiB7 and mutations. The locations of mutations identified in spontaneous mutants are denoted by arrowheads; deletions by solid arrowheads, insertions by an open arrowhead, and transversions by gray arrowheads. The whiB7 start codon is boxed. A predicted IdeR binding region is underlined, and an inverted repeat element is denoted by arrows.
Mutations in the 5′ UTR of whiB7 cause enhanced whiB7 expression and low-level KAN resistance.
Based on the location of the whiB7 mutations, we hypothesized that these mutations could affect whiB7 expression. Using quantitative RT-PCR (qRT-PCR), we compared whiB7 expression levels in the spontaneous mutants to those in the parental strains CDC1551 and H37Rv (Fig. 2A). When these results were normalized to the housekeeping sigma factor sigA mRNA in each strain, the relative expression levels of whiB7 increased between 23- and 145-fold in the spontaneous mutants (Fig. 2A). Comparison of sigA and whiB7 mRNA transcripts relative to 16S rRNA in CDC1551 and H37Rv revealed no significant difference in basal-level gene expression between the parental strains, indicating that the increased amount of whiB7 transcript in the mutants is not likely affected by the genetic background (see Fig S1a in the supplemental material). Several of these mutants were reverted back to the wild-type sequence through allelic exchange using the specialized transducing phage phAZ70 to investigate whether these mutations are sufficient to confer whiB7 overexpression and low-level KAN resistance in M. tuberculosis (see Table S1 in the supplemental material). Three spontaneous mutants were chosen that represented each type of mutation identified: the CDC1551 mutant, K301 harboring the C deletion at position +133 (ΔC+133), and the H37Rv mutants K209 harboring the A-to-G transversion at position +237 (A+237G) and K205 harboring the C insertion at position +133 (+C+133). The allelic exchange strains displayed wild-type levels of whiB7 mRNA expression compared to CDC1551 and H37Rv, confirming that the presence of any of these point mutations leads to highly increased whiB7 expression and suggesting that the mutations are located in a regulatory region for whiB7 (Fig. 2B). Additionally, the allelic exchange strains were susceptible to KAN, confirming the role of these mutations located upstream of whiB7 in conferring low-level KAN resistance (Table 1 and Fig. 3A). The slightly higher MIC (4 μg/ml) observed for the K205 revertant (K205-WT) suggests that additional genetic factors may be acting in this strain that prevent complete complementation back to intrinsic resistance levels.
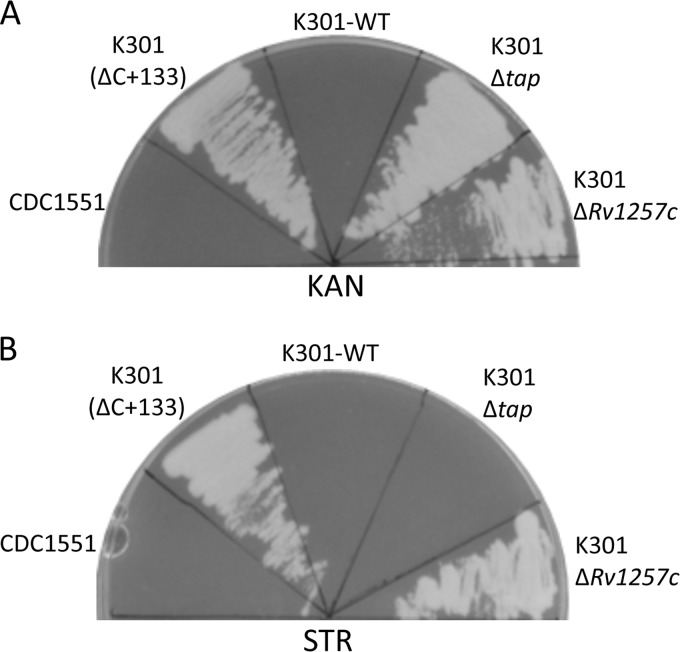
Fig 3.
Antibiotic resistance patterns of CDC1551 deletion strains. M. tuberculosis strains were streaked on 7H10 agar containing either 5 μg/ml KAN (A) or 2 μg/ml STR (B) and incubated for 28 days at 37°C.
KAN resistance in whiB7 mutants is due to increased expression of eis.
Previous microarray analysis indicated that eis is a member of the whiB7 regulon and that increased expression of whiB7 leads to enhanced levels of eis mRNA (10). Additionally, we recently showed that overexpression of the chromosomally encoded aminoglycoside acetyltransferase Eis (Rv2416c) confers low-level KAN resistance in M. tuberculosis through acetylation and inactivation of the drug (3). Taken together, these studies suggested that overexpression of whiB7 could lead to the subsequent upregulation of eis, which could explain the observed KAN resistance. Indeed, when we measured eis mRNA levels in the spontaneous mutants by qRT-PCR, we found that eis expression increased by 36- to 150-fold (Fig. 2A). The ratio of eis transcripts to sigA mRNA in the mutants is comparable to those of eis promoter mutants that are known to be resistant to KAN, such as K204, which harbors an eis C−14T allele (Table 1 and Fig. 2A). Additionally, cell lysates generated from the mutant strains with increased expression of whiB7 exhibited acetyltransferase activity against both KAN and AMK, while activity in the parental strains, CDC1551 and H37Rv, was either very low or undetectable (Fig. 2C). The amount of activity detected when KAN was used as a substrate was three times higher than that with AMK, which is characteristic of Eis protein activity (Fig. 2C) (3). In the whiB7 allelic exchange strains, eis mRNA expression and acetyltransferase activity reverted to wild-type levels, indicating that the mutations are sufficient to increase expression of eis (Fig. 2B and D). To confirm the acetyltransferase activity was due to Eis, we performed immunoblots on the cell lysates of each mutant and allelic exchange isolate with an anti-Eis antibody. As expected, Eis was more abundant in lysates generated from strains harboring mutations upstream of whiB7, suggesting that the mechanism of KAN resistance in these mutants is due to acetylation of the drug by Eis (Fig. 2E and F).
Increased expression of whiB7 also leads to STR resistance.
WhiB7 is thought to regulate expression of several genes involved in intrinsic antibiotic resistance of mycobacteria (10). It is possible that additional genes in the whiB7 regulon may be overexpressed in the mutants, and this modulation in gene expression could contribute to resistance to additional antibiotics. The MIC was determined for wild-type and mutant strains for several clinically used anti-TB drugs: streptomycin (STR), capreomycin (CAP), rifampin (RIF), isoniazid (INH), and ofloxacin (OFX). While there was no difference in MICs of CAP, RIF, INH, or OFX (data not shown), the MIC of STR increased 16-fold to 8 μg/ml for strains harboring mutations upstream of whiB7 (Table 1). Sequence analysis of the rrs, rpsL, and gidB loci in each mutant confirmed that no second site mutations were present that would account for the STR resistance (17–19). The Clinical and Laboratory Standards Institute document currently recommends testing STR susceptibility at two levels (2 μg/ml and 10 μg/ml) to distinguish between high- and low-level STR-resistant (Strr) isolates (11). Since the MIC is below 10 μg/ml, these strains are considered low-level Strr (11). The exception was K205, which harbors a +C+133 whiB7 allele and a slightly higher STR MIC of 10 μg/ml (Table 1). The whiB7 allelic exchange strains were susceptible to STR (MIC of ≤2 μg/ml) (Table 1 and Fig. 3B), confirming that mutations in the 5′ UTR of whiB7 confer cross-resistance to both KAN and STR and suggesting that a component of the whiB7 regulon is responsible for the STR resistance.
STR resistance is due to overexpression of tap.
Eis was initially ruled out as a contributing factor to STR resistance because STR is not a substrate of Eis (3), and eis promoter mutants such as K204 that overexpress the eis gene are susceptible to STR (Table 1). Instead, examination of the previously characterized whiB7 regulon revealed the tap (Rv1258c) gene encoding an efflux pump as a strong candidate that could contribute to STR resistance (10, 20). Numerous studies have indicated a role for efflux pumps in antibiotic resistance in M. tuberculosis, and the Tap homolog from the related mycobacterium M. fortuitum is involved in efflux of macrolides and aminoglycosides, including STR (20, 21). Therefore, we hypothesized that STR could also be a substrate of the M. tuberculosis Tap efflux pump. We confirmed that tap mRNA levels were highly elevated in the mutants by 30- to 175-fold (Fig. 2A). Also, we confirmed that mutations upstream of whiB7 confer increased tap expression, given that reversion of the mutations back to wild-type sequence in the allelic exchange strains yields tap expression levels similar to those of parental strains CDC1551 and H37Rv (Fig. 2B). To determine whether the Tap efflux pump was responsible for conferring STR resistance, we constructed tap deletion strains in both a wild-type and K301 (ΔC+133 mutation) background. However, tap is the first gene in a two-gene operon that includes the cotranscribed gene Rv1257c, which is annotated as coding for a putative oxidoreductase with no known role in antibiotic resistance in M. tuberculosis (10). We constructed deletions of tap and Rv1257c individually and then assessed the STR and KAN resistance phenotypes to circumvent the possibility that polar effects on Rv1257c due to disruption of tap could be responsible for the STR resistance phenotype. Deletion of the Rv1257c gene in either CDC1551 (wild type) or K301 (ΔC+133 mutation) had no effect on the STR or KAN MIC, and disruption of the tap gene did not significantly affect KAN resistance (Table 1 and Fig. 3A). However, the deletion of tap from either background caused the knockouts to become hypersusceptible to STR, decreasing the MIC 5-fold compared to the wild-type strains (Table 1 and Fig. 3B). These data confirm that the STR resistance in K301 is due to enhanced expression of tap, which probably results in increased efflux of STR. Overall, these data suggest that the cross-resistance between KAN and STR in the mutant strains is due to two distinct mechanisms: inactivation of KAN by the Eis protein and efflux of STR from the bacterial cell by the Tap efflux pump.
whiB7 contributes to the intrinsic resistance of M. tuberculosis to STR, but not KAN.
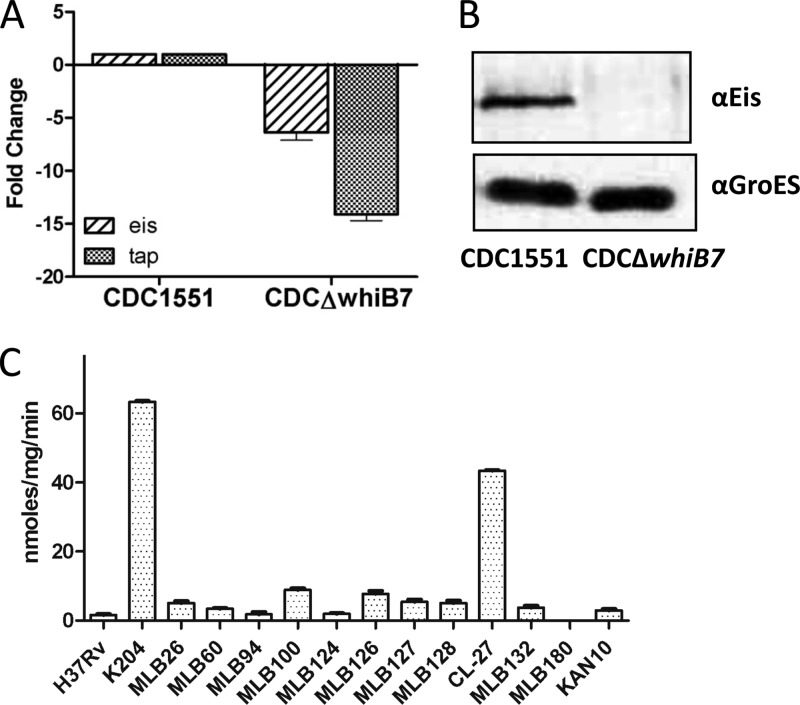
In agreement with a previous report (10), our results found that whiB7 is required for the intrinsic STR resistance in M. tuberculosis (MIC of 0.5 μg/ml) since a CDC1551ΔwhiB7 deletion strain is 2-fold more susceptible to STR than the wild type (Table 1). The loss of whiB7 leads to a 15-fold reduction in basal-level tap mRNA, which likely contributes to the reduced MIC of STR by decreasing efflux of STR from the cell (Fig. 4A). Similarly, in the absence of WhiB7, eis transcripts are reduced by 7.5-fold and the Eis protein is no longer detectable by immunoblotting (Fig. 4A and B). However, this reduction of Eis does not affect susceptibility to KAN, indicating that neither whiB7 nor eis is a limiting factor in the intrinsic resistance of M. tuberculosis to KAN (CDC1551ΔwhiB7) (Table 1). The observation that Eis can be completely removed from a cell (such as in an H37RvΔeis deletion strain) with no resulting deviation from wild-type KAN MIC levels supports this hypothesis (Table 1). Interestingly, disruption of whiB7 in the eis promoter mutant K204 (which harbors a C−14T mutation) has no effect on eis expression, indicating that expression from the mutant eis promoter is independent of WhiB7 (see Fig S1b in the supplemental material).
Fig 4.
eis and tap expression in a whiB7 knockout and acetyltransferase activity in Kanr clinical isolates. (A) The relative expression of gene transcripts was determined by qRT-PCR and normalized to sigA in H37Rv. Error bars represent the SEM from at least 3 experiments. (B) Immunoblot analysis of cell lysates. Twenty micrograms of cell lysate was probed with anti-Eis antibodies to ensure that basal-level Eis expression could be detected. (C) Sixteen clinical isolates with unexplained KAN resistance were analyzed by sequencing the eis, rrs, and whiB7 loci. With the exception of isolate CL-27 (which harbors a +C+133 whiB7 mutation), all of the loci were wild type. Acetyltransferase activity due to Eis was measured from crude cell lysates from 12 of the isolates and is expressed in nmol/mg/min. However, the remaining four clinical isolates grew very poorly in 7H9 medium and did not reach an OD600 sufficient to perform the acetyltransferase assays. Isolates CL-27, MLB100, and MLB126 displayed low-level cross-resistance to KAN and STR. H37Rv is a wild-type laboratory strain, and K204 harbors an eis C−14T mutation.
Increased whiB7 expression leads to increased frequency of emergence of high-level Strr mutants.
The use of STR as a chemotherapeutic for TB patients has been hindered by the high frequency of emergence of Strr strains (22). STR resistance in M. tuberculosis is commonly associated with the binding site of STR at the ribosome (9, 19). Point mutations in either rrs or rpsL have been shown to confer high-level STR resistance, with MICs of 10 μg/ml or greater (9, 19). Additionally, mutations throughout the open reading frame of the rRNA methyltransferase gene gidB confer low-level STR resistance (2 μg/ml ≤ MIC < 10 μg/ml), presumably by inhibiting efficient STR binding to the ribosome (18, 23). Intriguingly, gidB mutants are also 2,000 times more likely to acquire high-level STR resistance than a strain with a wild-type gidB locus (18). This observation led us to hypothesize that low-level Strr strains harboring mutations upstream of whiB7 may demonstrate a similar phenomenon. By plating CDC1551 (wild type) and K301 (ΔC+133 mutation) on plates containing a high concentration of STR (10 μg/ml), we compared the frequencies of emergence of high-level Strr isolates in the different backgrounds. Similar to a previous report that investigated gidB mutants (18), high-level Strr mutants arose in the K301 mutant at an average frequency greater than 1,100-fold higher than that of wild-type cells (Table 2). However, in contrast to the previous study that found numerous second site mutations at the rpsL locus (18), high-level Strr mutants derived from K301 primarily harbored mutations in gidB. Among 16 mutants analyzed, 12 had acquired a Gly37Stop mutation in gidB, while two harbored Pro78Ser missense mutations. No mutations were identified in the remaining two isolates in either gidB, rpsL, or rrs. The high mutation frequency was specific to STR as the emergence of high-level Kanr mutants (frequency of <8.40 × 10−9) remained the same in either strain background, indicating that the overall mutation rate of K301 was not affected.
Table 2.
Effect of the whiB7 ΔC+133 mutation on emergence of high-level Strr mutants
| Strain | STR (μg/ml) | Mutation frequency in: |
|
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| CDC1551 | 2 | 6.18 × 10−7 | 1.79 × 10−6 |
| K301 | 2 | NAa | NA |
| CDC1551 | 10 | <8.40 × 10−9 | <8.40 × 10−9 |
| K301 | 10 | 4.86 × 10−6 | 1.39 × 10−5 |
NA, not applicable.
Identification of a 5′ UTR whiB7 mutation in a clinical isolate of M. tuberculosis.
We sequenced whiB7 and its upstream region in 16 clinical isolates with unexplained low-level KAN resistance and identified one clinical isolate (CL-27) that harbored a mutation in the 5′ UTR of whiB7 and demonstrated acetyltransferase activity (Table 1 and Fig. 4C). Notably, CL-27 was one of four isolates (25%) that also demonstrated low-level Strr. A single cytosine insertion at position +133 was mapped to the whiB7 locus in isolate CL-27 (Table 1). This isolate was cross-resistant to both KAN (MIC of 20 μg/ml) and STR (MIC of 10 μg/ml), with a drug resistance profile that matched that of the spontaneous mutant K205, which also harbors a +C+133 whiB7 mutation (Table 1). The remaining unexplained Kanr isolates contained either very low or undetectable acetyltransferase activity (Fig. 4C).
DISCUSSION
In M. tuberculosis, resistance to multiple drugs is often due to the sequential acquisition of mutations at loci involved in drug resistance. However, cross-resistance occurs when a single-base-pair mutation (such as rrs A1401G) confers resistance to several members of a class of antibiotics, rendering several treatment options unviable simultaneously. Understanding the molecular mechanisms of cross-resistance should facilitate faster detection of drug-resistant M. tuberculosis isolates, a prospect that is especially critical for patients infected with MDR or XDR TB, who have limited treatment options. Here we characterize novel mutations located in the 5′ UTR of whiB7 that lead to KAN and STR cross-resistance in M. tuberculosis. As this study investigates a limited sample of clinical isolates, the frequency of whiB7 5′ UTR mutations in aminoglycoside cross-resistant clinical isolates remains unknown. However, given that these mutations cluster at a single locus, rapid molecular tests could be developed to detect these mutations and resistance pattern, ultimately leading to improved treatment for TB patients.
An increase in whiB7 expression leads to upregulation of at least two different antibiotic resistance genes in the whiB7 regulon—eis and tap. This induction is likely due to the direct recruitment and binding of WhiB7 to conserved putative WhiB7 binding sites located immediately upstream of each promoter (16). Similar to the closely related transcriptional regulator, WhiB3, WhiB7 may additionally regulate genes by interacting with the principal sigma factor SigA at promoters (24).
We recently demonstrated that Eis acetylates KAN, and mutations in the eis promoter that cause eis overexpression are a major mechanism of low-level KAN resistance in M. tuberculosis (3). Strains harboring mutations in the 5′ UTR of whiB7 demonstrate KAN MICs, eis expression levels, and acetyltransferase activities that are equivalent to those of eis promoter mutants, verifying that the KAN resistance in the mutants is due to inactivation of the drug by Eis. Notably, enhanced expression of the Tap efflux pump due to whiB7 5′ UTR mutations is a novel mechanism of low-level STR resistance in M. tuberculosis. Our genetic data provides strong evidence that STR is a substrate of the Tap efflux pump and that modulated expression of tap can confer either increased STR resistance or susceptibility. Given that the deletion of tap from either a wild-type or mutant background caused hypersusceptibility to STR suggests that Tap plays a key role in the intrinsic resistance of M. tuberculosis to STR. Previous studies have indicated a role for Tap in clinical STR resistance. A study investigating Strr isolates found 22% of clinical isolates showed increased susceptibility to STR in the presence of proton motive force inhibitors, suggesting an efflux pump, such as Tap, contributed to the resistance (17). Tap was also proposed to be involved in the efflux of INH, RIF, and OFX (25–27). However, we found no evidence of increased resistance to any of these compounds or CAP when tap was overexpressed, suggesting that none of these drugs are Tap substrates in M. tuberculosis.
An intriguing observation from this work is that whiB7 mutants progressed to high-level Strr strains at a frequency much higher than that of their wild-type counterparts. This phenomenon was previously described in strains that harbor gidB mutations (18). In both instances, the emergence of high-level resistance is specific to STR and not other antibiotics. In the case of K301, high-level STR resistance emerged at an average frequency of 9.31 × 10−6, similar to the mutation frequency reported for single-step gidB mutations (2.8 × 10−6) (18). Indeed, greater than 85% of the high-level Strr mutants harbored gidB mutations, suggesting that the frequency of emerging high-level Strr strains is largely a reflection of the spontaneous mutation rate at the gidB locus. These data show that a mutation at either the whiB7 or gidB locus leads to the acquisition of high-level STR resistance at an elevated frequency, which may partly explain why STR resistance can develop so quickly in the host.
It is interesting that the variety of mutations found in the 5′ UTR of whiB7 (insertions, deletions, and transversions) all confer overexpression of whiB7, suggesting that each causes whiB7 misregulation. whiB7 expression is modulated in response to diverse types of environmental stresses, including antibiotics, fatty acids, lung surfactant, heat shock, low iron, and upon entry into macrophages (10, 28–33). Additionally, at least seven proteins are predicted to interact at the promoter region of whiB7, although none of the residues that interact with the whiB7 promoter region are known (29, 30). One study postulated that a second small open reading frame could be present in the 5′ UTR of whiB7, although there is currently no evidence to suggest a hypothetical protein is produced from this DNA region (10). More likely, the recent identification of cyclic AMP receptor protein (Crp) binding sites in the 5′ UTR of several whiB family members suggests that this region could be an important regulatory feature for whiB7 as well (32). Consistent with this hypothesis, the 5′ UTR of whiB7 appears to contain several regulatory elements, including an iron-responsive IdeR binding consensus sequence (30) and an inverted repeat of unknown function (Fig. 1). Additionally, several of the mutations identified in this study cluster in the region between +124 and +133 bp, suggesting a regulatory element could interact with this sequence. Therefore, the mutations identified in the whiB7 5′ UTR in this study likely represent an uncharacterized region important for whiB7 regulation.
Both eis and tap likely serve other physiological or metabolic functions in the cell. Increased expression of eis was shown to confer a survival advantage in monocytes in a Beijing clinical isolate of M. tuberculosis, suggesting that strains which overexpress whiB7 could have an enhanced survival phenotype (34). Although the true substrate of Eis is still unknown, several studies demonstrate that Eis modulates cytokine expression in immune cells to suppress innate immunity and regulate autophagy and cell death during an M. tuberculosis infection (35, 36). This modulation was recently shown to occur in a redox-dependent manner (37). Given that the WhiB family of proteins is predicted to respond to changes in the redox state of the cell through an iron-sulfur cluster in the structure, it is possible that WhiB7 contributes to Eis-mediated immune suppression (24). Likewise, it is unlikely that the primary function of Tap is drug efflux. Tap was recently shown to play a key role in the ability of M. tuberculosis to grow in macrophages and the development of antibiotic tolerance during an infection, suggesting whiB7 mutants may have increased drug tolerance (27). The variety of stresses that WhiB7 responds to suggests that Tap, and perhaps other stress-induced efflux pumps, may work to alleviate stresses such as those encountered upon entering a macrophage. Additionally, antibiotics that target the 16S rRNA, including KAN, STR, and tetracycline, are potent inducers of whiB7, suggesting that whiB7 responds to ribosomal stress signals in the cell (10, 31). An intriguing aspect of our data is that the WhiB7-mediated response to ribosomal stress upregulates at least two genes that specifically alleviate aminoglycoside-induced ribosomal stress due to drug inactivation or extrusion. These observations raise the question of whether exposure to stressful conditions such as antibiotic treatment can induce phenotypic antibiotic resistance on M. tuberculosis. If so, this may have implications for designing treatment for tuberculosis.
Prior to this work, resistance to KAN and STR within a single strain was thought to be due to unlinked chromosomal mutations. However, we have identified point mutations in the upstream region of the transcriptional activator whiB7 that confer cross-resistance to both drugs. Identification of the molecular mechanisms that are responsible for conferring resistance and cross-resistance is critical for the future development of accurate and rapid molecular tests to diagnose drug-resistant TB strains. Importantly, strains harboring mutations in the 5′ UTR of whiB7 could have several phenotypes and consequences: low-level cross-resistance to the second-line drugs KAN and STR, a high frequency of progression to high-level Strr, a modulated stress response system which could alter the immune response to the infection, and possibly increased antibiotic tolerance.
ACKNOWLEDGMENTS
We acknowledge Richard Friedmann for providing the anti-Eis antibody used in this study, David Sikes for technical assistance, and Melisa Willby and Patrick M. Reeves for critical reading of the manuscript.
Footnotes
Published ahead of print 4 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02191-12.
REFERENCES
- 1. World Health Organization 2009. Global tuberculosis control—epidemiology, strategy, financing. World Report 2009 World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Centers for Disease Control and Prevention 2006. Notice to readers: revised definition of extensively drug-resistant tuberculosis. MMWR Morb. Mortal. Wkly. Rep. 55:1176 [Google Scholar]
- 3. Zaunbrecher MA, Sikes RD, Jr, Metchock B, Shinnick TM, Posey JE. 2009. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 106:20004–20009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Magnet S, Blanchard JS. 2005. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105:477–498 [DOI] [PubMed] [Google Scholar]
- 5. Alangaden GJ, Kreiswirth BN, Aouad A, Khetarpal M, Igno FR, Moghazeh SL, Manavathu EK, Lerner SA. 1998. Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1295–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maus CE, Plikaytis BB, Shinnick TM. 2005. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:3192–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suzuki Y, Katsukawa C, Tamaru A, Abe C, Makino M, Mizuguchi Y, Taniguchi H. 1998. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J. Clin. Microbiol. 36:1220–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taniguchi H, Chang B, Abe C, Nikaido Y, Mizuguchi Y, Yoshida SI. 1997. Molecular analysis of kanamycin and viomycin resistance in Mycobacterium smegmatis by use of the conjugation system. J. Bacteriol. 179:4795–4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Via LE, Cho SN, Hwang S, Bang H, Park SK, Kang HS, Jeon D, Min SY, Oh T, Kim Y, Kim YM, Rajan V, Wong SY, Shamputa IC, Carroll M, Goldfeder L, Lee SA, Holland SM, Eum S, Lee H, Barry CE., III 2010. Polymorphisms associated with resistance and cross-resistance to aminoglycosides and capreomycin in Mycobacterium tuberculosis isolates from South Korean patients with drug-resistant tuberculosis. J. Clin. Microbiol. 48:402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morris RP, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, Schoolnik G, Thompson CJ. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:12200–12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. NCCLS 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA: [PubMed] [Google Scholar]
- 12. Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, Hooks DP, Cowan LS, Plikaytis BB, Posey JE. 2011. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55:2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nusbaum C, Ohsumi TK, Gomez J, Aquadro J, Victor TC, Warren RM, Hung DT, Birren BW, Lander ES, Jaffe DB. 2009. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat. Methods 6:67–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017 [DOI] [PubMed] [Google Scholar]
- 16. Burian J, Ramon-Garcia S, Sweet G, Gomez-Velasco A, Av-Gay Y, Thompson CJ. 2012. The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J. Biol. Chem. 287:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spies FS, da Silva PE, Ribeiro MO, Rossetti ML, Zaha A. 2008. Identification of mutations related to streptomycin resistance in clinical isolates of Mycobacterium tuberculosis and possible involvement of efflux mechanism. Antimicrob. Agents Chemother. 52:2947–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K. 2007. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 63:1096–1106 [DOI] [PubMed] [Google Scholar]
- 19. Meier A, Sander P, Schaper KJ, Scholz M, Bottger EC. 1996. Correlation of molecular resistance mechanisms and phenotypic resistance levels in streptomycin-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 40:2452–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ainsa JA, Blokpoel MC, Otal I, Young DB, De Smet KA, Martin C. 1998. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J. Bacteriol. 180:5836–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Louw GE, Warren RM, Gey van Pittius NC, McEvoy CR, Van Helden PD, Victor TC. 2009. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53:3181–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Medical Research Council 1948. Streptomycin treatment of pulmonary tuberculosis. Br. Med. J. 2:769–782 [PMC free article] [PubMed] [Google Scholar]
- 23. Wong SY, Lee JS, Kwak HK, Via LE, Boshoff HI, Barry CE., III 2011. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55:2515–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. 2009. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5:e1000545 doi:10.1371/journal.ppat.1000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang X, Zhang W, Zhang Y, Gao F, Lu C, Zhang X, Wang H. 2008. Assessment of efflux pump gene expression in a clinical isolate Mycobacterium tuberculosis by real-time reverse transcription PCR. Microb. Drug Resist. 14:7–11 [DOI] [PubMed] [Google Scholar]
- 26. Siddiqi N, Das R, Pathak N, Banerjee S, Ahmed N, Katoch VM, Hasnain SE. 2004. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection 32:109–111 [DOI] [PubMed] [Google Scholar]
- 27. Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145:39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwab U, Rohde KH, Wang Z, Chess PR, Notter RH, Russell DG. 2009. Transcriptional responses of Mycobacterium tuberculosis to lung surfactant. Microb. Pathog. 46:185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo M, Feng H, Zhang J, Wang W, Wang Y, Li Y, Gao C, Chen H, Feng Y, He ZG. 2009. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 19:1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gold B, Rodriguez GM, Marras SA, Pentecost M, Smith I. 2001. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42:851–865 [DOI] [PubMed] [Google Scholar]
- 31. Geiman DE, Raghunand TR, Agarwal N, Bishai WR. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50:2836–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larsson C, Luna B, Ammerman NC, Maiga M, Agarwal N, Bishai WR. 2012. Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One 7:e37516 doi:10.1371/journal.pone.0037516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rohde KH, Abramovitch RB, Russell DG. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2:352–364 [DOI] [PubMed] [Google Scholar]
- 34. Wu S, Barnes PF, Samten B, Pang X, Rodrigue S, Ghanny S, Soteropoulos P, Gaudreau L, Howard ST. 2009. Activation of the eis gene in a W-Beijing strain of Mycobacterium tuberculosis correlates with increased SigA levels and enhanced intracellular growth. Microbiology 155:1272–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lella RK, Sharma C. 2007. Eis (enhanced intracellular survival) protein of Mycobacterium tuberculosis disturbs the cross regulation of T-cells. J. Biol. Chem. 282:18671–18675 [DOI] [PubMed] [Google Scholar]
- 36. Samuel LP, Song CH, Wei J, Roberts EA, Dahl JL, Barry CE, III, Jo EK, Friedman RL. 2007. Expression, production and release of the Eis protein by Mycobacterium tuberculosis during infection of macrophages and its effect on cytokine secretion. Microbiology 153:529–540 [DOI] [PubMed] [Google Scholar]
- 37. Shin DM, Jeon BY, Lee HM, Jin HS, Yuk JM, Song CH, Lee SH, Lee ZW, Cho SN, Kim JM, Friedman RL, Jo EK. 2010. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 6:e1001230 doi:10.1371/journal.ppat.1001230 [DOI] [PMC free article] [PubMed] [Google Scholar]