Abstract
Monte Carlo simulations (MCS) present a powerful tool to evaluate candidate regimens by determining the probability of target attainment. Although these assessments have traditionally incorporated variability in pharmacokinetic (PK) parameters and MICs, consideration of interstrain pharmacodynamic (PD) variability has been neglected. A population PK/PD model was developed for doripenem using murine thigh infection data based on 20 bacterial strains. PK data were fit to a linear two-compartment model with first-order input and elimination processes and an absorption lag time from a separate site (r2 > 0.96). PK parameters were utilized to simulate free-drug profiles for various regimens in PD studies, from which the percentage of the dosing interval for which free-drug concentrations exceed the MIC of the targeted strain (%fT>MIC) was calculated. Doripenem PD was excellently described with Hill-type models (r2 > 0.98); significant differences between mean PD estimates determined using a two-stage approach versus population analyses were not observed (P > 0.05); however, the variance in 50% effective concentration (EC50) and maximum effect (Emax) among strains was much greater using the two-stage approach. Even using the population approach, interstrain variability in EC50 (coefficient of variation expressed as a percentage [CV%] = 29.2%) and H (CV% = 46.1%) parameters was substantive, while the variability in Emax (CV% = 19.7%) was modest. This resulted in extensive variability in the range of %fT>MIC targets associated with stasis to those associated with a 2-log10 reduction in bacterial burden (CV% ∼ 50%). It appears that MCS, based on the assumption that PD variability is due to MIC alone, underestimates variability and may consequently underestimate treatment failures.
INTRODUCTION
The emerging threat posed by multidrug-resistant (MDR) bacterial strains has heightened, necessitating an urgent review of antimicrobial treatment strategies. Carbapenems are synthetic parenteral broad-spectrum β-lactam antibiotics, commonly employed as empirical therapy for the treatment of serious nosocomial infections. Doripenem (Doribax; Ortho-McNeil Pharmaceuticals, NJ) (1) is the most recent addition to the carbapenem class and boasts a broad spectrum of activity, providing excellent coverage across a wide range of Gram-positive and -negative bacteria (including MDR and β-lactamase-producing strains) as well as anaerobes (2). In comparison to the other carbapenems, doripenem has been credited with several advantages, including enhanced potency, superior stability when reconstituted in standard diluents for infusion (3, 4), and a lower potential to cause seizure-related toxicities in animal studies (5). The percentage of the dosing interval for which free-drug concentrations exceed the MIC of the targeted strain (i.e., %fT>MIC) is believed to be the pharmacokinetic (PK)/pharmacodynamic (PD) index most predictive of carbapenem efficacy. The extended stability of doripenem thus makes it a suitable candidate for administration in prolonged infusion regimens, allowing the %fT>MIC to be maximized.
Rational design of antimicrobial dosage regimens requires consideration of both PK and PD concepts in order to optimize drug exposure and enhance antimicrobial activity, translating to favorable clinical outcomes. For this purpose, Monte Carlo simulations (MCS) have been utilized as a powerful computer modeling tool to evaluate the probability of attaining predefined PK/PD targets following administration of candidate dosage regimens. This novel approach does not rely on mean PK parameter values but traditionally integrates intersubject variability in PK parameters together with the dispersion in microbiological susceptibility surveillance data (MICs) to determine the probability of target attainment across a large population of simulated subjects.
Evidently, the increasing body of literature accumulated from phase 1, 2, and 3 clinical studies of doripenem in recent years has seen the development and refinement of population PK models for doripenem (6–8), which incorporate relationships between PK parameters and key covariates in order to describe the nature of interpatient PK variability. These models have been applied to various MCS in attempts to optimize doripenem exposure across a range of MICs (6, 8–11). The existence of extensive PD variability among bacterial species has been previously demonstrated (particularly for β-lactam agents), with researchers reporting differences in the magnitude of PK/PD targets associated with activity for different organisms, as well as differences between and within drug classes (12). However, PD variability beyond that associated with MIC has been ignored in many MCS investigations to date, generating the potential for misleading evaluations of antimicrobial efficacy.
Consequently, as part of an intended MCS to evaluate doripenem regimens across a clinically relevant distribution of MICs while also accounting for interstrain PD variability, our objective here was to develop a population PD model to evaluate the extent of interstrain PD variability within our bacterial collection and determine the influence of this variability on the definition of PK/PD targets relating to activity. As a secondary objective, we further aimed to determine if (i) different measures of antimicrobial activity (compare equations 1 and 2 below) or (ii) different PD modeling methods (i.e., two-stage approach versus population approach) significantly influenced the results obtained.
MATERIALS AND METHODS
PK and PD data from neutropenic murine thigh infection models were provided by Andes et al. (13). Female Swiss ICR mice (Harlan Sprague Dawley; weight range, 24 to 27 g) were rendered neutropenic by 2 injections of cyclophosphamide at doses of 150 mg/kg of body weight and 100 mg/kg administered 4 days and 1 day before the study, respectively. Approval was obtained from the IACUC (William Middleton VA Hospital) prior to conducting animal experiments.
Murine PK studies and analyses.
Doripenem plasma concentrations were measured from groups of three thigh-infected neutropenic mice at each of the following time points: 0.25, 0.50, 0.75, 1, 1.5, 2, 3, and 4 h after subcutaneous administration (0.2-ml volume) of a single doripenem dose of 9.38, 37.5, or 150 mg/kg (8, 13). Blood was collected in heparinized capillary tubes, and each sample was analyzed individually. Concentrations were determined using a microbiologic assay with Staphylococcus aureus 6538P as the test organism. The lower limit of detection of doripenem was 0.12 mg/liter, and intraday variation was less than 10%. At each time point, the mean of the three samples was obtained, and data were weighted according to the standard deviation of the replicates. PK data were simultaneously comodeled in ADAPT 5 using a maximum-likelihood method (BMSR, CA) (14).
Murine PD studies and analyses.
Doripenem activity was assessed against 20 bacterial strains; these included strains of Pseudomonas aeruginosa (n = 1), Escherichia coli (n = 3), Klebsiella pneumoniae (n = 4), Enterobacter cloacae (n = 2), Staphylococcus aureus (n = 3), and Streptococcus pneumoniae (n = 7). MICs of doripenem against these strains were determined using standard Clinical and Laboratory Standards Institute (CLSI) methods and ranged from 0.004 to 0.5 mg/liter (see Table 2). Bacterial inocula were introduced into murine thighs (105.70 to 107.68 CFU/thigh) 2 h prior to therapy. Doripenem (powder; Shionogi and Co.) was prepared in 0.9% saline and administered subcutaneously at 6-h intervals over 24 h, with doses ranging between 0.018 and 4,800 mg/kg/day.
Table 2.
Bacterial strains and associated MICs, individual fitted population PD parameters, and %T>MIC values
| Organism | MIC (mg/liter) | Emaxa | EC50 (mg/liter)a | Hb | %fT>MIC (%) associated with: |
|
|---|---|---|---|---|---|---|
| Stasisb | 2-log reductionb,c | |||||
| E. cloacae 31-54A | 0.50 | 6.11 | 29.8 | 1.92 | 20.6 | 41.9 |
| E. cloacae 31-59A | 0.25 | 5.64 | 37.2 | 1.91 | 29.3 | 65.0 |
| E. coli 25922 | 0.015 | 4.36 | 53.1 | 3.77 | 54.4 | 152 |
| E. coli HBMS145 | 0.030 | 5.47 | 31.9 | 3.84 | 28.4 | 42.7 |
| E. coli HBMS154 | 0.060 | 5.55 | 34.4 | 2.33 | 27.1 | 52.2 |
| K. pneumoniae 43816 | 0.060 | 5.00 | 40.1 | 3.58 | 37.6 | 64.2 |
| K. pneumoniae 51504 | 0.060 | 4.60 | 49.3 | 2.94 | 47.7 | 108 |
| K. pneumoniae HBMS149 | 0.060 | 4.99 | 37.5 | 3.90 | 34.4 | 55.2 |
| K. pneumoniae HBMS152 | 0.060 | 5.34 | 43.6 | 1.38 | 31.6 | 103 |
| P. aeruginosa 27853 | 0.50 | 5.79 | 29.2 | 3.75 | 25.4 | 37.4 |
| S. aureus Smith | 0.015 | 5.94 | 25.3 | 3.92 | 21.2 | 30.3 |
| S. aureus 25923 | 0.015 | 5.27 | 39.0 | 2.38 | 33.1 | 66.9 |
| S. aureus 307192 | 4.0 | 5.49 | 27.6 | 5.66 | 25.1 | 33.0 |
| S. pneumoniae 10813 | 0.004 | 6.95 | 22.5 | 2.37 | 15.7 | 26.1 |
| S. pneumoniae 1293 | 0.25 | 6.27 | 30.3 | 2.02 | 22.3 | 43.0 |
| S. pneumoniae 1396 | 0.12 | 7.55 | 22.8 | 1.39 | 11.8 | 26.3 |
| S. pneumoniae 145 | 0.50 | 9.15 | 16.9 | 1.50 | 8.51 | 16.1 |
| S. pneumoniae 146 | 0.50 | 7.68 | 20.9 | 1.86 | 12.9 | 23.2 |
| S. pneumoniae 6301 | 0.004 | 7.49 | 26.1 | 0.67 | 5.23 | 29.6 |
| S. pneumoniae MNO418 | 0.004 | 5.97 | 30.9 | 1.94 | 21.6 | 44.3 |
S. pneumoniae is significantly different from all other species (Kruskal-Wallis test, P < 0.05).
S. pneumoniae is significantly different from K. pneumoniae, E. cloacae, and E. coli; K. pneumonia is significantly different from E. cloacae (Kruskal-Wallis test, P < 0.05).
Reduction in bacterial burden (representative of bactericidal activity).
The estimated murine PK parameters were used in conjunction with a previously reported murine doripenem plasma protein binding value of 25.2% (15) to simulate free-drug profiles for all regimens. Doripenem exposure was then expressed as the %fT>MIC (computed by numeric integration; ADAPT 5). The effect was described by the log10 difference in bacterial density (CFU/ml) for doripenem-treated mice at 24 h versus those for both (i) pre-antibiotic-exposed mice at 0 h (equation 1) and (ii) untreated control mice at 24 h (equation 2).
| (1) |
| (2) |
Using a standard two-stage PD modeling approach (SYSTAT version 13.00.05), doripenem exposure-activity relationships were individually fit to Hill-type models (equation 3) by nonlinear regression for each strain. A population PD analysis was also undertaken, whereby the exposure-activity relationships for all strains were simultaneously fit to Hill-type models using the MLEM algorithm in parallelized S-ADAPT (version 1.56) with the S-ADAPT Tran interface (16, 17); observations for each of the 20 bacterial strains were treated as 20 “subjects,” and all parameters were assumed to have a log normal likelihood distribution. Weighting of the observed log10 CFU/ml data for both PK/PD model analyses was assumed to be additive in the log domain. The %fT>MIC values associated with stasis and 1-log10- and 2-log10-unit changes in bacterial burden were subsequently calculated by solving equation 3 for the time above MIC (%fT>MIC) necessary to achieve the targeted effect (equation 4).
| (3) |
| (4) |
The significance of differences between selected PD parameters (EC50, Emax, and H) and targets (%fT>MIC) obtained from the two-stage approach versus a population approach was assessed by pairwise comparisons using a nonparametric Wilcoxon signed-rank test. The equivalence of PD parameters and targets was calculated for each strain using both modeling approaches by computing the (i) ratio (for the EC50 parameter and bacterial targets) or (ii) log ratio (for the H and Emax parameters) based on which transformation was Gaussian (SYSTAT version 13.00.05). Equivalence was concluded if the 90% confidence intervals for ratios and log ratios were within the limits of 80 to 125%. The significance of differences in variance between targets or parameter estimates derived from both methods was also determined using hypothesis testing (F test; SYSTAT version 13.00.05). Results from the population analysis were employed to test the significance of differences in targets and parameter estimates by bacterial species using a nonparametric Kruskal-Wallis one-way analysis of variance (ANOVA; P < 0.05) with the Dwass-Steel-Critchlow-Fligner test for all pairwise comparisons (SYSTAT version 13.00.05); bacterial species with <2 strains were excluded from this analyses.
RESULTS
PK analyses.
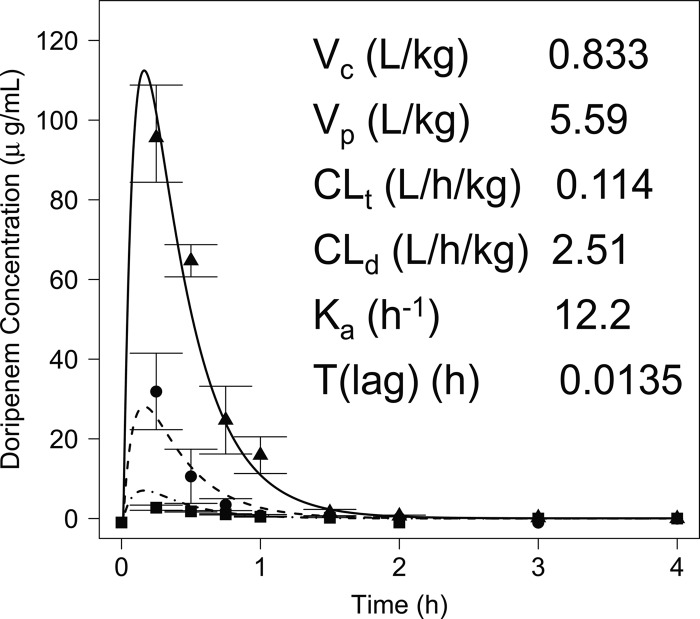
Concentration-time profiles of doripenem were well characterized by a linear two-compartment model with first-order input and elimination processes and an absorption lag time from a separate site (Fig. 1), as described by ordinary differential equations dX(c)/dt = ka × X(a) − CLd × [C(c) − C(p)] − CLt × C(c), dX(p)/dt = CLd × [C(c) − C(p)], and dX(a)/dt = −ka × X(a), where X(c) and X(p) are the amounts of doripenem (mg) in the central and peripheral compartments, respectively, X(a) is the amount of doripenem at the absorptive site following bolus administration, C(c) and C(p) are the concentrations of doripenem (mg/liter) in the central and peripheral compartments, respectively, ka is the first-order absorption rate constant, and CLd is the distributional clearance between the central and peripheral compartments, and CLt is the total clearance from the central compartment. Figure 2 illustrates the overlay of the observed and fitted plasma concentrations for each regimen.
Fig 1.

Two-compartment model used to model the PK characteristics of doripenem in mice. X(c), X(p), and X(a) represent the amounts of drug (mg) in the central, peripheral, and absorptive sites, respectively; ka is the first-order absorption rate constant (h−1); T(lag) is the absorption lag time (h); Vc and Vp are the volumes of distribution of the central and peripheral compartments, respectively (liters/kg); Cld is the distributional clearance between the central and peripheral compartments (liters/h/kg); and Clt is the total clearance from the central compartment (liters/h/kg).
Fig 2.
Plot of the observed doripenem concentrations (individual symbols) versus the derived PK model following administration of 150 (solid line), 37.5 (dashed line), or 9.38 (dot-dashed line) mg/kg in mice (r2 = 0.977, 0.961, and 0.968, respectively).
PD analyses.
Activity was quantified as the relationships between the log10 change in bacterial density (CFU/ml) in doripenem-treated mice at 24 h and preantibiotic-exposed mice at 0 h (equation 1) and between the log10 change in bacterial density in doripenem-treated mice and untreated control mice at 24 h (equation 2). As the EC50, Emax, and H parameters derived using both measures of activity were equivalent (data not shown), bacterial counts obtained from pre-antibiotic-exposed mice at 0 h (equation 1) were employed as the reference to measure activity for the remainder of this investigation.
Results obtained from both the population and two-stage analyses have been summarized in Table 1. Table 2 presents a listing of the PD parameters and targets by strain based on the population analysis, while Table 3 provides a statistical summary of these results by species (for species containing ≥3 strains). Using both modeling approaches, doripenem exposure-activity relationships were excellently described with Hill-type models for all 20 strains (Tables 1 and 2) (median r2 for two-stage analysis = 0.991, overall r2 for population analysis = 0.978). PD parameters were used to calculate the %fT>MIC targets associated with various log unit changes in bacterial burden for each strain. Since activity was defined in reference to pre-antibiotic-exposed mice, static activity was associated with a 0-log10 reduction in bacterial density, while bactericidal activity was associated with an ∼2-log10 reduction. For E. coli 25922, K. pneumoniae 51504, and K. pneumoniae HBMS152, calculated %fT>MIC targets associated with an ∼2-log reduction exceeded a value of 100%, implying that bactericidal activity was not theoretically achievable for these strains. No statistically significant differences between the selected PD parameters (EC50, Emax, and H) and targets obtained from the two-stage population analysis were observed (P > 0.2). In testing for equivalence, the 90% confidence interval ratio of EC50 ranged from 85.0 to 119%, while the ratios of Emax and H were 85.5 to 106% and 86.4 to 125%, respectively. The 90% confidence interval ratios of bacterial targets associated with stasis and a 2-log10-unit bacterial reduction were 95.4 to 102% and 90.8 to 107%, respectively. Thus, all parameters determined using the two-stage and population approaches were deemed to be equivalent. Hypothesis testing for differences in variances revealed that the variances observed in the two-stage estimates of EC50 and Emax for each strain were 3.74 times (F test; P < 0.05) and 2.30 times (F test; P < 0.05) greater, respectively, than the population estimates.
Table 1.
Summary of the individual fitted PD parameters determined using a two-stage approach and population approacha
| Analysis and type of value | E0 | Emax | H | EC50 (mg/liter) | r2 | %fT>MIC (%) associated with: |
||
|---|---|---|---|---|---|---|---|---|
| Stasis | Reductionb of: |
|||||||
| 1 log | 2 logs | |||||||
| Population | 0.978 | |||||||
| Minimum | 1.90 | 4.36 | 0.670 | 16.9 | 5.23 | 11.9 | 16.1 | |
| Maximum | 2.41 | 9.5 | 5.67 | 53.1 | 54.4 | 71.3 | 153 | |
| Median | 2.13 | 5.71 | 2.35 | 30.6 | 25.2 | 30.9 | 42.8 | |
| Mean | 2.12 | 6.03 | 2.65 | 32.4 | 25.7 | 34.9 | 53.0 | |
| SD | 0.118 | 1.19 | 1.22 | 9.45 | 12.3 | 16.3 | 33.8 | |
| CV% | 5.6 | 19.7 | 46.1 | 29.2 | 48.0 | 46.7 | 63.8 | |
| Two stage | ||||||||
| Minimum | 1.36 | 3.64 | 1.00 | 8.04 | 0.906 | 5.11 | 9.50 | 12.3 |
| Maximum | 3.00 | 10.0 | 5.63 | 69.1 | 1.00 | 58.7 | 83.8 | 131 |
| Median | 2.12 | 6.21 | 1.93 | 32.7 | 0.991 | 26.6 | 33.7 | 42.5 |
| Mean | 2.13 | 6.44 | 2.65 | 37.8 | 0.977 | 26.4 | 36.9 | 51.3 |
| SD | 0.441 | 1.81 | 1.56 | 19.3 | 0.027 | 13.4 | 19.2 | 29.8 |
| CV% | 20.7 | 28.0 | 58.6 | 48.2 | 2.8 | 50.7 | 51.8 | 58.2 |
r2 values were derived from the individual predicted values versus observations.
Reduction in bacterial burden.
Table 3.
Statistical summary of the individual fitted population PD parameters and calculated targets associated with static and 2-log bacterial reduction by species (n ≥ 3)
| Species and type of value | Emax | EC50 (mg/liter) | H | %fT>MIC (%) associated with: |
|
|---|---|---|---|---|---|
| Stasis | 2-log reductiona | ||||
| E. coli (n = 3) | |||||
| Minimum | 4.36 | 31.9 | 2.33 | 27.1 | 42.7 |
| Maximum | 5.50 | 53.1 | 3.84 | 54.4 | 152 |
| Median | 5.47 | 34.4 | 3.77 | 28.4 | 52.2 |
| Mean | 5.13 | 39.8 | 3.31 | 36.6 | 82.3 |
| CV% | 13.0 | 29.1 | 25.7 | 42.0 | 73.6 |
| K. pneumoniae (n = 4) | |||||
| Minimum | 4.60 | 37.5 | 1.38 | 31.6 | 55.2 |
| Maximum | 5.34 | 49.3 | 3.90 | 47.7 | 108 |
| Median | 5.00 | 41.9 | 3.26 | 36.0 | 83.6 |
| Mean | 4.98 | 42.6 | 2.95 | 37.8 | 82.6 |
| CV% | 6.10 | 12.0 | 38.0 | 18.6 | 32.4 |
| S. aureus (n = 3) | |||||
| Minimum | 5.27 | 25.3 | 2.38 | 21.2 | 30.3 |
| Maximum | 5.94 | 39.0 | 5.66 | 33.1 | 66.9 |
| Median | 5.49 | 27.6 | 3.92 | 25.1 | 33.0 |
| Mean | 5.57 | 30.6 | 4.00 | 26.5 | 43.4 |
| CV% | 6.10 | 23.9 | 41.2 | 22.9 | 47.0 |
| S. pneumoniae (n = 7) | |||||
| Minimum | 5.97 | 16.9 | 0.670 | 5.23 | 16.1 |
| Maximum | 9.15 | 30.9 | 2.37 | 22.3 | 44.3 |
| Median | 7.49 | 22.8 | 1.86 | 12.9 | 26.3 |
| Mean | 7.29 | 24.3 | 1.70 | 14.0 | 29.8 |
| CV% | 14.4 | 20.9 | 32.9 | 45.4 | 34.7 |
Reduction in bacterial burden (representative of bactericidal activity).
Although the variances in the population PD parameter estimates were reduced in comparison to those for the two-stage estimates, the presence of considerable interstrain variability in EC50 and H parameters determined using the population approach, together with modest variability in Emax, was still noted across the range of 20 strains (Table 1), as well as within a species (Table 3). Extensive variability in the %fT>MIC targets associated with static activity to those associated with a 2-log10 reduction in bacterial density was further highlighted (CV% = 48.0 to 63.8%) (Table 1). Comparisons of PD results by species (Tables 2 and 3) revealed statistically significant differences between Emax and EC50 population estimates for S. pneumoniae versus all other species (P < 0.05). Targets associated with static activity and 2 log units of activity, as well as the H parameter, were only significantly lower for S. pneumoniae than for K. pneumoniae, E. cloacae, and E. coli (Tables 2 and 3) (P < 0.05); significant differences in the same bacterial targets were also noted for K. pneumoniae versus E. cloacae (Tables 2 and 3) (P < 0.05).
DISCUSSION
Correlation of antimicrobial exposure with successful microbiological outcomes is dependent on the relationship between the PK parameters of the particular drug and the MIC of the pathogen of interest. For carbapenems, such as doripenem, the percentage of time during the dosing interval that the free-drug concentration remains above the MIC (%fT>MIC) is widely accepted as the PD index that best predicts activity (12). In vivo studies in preclinical models of infection have been used to simulate antibiotic exposure-activity relationships, which are often characterized using nonlinear regression with Hill-type models in a standard two-stage analysis (equation 3). From these relationships, PD exposure targets which correlate to a desired degree of activity against a particular strain may then be computed and have been applied to various MCS in order to evaluate the efficacy of candidate antimicrobial dosage regimens.
These analyses have been incorporated as part of clinical development initiatives for doripenem. Strategies that aim to optimize doripenem regimens by maximizing the %fT>MIC have particularly focused on the effects of prolonged or continuous infusion regimens (18). In this regard, the efficacy of simulated regimens of 500-mg, 1-g, and 2-g doses of doripenem administered every 8 h as a 1- or 4-h infusion has been evaluated in murine thigh infection models (19–21); the 500-mg dose administered as a 1-h infusion was reported to be sufficient to induce bactericidal activity against isolates with MICs ≤2 mg/liter, whereas enhanced activity against some isolates with MICs up to 4 mg/liter was observed using the 4-h infusion (21). It was also demonstrated that prolonged infusion regimens achieved greater effect following administration of 1 or 2 g against isolates with MICs ≤8 and 16 mg/liter, respectively, achieving a ≥2-log-unit reduction in bacterial density (19, 20). The ability of similar doripenem dosage regimens to attain optimal drug exposure against a range of strains with differing MICs has also been studied in various MCS (6, 8, 9, 11). However, in all of these studies, it must first be recognized that the human protein binding value of ∼8.5% was employed for data analyses, which is markedly different from the murine protein binding value of ∼25.2% applied in the present study (15). The influence of protein binding on the magnitude of the PK/PD parameter of interest has been previously acknowledged (12); thus, species differences in protein binding are likely to affect the accuracy of MCS results obtained. More importantly, the extent of variability in PD, besides MICs, was essentially ignored in the previous studies. Consequently, candidate dosage regimens were evaluated based on their ability to achieve target exposures, which were defined from a single averaged PD model constructed to describe the PD of several strains.
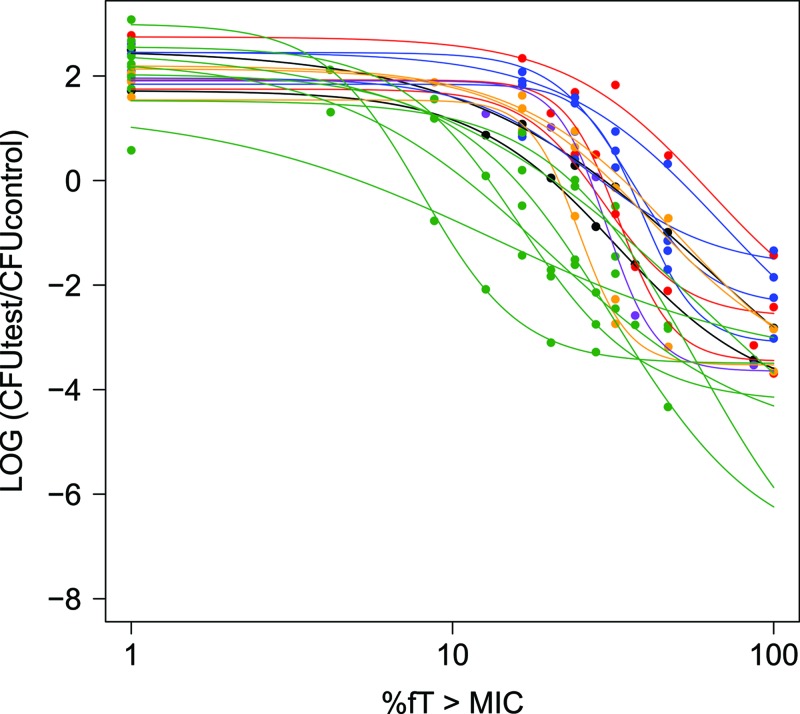
Differences in target magnitude for β-lactams both between drug classes and between organisms have been highlighted and may be attributed in part to differences in the rates of killing of specific “drug versus bug” combinations. These concepts may be similarly translated to the carbapenem class of antimicrobials. However, general definitions of “bacteriostatic” and “bactericidal” activity have been assigned to carbapenems at %fT>MIC values of greater than 20 and 40%, respectively (22, 23). Failure of these values to accurately predict activity against all bacterial strains and the inability of target exposures to be precisely determined from an “average” PD model have been reinforced by our present analyses of a broad range of 20 Gram-positive and -negative strains. Importantly, the presence of substantive interstrain variability in PD parameters was highlighted (Fig. 3; Tables 2 and 3), which translated to extensive variability in the range of calculated %fT>MIC targets associated with static activity to those associated with 2-log10-unit reductions in bacterial burden (CV% = 48.0 to 63.8%) (Tables 2 and 3). For example, in order to attain bacteriostatic activity, population estimates indicated that the required %fT>MIC ranged from 5.23 to 54.4%, while the targets associated with bactericidal activity (≥∼2-log10 bacterial reduction) ranged from 16.1 to 153%. Even within a species (Table 3), interstrain variability was still substantial, particularly for bacteriostatic (CV% = 25.7 to 45.4%) and bactericidal targets (CV% = 34.7 to 73.6%). The observed variability was thus demonstrated to be associated with bacterial species; in particular, Emax and EC50 estimates for S. pneumoniae were significantly different (P < 0.05) from those for all other species, while the calculated targets associated with bacteriostatic and bactericidal activity against S. pneumoniae were systematically lower than those obtained for K. pneumonia, E. cloacae, and E. coli (Table 2).
Fig 3.
Plots of the separate Hill models derived using population analyses for each of the 20 bacterial strains (lines) overlaid on the raw data points (individual symbols), illustrating the variability between bacterial species. Different colors correspond to the six different bacterial species included in the present investigation, namely, E. cloacae (black), E. coli (orange), K. pneumoniae (blue), P. aeruginosa (purple), S. aureus (red), and S. pneumoniae (green).
In this study, PD parameters were estimated using two approaches. First, using the population modeling approach, data from all strains were incorporated to concurrently estimate the population PD model, as well as the individual by-strain results and associated dispersion. Second, using the traditional standard two-stage modeling approach, parameter values for each strain were generated independent of the rest of the population, following which intersubject variability was determined with descriptive statistics; this enhances the potential for modeling artifacts to arise. Consequently, the population approach has been associated with more-precise results (24) and is supported by our data, which revealed larger variance in the two-stage estimates of EC50 and Emax than that for the population estimates (Table 1).
It is recognized that the apparent interstrain variability may certainly have originated from other sources. Arguably, the most significant source of variability may arise from measurement errors associated with the MIC, traditionally used to describe antimicrobial susceptibility (25). Indeed, there is a lack of precision achieved from multiple measurements of the MIC. The MIC further does not account for the existence of heterogenous bacterial populations within a single culture, each of which is likely to exhibit a different MIC. Additionally, the time course of activity is not explained by a fixed MIC, which rather reflects the net growth following a 24-h period during which bacteria are present at various growth phases due to continual death and regeneration.
Errors in PK estimates from in vivo animal studies can also contribute to the apparent variance in PD parameters. Accordingly, PK studies are usually conducted in groups of uninfected mice, which are likely to exhibit altered PK characteristics in comparison to infected mice employed in PD studies. Additionally, PK parameters are often derived from single-dose and dose ranging investigations; these are then extrapolated to simulate different multiple-dosage regimens in PD studies, which are designed to reflect clinically relevant doses. Finally, PK and PD studies are seldom conducted using the same animals, which poses a major source of variability.
In conclusion, the magnitude of PD variability between the array of 20 Gram-positive and -negative bacterial strains included in this study provided ample evidence to demonstrate that evaluations of candidate regimens in MCS based on the assumption that PD variability is attributable to MIC alone can substantially underestimate the extent of variability in responses, thus translating to mispredicted patient outcomes. Greater consideration of PD variability is necessary for the design of future MCS studies.
Footnotes
Published ahead of print 28 January 2013
REFERENCES
- 1. Ortho-McNeil Pharmaceutical 2007. Doribax package insert. Ortho-McNeil Pharmaceutical, Inc., Raritan, NJ [Google Scholar]
- 2. Livermore DM. 2009. Doripenem: antimicrobial profile and clinical potential. Diagn. Microbiol. Infect. Dis. 63:455–458 [DOI] [PubMed] [Google Scholar]
- 3. AstraZeneca 2007. Merrem package insert. AstraZeneca, Wilmington, DE [Google Scholar]
- 4. Merck & Co 2006. Primaxin IV package insert. Merck & Co., Inc., Whitehouse Station, NJ [Google Scholar]
- 5. Zhanel GG, Ketter N, Rubinstein E, Friedland I, Redman R. 2009. Overview of seizure-inducing potential of doripenem. Drug Saf. 32:709–716 [DOI] [PubMed] [Google Scholar]
- 6. Bhavnani SM, Hammel JP, Cirincione BB, Wikler MA, Ambrose PG. 2005. Use of pharmacokinetic-pharmacodynamic target attainment analyses to support phase 2 and 3 dosing strategies for doripenem. Antimicrob. Agents Chemother. 49:3944–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nandy P, Samtani MN, Lin R. 2010. Population pharmacokinetics of doripenem based on data from phase 1 studies with healthy volunteers and phase 2 and 3 studies with critically ill patients. Antimicrob. Agents Chemother. 54:2354–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van Wart SA, Andes DR, Ambrose PG, Bhavnani SM. 2009. Pharmacokinetic-pharmacodynamic modeling to support doripenem dose regimen optimization for critically ill patients. Diagn. Microbiol. Infect. Dis. 63:409–414 [DOI] [PubMed] [Google Scholar]
- 9. Ikawa K, Morikawa N, Ikeda K, Ohge H, Sueda T. 2008. Pharmacodynamic assessment of doripenem in peritoneal fluid against Gram-negative organisms: use of population pharmacokinetic modeling and Monte Carlo simulation. Diagn. Microbiol. Infect. Dis. 62:292–297 [DOI] [PubMed] [Google Scholar]
- 10. Ikawa K, Morikawa N, Uehara S, Monden K, Yamada Y, Honda N, Kumon H. 2009. Pharmacokinetic-pharmacodynamic target attainment analysis of doripenem in infected patients. Int. J. Antimicrob. Agents 33:276–279 [DOI] [PubMed] [Google Scholar]
- 11. Samtani MN, Flamm R, Kaniga K, Nandy P. 2010. Pharmacokinetic-pharmacodynamic-model-guided doripenem dosing in critically ill patients. Antimicrob. Agents Chemother. 54:2360–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Craig WA. 2003. Basic pharmacodynamics of antibacterials with clinical applications to the use of beta-lactams, glycopeptides, and linezolid. Infect. Dis. Clin. North Am. 17:479–501 [DOI] [PubMed] [Google Scholar]
- 13. Andes DR, Kiem S, Craig WA. 2003. In vivo pharmacodynamic activity of a new carbapenem, doripenem (DOR), against multiple bacteria in a murine thigh infection model, abstr A-308. Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., Chicago, IL, 14 to 17 September 2003 [Google Scholar]
- 14. D'Argenio D, Schumitzky A. 1997. ADAPT II user's guide. Biomedical Simulations Resource, Los Angeles, CA [Google Scholar]
- 15. Hori T, Nakano M, Kimura Y, Murakami K. 2006. Pharmacokinetics and tissue penetration of a new carbapenem, doripenem, intravenously administered to laboratory animals. In Vivo 20:91–96 [PubMed] [Google Scholar]
- 16. Bauer RJ, Guzy S, Ng C. 2007. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J. 9:E60–E83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bulitta JB, Bingolbali A, Shin BS, Landersdorfer CB. 2011. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J. 13:201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kotapati S, Kuti JL, Nightingale CH, Nicolau DP. 2003. Role of pharmacodynamics in designing dosage regimens for beta-lactams. Conn. Med. 67:265–268 [PubMed] [Google Scholar]
- 19. Bulik CC, Nicolau DP. 2010. In vivo efficacy of simulated human dosing regimens of prolonged-infusion doripenem against carbapenemase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:4112–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crandon JL, Bulik CC, Nicolau DP. 2009. In vivo efficacy of 1- and 2-gram human simulated prolonged infusions of doripenem against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:4352–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim A, Banevicius MA, Nicolau DP. 2008. In vivo pharmacodynamic profiling of doripenem against Pseudomonas aeruginosa by simulating human exposures. Antimicrob. Agents Chemother. 52:2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lodise TP, Lomaestro BM, Drusano GL. 2006. Application of antimicrobial pharmacodynamic concepts into clinical practice: focus on beta-lactam antibiotics: insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy 26:1320–1332 [DOI] [PubMed] [Google Scholar]
- 23. Mouton JW, Touzw DJ, Horrevorts AM, Vinks AA. 2000. Comparative pharmacokinetics of the carbapenems: clinical implications. Clin. Pharmacokinet. 39:185–201 [DOI] [PubMed] [Google Scholar]
- 24. Tam VH, Preston SL, Drusano GL. 2003. Comparative pharmacokinetic analysis by standard two-stage method versus nonparametric population modeling. Pharmacotherapy 23:1545–1549 [DOI] [PubMed] [Google Scholar]
- 25. Muller M, dela Pena A, Derendorf H. 2004. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: distribution in tissue. Antimicrob. Agents Chemother. 48:1441–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]