Abstract
Dengue virus (DENV) is the predominant mosquito-borne viral pathogen that infects humans with an estimated 50 to 100 million infections per year worldwide. Over the past 50 years, the incidence of dengue disease has increased dramatically and the virus is now endemic in more than 100 countries. Moreover, multiple serotypes of DENV are now found in the same geographic region, increasing the likelihood of more severe forms of disease. Despite extensive research, there are still no approved vaccines or therapeutics commercially available to treat DENV infection. Here we report the results of a high-throughput screen of a chemical compound library using a whole-virus assay that identified a novel small-molecule inhibitor of DENV, ST-610, that potently and selectively inhibits all four serotypes of DENV replication in vitro. Sequence analysis of drug-resistant virus isolates has identified a single point mutation, A263T, in the NS3 helicase domain that confers resistance to this compound. ST-610 inhibits DENV NS3 helicase RNA unwinding activity in a molecular-beacon-based helicase assay but does not inhibit nucleoside triphosphatase activity based on a malachite green ATPase assay. ST-610 is nonmutagenic, is well tolerated (nontoxic) in mice, and has shown efficacy in a sublethal murine model of DENV infection with the ability to significantly reduce viremia and viral load compared to vehicle controls.
INTRODUCTION
The Flaviviridae family comprises a group of positive-sense single-stranded RNA viruses that contain three genera, Hepacivirus, Pestivirus, and Flavivirus. The Flavivirus genus contains several important human pathogens, including dengue virus (DENV), West Nile virus (WNV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV). DENV transmission occurs through the bite of an infected Aedes mosquito, primarily Aedes aegypti, which is a day feeding mosquito found in tropical and subtropical regions around the world. Aedes albopictus mosquitoes can also transmit the virus and are found in an expanding geographic range, including recent spread into Europe, the Americas, and Africa (1). DENV infection is a rapidly growing global health problem with a dramatic increase in the number of infections and cases of disease in recent years (2, 3). The spread of dengue disease is thought to be due to many factors, including population growth, urbanization, migration, international transportation, spread of the mosquito vectors, and lack of effective vector control (3, 4).
Dengue fever (DF) is an acute febrile disease caused by one of four serologically distinct virus serotypes (dengue fever virus serotype 1 [DENV-1], DENV-2, DENV-3, and DENV-4) (5). Infection with DENV is frequently asymptomatic to mildly symptomatic. After an average 3- to 8-day incubation period, DENV infection can result in DF, which is characterized on the basis of the severity of its clinical features into classical DF or severe dengue (2). Symptoms of classical DF range from fever, frontal or retro-orbital headache, myalgia, chills, backache, malaise, anorexia, nausea, lymphadenopathy, leukopenia, and the appearance of a generalized transient rash among other respiratory symptoms, including cough, sore throat, and rhinitis (5, 6). DF has often been referred to as breakbone fever due to the aches and pain associated with the disease, and recovery can be associated with depression and prolonged fatigue (5, 6). Cases of severe disease are referred to as dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) and begin with the same symptoms as classical DF but are followed by plasma leakage with or without hemorrhage and often hepatomegaly, thrombocytopenia, elevated hematocrit, and circulatory failure, which can lead to shock and sometimes death (6). The pathogenesis of severe dengue is thought to be due to several factors, including virus virulence, host genetic factors, activation of serotype-specific cross-reactive memory T cells, and subneutralizing antibodies that lead to an enhancement of virus replication termed antibody-dependent enhancement (ADE) (7–13).
Infection with one serotype of virus provides lifelong immunity to subsequent infection from that particular serotype, but it provides only limited short-lived protection to the other three serotypes (6). A second infection with a different serotype of DENV can lead to an increase in disease severity. This phenomenon provides a challenging hurdle for the development of safe and effective DENV vaccines. A link has been established between viral load and severity of disease with viral titers in the blood averaging 10-fold higher in patients with DHF compared to patients with DF and 100- to 1000-fold higher in patients with DSS compared to those with DF (14–16). An antiviral drug administered early during the course of infection that inhibits viral replication and decreases the high viral load associated with the more severe forms of dengue disease would be an attractive strategy in the treatment and management of clinically apparent disease.
The DENV genome is approximately 11 kb in length and consists of a single-stranded, positive-sense RNA that is translated as a long polyprotein (17). This polyprotein is co- and posttranslationally cleaved by host and viral proteases into three structural and seven nonstructural proteins. The structural proteins, capsid (C), membrane (M), and envelope (E) are primarily involved in viral particle formation (18), while the nonstructural proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5, are involved in viral RNA replication and viral assembly and play a role in modulating the host immune response to infection (5, 17–19). The NS3 protein functions as a protease, helicase, and nucleoside triphosphatase and is essential to flavivirus replication and polyprotein processing. The N-terminal 180 amino acids of NS3 make up the serine protease domain, while the C-terminal domain encodes the helicase activity (20). The NS3 helicase unwinds the RNA secondary structure in the 3′ untranslated region (3′ UTR) to assist in the initiation of RNA synthesis (21).
In this article, we describe the discovery and characterization of a small-molecule compound, ST-610, that selectively inhibits DENV replication in vitro and in vivo. A virus variant with reduced compound susceptibility was found to contain a mutation in the gene encoding the helicase protein. We also demonstrate that ST-610 inhibits helicase RNA unwinding through the use of a molecular-beacon-based helicase assay.
MATERIALS AND METHODS
Compound synthesis.
ST-610 was purchased from ChemBridge (San Diego, CA), dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO) to a concentration of 10 mM, and stored at −20°C.
Cells and viruses.
Vero, A549, BSC40, BHK, C6/36, HeLa, L929, MDCK, and MDBK cells were acquired from American Type Culture Collection (ATCC) (Manassas, VA). Huh-7 cells were obtained from Japan Health Sciences Foundation (Tokyo, Japan). Vero, BSC40, BHK, HeLa, and L929 cells were maintained in minimal essential medium (MEM) (Invitrogen, Grand Island, NY) that was supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen), 2 mM l-glutamine (Invitrogen), and 10 μg/ml gentamicin (Invitrogen). C6/36 cells were maintained at 28°C with 5% CO2 in MEM supplemented as described above. MDBK cells were maintained in MEM supplemented as described above except 10% horse serum (HS) (Invitrogen) was used instead of FBS. A549 cells were maintained in F-12K medium (ATCC) supplemented with 10% FBS and 10 μg/ml gentamicin. MDCK cells were maintained in Eagle's modified essential medium (EMEM) (ATCC) with 10% FBS and 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen). DENV-1 strain TH-Sman (VR-344), DENV-2 strain New Guinea C (NGC) (VR-1255), DENV-3 strain H87 (VR-1256), and DENV-4 strain H241 (VR-1257) were acquired from ATCC. DENV-2 K0049 was a gift from Rebecca Rico-Hesse (Texas Biomedical Research Institute, San Antonio, TX). The DENVs were propagated on Vero or C6/36 cell cultures. Bovine viral diarrhea virus (BVDV) strain NADL (VR-534), Modoc virus strain M544 (VR-415), mouse hepatitis virus (MHV) strain A59 (VR-764), influenza virus A strain A/PR/8/34 (VR-1469), Sindbis virus strain AR-339 (VR-1585), and herpes simplex virus (HSV) strain KOS (VR-1493) were obtained from ATCC. Recombinant vaccinia virus expressing the green fluorescent protein (vvGFP) is described elsewhere (22). All cell culture incubations, except for C6/36 cell culture, were performed at 37°C with 5% CO2.
High-throughput screening (HTS) assay.
Vero cells were seeded on 96-well plates at 4.0 × 103 cells per well and incubated overnight. Individual compounds were added to the medium using a Multiprobe II liquid-handling robot (PerkinElmer, Waltham, MA) to a final concentration of 5 μM. The cell monolayers were infected with DENV-2 (strain NGC) at a multiplicity of infection (MOI) of 0.1 and incubated for 5 days. Each plate contained the following controls: four replicate wells containing virus-infected cells and four replicate wells containing uninfected cells. The dose-response curve was determined in duplicate for ribavirin (Sigma-Aldrich) at 500, 250, 125, and 62 μM as reference standards. Infected-cell monolayers were fixed with 5% glutaraldehyde (JT Baker, Central Valley, PA) in phosphate-buffered saline (PBS) (Fisher, Pittsburgh, PA) for 1 h and then stained with 0.1% crystal violet (Sigma) in 5% methanol for 30 min. To quantify virus-induced cytopathic effects (CPE), absorbance was measured at 595 nm on an Envision multilabel reader (PerkinElmer).
Inhibitory potency.
Vero cells were seeded in 96-well plates at 4.0 × 103 cells per well and incubated overnight. Compound dilutions prepared in DMSO were added at eight different concentrations (25, 8, 2.5, 0.8, 0.25, 0.08, 0.025, and 0.008 μM) in order to generate a dose-response curve. The final DMSO concentration in all samples was 0.5%. Cell monolayers with compound were infected with DENV-2 at a MOI of 0.1. At 5 days postinfection (dpi), the cells were fixed and stained as described above. The level of CPE was quantified by measuring absorbance at 595 nm. The effective concentration of compound that inhibited virus-induced CPE by 50% (EC50) was calculated by fitting the data to a four-parameter logistic model (variable-slope, nonlinear regression model) to generate a dose-response curve using XLfit 4.1 (IBDS, Emeryville, CA).
Cytotoxicity assays.
To determine the cytotoxicity of ST-610, Vero, BSC40, Huh-7, L929, HeLa (1.0 × 104 or 3.5 × 103 cells per well), BHK (5.0 × 103 cells per well), and C6/36 (4.0 × 104 cells per well) cells were seeded in MEM supplemented with 5% FBS in 96-well plates. MDBK cells were seeded at 5.0 × 103 cells per well in MEM supplemented with 2% horse serum, and A549 cells were seeded at 2.0 × 104 cells per well in F-12K medium supplemented with 5% FBS. The plates were incubated overnight (28°C with 5% CO2 for C6/36 cells) and then incubated with various concentrations of ST-610 for 48 h (96 h for MDBK cells). Resazurin (470 μM) (Sigma) was added at 10% of the total assay volume and incubated for 5 to 6 h, after which absorbance was read on an EnVision multilabel reader at 570/600 nm. A 5-day cytotoxicity assay was also performed where Vero cells were seeded at 4.0 × 103 cells per well in 96-well plates, incubated for 5 days after the addition of compound, and viability was measured using the CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI) according to the manufacturer's protocol. Fluorescence was read on an EnVision multilabel reader at 700 nm with a 0.1-s integration time.
Viral titer reduction assays.
To test the antiviral efficacy of ST-610 against DENV, Vero, Huh-7, and L929 cells were seeded at 1.0 × 105 cells per well, A549, BHK, and HeLa cells were seeded at 5.0 × 104 cells per well, and C6/36 cells were seeded at 3.0 × 105 cells per well in a 12-well plate and incubated overnight. For Modoc virus, Vero cells were seeded at 1.0 × 105 cells per well and incubated overnight. Cells were infected with DENV-1, DENV-2, DENV-3, DENV-4, or Modoc at a MOI of 0.1 in the presence of various concentrations of ST-610 and incubated for 1.5 h. After the incubation, the inoculum was removed and replaced with MEM supplemented with 2% FBS containing various concentrations of ST-610. After a 48-h incubation, supernatant was harvested and the titers of the virus were determined on Vero cells seeded overnight at 3.0 × 105 cells per well in a 6-well plate. The plates were overlaid with a 1:1 mix of 2.4% SeaPlaque agarose (Fisher) and 2× MEM after a 1.5-h incubation and removal of infection media. The plates were incubated for 5 days for Modoc virus, 7 days for DENV-2 and DENV-4, and 10 days for DENV-1 and DENV-3. The cells were fixed and stained as described above with the exception of a shorter crystal violet staining time of approximately 10 s. For MHV, L929 cells were seeded at 1 × 105 cells/well, infected at a MOI of 0.01, and incubated, and the titers of the virus were determined as described above. The plates were incubated for 3 days, and then the cells were fixed and stained as described above.
Antiviral specificity.
To determine the antiviral specificity of ST-610 in vitro, antiviral assays were performed with ST-610 against HSV, Sindbis virus, vvGFP, BVDV, and influenza virus, and an enzyme-linked immunosorbent assay (ELISA) was performed with DENV-2 K0049. The wells on 96-well plates were seeded at 2.0 × 104 cells per well with Vero cells (HSV) or BSC40 cells (Sindbis virus and vvGFP) or at 1.0 × 104 cells per well with MDBK cells (BVDV) in appropriate medium for each cell type supplemented with 2% FBS or HS (MDBK cells) and incubated overnight. The compound was serially diluted across the seeded plates, and the cells were infected with a tissue culture infectious dose causing ≥90% CPE (TCID90). The plates were incubated for 1.6 (Sindbis virus), 2 (HSV and vvGFP), or 4 (BVDV) days, and then the cells were fixed and stained as described above. Virus-induced CPE was quantified by measuring absorbance at 595 nm on an EnVision multilabel reader. For influenza virus, MDCK cells were seeded at 1.0 × 104 cells per well and incubated overnight. The cell culture medium (EMEM supplemented with 10% FBS) was replaced with UltraMDCK chemically defined serum-free renal cell medium with l-glutamine (Lonza, Allendale, NH) containing tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (5 μg/ml) (Amersham, Pittsburgh, PA). The compound and virus were added to the plates as described above, the plates were incubated for 72 h, and then the cells were fixed and stained as described above. For the DENV-2 K0049 ELISA, the wells on 96-well plates were seeded with Vero cells at 1.5 × 104 cells per well, and the compound was added as described above. The cells were infected at a titer that generated an ELISA signal of 2.5 at the end of the incubation period. The ELISA was performed using a 1:100 dilution of mouse monoclonal antibody to DENV prM glycoprotein (Abcam, Cambridge, MA) and 1:100 dilution of goat anti-mouse IgG (H+L)-horseradish peroxidase (HRP) conjugate (Bio-Rad, Hercules, CA) as a secondary antibody.
Time-of-drug-addition assay.
Vero cells were seeded in 24-well plates at 3 × 104 cells per well and incubated overnight before being infected with DENV-2 at a MOI of 1.0. After 1.5 h, the virus inoculum was replaced with MEM supplemented with 1% FBS. ST-610 at a concentration of 25 μM was added to the assay medium at 2 h prior to infection or at 0 (i.e., at the time of infection), 2, 4, 6, 8, 12, 24, or 48 h postinfection (hpi). At 48 hpi, the supernatant was harvested and plaque titrated on Vero cells as described above.
Ames assay for genotoxicity.
To evaluate the mutagenic potential of ST-610, the compound was tested in the bacterial reverse mutation assay (Ames assay) and the Muta-Chromo plate assay (EBPI, Mississauga, Canada). For the Ames assay, a top agar mixture (with 0.5% NaCl, 0.05 mM histidine, and 0.05 mM biotin) containing a culture of Escherichia coli tester strain WP-2 uvrA grown overnight in the presence or absence of 10% (vol/vol) Aroclor-1254-induced rat liver S9 (with 0.1 M phosphate buffer [pH 7.4], 4 mM NADP, 6 mM d-glucose-6-phosphate disodium salt hydrate, 33 mM MgCl2, and 8 mM KCl) and containing various concentrations of ST-610 with appropriate controls was overlaid onto plates containing minimal agar and glucose (Vogel-Bonner medium E with 2% glucose). The plates were allowed to harden and then incubated at 37°C for 48 to 72 h. Following the incubation, any revertant colonies were scored. The Muta-Chromo plate assay was conducted according to the manufacturer's protocol using Salmonella enterica serotype Typhimurium tester strains TA-98, TA-100, TA-1535, and TA-1537, Aroclor-1254-induced rat liver S9 with cofactors, and ST-610 at various concentrations. All bacterial tester strains were purchased from Molecular Toxicology, Inc. (Boone, NC).
Selection of drug-resistant virus variants.
By serially passaging wild-type DENV-2 in the presence of 5 μM ST-610, DENV isolates with reduced susceptibility to ST-610 were generated. The plates were seeded with Vero cells and infected with DENV-2 at a MOI of 0.01 in the presence of ST-610. After a 2-h incubation, the inoculum was removed and replaced with medium containing ST-610 and 2% FBS. At 6 dpi, the supernatant was harvested and used to infect a Vero cell monolayer in the presence of ST-610. The infection and incubation were repeated as described above until isolates with resistance to ST-610 were recovered. Virus isolates exhibiting reduced susceptibility to ST-610 were plaque purified three times in the presence of the compound. The purified virus isolates were amplified, and viral RNA was extracted using the QIAamp viral RNA minikit (Qiagen, Valencia, CA). Viral RNA was reverse transcribed into cDNA using the SuperScript III one-step reverse transcription-PCR (RT-PCR) system with Platinum Taq (Invitrogen), and the cDNA genome was sequenced at the Oregon State University Center for Genome Research and Biocomputing Core Laboratories (Corvallis, OR) and analyzed for mutations. Viral titer reduction assays with each isolate were performed as described above on Vero cells.
Plaque reduction assay.
To display the level of resistance of wild-type DENV-2 and the vA263T isolate against ST-610, a plaque reduction assay was performed. Vero cells were seeded at 3 × 105 cells per well in 6-well plates and incubated overnight. The growth medium was removed, and the cells were infected with 100 PFU per well of DENV-2 or vA263T in MEM supplemented with 1% FBS in the presence of 5 μM ST-610 or an equal volume of DMSO. The plates were incubated for 1.5 h, and after removal of the inoculum, the plates were overlaid with 2× MEM–agarose as described above in the presence of 5 μM ST-610 or an equal concentration of DMSO. The plates were fixed and stained at 7 dpi as described above with the exception of a shorter crystal violet stain time of approximately 10 s.
Multicycle growth curve.
To compare the replication fitness of the vA263T isolate and wild-type DENV-2, a multicycle growth curve was done. Vero cells were seeded at 1.0 × 105 cells per well in 12-well plates and incubated overnight before being infected with DENV-2 or vA263T at a MOI of 0.1. After 1.5-h incubation, the inoculum was removed, replaced with fresh medium containing 2% FBS, and incubated. The supernatant was harvested at various time points, and virus levels were quantified by plaque assay on Vero cells as described above.
Reverse genetics.
DENV-2 cDNA-Tonga/74 and Escherichia coli BD1528 ung-1 nadB7 (BD1528) cells were provided by Steve Whitehead at the National Institute of Allergy and Infectious Diseases (Bethesda, MD). To confirm the presence of the resistance-generating mutation A263T, the mutation was reverse engineered into DENV-2 cDNA and transfected into Vero cells to obtain infectious virus (vcDNA-A263T). Using primers DenMut3 (Den stands for dengue virus, and Mut stands for mutant) (5′ GATTGTAGACTTAATGTGTCATACCACATTTACCATGAGGCTG) and DenMut4 (5′ GATTGTAGACTTATGTGTCATACCACATTTACCATGAGGCTG), the A263T mutation was inserted into cDNA-Tonga/74 via site-directed mutagenesis using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) following the manufacturer's instructions. The mutant cDNA was transformed into competent E. coli BD1528 cells and plated on LB plates containing 10 μg/ml tetracycline. Colonies were picked and cultured, and cDNA with the A263T mutation (cDNA-A263T) was extracted from the cells using the QIAquick miniprep kit (Qiagen) and sequenced to confirm the presence of the mutation of interest. Using the AmpliCap SP6 high-yield message maker kit (Epicentre, Madison, WI), cDNA-A263T was linearized, transcribed into RNA, and purified using the RNeasy minikit (Qiagen) according to the manufacturer's protocol. Vero cells were split 1:2 in 1 ml of Opti-Pro serum-free medium (Invitrogen) 24 h prior to transfection. One microgram of transcribed RNA was diluted in 20 mM HEPES–saline, added to diluted DOTAP liposomal transfection reagent (Roche, Indianapolis, IN) according to the manufacturer's protocol, and incubated for 10 min at room temperature. The DOTAP mixture was added to the 1 ml of Opti-Pro serum-free medium on seeded Vero cells and incubated for 18 h, after which an additional 2 ml of Opti-Pro serum-free medium was added. Six days posttransfection, the supernatant from the transfected cells was serially diluted on Vero cells as described above and incubated for 7 days, and the cells were fixed and stained as described above to quantify the amount of infectious virus produced from the transfection. Viral titer reduction assays with vcDNA-A263T were performed as described above on Vero cells.
Construction of recombinant NS3 helicase expression plasmid.
The DENV NS3 helicase domain (positions 171 to 618) was amplified by PCR using cDNA-Tonga/74 (23) as a template and primers A (5′-CGATCATATGAGCATTGAAGCAATCC; the primer contains an engineered NdeI site, which is underlined, and the N-terminal sequence of NS3 helicase) and B (5′-CGATGGATCCCTACTTTCTTCCGGCTGCAAA; the primer contains an engineered BamHI site, which is underlined, and the C-terminal sequence of NS3). The PCR product was double digested with NdeI and BamHI and cloned into the pET-15b vector (EMD Millipore, Billerica, MA) to yield pET-16b-Den-NS3-helicase. The identity of the PCR-amplified region was verified by DNA sequencing.
Protein purification of NS3 helicase.
The DENV NS3 helicase was purified from E. coli at Aldevron (Fargo, ND) using a modified method based upon a previously published protocol (20). A 1-liter culture of E. coli containing a plasmid with the gene encoding the DENV NS3 helicase under the control of the lacZ promoter was grown at 37°C until the optical density reached 0.8. The culture was incubated at 14°C, and expression of the helicase protein was induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the culture medium for 20 h. The bacteria were concentrated by centrifugation, and the concentrated cell paste (10 g) was stored at −80°C. The cell paste was thawed and resuspended in IMAC lysis/bind buffer containing 50 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole, and 5% glycerol, pH 7.5. One tablet of protease inhibitor (Roche) was added per 50 ml of cell suspension. The cells were lysed by sonication and clarified by centrifugation at 16,000 × g for 20 min at 4°C. The clarified cell lysate (100 ml) was loaded on a 5-ml HisTrap FF crude column (GE Healthcare, Waukesha, WI) and washed with 10 ml of lysis buffer containing 40 mM imidazole followed by 10 ml of lysis buffer containing 60 mM imidazole. The helicase protein was eluted with lysis buffer containing 250 mM imidazole. The pooled peak fractions (15 ml) were dialyzed against 4 liters of buffer containing 10 mM HEPES (pH 8.0), 1.0 mM EDTA, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP), 100 mM trehalose, and 150 mM NaCl. The final protein concentration was 1.0 mg/ml with an estimated purity of 77% based upon SDS-PAGE and Coomassie blue staining.
Molecular-beacon-based helicase assay.
Molecular-beacon technology was used to measure the ATP-dependent duplex RNA unwinding activity of purified DENV NS3 helicase (24). An RNA duplex was prepared by mixing 20 μM concentration of the top strand RNA (Integrated DNA Technologies, Coralville, IA) containing a TYE665 fluorophore on the 5′ end and an Iowa Black RQ-Sp Quencher (IAbRQSp) on the 3′ end with 20 μM complementary bottom strand RNA (Integrated DNA Technologies) in buffer containing 10 mM Tris-HCl and 20 mM NaCl, pH 8.0. The bottom strand contains a 20-nucleotide poly(U) 3′ overhang to provide a binding site for helicase enzyme. The top strand RNA is self-complementary and forms a hairpin that brings the fluorescent TYE665 probe in proximity to the IAbRQSp quencher. The helicase substrate was heated to 95°C for 5 min and then allowed to cool to room temperature for 1 h to facilitate annealing of the top and bottom strands. Template RNA was stored at −20°C wrapped in foil to protect the fluorophore from degradation.
Helicase assays were conducted in quadruplicate in white 96-well polystyrene plates (Nunc, Rochester, NY). Reaction mixtures contained 10 mM morpholinepropanesulfonic acid (MOPS) (pH 6.5), 2 mM MgCl2, 2.5 nM TYE665-labeled duplex RNA substrate, 9.75 nM DENV NS3 helicase, and various concentrations of compounds distributed by an HP D300 digital dispenser (Palo Alto, CA). The fluorescence intensities of the reaction mixtures were measured five times in 60-s intervals using an EnVision multilabel reader with an excitation wavelength of 620 nm and an emission wavelength of 685 nm. The unwinding reaction was then initiated by the addition of ATP (Invitrogen) to a final concentration of 3 mM per reaction mixture using a multichannel pipette. The fluorescence intensity of the reaction mixtures were then measured at 60-s intervals for 15 min. Reactions were performed in quadruplicate.
ATPase assay.
ATP hydrolysis was measured by quantifying the release of free phosphate using a malachite green assay. DENV NS3 helicase was diluted in enzyme dilution buffer (25 mM MOPS, 0.2% Tween 20, 0.1 mg/ml bovine serum albumin [BSA], 1 mM dithiothreitol [DTT]). The reaction mixtures were prepared in phosphate-free reaction buffer (25 mM MOPS [pH 6.5], 5 mM MgCl2). Polyuridylic acid potassium salt (PolyU) (Sigma) RNA substrate diluted in H2O was added at various concentrations to the reaction mixture followed by 0.25 mM ATP. Reaction mixtures were prepared in clear 96-well round-bottom plates (Costar, Tewksbury, MA). An HP D300 dispenser was used to dispense drug into wells from a 10 mM stock solution diluted in DMSO. The ATPase reactions were initiated by adding 1 nM enzyme at 10% of the reaction volume. At selected time intervals, 20 μl malachite green reagent (BioAssay Systems, Hayward, CA) was added to each reaction mixture to form a complex with molybdate and free orthophosphate that is quantified by measuring absorbance at 620 nm. The plates were incubated for 30 min and read on a SpectraFluor Plus microplate reader (TECAN, Durham, NC). Standard curves were created by diluting phosphate buffer to known phosphate concentrations in duplicate. Sample absorbance values were then fitted to the linear portion of the standard curve to extrapolate the molar concentration of released phosphate in each sample. Phosphate-free reaction buffer was used as a background and subtracted from both the standard and the sample absorbance values. Reactions were performed in triplicate.
In vivo studies.
All studies involving vertebrate animals were approved by the Institutional Animal Care and Use Committee of Oregon State University, a fully AAALAC-certified facility, and followed all federal guidelines as outlined in the Guide for the Care and Use of Laboratory Animals (25). Strain 129 (129S2/SvPasCrl) mice were acquired from Charles River Laboratories (Wilmington, MA) and were approximately 6 weeks of age for the pharmacokinetic studies. AG129 mice (alpha/beta interferon [IFN-α/β] and gamma interferon [IFN-γ] receptor deficient) were expanded from breeding pairs originally purchased from B & K Universal (Hull, United Kingdom) and were 5 to 6 weeks of age for antiviral efficacy studies.
Pharmacokinetic analysis.
ST-610 was formulated for oral (p.o.), intraperitoneal (i.p.), and intravenous (i.v.) dosing of mice as an aqueous solution in 32% hydroxypropyl-beta-cyclodextrin (HPβCD) (Roquette, Lestrem, France). Plasma was separated (lithium heparin) from blood that was collected from strain 129 mice at various time intervals over a 24-h period after p.o., i.p., and i.v. administration of formulated compound. ST-610 concentrations in plasma were measured by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a 3200 Q TRAP LC-MS/MS system (Applied Biosystems/MDS Sciex, Foster City, CA). LC was performed with a Phenomenex Synergi JAX-RP column (4-μm particle size; 50 mm by 2 mm) at a flow rate of 0.5 ml/min with a mobile phase containing 20% acetonitrile and 80% 10 mM ammonium formate (pH 3.5) in water. WinNonlin (Pharsight, Sunnyvale, CA) was used to estimate pharmacokinetic values.
Antiviral efficacy in dengue murine model.
AG129 mice were housed in labeled cages (4 mice per cage), identified by ear notching, and weighed prior to any other procedures. The mice were injected i.p. with 1 × 106 PFU of DENV-2 strain NGC. Immediately postchallenge, either ST-610 formulated as a 10-, 6-, or 0.6-mg/ml solution in 32% HPβBD or vehicle as a control was administered by oral gavage or i.p. injection. The mice were treated once or twice daily for 3 days. Clinical observations and weight measurements were made daily. The mice were euthanized by CO2 asphyxiation on day 3 postinfection. Spleen, liver, and brain samples were taken from each animal and transferred to preweighed homogenization tubes (FastPrep-24 lysing matrix C with additional 1/4-inch ceramic spheres added [MP Biomedical, Solon, OH]) with 0.5 ml of Dulbecco's PBS (DPBS). Blood was collected into microcentrifuge tubes containing sodium citrate for plasma isolation. The virus titers in plasma and homogenized tissues were determined as described above to determine viremia and viral load. Cytokine levels in plasma were assessed using the BD cytometric bead array (CBA) mouse inflammation kit per the manufacturer's instructions (BD Biosciences, San Jose, CA).
Statistical analysis.
The statistical significance of differences in viral titers generated during efficacy studies was determined using one-way analysis of variance (ANOVA), with Holm-Sidak pairwise post hoc analysis. Data groups that did not pass the normality test (Shapiro-Wilk test; P < 0.05) were analyzed by ranks with a Tukey pairwise post hoc analysis. P values of ≤0.05 were considered statistically significant. Tests were performed using SigmaPlot version 12.0 (build 12.0.0.182; Systat Software, San Jose, CA).
RESULTS
ST-610 is a potent inhibitor of DENV in vitro.
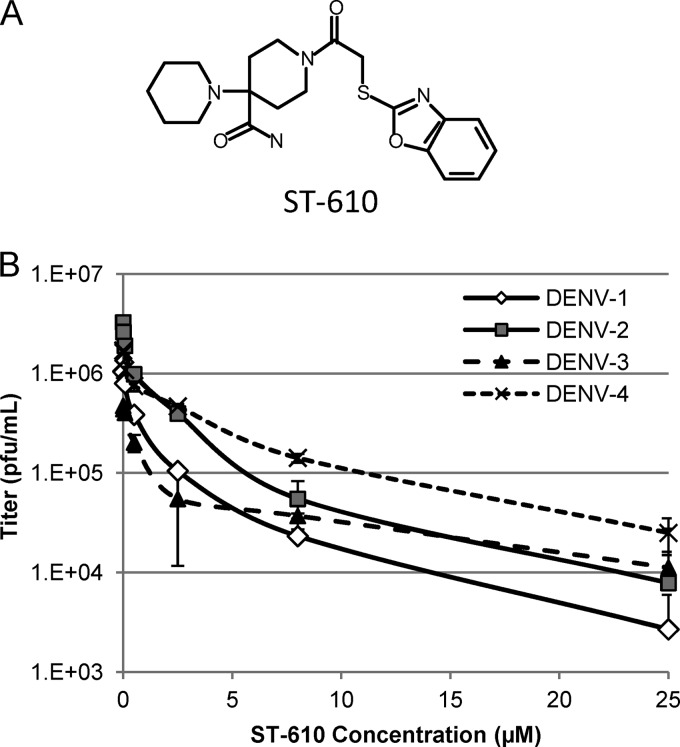
A high-throughput screening (HTS) assay was developed to identify inhibitors of DENV replication from a library of approximately 200,000 chemically diverse compounds. Hits from this screen were defined as compounds that inhibit virus-induced CPE by more than 50 percent (EC50) at an initial concentration of 5 μM. ST-610 (Fig. 1A) was identified from this screen. ST-610 inhibited DENV-2 in a viral titer reduction assay with an EC50 of 0.272 μM and an EC90 value of 3.59 μM (Fig. 1B). ST-610 inhibited DENV replication in multiple cell lines, including Vero, L929, A549, HeLa, BHK, Huh-7, and C6/36 cells with similar potencies (Table 1). The compound was also active against a clinical isolate of DENV-2, K0049. The compound was not cytotoxic in a 2-day assay in Vero, BSC40, C6/36, Huh-7, BHK, MDBK, L929, HeLa, and A549 cell lines or in a 5-day assay in Vero cells, with 50% cytotoxic concentration (CC50) values all >100 μM. ST-610 produced a dose-dependent decrease in the steady-state level of viral RNA at 48 hpi (data not shown).
Fig 1.
(A) Structure of ST-610 (27). (B) Antiviral activity of ST-610 against DENV. Vero cells were infected with the indicated viruses at a MOI of 0.1 and treated with ST-610. The culture medium was collected at 48 hpi, and the viral titers were determined by plaque assay. Data are mean values ± standard deviations (SD) (error bars) for 3 independent experiments.
Table 1.
Antiviral activity of ST-610 against DENVa
| Cell type | Virusb | EC50 (μM)c |
|---|---|---|
| Vero | DENV-1 | 0.236 ± 0.06 |
| DENV-2 | 0.272 ± 0.05 | |
| DENV-3 | 0.256 ± 0.17 | |
| DENV-4 | 0.203 ± 0.09 | |
| DENV-2 K0049 | 0.255 ± 0.24 | |
| vA263T | 13.736 ± 3.10 | |
| vcDNA-A263T | 11.640 ± 3.94 | |
| C6/36 | DENV-2 | 0.377 ± 0.07 |
| L929 | DENV-2 | 0.045 ± 0.01 |
| A549 | DENV-2 | 0.332 ± 0.13 |
| HeLa | DENV-2 | 0.300 ± 0.24 |
| Huh-7 | DENV-2 | 0.112 ± 0.01 |
| BHK | DENV-2 | 0.234 ± 0.03 |
Cells were infected with the indicated viruses at a MOI of 0.1 and treated with ST-610. Culture medium was collected at 48 hpi, and the viral titers were determined by plaque assay. For DENV-2 strain K0049, activity was measured by an ELISA.
vA263T is the drug-resistant DENV-2 isolate, and vcDNA-A263T is the virus recovered from the engineered cDNA clone.
The data are mean values ± standard deviation (SD) for 3 independent experiments.
ST-610 selectively inhibits flaviviruses.
The antiviral potency and selectivity of ST-610 was measured against a panel of viruses representing different virus families to determine the spectrum of activity. As shown in Fig. 1B and Table 2, ST-610 was active against all four serotypes of DENV, but less active against other members of Flaviviridae. ST-610 exhibited weak activity against yellow fever virus with an EC50 of 7.4 μM but was not active against hepatitis C virus, West Nile virus, bovine viral diarrhea virus, Japanese encephalitis virus, or Modoc virus. ST-610 did not inhibit viruses from families outside Flaviviridae, with the exception of Venezuelan equine encephalitis virus (VEEV) which showed weak inhibition. These results indicate that ST-610 specifically inhibits DENV within the Flavivirus genus.
Table 2.
ST-610 antiviral spectrum of activity and selectivity
| Virus | Family | Classificationa | EC50 (μM) | Assay type |
|---|---|---|---|---|
| Modoc virus | Flaviviridae | +ssRNA | >25 | Plaque titration |
| Bovine viral diarrhea virus | Flaviviridae | +ssRNA | >25 | Plaque titration |
| West Nile virusb | Flaviviridae | +ssRNA | >25 | Neutral red |
| Yellow fever virusb | Flaviviridae | +ssRNA | 7.4 | Neutral red |
| Hepatitis C virusc | Flaviviridae | +ssRNA | >50 | Luciferase |
| Japanese encephalitis virusd | Flaviviridae | +ssRNA | >50 | CPE |
| Mouse hepatitis virus | Coronaviridae | +ssRNA | >25 | Plaque titration |
| SARS-CoVb,e | Coronaviridae | +ssRNA | >25 | Neutral red |
| Sindbis virus | Togaviridae | +ssRNA | >25 | CPE |
| Venezuelan equine encephalitis virusb | Togaviridae | +ssRNA | 10.1 | Neutral red |
| Influenza virus | Orthomyxoviridae | −ssRNA | >25 | CPE |
| Vaccinia virus | Poxviridae | dsDNA | >25 | CPE |
| Herpes simplex virus | Herpesviridae | dsDNA | >25 | CPE |
+ssRNA, positive-sense single-stranded RNA virus; −ssRNA, minus-sense single-stranded RNA virus; dsDNA, double-stranded DNA virus.
In vitro testing done by NIAID screening program.
In vitro testing done by the laboratory of J. Neyts at the Rega Institute using a subgenomic HCV replicon (genotype 1b) in Huh 5.2 cells (26).
In vitro testing done by Integrated BioTherapeutics via a cytopathic effect (CPE)-based assay in Vero cells.
SARS-CoV, severe acute respiratory syndrome coronavirus.
ST-610 is not mutagenic.
ST-610 was tested by the Ames assay and the Muta-Chromo plate assay to evaluate the mutagenic potential of the compound. The assay used Salmonella Typhimurium tester strains TA-98, TA-100, TA-1535, and TA-1537 as well as Escherichia coli tester strain WP-2 uvrA with or without Aroclor-1254-induced rat liver S9. No mutagenic potential was observed under any of these conditions (data not shown).
Time-of-drug-addition studies.
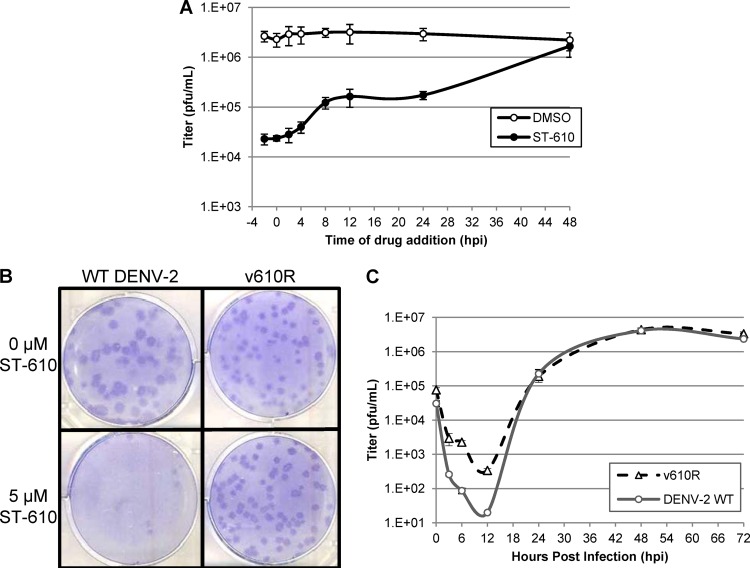
A time-of-drug-addition experiment was conducted to explore which stage of the viral life cycle is affected by ST-610. ST-610 was added at a concentration of 25 μM to DENV-2-infected Vero cells either before the time of infection or at several time points after infection. At 48 hpi, viral yield was quantified by plaque assay. The data indicate that ST-610 was most effective when added within 4 hpi but that virus yield was significantly reduced even when added at 24 hpi. This is consistent with a postentry mechanism of action (Fig. 2A).
Fig 2.
(A) Time-of-drug-addition study. DENV-2-infected Vero cells were treated with ST-610 at various time points, and virus levels were quantified by plaque assay at 48 hpi. (B) Plaque formation of wild-type (WT) DENV-2 and the ST-610-resistant virus variant vA263T (v610R) in the presence and absence of ST-610. Vero cell monolayers were infected with approximately 100 PFU of DENV-2 or vA263T in the presence and absence of 5 μM ST-610 and covered with an agarose overlay. At 7 dpi, the cultures were fixed and stained. (C) Multicycle growth curve of wild-type DENV-2 and vA263T in the absence of ST-610. Vero cell monolayers were infected with virus, harvested at various time points, and plaque titrated. The data are mean values ± SD for three independent experiments.
Characterization of a drug-resistant virus variant.
Virus variants that are resistant to the inhibitory effects of a compound provide evidence that the compound is acting on a virus-specific target and not on a cellular target. Moreover, mapping the drug susceptibility locus can facilitate target identification. In order to generate compound-resistant virus, wild-type DENV-2 was serially passaged eight times on Vero cells in the presence of 5 μM ST-610. Virus capable of growing in the presence of ST-610 was isolated and plaque purified three times in the presence of compound. Five separate drug-resistant virus isolates were selected and further characterized. Nucleotide sequencing of these isolates identified a single point mutation (A263T) in the NS3 gene in each isolate that correlated with reduced compound susceptibility. To measure the susceptibility of these virus isolates to ST-610, a plaque reduction assay was conducted with wild-type virus and the drug-resistant virus variants in the presence and absence of ST-610 (Fig. 2B and data not shown). After the cells were fixed, crystal violet stain was applied for approximately 3 to 5 s, resulting in a lighter cell monolayer with dark plaques (27). In the presence of ST-610, plaque formation of wild-type virus was greatly reduced relative to DMSO control wells, while a similar number of plaques was observed for the drug-resistant virus isolate vA263T in both the presence and absence of compound. The plaque size of vA263T appears slightly smaller in the presence and absence of ST-610 compared to wild-type virus in the absence of compound. In multicycle growth curve experiments, the replication rate and peak titer of vA263T in the absence of compound were comparable to those of wild-type virus, which suggests that the resistance mutation had no effect on viral fitness (Fig. 2C). A viral titer reduction assay was used to determine the level of susceptibility of the resistant virus to ST-610. The EC50 for inhibition of the resistant variant (vA263T) (13.736 μM) was 50-fold higher than the EC50 for inhibition of wild-type DENV-2 (0.272 μM) (Table 1).
ST-610 resistance maps to the NS3 gene.
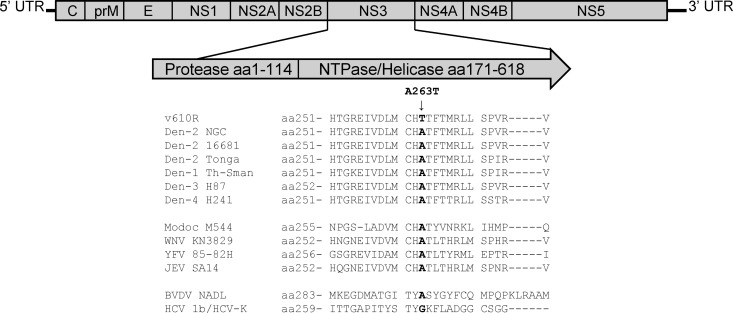
To determine the locations of the changes in the viral genome that correlate with reduced drug susceptibility, genomic RNA from pools of plaque-purified resistant virus were isolated and amplified by reverse transcription-PCR to generate cDNA, which was then sequenced in overlapping segments. A single nucleotide change was observed in five separate resistant virus genomes that resulted in an alanine (Ala)-to-threonine (Thr) change at amino acid position 263 in the helicase domain of the NS3 gene (Fig. 3). To confirm that this mutation was responsible for conferring resistance to ST-610, the Ala-to-Thr change was reengineered back into a wild-type DENV-2 cDNA. Infectious virus recovered from this engineered cDNA (vcDNA-A263T) showed reduced susceptibility to ST-610 (EC50 of 11.640 μM) which is 43-fold higher than the EC50 for inhibition of wild-type DENV-2 (0.272 μM), supporting the conclusion that this mutation is sufficient to confer reduced compound susceptibility (Table 1).
Fig 3.
Sequence comparison of NS3 orthologs. Graphic representation of the DENV genome, illustrating the location of the ST-610-resistant allele (NS3 A263T) and an amino acid sequence comparison of NS3 orthologs from the helicase region of other flaviviruses showing the degree of sequence conservation. aa, amino acids.
ST-610 inhibits NS3 helicase activity.
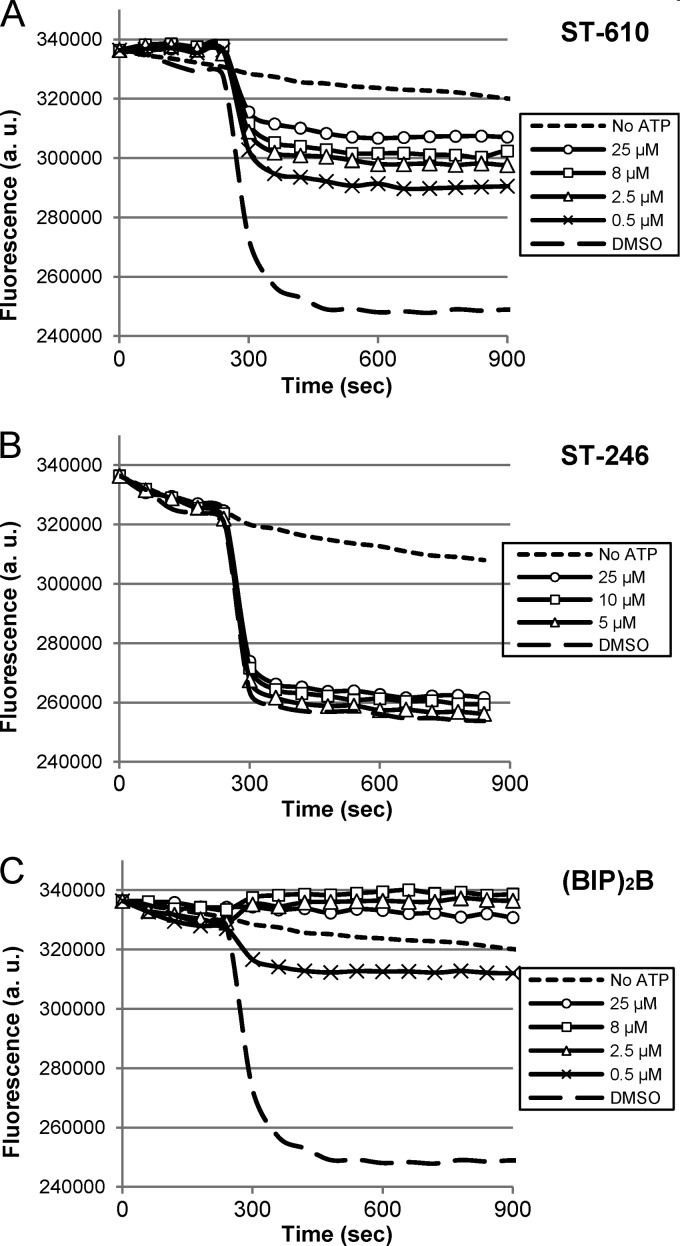
Although resistance mapped to the helicase domain, definitive evidence that ST-610 altered NS3 helicase activity was required to support the hypothetical mechanism of action of the compound. The ATP-dependent helicase activity was measured using a molecular-beacon-based assay (24). This assay measures the reduction in fluorescence of a 5′-TYE665-labeled RNA oligomer that forms a hairpin upon release from the duplex substrate bringing into proximity a fluorescence quencher located on the 3′ end of the molecule. Purified DENV NS3 helicase was incubated with duplex substrate in the presence and absence of various concentrations of ST-610 (Fig. 4A); an inhibitor of orthopoxviruses, ST-246 (tecovirimat), as a negative control (Fig. 4B); and a benzimidazole-phenyl-carboxamide compound, 1-N,4-N-bis[4-(1H-benzimidazol-2-yl)phenyl]benzene-1,4-dicarboxamide [(BIP)2B], that has been reported to inhibit flavivirus helicase activity as a positive control (28) (Fig. 4C). The reaction mixtures were incubated for 5 min at room temperature, and helicase activity was initiated by the addition of ATP. Upon the addition of ATP, a decrease in fluorescence intensity was observed in the absence of compound (DMSO samples), consistent with unwinding of the duplex substrate by active helicase. The unwinding activity did not reach completion based upon the fluorescence intensity of reaction mixtures containing top strand 5′TYE665-labeled RNA oligomer alone (data not shown). Furthermore, control experiments demonstrated that helicase activity could be observed only under single-turnover conditions with enzyme concentrations that were higher than the substrate concentrations (data not shown). This observation is consistent with the observations of Belon et al., using this experimental design to characterize inhibition of hepatitis C virus (HCV) helicase (28).
Fig 4.
Molecular-beacon-based helicase unwinding assay. Purified DENV helicase was incubated with duplex substrate in the presence and absence of ST-610, ST-246, or (BIP)2B. The unwinding reaction was initiated by the addition of 3 μM ATP, and the fluorescence intensity of the reaction mixtures was measured in 60-s intervals. Fluorescence intensity values were measured in arbitrary units (a.u.). (A to C) DENV helicase with various concentrations of ST-610 (A), ST-246 (B), or (BIP)2B (C). Data are mean values of four independent experiments.
In the presence of ST-246, the decrease in fluorescence intensity was similar to that of DMSO-treated samples, suggesting that ST-246 did not inhibit helicase activity. In contrast, ST-610 and (BIP)2B reduced the level of fluorescence quenching in a dose-dependent manner consistent with inhibition of helicase activity. The compounds appeared to inhibit the extent of the helicase reaction; however, due to the high rate of fluorescence quenching upon the addition of ATP, it was not possible to determine whether they inhibited the reaction rate. (BIP)2B showed the highest levels of helicase inhibition, with the highest concentrations of compounds inhibiting 100% of the activity. ST-610 showed intermediate levels of inhibition. While DENV helicase was found to unwind DNA as well as RNA substrates, ST-610 inhibited only ATP-dependent unwinding activity in the presence of the RNA substrate (data not shown).
ST-610 does not inhibit the RNA-dependent ATPase activity of the DENV NS3 helicase.
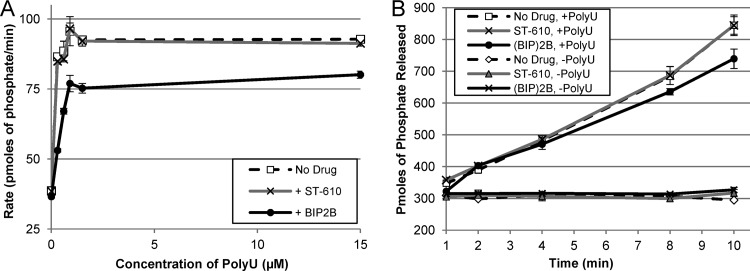
A malachite green assay was used to measure the effects of ST-610 on the RNA-dependent ATPase activity of the DENV NS3 helicase. A purified fragment of the DENV NS3 helicase (amino acids 171 to 610) was incubated with polyuridylic acid (PolyU) in the presence or absence of ST-610, and the reaction was initiated by the addition of ATP. PolyU is an allosteric activator of the DENV NS3 helicase ATPase activity, increasing the rate of ATP hydrolysis by 5- to 10-fold. The results show that the ATPase activity of the DENV NS3 helicase was not inhibited by ST-610 in the presence or absence of PolyU (Fig. 5). (BIP)2B showed slight inhibition of ATPase activity at low concentrations of PolyU (≤15 μM), consistent with published results (28). Although ST-610 inhibited the ATP-dependent helicase activity of the DENV helicase, the compound does not appear to have a measurable effect on the RNA-dependent ATPase activity of the DENV NS3 protein.
Fig 5.
RNA-dependent ATPase activity assay. ATP hydrolysis was measured by quantifying the release of free phosphate using a malachite green assay. (A) Rate of phosphate released (pmol/min) with 5 μM ST-610 (+ ST-610) and 5 μM (BIP)2B (+ BIP2B) in the presence of various concentrations of PolyU after a 10-min reaction. (B) Amount of phosphate released (pmol) at various reaction times in the presence (+) and absence (−) of 5 μM ST-610, 5 μM (BIP)2B, and 15 μM PolyU. Data are mean values ± SD of three independent experiments.
Pharmacokinetic analyses and tolerability.
Prior to conducting in vivo efficacy studies, the tolerability and pharmacokinetic parameters of ST-610 in mice were determined. The compound was formulated as an aqueous solution in 32% hydroxypropyl-beta-cyclodextrin (HPβCD). Oral (p.o.) and intraperitoneal (i.p.) administration of ST-610 at 100 mg/kg of body weight in strain 129 mice generated peak plasma compound concentrations of 536 and 8,390 ng/ml, respectively (Table 3), which are 4- and 64-fold above the EC50, respectively. Intravenous (i.v.) administration of ST-610 at 10 mg/kg generated a peak plasma drug concentration of 4,170 ng/ml, 31-fold above the EC50. The area under the plasma drug concentration-time curve (AUC) was greatest in mice administered ST-610 via the i.p. route. The absolute oral bioavailability of ST-610 was 7%. While the oral bioavailability of the compound was relatively low, the compound was rapidly absorbed after oral dosing, with the time to maximum plasma drug concentration measured at 1.33 h. Intraperitoneal administration produced higher plasma drug concentrations with rapid absorption, peak plasma drug concentrations at 1.00 h, and bioavailability of 56%. There was no overt toxicity, or changes in physical signs, including body weights, observed in mice after repeated p.o. or i.p. ST-610 administration of 100 mg/kg/day for 3 days. These data indicate that ST-610 has good systemic availability following i.p. administration, although it shows limited oral bioavailability and fairly rapid clearance.
Table 3.
Pharmacokinetic parameters of ST-610 in micea
| Route | Dosage (mg/kg) | Tmax (h) | Cmax (ng/ml) | AUC (h · ng/ml) |
|---|---|---|---|---|
| p.o. | 100 | 1.33 | 536 | 1,146 |
| i.v. | 10 | 0.08 | 4,170 | 1,688 |
| i.p. | 100 | 1.00 | 8,390 | 9,382 |
Plasma drug concentrations were measured by LC-MS/MS over a 24-h time period. Tmax, time to maximal concentration of drug; Cmax, maximal concentration of drug; AUC, area under the plasma drug concentration-time curve.
In vivo antiviral efficacy.
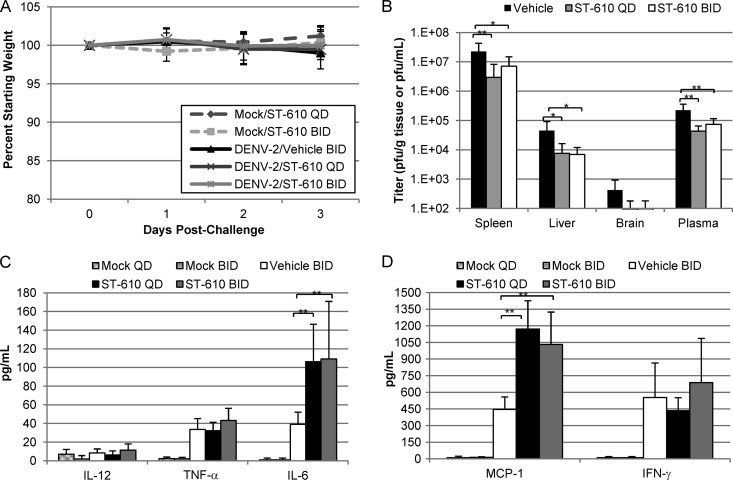
AG129 mice, which are deficient in alpha/beta interferon (IFN-α/β) and gamma interferon (IFN-γ) receptors, are permissive for DENV infection (29–33). Intraperitoneal inoculation of mice with 1.0 × 106 PFU of DENV-2 strain NGC produces a nonlethal infection characterized by a transient viremia with peak titers observed by day 3 postinoculation that fall below the limit of quantification by day 9 postinoculation. This model was used to evaluate the antiviral efficacy of ST-610 (Fig. 6). The mice were infected with DENV-2 via i.p. injection, and vehicle or ST-610 at 100 mg/kg/dose was administered i.p. either once or twice daily for 3 days. None of the vehicle- or ST-610-treated mice lost weight during the course of the experiment (Fig. 6A). Once-daily treatment with ST-610 reduced peak viremia by 5.2-fold and reduced viral load in the spleen, liver, and brain by 7.6-, 6.0-, and 4.4-fold, respectively (Fig. 6B). A comparable reduction was seen with twice-a-day dosing. This difference in viremia relative to vehicle-treated mice was statistically significant as determined by one-way ANOVA. Similar to what is observed in the blood of patients who have DF (11), levels of inflammatory cytokines (tumor necrosis factor alpha [TNF-α], interleukin 6 [IL-6], macrophage chemoattractant 1 [MCP-1], and IFN-γ) are elevated in the AG129 model of dengue disease. DENV-infected mice treated with ST-610 showed no statistically significant difference in the levels of IL-12, TNF-α, or IFN-γ, but they did exhibit an increase in the levels of IL-6 and MCP-1 that was statistically significant (Fig. 6C and D).
Fig 6.
In vivo efficacy of ST-610. AG129 mice (8 mice/group) were infected with 1.0 × 106 PFU DENV-2 (strain NGC) i.p. on day 0. The mice were treated with 100 mg/kg ST-610 i.p. either once a day (QD) or twice a day (BID). On day 3 postinfection (the peak of viremia), the viral load and viremia were quantified by plaque assay, and inflammatory cytokine levels were determined by using the BD CBA mouse inflammation kit (BD Biosciences). (A) Weight of the mice compared to their starting weight during each day of the study. (B) Viral load and viremia. (C and D) Inflammatory cytokine levels in plasma. The lower limit of quantification for the BD CBA mouse inflammation kit assay is 20 pg/ml. Bars represent the arithmetic means plus SD (error bars). Values that are significantly different are indicated by brackets and asterisks as follows: *, P value < 0.05; **, P value < 0.01.
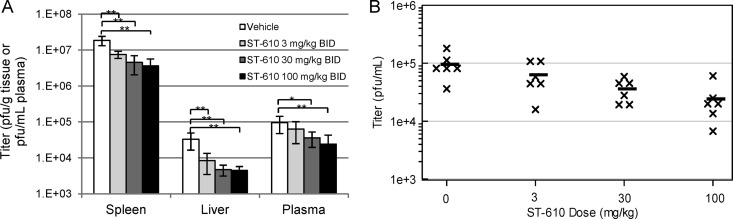
To determine the optimal dose of ST-610, infected mice were treated i.p. with increasing amounts of compound. Twice-daily treatment of infected mice with ST-610 at 3, 30, and 100 mg/kg reduced peak viremia by 1.5-, 2.7-, and 4.0-fold, respectively (Fig. 7).
Fig 7.
Dose-dependent in vivo efficacy of ST-610. AG129 mice were infected with 1.0 × 106 PFU DENV-2 (strain NGC) i.p. on day 0. The mice were treated with the indicated doses of ST-610 i.p. On day 3 postinfection (the peak of viremia), the viral load and viremia were quantified by plaque assay. (A) Average virus titer with BID ST-610 treatment. Bars represent the arithmetic means ± SD. Values that are significantly different are indicated by brackets and asterisks as follows: *, P value < 0.05; **, P value < 0.01. (B) Viremia level per mouse with BID ST-610 treatment. Each symbol represents the value for an individual mouse. The black bars represent the arithmetic means for the group of mice.
DISCUSSION
A novel benzoxazole inhibitor (ST-610) of DENV replication was identified by high-throughput screening of a chemical library for compounds that inhibit virus-induced cytopathic effects. ST-610 was found to be potent (EC50 = 0.045 to 0.377 μM) and nontoxic (CC50 > 100 μM in both 2- and 5-day assays) with activity against all 4 DENV serotypes in cell culture. The antiviral activity was independent of cell type, suggesting that ST-610 targets a virus-specific function. Analysis of resistant variants identified an amino acid substitution (A263T) in the dengue virus NS3 helicase domain that correlated with reduced compound susceptibility. ST-610 showed modest inhibition of YFV replication (EC50 = 7.4 μM) and no activity against other flaviviruses. The weak activity against YFV could not be explained by the degree of sequence conservation of the putative target, since there was noticeable sequence divergence in this region of the virus genome among flaviviruses (Fig. 3). Moreover, there was no antiviral activity observed against other species of virus from different virus families with the exception of VEEV (EC50 = 10.1 μM). However, there is very little sequence homology between VEEV and DENV, and this result may be an artifact of the testing system which needs to be confirmed with further testing. These results suggest that ST-610 is a potent and relatively specific inhibitor of DENV replication.
ST-610 was nonmutagenic in bacterial Ames tests and was well tolerated in strain 129 mice and the derivative AG129 mice when administered via intraperitoneal injection. Peak plasma drug levels were 64-fold above the in vitro EC50s following compound administration. Intraperitoneal injection resulted in better exposure of ST-610 relative to oral delivery with peak plasma drug concentrations (Cmax) and plasma drug exposure levels (AUC [h · ng/ml]) 15- and 8-fold higher relative to oral delivery of ST-610, respectively. While intraperitoneal injection is not a conventional route of clinical delivery for anti-infective drugs, for proof-of-concept efficacy studies, it provided a means of assessment of antiviral activity. Chemical optimization of the ST-610 series will focus on identification of compound analogues with improved oral bioavailability.
Intraperitoneal inoculation of interferon receptor-deficient AG129 mice with DENV produces a transient viremia that can be quantified by plaque assay of blood and tissue samples from infected animals. The peak of virus replication occurs by day 3 postinoculation with virus yields approximately 10- to 100-fold higher than virus yields measured on day 0 after inoculation. Virus yields begin to decline after day 3 postinoculation, and mice ultimately recover from infection. Initiating ST-610 treatment at the time of infection and continuing ST-610 treatment once or twice a day for 3 consecutive days reduced virus yield at peak times by 5-fold compared to animals treated with vehicle alone. One-way ANOVA showed that this decrease in virus yield was statistically significant relative to vehicle-treated animals. The compound appeared to be well tolerated over the course of the study, and no adverse effects were observed. These results demonstrate that ST-610 can reduce virus replication in AG129 mice infected with DENV. However, an increase in the levels of MCP-1 and IL-6 were observed in ST-610-treated mice. It is possible that the inhibition of double-stranded RNA (dsRNA) unwinding activity of the viral NS3 helicase by ST-610 results in the intracellular accumulation of DENV dsRNA intermediates in infected cells. This accumulation in return may result in enhanced triggering of the innate immune system's dsRNA sensing and signaling pathways and lead to the upregulated expression of cytokines and chemokines (34).
Drug resistance mapped to amino acid position 263 in the helicase domain of the DENV NS3 protein. An alanine-to-threonine change at this position appeared to correlate with reduced compound susceptibility. Reengineering this mutation back into wild-type DENV resulted in a virus recombinant that was resistant to ST-610, suggesting that this amino acid change was responsible for the reduced compound susceptibility. On the basis of the crystal structure of the DENV helicase, the alanine residue at position 263 is located within a basic patch of amino acids within a channel in the protein believed to be involved in nucleic acid binding. In fact, a threonine residue at position 264 has been purported to hydrogen bond with the hydroxyl of the ribose moiety of the RNA substrate (35). We hypothesize that binding of ST-610 to this region of the molecule might interfere with nucleic acid substrate binding or translocation of single-stranded RNA through this channel. The amino acid change from alanine to threonine at position 263 could block ST-610 binding to this region of the protein or perhaps help stabilize nucleic acid binding such that ST-610 could not inhibit this process.
The ability of ST-610 to inhibit the biochemical activity associated with purified DENV NS3 helicase was measured with a molecular-beacon-based helicase assay and a malachite green ATPase assay. ST-610 appeared to have a dose-dependent inhibitory effect on the RNA unwinding activity, but not the RNA-dependent ATPase activity of purified NS3. Moreover, a benzimidazole-phenyl-carboxamide compound, (BIP)2B, partially inhibited both DENV NS3 helicase unwinding and ATPase activity. This molecule has been shown to inhibit unwinding activity from a variety of flavivirus helicase enzymes using both RNA and DNA duplex substrates (28). Molecular modeling studies using a flavivirus NS3 helicase from hepatitis C virus suggest that (BIP)2B binds to the basic patch of amino acids within the helicase that are involved in RNA binding. Kinetic studies conducted with hepatitis C virus NS3 helicase and (BIP)2B are consistent with this mode of action. Resistance to ST-610 maps to this region of the molecule and is able to inhibit some of the enzymatic activities associated with purified protein. It is also possible that ST-610 interferes with another function of the helicase protein or some other target within the replication complex. ST-610 binding specificity may require intact replication complex for optimal activity. Future studies with purified replication complexes and purified DENV NS3 helicase are needed to understand the molecular mechanism of ST-610 action. Medicinal chemistry efforts are also required to improve the in vitro and in vivo activity of this series of compounds.
ACKNOWLEDGMENTS
This work was supported by NIH grant 1R43AI079937-01A1.
This work was performed by employees of SIGA Technologies who hold stock in the company.
We thank Steve Whitehead for the cDNA and Rebecca Rico-Hesse for the K0049 strain of virus. We also thank Johan Neyts for testing ST-610 against HCV and confirming activity against DENV-2 in vitro. Thanks to Kady Honeychurch for DENV-specific primer design, Andrew Wieczorek and Thuan Tran for HTS support, and Yali Chen for carrying out the Ames assay.
Footnotes
Published ahead of print 12 February 2013
REFERENCES
- 1. Weaver SC, Reisen WK. 2010. Present and future arboviral threats. Antiviral Res. 85:328–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization 2009. Dengue: guidelines for diagnosis, treatment, prevention and control, new edition World Health Organization, Geneva, Switzerland: [PubMed] [Google Scholar]
- 3. Monath TP. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. U. S. A. 91:2395–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gubler DJ. 2002. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10:100–103 [DOI] [PubMed] [Google Scholar]
- 5. Henchal EA, Putnak JR. 1990. The dengue viruses. Clin. Microbiol. Rev. 3:376–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. World Health Organization 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control, 2nd ed World Health Organization, Geneva, Switzerland [Google Scholar]
- 7. Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos de Chacon I, Ramos C, Rico-Hesse R. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 73:4738–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martina BE, Koraka P, Osterhaus AD. 2009. Dengue virus pathogenesis: an integrated view. Clin. Microbiol. Rev. 22:564–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rothman AL. 2003. Immunology and immunopathogenesis of dengue disease. Adv. Virus Res. 60:397–419 [DOI] [PubMed] [Google Scholar]
- 10. Rico-Hesse R. 2007. Dengue virus evolution and virulence models. Clin. Infect. Dis. 44:1462–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halstead SB. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476–481 [DOI] [PubMed] [Google Scholar]
- 12. Fink J, Gu F, Vasudevan SG. 2006. Role of T cells, cytokines and antibody in dengue fever and dengue haemorrhagic fever. Rev. Med. Virol. 16:263–275 [DOI] [PubMed] [Google Scholar]
- 13. Halstead SB. 2009. Antibodies determine virulence in dengue. Ann. N. Y. Acad. Sci. 1171(Suppl 1):E48–E56 [DOI] [PubMed] [Google Scholar]
- 14. Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. 2000. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181:2–9 [DOI] [PubMed] [Google Scholar]
- 15. Libraty DH, Endy TP, Houng HS, Green S, Kalayanarooj S, Suntayakorn S, Chansiriwongs W, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. 2002. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J. Infect. Dis. 185:1213–1221 [DOI] [PubMed] [Google Scholar]
- 16. Murgue B, Roche C, Chungue E, Deparis X. 2000. Prospective study of the duration and magnitude of viraemia in children hospitalised during the 1996-1997 dengue-2 outbreak in French Polynesia. J. Med. Virol. 60:432–438 [DOI] [PubMed] [Google Scholar]
- 17. Schlesinger S, Schlesinger MJ. 1990. Replication of Togaviridae and Flaviviridae, p 697–710 In Fields BN, Knipe DM, Chanock RM, Hirsch MS, Melnick JL, Monath TP, Roizman B. (ed), Virology, 2nd ed, vol 1 Ravens Press, New York, NY [Google Scholar]
- 18. Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22 [DOI] [PubMed] [Google Scholar]
- 19. Bartenschlager R, Miller S. 2008. Molecular aspects of dengue virus replication. Future Microbiol. 3:155–165 [DOI] [PubMed] [Google Scholar]
- 20. Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J. 2005. Structure of the dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J. Virol. 79:10278–10288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, Cook S, Coutard B, Decroly E, de Lamballerie X, Gould EA, Grard G, Grimes JM, Hilgenfeld R, Jansson AM, Malet H, Mancini EJ, Mastrangelo E, Mattevi A, Milani M, Moureau G, Neyts J, Owens RJ, Ren J, Selisko B, Speroni S, Steuber H, Stuart DI, Unge T, Bolognesi M. 2010. Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res. 87:125–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Byrd CM, Bolken TC, Mjalli AM, Arimilli MN, Andrews RC, Rothlein R, Andrea T, Rao M, Owens KL, Hruby DE. 2004. New class of orthopoxvirus antiviral drugs that block viral maturation. J. Virol. 78:12147–12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blaney JE, Jr, Hanson CT, Hanley KA, Murphy BR, Whitehead SS. 2004. Vaccine candidates derived from a novel infectious cDNA clone of an American genotype dengue virus type 2. BMC Infect. Dis. 4:39 doi:10.1186/1471-2334-4-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Belon CA, Frick DN. 2008. Monitoring helicase activity with molecular beacons. Biotechniques 45:433–440, 442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed National Academy Press, Washington, DC [Google Scholar]
- 26. Vrolijk JM, Kaul A, Hansen BE, Lohmann V, Haagmans BL, Schalm SW, Bartenschlager R. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201–209 [DOI] [PubMed] [Google Scholar]
- 27. Byrd CM, Dai D, Grosenbach DW, Berhanu A, Jones KF, Cardwell KB, Schneider C, Wineinger KA, Page JM, Harver C, Stavale E, Tyavanagimatt S, Stone MA, Bartenschlager R, Scaturro P, Hruby DE, Jordan R. 2013. A novel inhibitor of dengue virus replication that targets the capsid protein. Antimicrob. Agents Chemother. 57:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Belon CA, High YD, Lin TI, Pauwels F, Frick DN. 2010. Mechanism and specificity of a symmetrical benzimidazolephenylcarboxamide helicase inhibitor. Biochemistry 49:1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson AJ, Roehrig JT. 1999. New mouse model for dengue virus vaccine testing. J. Virol. 73:783–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, Harris E. 2004. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J. Virol. 78:2701–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kyle JL, Beatty PR, Harris E. 2007. Dengue virus infects macrophages and dendritic cells in a mouse model of infection. J. Infect. Dis. 195:1808–1817 [DOI] [PubMed] [Google Scholar]
- 32. Schul W, Liu W, Xu HY, Flamand M, Vasudevan SG. 2007. A dengue fever viremia model in mice shows reduction in viral replication and suppression of the inflammatory response after treatment with antiviral drugs. J. Infect. Dis. 195:665–674 [DOI] [PubMed] [Google Scholar]
- 33. Williams KL, Zompi S, Beatty PR, Harris E. 2009. A mouse model for studying dengue virus pathogenesis and immune response. Ann. N. Y. Acad. Sci. 1171(Suppl 1):E12–E23 [DOI] [PubMed] [Google Scholar]
- 34. Yu M, Levine SJ. 2011. Toll-like receptor, RIG-I-like receptors and the NLRP3 inflammasome: key modulators of innate immune responses to double-stranded RNA viruses. Cytokine Growth Factor Rev. 22:63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, Vasudevan SG, Lescar J. 2008. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 27:3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]