Abstract
Infection by human cytomegalovirus (CMV) elicits a strong humoral immune response and robust anti-CMV antibody production. Diagnosis of virus infection can be carried out by using a variety of serological assays; however, quantification of serum antibodies against CMV may not present an accurate measure of a patient's ability to control a virus infection. CMV strains that express green fluorescent protein (GFP) fusion proteins can be used as screening tools for evaluating characteristics of CMV infection in vitro. In this study, we employed a CMV virus strain, AD169, that ectopically expresses a yellow fluorescent protein (YFP) fused to the immediate-early 2 (IE2) protein product (AD169IE2-YFP) to quantify a CMV infection in human cells. We created a high-throughput cell-based assay that requires minimal amounts of material and provides a platform for rapid analysis of the initial phase of virus infection, including virus attachment, fusion, and immediate-early viral gene expression. The AD169IE2-YFP cell infection system was utilized to develop a neutralization assay with a monoclonal antibody against the viral surface glycoprotein gH. The high-throughput assay was extended to measure the neutralization capacity of serum from CMV-positive subjects. These findings describe a sensitive and specific assay for the quantification of a key immunological response that plays a role in limiting CMV dissemination and transmission. Collectively, we have demonstrated that a robust high-throughput infection assay can analyze the early steps of the CMV life cycle and quantify the potency of biological reagents to attenuate a virus infection.
INTRODUCTION
The coevolution of human herpesviruses with their hosts over the past millions of years has led to the development of complex strategies of immune evasion that allow persistent viral infection despite the presence of an active immune response (1). A comprehensive understanding of cytomegalovirus (CMV) infection is essential to delineate the molecular and cellular interactions necessary for priming a targeted humoral immune response and how a pathogen may coopt these processes to establish a persistent and lifelong infection (2). Furthermore, a more complete comprehension of CMV entry, replication, and immune evasion is paramount in developing strategies to diagnose and alleviate CMV disease in immunocompromised patients such as transplant recipients, AIDS patients, and neonates.
Analysis of viral protein expression during CMV infection can be useful in studying viral entry, cellular manipulation, and egress. The replication cycle of CMV is temporally controlled and regulated by different segments of the viral genome. The replicative cycle is divided into immediate-early (IE), early (E), and late (L) phases of replication. CMV IE proteins are produced first and appear within 6 h postinfection (hpi). IE proteins are potent transactivators that stimulate the transcription of E genes (3, 4). IE1 and IE2 are the best-characterized IE gene products and are essential for viral replication and controlling downstream transcription factors (5–8).
Recombinant green fluorescent protein (GFP)-expressing virus strains have contributed to our understanding of the contribution of viral genes to CMV replication and dissemination (9), including viral latency (10) and cell cycle manipulation (11). Such strains have also been used to examine the kinetics and localization of viral protein expression during CMV and herpes simplex virus 1 (HSV-1) infections (12). Viral strains that express fluorescent proteins alone or as a viral chimera are also useful for measuring virus infectivity, testing the antiviral properties of small molecules and the neutralizing capability of monoclonal anti-CMV antibodies, and elucidating early steps in viral binding and entry (13–15).
In this study, we employed a CMV strain that ectopically expresses a yellow fluorescent protein (YFP) fused to the IE2 transcript, AD169IE2-YFP, to establish a high-throughput cell-based assay that can quantify viral entry into human cells by measuring fluorescence of infected cells. The high-throughput format of the assay offers an easy and rapid approach for evaluating viral growth kinetics and, as we demonstrate, can be employed to test the virus-neutralizing capability of monoclonal antibodies and human serum from CMV-positive patients. The infection readout of YFP fluorescence can assist in elucidating the early steps of the CMV life cycle and can be utilized for identification of anti-CMV therapeutics.
MATERIALS AND METHODS
Cells and antibodies.
MRC5 lung fibroblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 8% fetal bovine serum, 1 mM HEPES, 100 U/ml penicillin, and 100 g/ml streptomycin at 37°C in a humidified atmosphere (95% air–5% CO2). The monoclonal anti-gH antibody (clone 14-4b) was purified from hybridoma culture supernatant (16). Monoclonal antibody HC-10 (anti-HC) (recognizes free major histocompatibility complex [MHC] class I heavy chains) (17) was purified from hybridoma culture supernatant. For HCMV IE1 and IE2 detection, a primary mouse monoclonal antibody against a shared epitope present in IE1 and IE2 was used (MAB810; Millipore). Actin was detected by using a primary goat polyclonal IgG against human actin (sc-2005; Santa Cruz Biotechnology, Inc.).
Cloning of recombinant viruses.
CMV AD169IE2-YFP was constructed in the CMV AD169 background (18) by inserting enhanced YFP (Clontech) into the 3′ end of IE2 exon 5 in the parent AD169 bacterial artificial chromosome (BAC), as previously described (19–21). The following IE2-targeting primers were used (sequences in uppercase type are the homology arms to the IE2 sequence): 5′-CTGAGCCTGGCCATCGAGGCAGCCATCCAGGACCTGAGGAACAAGTCTCAGgccggaagaagatggaaaaag-3′ (forward) and 5′-ACGGGGAATCACTATGTACAAGAGTCCATGTCTCTCTTTCCAGTTTTTCACcgtcgtggaatgccttcg-3′ (reverse). The CMVGFP control virus (22) encodes a simian virus 40 (SV40) promoter-enhanced GFP cassette. The CMV Δcrs IE2-YFP strain was constructed by BAC “recombineering” (23) of the CMV IE2-YFP strain, as previously described (24). The integrity of all BAC recombinant viruses was verified by (i) restriction digestion using HindIII and EcoRI to verify banding patterns (New England BioLabs), (ii) direct sequencing of the recombineered locus and 1,000 bp in each direction, (iii) replication kinetics of the resulting virus, and (iv) YFP or GFP expression. To propagate and purify virus, BAC DNA was electroporated (19) into MRC5 cells (American Type Culture Collection) by using a GenePulser Xcell electroporation system (Bio-Rad). Upon infection reaching a 100% viral cytopathic effect or 100% GFP, the culture supernatant was collected and filtered with a 0.2-μm filter (Corning, Inc.). Titers of virus stocks were determined by the 50% tissue culture infectious dose (TCID50) and converted to PFU/ml (25).
Replication kinetics.
MRC5 cells were seeded into cell culture plates and maintained for 1 to 2 days until cells reached confluence at ∼5 × 104 cells per well in 24-well tissue culture plates. The monolayers were then infected with recombinant CMV IE2-YFP or CMVGFP as a control. Cells were infected at a multiplicity of infection (MOI) of 1. Inoculum was prepared by using virus stock diluted in culture medium and adsorbed onto cells in a volume of 200 μl for 1 h at 37°C in a humidified CO2 incubator. The inoculum was then removed and replaced with 1 ml of fresh medium. The amount of infectious virus used to prepare the inoculum was based on plaque assay titrations (26) of virus stocks and is shown as time point zero in each figure. At each time point, three separate sample wells were collected. Infectious cell-free virus was collected by harvesting medium from infected culture wells at the indicated time points and stored at −80°C. To measure replication, samples were thawed and prepared as a 10-fold serial dilution series in culture medium and then analyzed by TCID50. The results were then converted to PFU/ml. Error ranges were calculated by using standard deviations.
Quantitative Western blot analysis.
MRC5 cultures at ∼60% confluence were infected at an MOI of 1 with previously described strains or recombinants of CMV. Adsorption was done at 4°C to synchronize entry of virus. After adsorption, cells were washed once in phosphate-buffered saline (PBS) (Mediatech, Inc.), and fresh medium was added at 1 ml per well. A zero-time-point sample was collected as a control for viral protein levels that could potentially be carried over from the inoculum. Cells were then placed into a 37°C incubator. Samples were collected every 2 h over 20 to 24 consecutive hours or every 1 h over 20 consecutive hours where indicated. For sample collection, cell monolayers were rinsed in PBS, and cells from each well were scraped into 50 μl of radioimmunoprecipitation assay (RIPA) buffer and stored at −80°C. For analysis, samples were thawed, and total protein content was quantitated by using a modified Lowry assay (Bio-Rad DC protein assay kit). Equivalent amounts of total protein (10 μg) were added to appropriate volumes of 4× sample loading buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 1% beta-mercaptoethanol, 12.5 mM EDTA, and 0.02% bromophenol blue) and heated at 95°C for 5 min. Protein samples were loaded and separated on precast SDS-PAGE 10% or 7.5% bisacrylamide gels (Bio-Rad). Protein transfer and blot preparation were handled as previously described (26). For detection of bands, the blot was incubated with the respective primary and secondary antibodies, followed by incubation with chemiluminescence substrate from the Western Lightning ECL detection kit (NEN/Perkin-Elmer).
Virus preparation and infection.
The AD169IE2-YFP virus was propagated in MRC5 cells and purified by density gradient centrifugation by spinning at 20,000 rpm at room temperature for 1.5 h over a 20% sorbitol cushion. Infectious-virus yield was assayed on human fibroblasts by TCID50. MRC5 cells were infected with AD169IE2-YFP for 1 h at the respective MOIs, with agitation every 15 min at 37°C.
Human serum.
Human blood was obtained from subjects from a study supported by an NIH-NIAID-funded contract (contract no. HHSN266200500028C) that studied immune changes during the course of pregnancy. All subjects gave informed consent, and the IRB included the use of samples for CMV research.
Confocal microscopy.
Neonatal human dermal fibroblast (NHDF) cells were seeded overnight, infected (MOI of 5), and then fixed at 7 hpi with BD Biosciences Cytofix/Cytoperm solution (45 min at 4°C). Cells were stained by using Hoechst reagent (25 μg/ml) and washed with PBS, and data were collected for nuclear staining and YFP fluorescence by using a Molecular Devices ImageXpress Ultra (IXU) plate-scanning confocal microscope. Images were analyzed by using MetaExpress software.
Flow cytometry analysis.
Flow cytometry was performed as described previously (27). In brief, MRC5 cells were plated at 250,000 MRC5 cells/well and incubated overnight at 37°C, followed by AD169IE2-YFP infection (MOI of 5). Cells were harvested by trypsinization, washed with PBS, and subjected to flow cytometry using a Cytomics FC 500 flow cytometer (Beckman Coulter). The data were quantified by using Flow Jo software (Tree Star, Inc.) and plotted as a normalized cell number versus the YFP fluorescence signal. The percentage of infected cells was determined by gating mock-infected cells as “negative” and all events with a fluorescent signal above mock as “positive.”
Neutralization assay.
AD169IE2-YFP was preincubated with or without the respective antibody (50-μl total inoculum volume for 2.5 h at 4°C). The inoculum was then added to MRC5 cells (5,000 to 15,000 cells/well) in a 96-well plate. After 1 h, the virus/antibody inoculum was removed and replaced with 100 μl DMEM. The plate was then read in an Acumen eX3 laser scanning fluorescence microplate cytometer to measure YFP fluorescence levels for up to 17 hpi. Serum samples were diluted with DMEM prior to virus incubation (total, 50 μl). For studies in which antibody was preincubated with cells, the anti-gH antibody was preincubated with cells for 2.5 h. Following incubation, cells were washed twice with DMEM and then exposed to virus inoculum.
Fluorometric analysis by use of an Acumen eX3 laser scanning fluorescence microplate cytometer.
Cells were excited with a 488-nm laser, and shot thresholding was used to determine average background levels (shot thresholding tracks the midpoint of the background noise and sets a dividing line above this to separate data from noise). Fluorescent emission above background levels by 2 standard deviations was registered as a positive signal. To delineate between autofluorescence caused by particulate from cells or cell culture medium and emission from YFP, any fluorescent signal larger than 5 μm, smaller than 300 μm, and separated from any other emission by at least 0.5 μm in both x and y axes was considered to be an “event.” One event was assumed to be equivalent to one infected cell, and thus, the number of infected cells in a given well was extrapolated from the fluorescence emission data. Background autofluorescence was adjusted for by subtracting the mean number of events in noninfected wells from that in each infected well. Percent infection was determined by dividing the number of events in each well by the number of events in AD169IE2-YFP-infected control wells (no antibody).
ELISA CMV diagnostic test.
Patient sera were banked and stored at −80°C for use in CMV enzyme-linked immunosorbent assays (ELISAs). CMV IgG and CMV IgM ELISAs (Calbiotech) were performed according to the manufacturer's instructions. Freshly thawed sera (10 μl) were diluted 1:20 and incubated on ELISA plates precoated with the CMV pp65 protein. Plates were washed before and after the addition of enzyme conjugate. TMB (3,3′,5,5″-tetramethylbenzidine) substrate and stop solution were added before reading at 450 and 600 nm in a plate reader (Bio Tek, Winooski, VT).
Statistical analysis.
Student's unpaired, two-tailed t tests and 2-way analysis of variance (ANOVA) were performed by using GraphPad Prism.
RESULTS
Expression kinetics and intracellular localization of IE2-YFP in AD169IE2-YFP-infected cells.
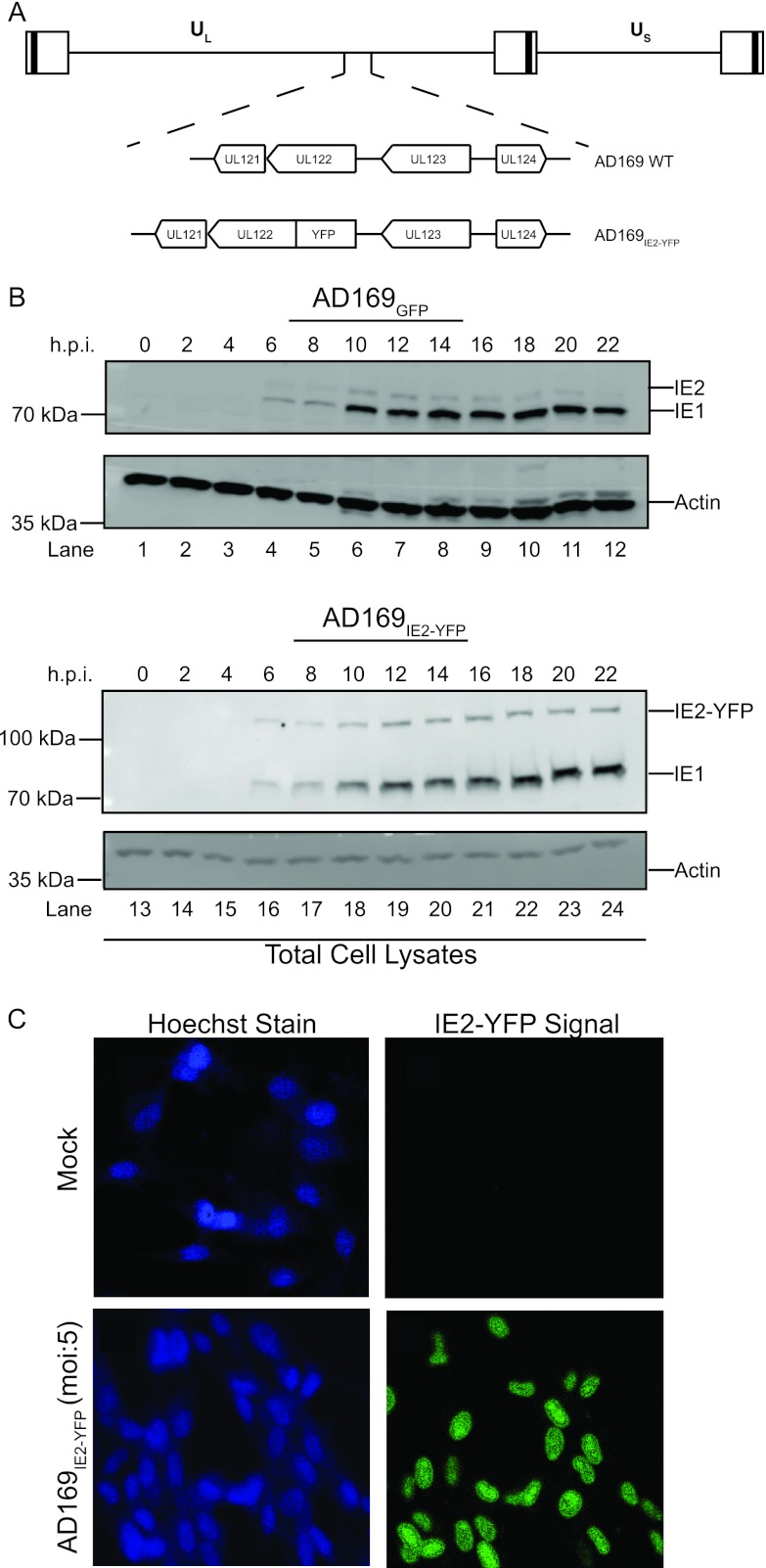
The expression of CMV gene products occurs in a tightly regulated cascade of immediate-early (IE), early (E), and late phases of replication. CMV immediate-early transcripts are produced within 6 h postinfection (hpi). IE proteins act as potent transactivators to stimulate transcription of E genes, which function primarily to replicate viral genomic DNA and alter host immune recognition (3, 4). The temporal nature of the CMV immediate-early phase of infection was exploited to study the events that permit virus entry. More specifically, a variant of the CMV AD169 strain that expresses a chimeric fusion of the IE2 protein product with enhanced yellow fluorescent protein (YFP) (AD169IE2-YFP) (Fig. 1A) was utilized to measure a CMV infection. We examined the expression kinetics of the IE2-YFP chimeric protein in cells infected with AD169IE2-YFP compared to an AD169 strain expressing GFP (AD169GFP) (Fig. 1B). Human fibroblast (MRC5) cells infected (multiplicity of infection [MOI] of 1) with AD169IE2-YFP and AD169GFP for up to 22 h were subjected to immunoblot analysis using antibodies against IE1/IE2 and actin. The IE1, wild-type IE2, and IE2-YFP fusion proteins were observed at 6 hpi, and their levels increased over 22 hpi (Fig. 1B, top). The expression kinetics of IE2-YFP were similar to those of the wild-type IE2 protein from AD169GFP-infected cells (Fig. 1B, top). More importantly, the IE2-YFP polypeptide migrated more slowly than the IE2 protein, confirming the expression of the chimeric IE2-YFP construct. Actin levels verified equivalent protein loading (Fig. 1B, bottom). Also, the IE2-YFP construct did not alter the growth kinetics of AD169IE2-YFP (see Fig. S1 in the supplemental material). Collectively, these results demonstrate that IE2-YFP is expressed with immediate-early kinetics similar to those of wild-type IE2 and does not interfere with viral replication.
Fig 1.
AD169IE2-YFP-infected cells express IE2-YFP as an immediate-early protein. (A) Diagram of the recombinant AD169IE2-YFP virus. CMV unique long (UL) and unique short (US) regions of the genome, the orientation of the UL121-124 genes, and the UL122-YFP chimera are indicated. (B) AD169GFP-infected (lanes 1 to 12) and AD169IE2-YFP-infected (lanes 13 to 24) (MOI of 1) MRC5 cells were harvested at between 0 and 22 hpi and subjected to immunoblot analysis for IE1 and IE2 polypeptides and actin protein. The respective polypeptides and relative molecular mass markers are indicated. (C) Mock- and AD169IE2-YFP-infected MRC5 cells (MOI of 5) were stained with Hoechst reagent and analyzed by using confocal microscopy (magnification, ×20). The Hoechst reagent and IE2-YFP signal localized exclusively to the nucleus.
We next analyzed the intracellular localization of IE2-YFP signal in MRC5 cells infected with AD169IE2-YFP using confocal microscopy (Fig. 1C). MRC5-infected cells (MOI of 5) were fixed at 7 hpi and stained with Hoechst reagent to identify the nucleus. The YFP fluorescent signal was observed exclusively in the nucleus of cells and coincided with Hoechst staining. This exclusive nuclear localization pattern of IE2-YFP is consistent with previous reports characterizing the nuclear localization of IE2 (28–30). Together, the data demonstrate that IE2-YFP expression acts like its wild-type immediate-early counterpart.
Analysis of fluorescent signal from AD169IE2-YFP-infected cells.
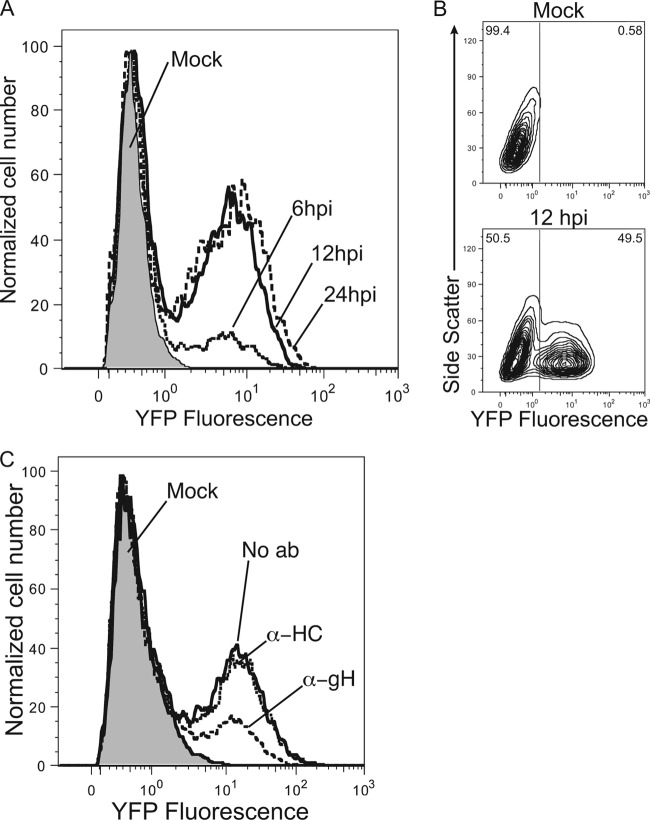
To further characterize IE2-YFP expression, the kinetics of the IE2-YFP fluorescent signal from AD169IE2-YFP-infected cells were examined by using flow cytometry (Fig. 2A). Cells were infected (MOI of 5) and analyzed for fluorescence signals at 6, 12, and 24 hpi. YFP expression peaked by 12 hpi and remained through 24 hpi. Two distinct peaks were observed in the flow cytometry plot, allowing quantification of virus-infected and noninfected cells. An increase of the fluorescence signal was observed for AD169IE2-YFP-infected cells at between 6 hpi and 12 hpi, indicating that about 50% of the cells were IE2-YFP positive (Fig. 2B). The results demonstrate that AD169IE2-YFP infections can be quantified through the fluorescent signal from IE2-YFP expression.
Fig 2.
AD169IE2-YFP-infected cells emit a robust fluorescent signal. (A and B) Mock- and AD169IE2-YFP-infected MRC5 cells (MOI of 5) analyzed by flow cytometry at 6, 12, and 24 hpi for YFP fluorescent signal are represented by a histogram (A) or a contour plot (12 hpi only) (B). (C) AD169IE2-YFP-infected cells (MOI of 3) preincubated with 4.0 μg/ml anti-gH, anti-MHC class I heavy chain (HC), or no antibody (No ab) were analyzed by flow cytometry (20 hpi) for YFP signal. Mock-infected cells are indicated by a gray peak.
We next determined whether AD169IE2-YFP-infected cells could be employed in an antibody-mediated neutralization assay. Virus was preincubated with a well-characterized neutralizing antibody against CMV glycoprotein gH (anti-gH) (mAb14-4b) (16). As a control, we utilized a monoclonal antibody against the human MHC class I heavy chains (anti-HC) (17). AD169IE2-YFP preincubated with antibodies (4 μg/ml) was used to infect MRC5 cells, which were subsequently examined by flow cytometry. The flow cytometry results demonstrated that preincubation with anti-gH caused a significant decrease in the fluorescent signal of virus-infected cells compared to virus preincubated with anti-HC (Fig. 2C). Quantification of the fluorescent signal from virus-infected cells revealed that preincubation of anti-gH antibody exhibited a dose-dependent neutralization and showed maximal neutralization at 8.0 μg/ml, while the anti-HC antibody did not exhibit a significant dose-dependent neutralization (data not shown). These data provide the proof of concept that AD169IE2-YFP-infected cells can be utilized to measure the neutralizing capacity of anti-CMV antibodies.
Quantification of CMV neutralization using an anti-CMV monoclonal antibody in a high-throughput assay.
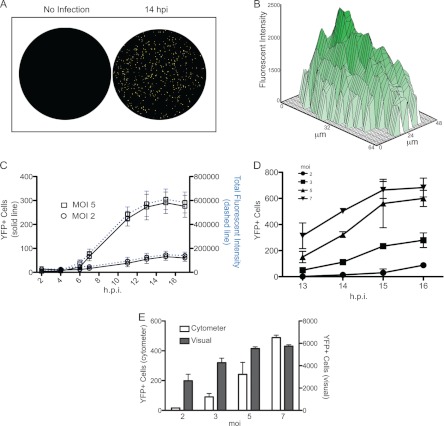
We next established conditions to quantify AD169IE2-YFP-infected cells using a laser scanning fluorescence microplate cytometer in a high-throughput format. The YFP fluorescence was measured from AD169IE2-YFP-infected (MOI of 2 or 5) MRC5 cells (5,000 cells/well) (Fig. 3). Numerous cells emitting a fluorescent signal were visualized in virus-infected wells (Fig. 3A). Analysis of the fluorescent intensity of infected cells revealed a Gaussian distribution of fluorescent signals that coalesced within an infected cell, demonstrating an intense fluorescent signal throughout the nucleus (Fig. 3B). This punctate localization corresponded well with the nuclear localization observed in confocal microscopy images of AD169IE2-YFP-infected cells (Fig. 1C). The quantification of the fluorescent signal was extrapolated to determine the number of infected cells/well. YFP signal increased above background levels by 6 hpi and continued to increase until 14 hpi. To confirm that measuring the number of virus-infected cells by fluorescence events was valid, the total fluorescence intensity per well was determined (Fig. 3C, right axis). Remarkably, a dramatic increase in the number of YFP-positive cells and total fluorescence intensity was quite consistent and dependent on the MOI. We then conducted infections in wells seeded at 15,000 cells per well at various MOIs (2 to 7) (Fig. 3D). The number of YFP-positive cells corresponded well with MOIs. To validate the number of fluorescent cells detected by the cytometer, we compared the number of YFP-positive cells visible by wide-field microscopy to the cytometer value (Fig. 3E). As with the cytometer readout, the number of cells with visible YFP expression corresponded well with the MOI. Thus, quantification of the fluorescent signal from AD169IE2-YFP-infected cells provides a robust readout to measure CMV infection that is an accurate proxy for visual assessment of infection.
Fig 3.
Characterization of AD169IE2-YFP-infected cells. (A and B) Mock- or AD169IE2-YFP-infected MRC5 cells (MOI of 5) analyzed with an Acumen eX3 laser scanning fluorescence microplate cytometer demonstrated points of intense fluorescence signal (A) distributed throughout the infected-cell nucleus (B). (C) The fluorescence signal from AD169IE2-YFP-infected cells (MOI of 2 or 5) up to 17 hpi was measured by using a microplate cytometer. The number of YFP-positive cells per well (left axis) and the total fluorescence intensity in each well (right axis) were plotted over the course of infection. (D) Furthermore, the number of YFP-positive cells per well from AD169IE2-YFP-infected cells at between 13 and 16 hpi at an MOI of 2, 3, 5, or 7 was measured by using a microplate cytometer. The 2-way ANOVA test showed a significant effect at all MOIs and time points tested (P < 0.05). (E) The number of YFP-positive cells from AD169IE2-YFP-infected cells (MOIs of 2, 3, 5, and 7) at 14 hpi was measured by using a fluorescence cytometer (left axis) or by visual counting using a wide-field fluorescence microscope (right axis). Error bars represent standard errors of means.
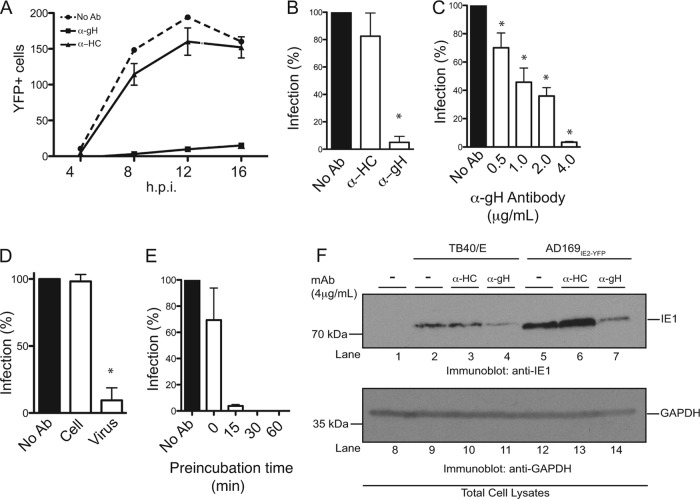
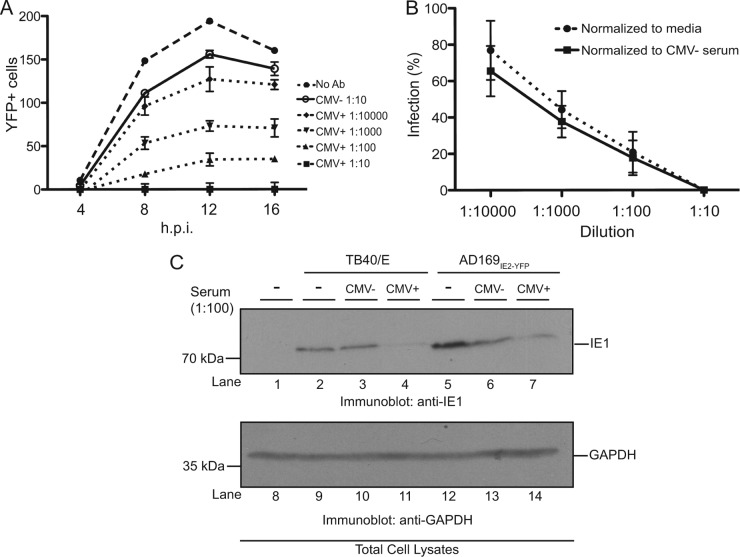
Furthermore, we utilized the newly developed high-throughput AD169IE2-YFP infection assay to measure the ability of anti-CMV antibodies to neutralize virus infection. MRC5 cells infected with AD169IE2-YFP alone or preincubated with the anti-gH and anti-HC antibodies were analyzed for YFP-positive cells for up to 16 hpi (Fig. 4A). Cells infected with the AD169IE2-YFP/anti-gH inoculum exhibited very low YFP fluorescence postinfection compared to cells infected with AD169IE2-YFP alone or AD169IE2-YFP/anti-HC antibody (Fig. 4A). Quantification of the fluorescence signal at 12 hpi revealed that the AD169IE2-YFP/anti-gH inoculum decreased the infection by >95% compared to the slight decrease in the fluorescent signal observed for AD169IE2-YFP/anti-HC-infected cells (Fig. 4B). We verified the dose dependency of anti-gH-mediated neutralization in the high-throughput assay. As expected, the anti-gH antibody exhibited dose-dependent neutralization at concentrations of between 0.5 and 4.0 μg/ml (Fig. 4C). To validate the specificity of the high-throughput neutralization assay, cells were preincubated with anti-gH antibody, washed thoroughly, and then infected with AD169IE2-YFP (Fig. 4D). The anti-gH antibody reduced infection only when preincubated with virus and not cells. Finally, we investigated the effect of anti-gH preincubation times on AD169IE2-YFP infection (Fig. 4E). Anti-gH was incubated with a virus inoculum between 0 and 60 min prior to infection (Fig. 4E). Strikingly, a significant reduction was observed with only a 15-min preincubation, and a complete block of infection was observed with 30- and 60-min preincubations. We further validated the fluorescence-based neutralization assay by immunoblotting. Cells were infected with two CMV strains that were incubated with anti-HC or anti-gH antibody as described above and harvested at 12 hpi (Fig. 4F). For both virus strains, expression of the CMV gene product IE1 was significantly reduced when virus inoculum was incubated with anti-gH antibody but not anti-HC antibody. Collectively, these results demonstrate that we have established a robust, high-throughput, fluorescence-based neutralization assay to quantify the neutralizing ability of anti-CMV antibodies and possibly other biological reagents.
Fig 4.
Establishment of a high-throughput HCMV neutralization assay. (A) MRC5 cells infected with AD169IE2-YFP (MOI of 5) preincubated with anti-gH, anti-MHC class I heavy chain (HC), or medium were analyzed at 4, 8, 12, and 16 hpi for the number of YFP-positive cells per well by using a microplate cytometer. (B to E) The percentage of infected cells (12 hpi) was calculated by using the number of YFP-positive cells from AD169IE2-YFP-infected cells as 100%. Student's t test confirmed statistical significance (*, P < 0.05), and error bars represent standard errors of means in all experiments (B to E). (C) AD169IE2-YFP-infected cells preincubated with various concentrations (0.5 to 4.0 μg/ml) of anti-gH or anti-HC antibody or medium were analyzed (12 hpi) to determine the optimal neutralization concentration of anti-gH antibody. (D) MRC5 cells infected with AD169IE2-YFP (MOI of 5) and pretreated with anti-gH antibody or cells pretreated with anti-gH prior to infection were analyzed (12 hpi) to validate the specificity of the anti-gH antibody. (E) MRC5 cells infected with AD169IE2-YFP (MOI of 5) and pretreated with anti-gH antibody for 0, 15, 30, and 60 min were analyzed (12 hpi) to measure the minimal time required for neutralization. (F) MRC5 cells infected with TB40/E or AD169IE2-YFP (MOI of 2) preincubated with anti-gH or anti-HC antibody (4 μg/ml) were subjected to immunoblot analysis (12 hpi) for IE1 (lanes 1 to 7) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (lanes 8 to 14) proteins. The respective polypeptides and relative molecular mass markers are indicated.
Analysis of CMV neutralization by human serum using a high-throughput-based fluorescent assay.
We examined the efficacy of our assay to measure the neutralization capability of human serum from CMV-positive donors. CMV infection elicits a robust anti-CMV antibody response by the humoral branch of the immune system. Neutralizing antibodies to CMV are important for controlling CMV disease in bone marrow transplant recipients (31) and solid-organ transplant recipients (32, 33) as well as for potentially reducing virus transmission in newborn infants exposed to infectious CMV (34). To analyze the presence of CMV-reactive antibodies in human serum, a CMV-positive donor and a CMV-negative donor were identified by using an ELISA-based assay that employed a virus-coated plate and colorimetric IgG detection (data not shown). Sera from these individuals were tested in our neutralization assay (Fig. 5A). Four serum dilutions of between 1:10 and 1:10,000 were tested. Serum from the CMV-positive donor exhibited effective neutralizing capability at all dilutions in a dose-dependent manner, while serum from the CMV-negative donor only slightly decreased YFP fluorescence at the highest concentration. To ensure that the observed neutralization capacity of the CMV-positive serum was not due to nonspecific interactions, the number of infected cells from the AD169IE2-YFP/CMV-positive reaction was normalized to the numbers of AD169IE2-YFP-infected cells incubated with medium and CMV-negative serum, respectively (Fig. 5B). In both cases, incubation with CMV-positive serum was found to significantly and specifically reduce infection in a dose-dependent manner. We validated these results by immunoblotting. Cells were infected with either TB40/E or AD169IE2-YFP inoculum that was incubated with serum from a CMV-negative donor or serum from a CMV-positive donor. Infected cells were harvested at 12 hpi (Fig. 5C). In cells infected with either virus strain, the expression level of the CMV gene product IE1 was significantly reduced when virus inoculum was incubated with CMV-positive serum but not CMV-negative serum. We conclude that our neutralization assay can be employed to measure the CMV-neutralizing activity of human serum.
Fig 5.
Serum from a CMV-positive donor neutralizes AD169IE2-YFP infection. (A) MRC5 cells infected with AD169IE2-YFP (MOI of 5) and pretreated with medium or human serum at various dilutions were analyzed with a microplate cytometer at 4, 8, 12, and 16 hpi for the number of YFP-positive cells. The 2-way ANOVA test showed significance at all dilutions compared to CMV-negative serum (P < 0.05). Error bars represent standard errors of means. (B) The percentage of AD169IE2-YFP-infected cells treated with CMV-positive serum (12 hpi) was expressed as percent infection compared to cells infected with inoculum incubated with medium or inoculum incubated with CMV-negative serum. (C) MRC5 cells infected with TB40/E or AD169IE2-YFP (MOI of 2) preincubated with CMV-positive serum were subjected to immunoblot analysis (12 hpi) for IE1 (lanes 1 to 7) and GAPDH (lanes 8 to 14) proteins. The respective polypeptides and relative molecular mass markers are indicated.
How does the CMV-neutralizing capability of human serum compare to anti-CMV immunoglobulin levels? Serum samples from four CMV-positive donors and one CMV-negative donor were tested for the presence of anti-CMV pp65 IgG and IgM levels by using a commercially available diagnostic kit (Fig. 6A). Sera from three donors were found to contain high titers of anti-CMV IgG that ranked above the cutoff for a “CMV-positive” designation. Donor 4 ranked just below the cutoff for the CMV-positive designation. To determine whether the various levels of IgG were due to a primary or recurrent infection, we measured the levels of anti-CMV IgM (Fig. 6A). Interestingly, donor 4 exhibited IgM levels above the cutoff for the CMV-positive designation, possibly suggesting that donor 4 recently experienced a primary or recurrent infection. Collectively, the data provide an overall “snapshot” of the anti-CMV immunoglobulins from various donors.
Fig 6.
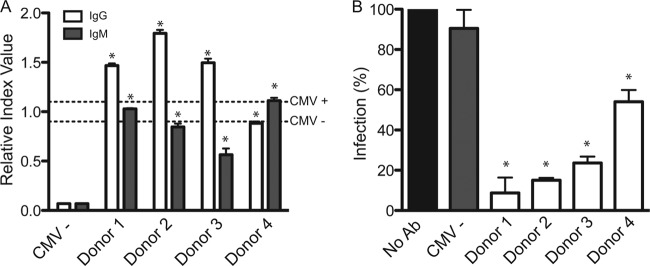
Analysis of the ability of sera from different donors to neutralize AD169IE2-YFP infection. (A) Anti-CMV IgG and IgM levels from sera of five human donors were determined by using a commercial ELISA-based anti-CMV kit. Cutoff values for CMV-positive and CMV-negative donors, as recommended by the manufacturer, are indicated with dashed lines. Student's t test showed significance of IgG and IgM levels in all samples compared to the CMV-negative control (*, P < 0.05). (B) AD169IE2-YFP-infected MRC5 cells (MOI of 5) preincubated with serum (1:100 dilution) from all five donors were analyzed (14 hpi) by using a microplate cytometer to determine YFP-positive cells. The percentage of infected cells was calculated by using AD169IE2-YFP-infected cells as 100%. Student's t test showed significance in all samples compared to the CMV-negative control (*, P < 0.05). Error bars represent standard errors of means.
Sera from the four donors and one CMV-negative donor were preincubated with the AD169IE2-YFP inoculum at a 1:100 dilution, and the inoculum was then used to infect MRC5 cells. The fluorescence signal from AD169IE2-YFP-infected cells was quantified by using nontreated AD169IE2-YFP-infected cells as 100% (Fig. 6B). Virus-neutralizing activity was detected in the sera from all CMV-positive donors, resulting in 8.73% ± 13.5%, 15.01% ± 0.82%, 23.63% ± 2.22%, and 54.03% ± 4.12% infection for donors 1 to 4, respectively. The relative levels of CMV-reactive antibodies corresponded well with the neutralizing capability of serum from donors 1 to 3. Importantly, the neutralization assay identified donor 4 to be capable of substantially neutralizing a CMV infection despite the ELISA results that ranked donor 4 as borderline negative for anti-CMV antibodies. These results suggest that the high-throughput AD169IE2-YFP neutralization assay would provide a robust readout that directly measures CMV-neutralizing antibodies in human serum. In summary, the neutralization assay offers a rapid and quantitative assessment of the neutralizing capability of both monoclonal antibodies directed at CMV and human serum. The assay was optimized for use in a high-throughput format that requires minimal material and allows thorough time course analysis of viral infection, as measured by IE2-YFP signals. Our assay can provide an immunological parameter that assesses a patient's capacity to control CMV proliferation.
DISCUSSION
In this study, we employed a recombinant CMV virus, AD169IE2-YFP, that expresses a fluorescently tagged IE2-YFP protein to visualize fluorescent expression and quantify virus infectivity. We established conditions for an antibody neutralization assay that uses YFP expression as a quantitative readout. Finally, we adapted our assay for use in a high-throughput format that uses minimal material to quantify virus infection with a rapid turnover. There are many potential applications in which the fluorescent readout of AD169IE2-YFP infection could be employed. One such example is high-throughput screening for identifying compounds or antibodies that attenuate or block infection as well as for diagnostic purposes in which the neutralizing capability of human serum can be tested in a rapid and accurate assay. Indeed, a recent report demonstrated the usefulness of CMV strains that express fluorescent fusion proteins for such applications (13).
The high-throughput AD169IE2-YFP neutralization assay could be utilized to evaluate the neutralizing capability of human serum from patients at high risk for CMV-associated diseases. For example, determining the neutralizing ability of serum from an immunocompromised patient may indicate whether an individual has CMV-neutralizing immunoglobulins, possibly due to a bout of CMV reactivation. Indeed, neutralizing antibodies have been proposed to play critical roles in protective immunity in the case of CMV pneumonitis (35) and may thus serve as critical indicators of a reactivated CMV infection. Remarkably, the assay (Fig. 5A and 6B) requires only 0.5 μl of human serum, which could likely be obtained from a finger prick. Sera from patients who had tested positive for CMV by ELISA displayed significant neutralizing capability compared to serum from a CMV-negative donor. Relative levels of CMV neutralization corresponded to levels of CMV-reactive IgG, as measured by a commercial diagnostic ELISA kit. One donor (donor 4) displayed approximately 60% infection (or 40% neutralizing capability), while the ELISA-based diagnostic kit ranked the same donor as just below the cutoff for CMV positivity. Thus, our assay is a sensitive evaluation of the ability of an individual's serum to neutralize a CMV infection and a method to survey the overall health of the patient's humoral immunity. A potential extension of our assay would be to test the neutralizing capability of antibodies found in saliva and urine. For example, a recent report demonstrated the presence of CMV-neutralizing antibodies in saliva and indicated an important distinction in neutralizing capability between fibroblast and epithelial cells (36).
One limitation of the assay developed here is that it employs a laboratory CMV strain that lacks the UL128-131 region, known to be essential for pH-dependent membrane fusion in endothelial and epithelial cells (37–40). Our assay could thereby “miss” the detection of neutralizing antibodies against this glycoprotein complex. Indeed, anti-UL130 and -UL131 antibodies can block CMV entry into mucosal epithelial cells (41). However, CMV infects fibroblasts, such as those utilized in this assay, by fusion at the cell surface in a gH/gL/UL128-131-independent fashion (42). Additionally, serum-mediated neutralization measures a polyclonal antibody response that is unlikely to be directed against only one glycoprotein complex (43, 44). Furthermore, neutralizing antibodies against the gB glycoprotein, present in AD169, are very common in CMV-seropositive individuals. One study found neutralizing antibodies to gB in 86% of 600 CMV-positive individuals (45). As several CMV vaccine strategies utilize the gB protein as an immunogen, our assay could be ideal for assessing vaccine efficacy (46). We show here (Fig. 5C) that serum from a CMV-positive individual is capable of neutralizing both a clinical (TB40/E) and a laboratory (AD169IE2-YFP) CMV strain. A clinical CMV strain that employs a similar YFP chimera or a reporter cell line capable of infection by various CMV strains (47) may be ideal for quantifying infection under a variety of conditions.
The transmission of CMV from mother to child is a result of a primary or recurrent maternal CMV infection and carries a risk of transmission of between 24 and 75% (48–50). Anti-CMV IgG avidity tests are a reliable determinant of primary versus secondary CMV infection (51–54). Combining the results from avidity and neutralization tests could strengthen the diagnostic value of these assays alone and identify patients with a higher risk of CMV transmission, potentially secondary to poor virus-neutralizing activity. A longitudinal study that could correlate virus-neutralizing antibodies with the postulated gestational age at which intrauterine transmission occurred could identify parameters that result in a high risk of transmission (55). Naturally, these studies could extend beyond maternal and congenital CMV infections to transplant recipients and patients who suffer from chronic immune-regulatory disorders such as vascular and autoimmune diseases as well as chronic immunosuppression (56).
CMV can produce noninfectious particles termed dense bodies (DB) and noninfectious enveloped particles (NIEPs) (57–60). Importantly, CMV-positive human sera react to these noninfectious particles (61), thereby demonstrating their antigenic properties. In fact, most commercially available test kits for detection of anti-CMV antibodies contain large amounts of DB in the antigen preparation (62). Previous studies indicated that the neutralization capacity of human sera is not influenced by the presence of noninfectious envelope glycoproteins, even when added at a 100-fold excess (63). Thus, only virus-neutralizing antibodies and not ELISA-reactive antibodies should be associated with protection. Our assay offers the unique advantage of measuring the protective quality of a patient's antibody response. Furthermore, the high-throughput format will allow investigators to assay large numbers of patient samples to quantify the protective response with sufficient numbers of patients to allow statistically meaningful results.
In conclusion, we developed a high-throughput assay utilizing a recombinant CMV that expresses a fluorogenic viral protein to measure the neutralizing capability of antibodies directed against the CMV virion and human serum. The assay significantly reduces the amount of material needed for analysis and measures data from multiple time points during viral replication to demonstrate IE2 expression kinetics during a lytic CMV infection. Furthermore, our high-throughput, fluorescence-based assay will be useful in the future for characterizing the neutralizing capability of human patient cohorts as well as investigating the biology of the humoral immune response toward a CMV infection in various disease states.
ACKNOWLEDGMENTS
This work was supported in part by the NIH grants AI060905 and GM083395, the Irma T. Hirschl Trust, and the American Heart Association. T.J.G. is supported by a predoctoral trainee award supported in part by USPHS institutional research training award T32-AI07647 and a Helmsley Trust fellowship, and M.W.T. is supported by a graduate research fellowship from the National Science Foundation.
We give special thanks to Scott Terhune, Nat Moorman, and Thomas Shenk, who were critical in generating the AD169IE2-YFP variant. We thank Vanessa Noriega, Sui Lee-Arteaga, Sharon Czelusniak, and Joseph Castiglione for their technical assistance and helpful discussions.
Footnotes
Published ahead of print 6 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00644-12.
REFERENCES
- 1.Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW., IV 2007. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447:326–329 [DOI] [PubMed] [Google Scholar]
- 2.Noriega V, Redmann V, Gardner T, Tortorella D. 2012. Diverse immune evasion strategies by human cytomegalovirus. Immunol. Res. 54:140–151 [DOI] [PubMed] [Google Scholar]
- 3.Cherrington JM, Mocarski ES. 1989. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meier JL, Stinski MF. 1997. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J. Virol. 71:1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heider JA, Yu Y, Shenk T, Alwine JC. 2002. Characterization of a human cytomegalovirus with phosphorylation site mutations in the immediate-early 2 protein. J. Virol. 76:928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White EA, Clark CL, Sanchez V, Spector DH. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reeves M, Murphy J, Greaves R, Fairley J, Brehm A, Sinclair J. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 80:9998–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass M, Busche A, Wagner K, Messerle M, Borst EM. 2009. Conditional and reversible disruption of essential herpesvirus proteins. Nat. Methods 6:577–579 [DOI] [PubMed] [Google Scholar]
- 10.Goodrum F, Reeves M, Sinclair J, High K, Shenk T. 2007. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110:937–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy EA, Streblow DN, Nelson JA, Stinski MF. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai P, Person S. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Straschewski S, Warmer M, Frascaroli G, Hohenberg H, Mertens T, Winkler M. 2010. Human cytomegaloviruses expressing yellow fluorescent fusion proteins—characterization and use in antiviral screening. PLoS One 5:e9174 doi:10.1371/journal.pone.0009174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibig-Rehm Y, Gotte M, Gabriel D, Woodhall D, Shea A, Brown NE, Compton T, Feire AL. 2011. High-content screening to distinguish between attachment and post-attachment steps of human cytomegalovirus entry into fibroblasts and epithelial cells. Antiviral Res. 89:246–256 [DOI] [PubMed] [Google Scholar]
- 15.Dal Pozzo F, Andrei G, Daelemans D, Winkler M, Piette J, De Clercq E, Snoeck R. 2008. Fluorescence-based antiviral assay for the evaluation of compounds against vaccinia virus, varicella zoster virus and human cytomegalovirus. J. Virol. Methods 151:66–73 [DOI] [PubMed] [Google Scholar]
- 16.Simpson JA, Chow JC, Baker J, Avdalovic N, Yuan S, Au D, Co MS, Vasquez M, Britt WJ, Coelingh KL. 1993. Neutralizing monoclonal antibodies that distinguish three antigenic sites on human cytomegalovirus glycoprotein H have conformationally distinct binding sites. J. Virol. 67:489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stam NJ, Spits H, Ploegh HL. 1986. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 137:2299–2306 [PubMed] [Google Scholar]
- 18.Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Chee MS, Hutchison CA, III, Kouzarides T, Martignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston KM, Barrell BG. 1991. The DNA sequence of the human cytomegalovirus genome. DNA Seq. 2:1–12 [DOI] [PubMed] [Google Scholar]
- 19.Yu D, Smith GA, Enquist LW, Shenk T. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng MW, Bolovan-Fritts C, Dar RD, Womack A, Simpson ML, Shenk T, Weinberger LS. 2012. An endogenous accelerator for viral gene expression confers a fitness advantage. Cell 151:1569–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36 doi:10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuevas-Bennett C, Shenk T. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82:9525–9536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevels M, Brune W, Shenk T. 2004. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 78:7803–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolovan-Fritts C, Wiedeman JA. 2001. Human cytomegalovirus strain Toledo lacks a virus-encoded tropism factor required for infection of aortic endothelial cells. J. Infect. Dis. 184:1252–1261 [DOI] [PubMed] [Google Scholar]
- 27.Noriega VM, Hesse J, Gardner TJ, Besold K, Plachter B, Tortorella D. 2012. Human cytomegalovirus US3 modulates destruction of MHC class I molecules. Mol. Immunol. 51:245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pizzorno MC, Mullen MA, Chang YN, Hayward GS. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otto SM, Sullivan-Tailyour G, Malone CL, Stinski MF. 1988. Subcellular localization of the major immediate early protein (IE1) of human cytomegalovirus at early times after infection. Virology 162:478–482 [DOI] [PubMed] [Google Scholar]
- 30.Ahn JH, Hayward GS. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoppel K, Schmidt C, Einsele H, Hebart H, Mach M. 1998. Kinetics of the antibody response against human cytomegalovirus-specific proteins in allogeneic bone marrow transplant recipients. J. Infect. Dis. 178:1233–1243 [DOI] [PubMed] [Google Scholar]
- 32.Falagas ME, Snydman DR, Ruthazer R, Griffith J, Werner BG, Freeman R, Rohrer R. 1997. Cytomegalovirus immune globulin (CMVIG) prophylaxis is associated with increased survival after orthotopic liver transplantation. The Boston Center for Liver Transplantation CMVIG Study Group. Clin. Transplant. 11:432–437 [PubMed] [Google Scholar]
- 33.Werner BG, Snydman DR, Freeman R, Rohrer R, Tilney NL, Kirkman RL. 1993. Cytomegalovirus immune globulin for the prevention of primary CMV disease in renal transplant patients: analysis of usage under treatment IND status. The Treatment IND Study Group. Transplant. Proc. 25:1441–1443 [PubMed] [Google Scholar]
- 34.Snydman DR, Werner BG, Meissner HC, Cheeseman SH, Schwab J, Bednarek F, Kennedy JL, Jr, Herschel M, Magno A, Levin MJ, Ron J, Valaes T, Berkman E, McIver J, Leszczynski J, Griffith J, Grady GF. 1995. Use of cytomegalovirus immunoglobulin in multiply transfused premature neonates. Pediatr. Infect. Dis. J. 4:34–40 [DOI] [PubMed] [Google Scholar]
- 35.Grundy JE, Shanley JD, Griffiths PD. 1987. Is cytomegalovirus interstitial pneumonitis in transplant recipients an immunopathological condition? Lancet ii:996–999 [DOI] [PubMed] [Google Scholar]
- 36.Saccoccio FM, Gallagher MK, Adler SP, McVoy MA. 2011. Neutralizing activity of saliva against cytomegalovirus. Clin. Vaccine Immunol. 18:1536–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patrone M, Secchi M, Fiorina L, Ierardi M, Milanesi G, Gallina A. 2005. Human cytomegalovirus UL130 protein promotes endothelial cell infection through a producer cell modification of the virion. J. Virol. 79:8361–8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Shenk T. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 79:10330–10338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80:710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saccoccio FM, Sauer AL, Cui X, Armstrong AE, Habib E-SE, Johnson DC, Ryckman BJ, Klingelhutz AJ, Adler SP, McVoy MA. 2011. Peptides from cytomegalovirus UL130 and UL131 proteins induce high titer antibodies that block viral entry into mucosal epithelial cells. Vaccine 29:2705–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Compton T, Nepomuceno RR, Nowlin DM. 1992. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 191:387–395 [DOI] [PubMed] [Google Scholar]
- 43.Britt WJ, Vugler L, Butfiloski EJ, Stephens EB. 1990. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J. Virol. 64:1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerna G, Sarasini A, Patrone M, Percivalle E, Fiorina L, Campanini G, Gallina A, Baldanti F, Revello MG. 2008. Human cytomegalovirus serum neutralizing antibodies block virus infection of endothelial/epithelial cells, but not fibroblasts, early during primary infection. J. Gen. Virol. 89:853–865 [DOI] [PubMed] [Google Scholar]
- 45.Marshall GS, Li M, Stout GG, Louthan MV, Duliege AM, Burke RL, Hunt LA. 2000. Antibodies to the major linear neutralizing domains of cytomegalovirus glycoprotein B among natural seropositives and CMV subunit vaccine recipients. Viral Immunol. 13:329–341 [DOI] [PubMed] [Google Scholar]
- 46.Sung H, Schleiss MR. 2010. Update on the current status of cytomegalovirus vaccines. Expert Rev. Vaccines 9:1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukui Y, Shindoh K, Yamamoto Y, Koyano S, Kosugi I, Yamaguchi T, Kurane I, Inoue N. 2008. Establishment of a cell-based assay for screening of compounds inhibiting very early events in the cytomegalovirus replication cycle and characterization of a compound identified using the assay. Antimicrob. Agents Chemother. 52:2420–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alford CA, Stagno S, Pass RF, Britt WJ. 1990. Congenital and perinatal cytomegalovirus infections. Rev. Infect. Dis. 12(Suppl 7):S745–S753 [DOI] [PubMed] [Google Scholar]
- 49.Fowler KB, Stagno S, Pass RF, Britt WJ, Boll TJ, Alford CA. 1992. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N. Engl. J. Med. 326:663–667 [DOI] [PubMed] [Google Scholar]
- 50.Kenneson A, Cannon MJ. 2007. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev. Med. Virol. 17:253–276 [DOI] [PubMed] [Google Scholar]
- 51.Grangeot-Keros L, Mayaux MJ, Lebon P, Freymuth F, Eugene G, Stricker R, Dussaix E. 1997. Value of cytomegalovirus (CMV) IgG avidity index for the diagnosis of primary CMV infection in pregnant women. J. Infect. Dis. 175:944–946 [DOI] [PubMed] [Google Scholar]
- 52.Lazzarotto T, Spezzacatena P, Pradelli P, Abate DA, Varani S, Landini MP. 1997. Avidity of immunoglobulin G directed against human cytomegalovirus during primary and secondary infections in immunocompetent and immunocompromised subjects. Clin. Diagn. Lab. Immunol. 4:469–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eggers M, Bader U, Enders G. 2000. Combination of microneutralization and avidity assays: improved diagnosis of recent primary human cytomegalovirus infection in single serum sample of second trimester pregnancy. J. Med. Virol. 60:324–330 [DOI] [PubMed] [Google Scholar]
- 54.Mace M, Sissoeff L, Rudent A, Grangeot-Keros L. 2004. A serological testing algorithm for the diagnosis of primary CMV infection in pregnant women. Prenat. Diagn. 24:861–863 [DOI] [PubMed] [Google Scholar]
- 55.Kraus TA, Engel SM, Sperling RS, Kellerman L, Lo Y, Wallenstein S, Escribese MM, Garrido JL, Singh T, Loubeau M, Moran TM. 2012. Characterizing the pregnancy immune phenotype: results of the viral immunity and pregnancy (VIP) study. J. Clin. Immunol. 32:300–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soderberg-Naucler C. 2006. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J. Intern. Med. 259:219–246 [DOI] [PubMed] [Google Scholar]
- 57.Craighead JE, Kanich RE, Almeida JD. 1972. Nonviral microbodies with viral antigenicity produced in cytomegalovirus-infected cells. J. Virol. 10:766–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiala M, Honess RW, Heiner DC, Heine JW, Jr, Murnane J, Wallace R, Guze LB. 1976. Cytomegalovirus proteins. I. Polypeptides of virions and dense bodies. J. Virol. 19:243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irmiere A, Gibson W. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118–133 [DOI] [PubMed] [Google Scholar]
- 60.Sarov I, Abady I. 1975. The morphogenesis of human cytomegalovirus. Isolation and polypeptide characterization of cytomegalovirions and dense bodies. Virology 66:464–473 [DOI] [PubMed] [Google Scholar]
- 61.Forghani B, Schmidt NJ. 1980. Humoral immune response to virions and dense bodies of human cytomegalovirus determined by enzyme immunofluorescence assay. J. Med. Virol. 6:119–127 [DOI] [PubMed] [Google Scholar]
- 62.Pepperl S, Munster J, Mach M, Harris JR, Plachter B. 2000. Dense bodies of human cytomegalovirus induce both humoral and cellular immune responses in the absence of viral gene expression. J. Virol. 74:6132–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klein M, Schoppel K, Amvrossiadis N, Mach M. 1999. Strain-specific neutralization of human cytomegalovirus isolates by human sera. J. Virol. 73:878–886 [DOI] [PMC free article] [PubMed] [Google Scholar]