Abstract
Protection against the avian influenza (AI) H5N1 virus is suspected to be mainly conferred by the presence of antibodies directed against the hemagglutinin (HA) protein of the virus. A single electroporation delivery of 100 or 250 μg of a DNA vaccine construct, pCAG-HA, carrying the HA gene of strain A/Hanoi/30408/2005 (H5N1), in chickens led to the development of anti-HA antibody response in 16 of 17 immunized birds, as measured by a hemagglutination inhibition (HI) test, competitive enzyme-linked immunosorbent assay (cELISA), and an indirect ELISA. Birds vaccinated by electroporation (n = 11) were protected from experimental AI challenge with strain A/chicken/Pennsylvania/1370/1/1983 (H5N2) as judged by low viral load, absence of clinical symptoms, and absence of mortality (n = 11). In contrast, only two out of 10 birds vaccinated with the same vaccine dose (100 or 250 μg) but without electroporation developed antibodies. These birds showed high viral loads and significant morbidity and mortality after the challenge. Seroconversion was reduced in birds electroporated with a low vaccine dose (10 μg), but the antibody-positive birds were protected against virus challenge. Nonelectroporation delivery of a low-dose vaccine did not result in seroconversion, and the birds were as susceptible as those in the control groups that received the control pCAG vector. Electroporation delivery of the DNA vaccine led to enhanced antibody responses and to protection against the AI virus challenge. The HI test, cELISA, or indirect ELISA for anti-H5 antibodies might serve as a good predictor of the potency and efficacy of a DNA immunization strategy against AI in chickens.
INTRODUCTION
The Eurasian H5N1 strain of the avian influenza (AI) virus can be fatal in humans following infection and might result in a pandemic if viral genetic reassortment or mutation produces a virus with efficient human-to-human transmission properties. Vaccination has been proposed as a viable tool for controlling epizootic or panzootic influenza in poultry and has been used in a number of countries in Asia and Central America and in Mexico to afford protection against H5N1 and H5N2 AI (1, 2). Protection against AI viruses has been observed with little or undetectable anti-AI antibodies in chickens (3, 4), ducks (5, 6), and turkeys (7), suggesting some contribution to protection by cell-mediated immunity. In an overwhelming majority of cases, however, the production of hemagglutination inhibition (HI) antibodies directed against the hemagglutinin (HA) protein, which is a critical pathogenicity determinant for the AI virus, has been the most reliable indicator of immunity against the H5N1 AI virus among many avian and nonavian species (4, 8, 9, 10). In one study, an HI titer of 1:40 was found to be an excellent predictor of protection from death and of reduced viral shedding in chickens (11). Accordingly, the HI test, as a better correlate of host protection against influenza, has surpassed the former standard serological test for AI, the agar gel immunodiffusion test (10). In addition, the competitive enzyme-linked immunosorbent assay (cELISA) is beginning to gain acceptance. Recently, a cELISA for the hemagglutinin protein of H5N1 was developed using an insect cell-expressed recombinant antigen (12), obviating the need to handle live AI virus and removing the associated biocontainment constraints required for HI testing. Potentially, the cELISA could have a wide usage in assessing the host response to vaccination. Thus, there is a need to compare available serological tests so as to understand their limitations and to assess their performance before they are used as tools for evaluating newly developed vaccine products.
The stability and ease of the production of DNA vaccines for mass distribution make them a suitable vaccine candidate as part of the strategy for pandemic preparedness. Unlike live vectors, where preexisting immunity might interfere with vaccine efficacy, multiple vaccinations can be done with a DNA vaccine if required. Similarly, DNA vaccines have been shown to overcome interference by maternal antibodies, which is a barrier to successful vaccination seen in some other types of vaccine platforms (13). Recent improvements to vaccine vector constructs and efficient delivery methodologies have been timely in addressing most of the early concerns about DNA vaccine potency in species other than laboratory rodents (14, 15). Intramuscular injection of DNA followed by the application of electrical charges or stimuli, known as electroporation, has resulted in a marked improvement in the potency of DNA vaccines (16, 17). DNA vaccine technology is now seen as a next-generation vaccine platform for use in animals and humans (18). Commercial application of DNA vaccines was first used in the veterinary market, and this has resulted in the availability of at least three DNA-based vaccines, including vaccines for West Nile virus in horses, infectious hematopoietic necrosis virus in salmon, and melanoma in dogs.
The objectives of this study were to (i) measure the anti-H5 antibody response in chickens immunized with a DNA vaccine carrying the hemagglutinin protein of H5N1 delivered with or without electroporation, (ii) assess the correlation between three anti-H5 antibody tests, the indirect ELISA, cELISA, and the HI test, and (iii) assess the usefulness of the tests in predicting DNA vaccine potency and/or efficacy.
MATERIALS AND METHODS
Animals.
Three-week-old White Leghorn chickens were obtained from the animal care unit of the Ottawa Laboratory Fallowfield (OLF), Canadian Food Inspection Agency (CFIA). The OLF stock was developed from Charles River SPAFAS birds (Storrs, CT). Animal care and handling procedures were done according to the Canadian Council on Animal Care guidelines and as outlined in the protocols approved by the institute.
Antibodies.
Antiserum against H5N2 AI virus was produced in a rabbit by immunization with binary ethylenimine-inactivated strain A/chicken/Pennsylvania/1370/1983 (H5N2) and bled 86 days after immunization. Monoclonal mouse IgG1 antibody M3095, produced using the hemagglutinin of the H5N1 AI virus (monoclonal antibody [MAb] anti-H5 antibody), a gift of Klaus Nielsen, was used in the affinity purification of the recombinant hemagglutinin protein of the A/turkey/Ontario/7732/1966 H5N9 virus that was used to coat indirect ELISA plates. The monoclonal anti-hemagglutinin antibody (IgG1 MAb no. 9) used in the cELISA was generated against the hemagglutinin protein of strain A/turkey/Ontario/6213/1966 (H5N1) at the CFIA National Centre for Foreign Animal Diseases (NCFAD), Winnipeg, Canada.
Antigen.
A 927-bp fragment (bases 55 to 981) of the hemagglutinin gene of strain A/turkey/Ontario/7732/1966 (H5N9), encoding a 40-kDa protein, was cloned into the BamHI-HindIII site of the pMelBac transfer vector (Invitrogen, CA) and was cotransfected with the linearized baculovirus Autographa californica multiple nuclear polyhedrosis virus into the insect cell line Spodoptera frugiperda (Sf9). The baculovirus stock containing the cloned H5 gene was released into the medium over a period of 72 to 168 h, and was harvested and plaque purified based on a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-mediated color selection procedure. The presence of an insert in the baculovirus stock was confirmed by PCR analysis using the primers flanking the polyhedrin gene present in the backbone of the pMEL vector (forward primer, 5′-TTTACTGTTTTCGTAACAGTTTTG-3′, and reverse primer, 5′-CAACAACGCACAGAATCTAGC-3′). Plaque-purified baculovirus stock containing a PCR-confirmed insert was used to generate a high-titer viral stock by infecting Sf9 cells in a suspension flask. The viral titer of the supernatant was determined to be 6.9 × 107 CFU. The cloned H5 protein was expressed in the insect cell line (High Five; Invitrogen, Carlsbad, CA) at a multiplicity of infection (MOI) of 10. Culture supernatants were harvested at 72 h postculture and used for downstream applications as described below, sometimes without further processing. At other times, the recombinant hemagglutinin protein was purified by sequential ammonium sulfate precipitation and affinity chromatography developed with monoclonal IgG1 anti-hemagglutinin antibody M3095, conjugated to CNBr-activated Sepharose (GE Healthcare, Baie d'Urfe, Quebec, Canada).
Construction of pCAG-HA DNA vaccine.
The HA gene of A/Hanoi/30408/2005 (H5N1) was inserted into the pCAGα vector under the control of a chicken β-actin promoter as described previously (19). The plasmid was grown in Escherichia coli and purified using endotoxin-free Giga-Preps (Qiagen, Mississauga, Ontario, Canada). The control plasmid consisted only of the pCAGα vector. The DNA content of the purified plasmid preparation was determined using a UV spectrophotometer (Beckman Coulter Canada, Mississauga, Ontario, Canada). The endotoxin level of each batch of DNA was determined by the Limulus amebocyte lysate test (Lonza, Walkersville, MD) and found to be sufficiently low (2.62 to 13.53 endotoxin units [EU]/mg DNA).
Hemagglutination inhibition assay.
An HI assay was performed following an established protocol (20). Briefly, sera were prescreened for the presence of natural hemagglutinins and all were found to be negative. Four HA units of strain A/chicken/Vietnam/14/2005 (H5N1) was then combined with 2-fold serial dilutions of each serum sample beginning with 1:2. Chicken red blood cells (0.5% [vol/vol] suspension) were then added and HI endpoints read. Chicken antiserum to strain A/duck/British Columbia/26-2/2005 (H5N2) was used as a positive control, producing endpoint titers of 1:80 to 1:160, while the negative-control serum did not inhibit the virus-induced hemagglutination (titer, <1:4). A serum HI titer of ≥1:4 was considered positive.
cELISA.
The cELISA was performed as described recently (12), with minor modifications. Briefly, microtiter plates (Nunc-Immuno plates, Roskilde, Denmark) were coated with 1 μg of recombinant baculovirus H5 protein (A/turkey/Ontario/7732/1966 [H5N9]) in carbonate buffer (pH 9.6) at 100 μl/well and plates were incubated overnight at 4°C. After washing, equal volumes (50 μl) of diluted test serum (1:5) and hybridoma culture supernatants containing monoclonal anti-hemagglutinin (H5N1) IgG1 MAb no. 9 (1:200) were added to the plates and incubated at 37°C for 1 h with agitation. Then horseradish peroxidase (HRP)-conjugated anti-mouse IgG was added and incubated for 1 h at 37°C with subsequent washing. Afterwards, the enzyme substrate 3,3′,5,5′-tetramethylbenzidine (TMB) (Sigma-Aldrich) was added and color development was stopped after 15 min with 50 μl/well of 2.0 M sulfuric acid. The optical density (OD) at 450 nm was determined on an automated plate reader (ThermoMax, Molecular Devices, Sunnyvale, CA). Results were expressed as a percentage of inhibition as derived using the following formula: percentage of inhibition (PI) = [(negative reference serum OD − test sample OD)/(negative reference serum OD − positive reference serum OD)] × 100%. A test serum sample conferring ≥50% inhibition of the hemagglutinin protein-monoclonal antibody binding was considered positive).
Indirect enzyme-linked immunosorbent assay.
Supernatant from baculovirus-infected High Five cells containing recombinant HA protein derived from A/turkey/Ontario/7733/1966 H5N9 (see “Antigen”) was suspended in 50 mM carbonate buffer (pH 9.6) at a 1:25 dilution, applied to the wells of a microtiter plate (0.2 μg/well), and incubated overnight at 4°C. Unbound antigen was discarded and wells were washed (10 mM Tris-HCl [pH 8.0] containing 150 mM NaCl and 0.05% Tween 20). The microtiter wells were blocked by the addition of 2% milk in sample diluent (10 mM Tris-HCl [pH 8.0] containing 150 mM NaCl, 0.05% Tween 20, and 0.02% NaN3). A 2-fold dilution of each serum sample was carried out starting with 1:25 until a dilution of 1:200, applied to the wells, and incubated. For the conjugate binding step, alkaline phosphatase rabbit anti-chicken IgG (heavy and light chains) (Jackson ImmunoResearch Laboratories, West Grove, PA) was diluted in assay buffer to 1:5,000 and incubated. Unless specified, all incubations were carried out at room temperature for 60 min, followed by a wash step using an automated washer (BioTek ELx405, Winooski, VT). The binding of chicken anti-influenza virus IgG to the 40-kDa HA protein was revealed by the addition of p-nitrophenyl phosphate (Sigma, St. Louis, MO) during a final incubation step at room temperature for 10 min. Optical density values were determined using an ELISA reader (ThermoMax, Molecular Devices, Menlo Park, CA) and analyzed by determining the ELISA index against a single negative and a single positive serum as described previously (22). To determine the cutoff ELISA index value for the samples tested in this study, another set of experimental chicken serum samples shown previously to be positive (n = 33) or negative (n = 66) for anti-hemagglutinin (H5N1) antibodies by cELISA and/or HI assays was tested in the indirect ELISA. The negative group consisted of chickens that were electroporated with saline (n = 35), electroporated with pCAG (empty vector) at 10 μg per chicken (n = 9) or 100 μg per chicken (n = 15), or electroporated with 100 μg pSLKIA empty vector (n = 7). The positive group consisted of chickens immunized with a commercially available H5N3 AI killed virus vaccine following the manufacturer's recommendations (FluFend; Fort Dodge Animal Health, Fort Dodge, IA). The cutoff value was determined to be 10% by means of a two-graph receiver operating characteristic curve (23).
Electroporation delivery of DNA vaccine in chickens and postvaccination bleeding.
Chickens were anesthetized with isoflurane delivered using an anesthetic machine (Matrix Medical, Inc., Minneapolis, MN) and were vaccinated with different doses of pCAG-HA DNA vaccine or the empty pCAG vector (0, 10, 100, or 250 μg plasmid DNA). Vaccine or control DNA was aspirated into a 0.3-ml syringe (Becton, Dickinson, Franklin Lakes, NJ) and fitted into the center of the TriGrid electrode (Ichor Medical Systems, San Diego, CA), comprised of four electrodes arranged in two adjacent equilateral triangles with 4.5-mm spacing between the apexes. Vaccine or control DNA was delivered intramuscularly into the anterior region of the pectoral muscle, about 1 cm anterior-lateral to the cranial apex of the sternum, and was immediately followed by five electric pulses of 112 V delivered around the immunization sites of the anesthetized chickens at intervals of 100 ms, each lasting 20 ms (Gene Pulser Xcell electroporation system; Bio-Rad, Mississauga, Ontario, Canada). Serum samples were obtained on 14, 28, 42, and 56 days postvaccination.
Highly pathogenic avian influenza (HPAI) H5N2 virus challenge.
Chickens immunized with the pCAG-HA DNA vaccine, as well as their age-matched controls vaccinated with the empty pCAGα control vector, were shipped from OLF, Ottawa, Canada, to the National Centre for Foreign Animal Diseases Laboratory, Winnipeg, Canada, and housed in biological safety level 3 (BSL3+) animal cubicles where they were allowed to acclimate for 7 days. Seventy days postvaccination, the birds were inoculated via the oronasal route with 1 × 106 ELD50 (50% egg lethal dose) of the highly pathogenic A/chicken/Pennsylvania/1370/1/1983 H5N2 AI strain (24). Following a virus challenge, the birds were monitored twice daily for clinical signs of AI. Oropharyngeal and cloacal swab specimens were collected from all animals at predetermined intervals postinoculation. To avoid death as an endpoint, birds that became moribund were humanely euthanized. In all, a total of 28 pCAG-HA-immunized birds (19 electroporated and 9 nonelectroporated, 10 to 250 μg plasmid DNA) were challenged out of a total of 50 vaccinated birds (Tables 1, 2, and 3). The remaining birds were sacrificed and their spleens and bursas were harvested for cell culture and cytokine determination, the results of which will be the subject of another report.
Table 1.
HIa antibody titers in chickens immunized with pCAG-HA DNA vaccine carrying the hemagglutinin gene of avian influenza virus strain A/Hanoi/30408/2005 (H5N1)
| Vaccine dose (μg) | + Electroporation |
− Electroporation |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vaccinated birds at 56 dpvb,c |
Seroconversion during course of expt |
Vaccinated birds at day 56 dpv |
Seroconversion during the course of expt |
|||||||||
| Total no. | GMT | Range of endpoint titer | No. of seropositive birds/total no. of immunized | Earliest detection (avg dpv) | Antibody persistence (avg no. of days)d | Total no. | GMT | Range of endpoint titer | No. of seropositive birds/total no. of immunized | Earliest detection (avg dpv) | Antibody persistence (avg no. of days) | |
| 0 | 5 | 1 | 1 | 0/5 | Not detectede | 0 | 10 | 1 | 1 | 0/10 | Not detectede | 0 |
| 10 | 13 | 9.2 | 1–128 | 7/13 | 36.4 | 13.4 | 13 | 1 | 1 | 2/13 | 32.5 | 1 |
| 100 | 12 | 48.5 | 1–128 | 11/12 | 29.1 | 27.9 | 12 | 1 | 1–8 | 1/12 | 56 | 1 |
| 250 | 5 | 42.2 | 16–128 | 5/5 | 20.2 | 23.4 | 5 | 1 | 1–8 | 1/5 | 44 | 1 |
HI, hemagglutination inhibition.
dpv, days postvaccination; GMT, geometric mean titer.
Samples testing negative by the HI test were ascribed a titer of 1 for the purpose of calculating the GMT. An HI titer of ≥4 (i.e., 1:4) was considered positive. Endpoint dilution values were converted to log2, and the GMT was determined and expressed as the endpoint mean titer after a reconversion.
The study design allowed only for measuring antibody persistence to a maximum of 43 days before termination of the experiment.
Not detected indicates that none of the chickens tested positive at any of the sampling times.
Table 2.
Serum anti-hemagglutinin antibody in chickens immunized with pCAG-HA DNA vaccine carrying the hemagglutinin gene of avian influenza virus strain A/Hanoi/30408/2005 (H5N1) as measured by a competitive ELISA
| Vaccine dose (μg) | + Electroporation |
− Electroporation |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vaccinated birds |
Seroconversion during the course of expt |
Vaccinated birds |
Seroconversion during the course of expt |
|||||||||
| Total no. | Median % inhibition | Range of inhibition (%) | No. of seropositive birds/total no. immunized | Earliest detection, (avg dpv)a | Antibody persistence (avg no. of days)b | Total no. | Median % inhibition | Range of inhibition (%) | No. of seropositive birds/total no. immunized | Earliest detection, (avg dpv)a | Antibody persistence (avg no. of days) | |
| 0 | 5 | 3 | 0–9 | 0/5 | Not detected | 0 | 10 | 0 | 0 | 0/10 | Not detected | 0 |
| 10 | 13 | 48 | 3–99 | 6/13 | 42 | 10 | 13 | 9 | 0–36 | 0/13 | Not detected | 0 |
| 100 | 12 | 85 | 29–101 | 11/12 | 31 | 26 | 12 | 12 | 0–47 | 0/12 | Not detected | 0 |
| 250 | 5 | 96 | 58–99 | 5/5 | 29 | 26 | 5 | 17 | 1–24 | 0/5 | Not detected | 0 |
Not detected: none of the chickens tested positive at any of the sampling times. Serum with a cELISA inhibition value of ≥50% was considered positive.
The study design allowed only for measuring antibody persistence to a maximum of 43 days before termination of experiment.
Table 3.
Serum anti-hemagglutinin IgG antibody in chickens immunized with pCAG-HA DNA vaccine carrying the hemagglutinin gene of avian influenza virus strain A/Hanoi/30408/2005 (H5N1) as measured by an indirect ELISA
| Vaccine dose (μg) | + Electroporation |
− Electroporation |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vaccinated birds at 56 dpv |
Seroconversion during the course of expt |
Vaccinated birds at 56 dpv |
Seroconversion during the course of expt |
|||||||||
| Total no. | Median ELISA index (%) | Range of ELISA index (%) | No. of seropositive birds/total no. of immunized | Earliest detection, (avg dpv)a | Antibody persistence (avg no. of days)b | Total no. | Median ELISA index (%) | Range of ELISA index (%) | No. of seropositive birds/total no. of immunized | Earliest detection, (avg dpv)a | Antibody persistence (avg no. of days) | |
| 0 | 5 | 3 | 1–6 | 0/5 | Not detected | 0 | 10 | 3 | 2–8 | 0/10 | Not detected | 0 |
| 10 | 13 | 10 | 1–61 | 10/13 | 30.4 | 24.8 | 13 | 4 | 0–15 | 2/13 | 42 | 1 |
| 100 | 12 | 63 | 1–105 | 11/12 | 27.2 | 31.2 | 12 | 4 | 1–8 | 0/12 | Not detected | 0 |
| 250 | 5 | 36 | 29–79 | 5/5 | 28.6 | 35.4 | 5 | 59 | 1–81 | 0/5 | Not detected | 0 |
Not detected: none of the chickens tested positive at any of the sampling times. Serum with an indirect ELISA score of ≥10% was considered positive.
The study design allowed only for measuring antibody persistence to a maximum of 43 days before termination of the experiment.
Reverse transcriptase PCR (RT-PCR) for influenza A virus.
Total RNA was extracted from 0.5 ml of swab specimen from the oropharynx and cloaca of H5N2 AI virus-inoculated chickens by means of an RNeasy mini kit (Qiagen, Mississauga, Ontario, Canada). A semiquantitative real-time RT-PCR assay (RRT-PCR), which targets the M1 gene of influenza A virus segment 7, was carried out as described previously (25). Full-length in vitro-transcribed segment 7 RNA, serially diluted in buffer, was run with each assay in order to give a semiquantitative estimate of the viral load in the sample, expressed as the cycle threshold (CT).
Relatedness of the amino acid sequences of hemagglutinin proteins used in this study.
The four different H5 hemagglutinins used in this study (i.e., DNA vaccine, challenge virus, ELISA antigen, and HI assay virus) provide a means of assessing both heterologous protection of H5N1 vaccines and the cross-reactive antibody responses among different clades of H5 AI types. To that end, we assessed the degrees of identity among the HA amino acid sequences of all four virus isolates employed in this study using sequence alignment software (Clone Manager v9; Scientific & Educational Software, Cary, NC). To permit a broader interpretation of our experimental results (i.e., serological and challenge experiments), these four HA sequences were included in a larger sequence analysis involving another 50 isolates of both H5N1 and H5N2 types that were selected based on the following criteria: (i) a wide geographical representation of many continents, including Asia, North America, Africa, and Europe, (ii) a mix of older and newer isolates spanning a considerable period of time (1959-2012), (iii) a variety of clades, including those currently in circulation and those that have been detected previously but are currently noncirculating, and (iv) different hosts, including humans, domestic birds, wild birds, domestic animals (a cat), and livestock (a horse). The amino acid residues of the H5 protein of 54 AI isolates were compared by assessing the percent match of the other proteins with the A/Hanoi vaccine strain (Clone Manager software) and by constructing a phylogenetic tree employing the neighbor-joining method (Geneious software version 6.04; Biomatters, Auckland, New Zealand). By aligning the amino acid resides, we carried out a detailed comparison of the amino acid residues present at the antigenic sites A (amino acid positions 136 to 141) and B (124 to 129 and 152 to 153), which are considered to be critical sites for antibody-mediated virus neutralization (26–28). Amino acid sequences of the HA proteins were obtained from GenBank or at http://H5N1.flugenome.org/show_subtypes.php. The vaccine strain was used as the reference sequence for the alignment analysis.
Statistical and arithmetic analysis.
The geometric mean of the HI titers and the correlation coefficient of antibody test results were determined with the aid of a statistical software program (MedCalc software; Mariakerke, Belgium). In assessing the geometric mean of each HI titer, a serum sample testing negative at the highest concentration tested (i.e., dilution of 1:2) was assessed to be <2 HI units, and an arbitrary value of 1 unit was assigned. Significant differences between mean values (P < 0.05) were determined by t test on the data shown to follow a normal distribution as analyzed by the Kolmogorov-Smirnov test (MedCalc software).
The 2-week interval between the samplings of vaccinated chickens (see “Electroporation delivery of DNA vaccine in chickens and postvaccination bleeding”) inherently imposed a limitation on the estimates of the period of seroconversion and of antibody persistence in the vaccinated chickens. The study design allowed only for measuring antibody persistence at a maximum of 43 days. Agreement between the results of two serological tests was determined (MedCalc software) by calculating the weighted kappa value (κ) on the results scored as positive or negative based on the cutoff value of each test.
RESULTS
The effect of electroporation on antibody responses.
Sixteen of 17 birds (94%) receiving 100 μg or 250 μg of DNA vaccine carrying the HA gene of H5N1 AI virus (strain Hanoi 2005) delivered by electroporation had positive antibody titers at day 56 as measured by three serological tests, namely, HI, cELISA, and indirect ELISA (Tables 1 to 3). In contrast, only two of 17 birds (12%) that received identical doses of vaccine without electroporation responded by day 56 with a very transient and weak antibody response that was positive only by HI test. Of the 13 birds that received 10 μg of the DNA vaccine by electroporation, six showed a sustained positive antibody response in all three tests; however, as many as 10 birds actually mounted an antibody response. Among the 13 birds that received 10 μg of the DNA vaccine without electroporation, two had very weak and transient antibody responses detected only by the indirect ELISA. Birds injected with 10 μg (n = 12), 100 μg (n = 12), or 250 μg (n = 5) of the empty pCAG vector delivered by electroporation did not develop an anti-HA antibody response in any of the three serological tests.
The effect of vaccine dose on antibody responses as measured by three serological tests.
Among birds that received the highest DNA vaccine dose, i.e., 250 μg, delivered by electroporation, HI and cELISA antibody responses developed more quickly than in those receiving lower doses under identical conditions (Tables 1 and 2). The magnitude and persistence of antibody responses in birds receiving 100 μg or 250 μg of electroporation-delivered vaccine were generally similar (Mann-Whitney test for differences in the magnitude of antibody response between the two groups at day 56: HI, P = 0.49; cELISA, P = 0.24; indirect ELISA, P = 0.95; Mann-Whitney test for antibody persistence: HI, P = 0.55; cELISA, P = 0.69; indirect ELISA, P = 0.42). In general, DNA vaccines induced an antibody response in birds starting 3 to 6 weeks after vaccination and tended to persist for a longer period in chickens that received the higher vaccine doses. The longer persistence of antibodies in chickens electroporated with ≥100 μg DNA vaccine over chickens electroporated with 10 μg DNA vaccine attained statistical significance for HI and cELISA (t test, P = 0.02 for both serological tests) but not for the indirect ELISA (P = 0.07). When chickens receiving different doses of the electroporated-delivered vaccine were considered separately, the antibody profiles obtained using the different tests showed the greatest disparities in the group that received the lowest dose of vaccine, i.e., 10 μg (n = 13). Ten of the birds (77%) in this group had antibody titers that were positive by indirect ELISA, in comparison to 54% and 46% that were positive by cELISA and HI, respectively. Nevertheless, the weighted kappa values obtained by comparing test results from any pair of serological tests on the DNA-vaccinated chickens, electroporated and nonelectroporated, were >0.8 (Table 4), which indicated that the serological tests showed very good agreement.
Table 4.
Test agreement between serological assays for detecting anti-hemagglutinin antibodies in chickens following electroporation and nonelectroporation delivery of pCAG-HA DNA vaccine constructed using the hemagglutinin gene of strain A/Hanoi/30408/2005 (H5N1)a
| Tests comparedb | κ | SE | Agreement |
|---|---|---|---|
| HI test vs cELISA | 0.86 | 0.13 | Very good |
| HI test vs indirect ELISA | 0.87 | 0.12 | Very good |
| cELISA vs indirect ELISA | 0.86 | 0.13 | Very good |
The serological test results of electroporated (n = 30) and nonelectroporated chickens (n = 30) were used in the analysis after determining whether the serum sample tested positive or negative on day 56 postvaccination (Tables 1 to 3). The strength of test agreement was judged as poor (κ < 0.2), fair (κ = 0.21 to 0.40), moderate (κ = 0.41 to 0.61), good (κ = 0.61 to 0.80), or very good (κ = 0.81 to 1).
HI, hemagglutination inhibition test; cELISA, competitive enzyme-linked immunosorbent assay.
Protective effect of DNA vaccine against H5N2 virus challenge.
Following challenge with the A/chicken/PA/1370/1983 (H5N2) virus, chickens that were immunized previously with pCAG-HA vaccine delivered by electroporation exhibited significant protection against infection as assessed by viral burden on day 6 postinoculation, presence of clinical signs of the virus, and mortality (Table 5). All 11 chickens challenged with the AI virus following electroporation with 100 μg (n = 8) or 250 μg (n = 3) of the DNA vaccine survived infection (0% mortality). None showed a clinical sign (0% morbidity), and all of them, with the exception of one bird electroporated with 100 μg, failed to excrete detectable amounts of the virus in their feces by day 6. Although most of the birds still harbored the virus in the oropharynx by day 6, in time, they eliminated their viral load, with the exception of two birds (given 100 μg of DNA vaccine) that retained very small amounts in their oral cavities by day 9 (CT values, 35.6 and 37.8). In contrast, three of the five chickens injected with the 100 μg of pCAG-empty vector delivered by electroporation showed clinical signs (60% morbidity), two died (40% mortality), and all had considerable amounts of the virus in both the oropharynx and cloaca (CT values, 20.7 and 32.1), with the exception of one bird that did not shed virus in feces. Furthermore, birds receiving pCAG-HA vaccine without electroporation showed a similar pattern of susceptibility as those injected with the control plasmid. Three birds immunized with 100 μg of pCAG-HA without electroporation harbored the virus until day 6 and beyond, two of them showed clinical signs, and one had to be euthanized. Similarly, two of three birds injected with 250 μg of the vaccine without electroporation developed clinical signs following viral challenge and they all harbored the virus, albeit in limited amounts, in the oropharynx (range of CT values, 35.6 to 39.1) and shed even more virus from the cloaca (range of CT values, 27.6 to 28.8). Without exception, all the birds inoculated with the virus became infected, as measured by the presence of virus in the oropharynx at day 3 postinoculation (range of CT values, 20.3 to 30.3, n = 35), but the spread of the virus was slowed or often abrogated in DNA-electroporated chickens to the extent that only a minority shed the virus in their feces. Along the same vein, morbidity was remarkably reduced and mortality was prevented in chickens that received electroporation-delivered DNA vaccine prior to the AI virus challenge.
Table 5.
Effect of DNA vaccination on the pathogenicity of H5N2 avian influenza (strain A/chicken/Pennsylvania/1370/1/1983) in chickens
| Vaccine delivery and type | Vaccine dose (μg/bird) | H5N2 viral challenge |
|||
|---|---|---|---|---|---|
| Viral load assessed by RT-PCR (CT value in individual birds 6 days postinoculation)a |
No. affected/total no. challenged with virus |
||||
| Oropharynx | Cloaca | Morbidityb | Mortality | ||
| Controlc | 10 | 27.3, 30.0, 31.0, 22.6, *d | 23.5, 25.7, 26.9 29.2, *d | 5/5 | 1/5 |
| Control | 100 | 26.6, 29.4, 25.5, 20.7, 33.4 | 25.4, 27.4 28.9, 32.1, ND | 3/5 | 1/5 |
| Control | 250 | Not tested | Not tested | Not tested | Not tested |
| DNA vaccine | 10 | 30.0, 30.0, 35.3, ▲e, 34.8, 30.9, 29.9, 31.7 | 37.2, ND, ND, ▲e, 32, ND, ND, 25.1 | 2/8 | 2/8 |
| DNA vaccine | 100 | 31.3, 37.4, 36.2, 36.3, 29.9, 34.3, 37.8, 39.1 | ND, ND, ND, ND, 24.2, ND, ND, ND | 0/8 | 0/8 |
| DNA vaccine | 250 | 35.6, 30.9, 20.8 | ND, ND, ND | 0/3 | 0/3 |
| Nonelectroporation delivery of vaccine | |||||
| DNA vaccine | 10 | 23.4, 25.4, 26.2 | 26.9, 27.1, 28.9 | 3/3 | 1/3 |
| DNA vaccine | 100 | 39.3, 22.3, 34.1 | 39.8, 24.3, 30.0 | 2/3 | 1/3 |
| DNA vaccine | 250 | 28.6, 27.6, not tested | 27.6, 28.9, not tested | 2/3 | 0/3 |
A CT value of ≥40 was considered negative and reported as ND (virus not detected). The CT values are inversely correlated with viral load. Amount of virus present in the oropharyngeal and cloacal swabs was determined for some of the birds (n = 18) using the CT values presented above analyzed against a linear curve, and the results are presented as median virus copy number (range of virus copy number; number of birds). For oropharynx swabs: control, 10 μg: 8.5 × 106 (1 × 105 to 1.8 × 107; n = 4); control, 100 μg: 2.6 × 105 (2.2 × 104 to 6.2 × 107; n = 5); DNA vaccine, 10 μg: 9.5 × 104 (6.7 × 103 to 1.8 × 105; n = 4); DNA vaccine, 100 μg: 3.8 × 103 (1.8 × 103 to 8.1 × 104; n = 5); for cloaca swabs: control, 10 μg: 2.0 × 106 (2.9 × 105 to 1.0 × 107; n = 4); control, 100 μg: 3.8 × 105 (0 to 3.2 × 106; n = 5); DNA vaccine, 10 μg: 1.1 × 103 (0 to 5.2 × 104, n = 4); vaccine, 100 μg: 0 (0 to 7.1 × 106; n = 5).
Clinical signs (morbidity) observed included depression, swollen hock, lameness, cyanosis of comb, and petechial hemorrhages on feet, joint, and comb.
Control DNA: empty pCAG-α vector.
An asterisk indicates that the bird died on day 5 and viral load was significant on day 3 in the oropharynx (CT = 27.2) and cloaca (CT = 26.1).
A ▲ indicates that the bird died on day 5 and virus was detected on day 3 in the oropharynx (CT = 28.5) and cloaca (CT = 37.7).
Correlation between anti-HA antibody seroconversion and protection against avian influenza virus.
All 11 chickens that survived the challenge with the H5N2 virus and had a history of electroporation with 100 μg (n = 8) or 250 μg (n = 3) of the DNA vaccine had anti-HA antibodies that were positive in all 3 serological tests at day 56 postelectroporation (Tables 1 and 5). Six of eight chickens that received 10 μg by electroporation and had positive antibody responses did not show any clinical signs upon virus challenge and survived (Tables 1 and 5). One chicken that succumbed to virus challenge was negative by HI but tested positive by both the cELISA and indirect ELISA. The last of the eight chickens, i.e., among those electroporated with 10 μg DNA vaccine and subsequently challenged with the virus, failed to develop an antibody response following vaccination and was not protected upon challenge (Tables 1, 2, 3, and 5). Chickens injected with the DNA vaccine but without electroporation were usually ill following virus challenge (10 μg, 3 sick out of 3; 100 μg, 2 sick out of 3; 250 μg, 2 sick out of 3) (Table 5). All the sick birds were negative by all three antibody tests, while one protected bird was positive by HI but negative by the other two serological tests. The other protected bird (250 μg) was negative by all three serological tests. Odds ratio (OR) analysis showed that positive anti-HA test results in each of the three serological tests significantly predicted immunity against AI (HI, OR = 144, P = 0.0008; cELISA, OR = 22.4, P = 0.0085; indirect ELISA, OR = 68, P = 0.0011) (Table 6).
Table 6.
Relationship between anti-hemagglutinin seropositivity in chickens immunized with pCAG-HA DNA vaccine and protection against avian influenza (strain A/chicken/Pennsylvania/1370/1/1983 [H5N2])a
| AI protection status | HI result |
cELISA result |
Indirect ELISA result |
|||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | |
| Protected | 18 | 1 | 14 | 5 | 17 | 2 |
| Not protected | 1 | 8 | 1 | 8 | 1 | 8 |
| Total | 19 | 9 | 15 | 13 | 18 | 10 |
Similarities and differences among the HA (H5) sequences of AI virus strains, including the DNA vaccine and challenge virus.
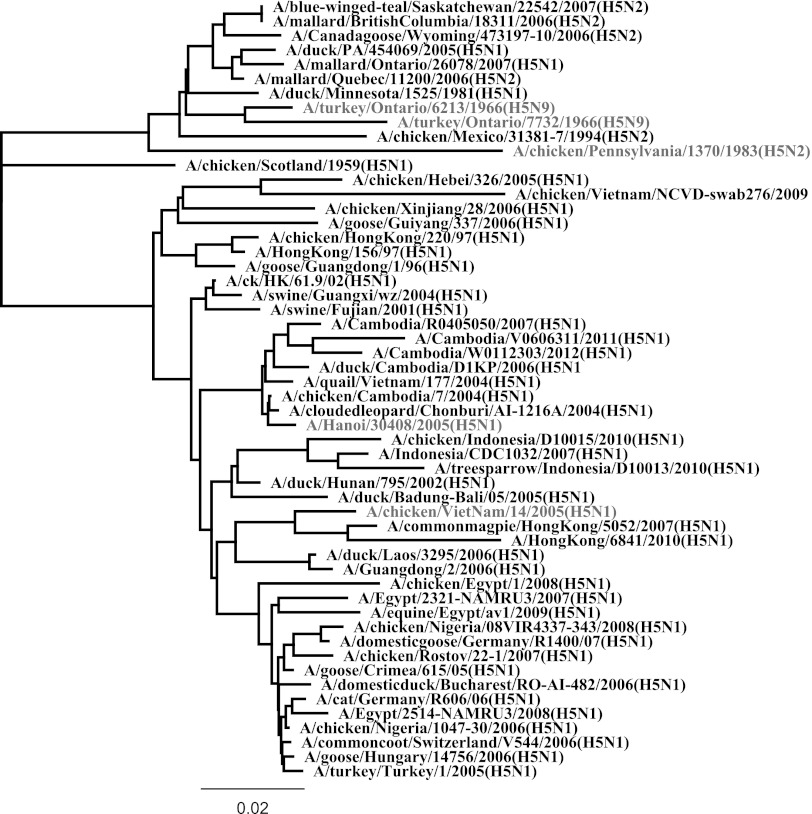
Comparison of the full HA (H5) protein sequence of the vaccine strain, A/Hanoi/30408/2005, with that of the challenge virus strain, A/chicken/Pennsylvania/1370/1/1983, indicated an identity of 86% at the amino acid level (Table 7 and Fig. 1). The vaccine HA amino acid sequence showed 87% identity with the A/turkey/ON/7732/1966 HA used as the ELISA coating antigen and 95% identity with A/chicken/Vietnam/14/2005 HA, which was used to assess the HI antibodies in immunized chickens (Table 7 and Fig. 1). A comparison of an additional 50 full H5 protein sequences with the vaccine strain indicates a high similarity among diverse H5 isolates (87 to 99% identity), although the North American strains were predictably less similar (87 to 89% identity) to the vaccine strain (A/Hanoi) compared to isolates from Asia and the rest of the world (90 to 99% identity). All the Asian isolates were similar to each other and to isolates from the other geographical locations, apart from North America. For example, A/Hanoi (Asian) showed 96% identity with A/chicken/Nigeria/1047-30/2006 (Africa) and 95% identity with A/turkey/Turkey/1/2005. Examination of all 54 aligned hemagglutinin protein sequences revealed considerable amino acid diversity at the recognized antigenic sites A and B of the H5 protein (10); we found significant variations at positions 136, 139, and 140 in the antigenic site A, and at position 124 in the antigenic site B. The residue S occupied position 136 in a vast majority of the isolates (48 out of 54), while the remaining sequences had N (4 isolates) or D (2 isolates) in that position. Similarly, position 139 was occupied by an S in most of the isolates (45 out of 54), while P was present in 6 isolates, and A, T, and L were present in 1 isolate each. The residue S was also commonly encountered at position 140 (10 isolates) but was not as frequent as D (24 isolates) or N (19 isolates) in that position. The residue E was present at position 140 only in the earliest isolate from North America (A/turkey/Ontario/7732/1966), which we used for the indirect and competitive ELISAs, whereas all the remaining 8 isolates from North America had N at position 140. The diversity at antigenic site B showed a remarkable contrast from the observations described above for antigenic site A in that there was a clear-cut dichotomy between Eurasian and North American isolates: residue I was consistently observed at position 124 of the antigenic site B among the Eurasian isolates, while the North American isolates had T at the same position. The preceding residue at position 123, which has not been previously linked with an antigenic site, also followed a similar pattern, where R was found in the Eurasian isolates compared to S in the North American isolates. The residues at positions 154 and 156, which are immediately distal to the recognized antigenic site B (152 and 153), also showed noticeable diversity: all the North American isolates had N at position 154, with the exception of A/chicken/Pennsylvania/1370/1983, our challenge virus, which had L in that position. The Eurasian isolates showed considerable diversity at position 154, as a majority had Q (33 of 43 isolates), some had L (9 of 43 isolates), and a single isolate had H (A/goose/Guangdong/1/1996). At position 156, N was present in all North American isolates studied, and although it also was the most frequently occurring residue among the Eurasian isolates (19 of 43), K was also fairly common at this position (15 of 43 isolates). The recognized antigenic sites and the adjoining positions explored in this analysis show obvious, and sometimes defining, differences between the North American and Eurasian strains, including those examined during this study. Despite these differences, the antibodies generated as a result of immunization with an HA DNA sequence of Eurasian origin generated serological cross-reactivity with North American HA proteins, as measured by the indirect and competitive ELISAs (Tables 2 to 4). The Eurasian HA sequence used in this study cross-protected against a North American-sourced H5N2 virus (Table 5).
Table 7.
Relationship among a wide selection of hemagglutinin proteins of H5N1 and H5N2 avian influenza virus strains, including four used in this studya
| Strain | Amino acid residues | Match | Nonmatch | % Matchb |
|---|---|---|---|---|
| A/Hanoi/30408/2005 (H5N1) | 567 | |||
| A/blue-winged teal/Saskatchewan/22542/2007 (H5N2) | 564 | 503 | 64 | 89 |
| A/Cambodia/R0405050/2007 (H5N1) | 565 | 555 | 13 | 98 |
| A/Cambodia/V0606311/2011 (H5N1) | 568 | 549 | 19 | 97 |
| A/Cambodia/W0112303/2012 (H5N1) | 568 | 553 | 15 | 97 |
| A/cat/Germany/R606/06 (H5N1) | 568 | 547 | 21 | 96 |
| A/chicken/Cambodia/7/2004 (H5N1) | 553 | 549 | 19 | 99 |
| A/chicken/Egypt/1/2008 (H5N1) | 568 | 537 | 31 | 95 |
| A/chicken/Hebei/326/2005 (H5N1) | 548 | 515 | 53 | 94 |
| A/Ck/HK/61.9/02 (H5N1) | 548 | 535 | 33 | 98 |
| A/Chicken/Hong Kong/220/97 (H5N1) | 568 | 542 | 26 | 95 |
| A/chicken/Nigeria/08VIR4337-343/2008 (H5N1) | 542 | 517 | 51 | 95 |
| A/chicken/Indonesia/D10015/2010 (H5N1) | 568 | 541 | 27 | 95 |
| A/chicken/Mexico/31381-7/1994 (H5N2) | 564 | 492 | 75 | 87 |
| A/chicken/Nigeria/1047-30/2006 (H5N1) | 568 | 547 | 21 | 96 |
| A/chicken/Pennsylvania/1370/1983 (H5N2) | 547 | 470 | 97 | 86 |
| A/chicken/Rostov/22-1/2007 (H5N1) | 568 | 544 | 24 | 96 |
| A/chicken/Scotland/1959 (H5N1) | 564 | 513 | 54 | 91 |
| A/chicken/Vietnam/14/2005 (H5N1) | 567 | 540 | 27 | 95 |
| A-chicken-Vietnam-NCVD-swab276/2009 | 550 | 495 | 76 | 90 |
| A/chicken/Xinjiang/28/2006 (H5N1) | 552 | 525 | 43 | 95 |
| A/clouded leopard/Chonburi/AI-1216A/2004 (H5N1) | 564 | 559 | 9 | 99 |
| A/common coot/Switzerland/V544/2006 (H5N1) | 568 | 545 | 23 | 96 |
| A/common magpie/Hong Kong/5052/2007 (H5N1) | 555 | 527 | 40 | 95 |
| A/domestic duck/Bucharest/RO-AI-482/2006 (H5N1) | 546 | 524 | 44 | 96 |
| A/duck/Badung-Bali/05/2005 (H5N1) | 529 | 505 | 63 | 95 |
| A/duck/PA/454069/2005 (H5N1) | 564 | 502 | 65 | 89 |
| A/duck/Cambodia/D1KP/2006 (H5N1) | 568 | 559 | 9 | 98 |
| A/duck/Hunan/795/2002 (H5N1) | 565 | 548 | 20 | 97 |
| A/duck/Laos/3295/2006 (H5N1) | 567 | 544 | 24 | 96 |
| A/duck/Minnesota/1525/1981 (H5N1) | 564 | 504 | 63 | 89 |
| A/Egypt/2321-NAMRU3/2007 (H5N1) | 568 | 544 | 24 | 96 |
| A/Egypt/2514-NAMRU3/2008 (H5N1) | 549 | 527 | 41 | 96 |
| A/equine/Egypt/av1/2009 (H5N1) | 567 | 536 | 32 | 95 |
| A/goose/Crimea/615/05 (H5N1) | 568 | 547 | 21 | 96 |
| A/domestic goose/Germany/R1400/07 (H5N1) | 568 | 542 | 26 | 95 |
| A/Goose/Guangdong/1/96 (H5N1) | 568 | 544 | 24 | 96 |
| A/goose/Guiyang/337/2006 (H5N1) | 556 | 527 | 41 | 95 |
| A/goose/Hungary/14756/2006 (H5N1) | 568 | 546 | 22 | 96 |
| A/Guangdong/2/2006 (H5N1) | 567 | 542 | 25 | 96 |
| A/Hong Kong/156/97 (H5N1) | 568 | 542 | 26 | 95 |
| A/Hong Kong/6841/2010 (H5N1) | 528 | 488 | 79 | 92 |
| A/Indonesia/CDC1032/2007 (H5N1) | 552 | 527 | 41 | 95 |
| A/mallard/British Columbia/18311/2006 (H5N2) | 564 | 503 | 64 | 89 |
| A/mallard/Ontario/26078/2007 (H5N1) | 564 | 501 | 66 | 89 |
| A/mallard/Quebec/11200/2006 (H5N2) | 551 | 493 | 74 | 89 |
| A/quail/Vietnam/177/2004 (H5N1) | 565 | 557 | 11 | 99 |
| A/swine/Fujian/2001 (H5N1) | 568 | 550 | 18 | 97 |
| A/swine/Guangxi/wz/2004 (H5N1) | 568 | 553 | 15 | 97 |
| A/tree sparrow/Indonesia/D10013/2010 (H5N1) | 568 | 536 | 32 | 94 |
| A/Canada goose/Wyoming/473197-10/2006 (H5N2) | 564 | 500 | 64 | 89 |
| A/turkey/Ontario/6213/1966 (H5N9) | 564 | 499 | 68 | 88 |
| A/turkey/Ontario/7732/1966 (H5N9) | 565 | 491 | 76 | 87 |
| A/turkey/Turkey/1/2005 (H5N1) | 568 | 545 | 23 | 96 |
A total of 54 hemagglutinin sequences obtained from sources indicated below were analyzed for similarities with the hemagglutinin sequence used for constructing the pCAG-HA DNA vaccine, A/Hanoi/30408/2005 (H5N1), as the reference sequence. Global sequence alignment analysis was performed using the BLOSUM 62 scoring matrix (Clone Manager Professional v9.2, Scientific & Educational Software, Cary, NC). Sources of hemagglutinin sequences: http://www.fludb.org/brc/home.do?decorator=influenza and http://www.ncbi.nlm.nih.gov/genomes/FLU/.
The % match is the proportion of matching amino acids compared to the total number of amino acid residues.
Fig 1.
Phylogenetic relationships among 54 H5-type avian influenza virus hemagglutinins, including those used in this study. Shown in light grey font are AI virus strains used as sources of DNA vaccine (A/Hanoi/30408/2005), challenge virus (A/chicken/Pennsylvania/1370/1983), hemagglutination inhibition assay virus (A/chicken/Vietnam/14/2005), and recombinant protein used in ELISA or cELISA (A/turkey/Ontario/7732/1966), as well as the virus used to produce the cELISA monoclonal antibody (A/turkey/Ontario/6213/1966) (see details in Materials and Methods).
DISCUSSION
Apart from the recent use of vaccines in commercial turkey operations, vaccination against AI is not widely practiced by the poultry industry in Canada. In other areas of the world, however, vaccination campaigns employing killed or inactivated virus preparations have been successfully mounted against outbreaks caused by the AI virus (29). Strategic vaccination against H7N1 was carried out in Italy to deal with the reemergence of a low pathogenic H7N1 virus (30). In the United States, vaccination of turkeys against low pathogenic strains has been done using commercially available vaccines under special licenses (31–34). Many authorities worldwide have now established an AI vaccine bank in anticipation of the use of vaccination to control the next AI pandemic.
It is difficult to predict which AI strain will cause the next epizootic or panzootic. As the recent experience with the pH1N1 2009 virus in humans has reinforced, it is imperative that scientists investigate vaccine platforms that are conducive to rapid production and deployment in adequate quantities in response to the emergence of a novel influenza virus strain with panzootic potential. A key advantage of the DNA vaccine platform is that candidate vaccine development can commence immediately following the sequencing of the viral genome, even if viral stocks are not yet available. In addition, initiation and scale-up of production can be rapidly and predictably carried out because manufacturing processes developed for one DNA vaccine candidate can be swiftly adapted to subsequent vaccine candidates. Our investigation revealed a significant degree of HA cross-reactivity among our isolates based on two sets of evidence. First, there is broad recognition of anti-HA antibodies developed by chickens immunized using the DNA vaccine construct from A/Hanoi in assays utilizing another Asian strain of a different clade (A/chicken/Vietnam) or a North American strain. Second, we demonstrated cross-protection between an Asian and a North American strain (86% identity) of HPAI. Based on this study, a wide and broad cross-reactivity might be inferred among the HA proteins based on their amino acid sequence profiles (82 to 98% identity). In the face of an H5N- AI pandemic, early vaccination using an existing DNA vaccine delivered properly might be of some benefit by producing significant protection against H5N- viruses in chickens, pending the availability, perhaps in no more than a few weeks, of a homologous DNA vaccine. The degree of divergence seen among the 54 isolates analyzed in this study as shown by the percent identity (Table 7) and a phylogenetic tree (Fig. 1) is similar to that observed for a tree drawn from 2,947 HA sequences of isolates believed to represent all currently circulating viral clades (http://www.who.int/influenza/gisrs_laboratory/201101_h5fulltree.pdf). We observed diversity among the amino acids at positions 123, 124, 154, and 156, all overlapping or adjoining the antigenic sites A and B. The differences in positions 123 and 124 were predictive of the geographical origin of the isolates (North America or Eurasia/Africa). In contrast, the differences in the other positions could not be used to determine the lineage of the AI viruses. Importantly, differences in the HA protein at all sites were not sufficient to affect the ability of a DNA vaccine developed with an HA gene sourced from Asia to protect birds infected with virus of a North American strain. We have now also demonstrated that our vaccination protocol, which is based on the HA gene of a clade 1 strain, produced 100% protection in chickens (n = 15) challenged with a Eurasian strain of the H5N1 virus, A/chicken/Vietnam/14/2005, belonging to clade 2.3.2 (unpublished data).
The HI test is widely used to assess the protection of birds. Our data show that birds with HI titers that are ≥1:8 consistently controlled AI viral replication following challenge with the HPAI H5N2 isolate. A disadvantage to using the HI test for assessing the protection of chickens vaccinated against HPAI is the requirement to handle the virus under strict biocontainment procedures (i.e., level 3) for routine serological testing. For that reason, we have developed two new tests based on the use of recombinant H5 antigen, which does not require a high biocontainment environment. The data show a significant correlation between the results from any pair of these tests. It appears that any of the tests might be used to detect general trends in the protection of birds against AI. When protection was assessed by a combination of viral load, clinical signs, and mortality, the induction of antibodies following DNA vaccination generally led to protection. All birds with HI titers that were ≥1:8, cELISA results of ≥50%, and indirect ELISA results of ≥10% prior to challenge did not show any significant clinical signs and eliminated their viral loads during the course of AI infection. One bird immunized with 10 μg of the DNA vaccine was positive by cELISA (59%) and indirect ELISA (26%), but was negative by HI before challenge and died on day 5 postinoculation. We also observed that out of the 3 birds injected with 100 μg of DNA vaccine without electroporation, the only one protected from virus challenge (i.e., no clinical signs postinoculation and no detectable viral load on day 6 postinoculation) had a positive HI titer (1:8) on day 56 postvaccination but was negative by cELISA (19%) and indirect ELISA (8%). Our data show that the HI test may be superior to the other tests as a predictor of vaccine efficacy (Table 6), which suggests that a functional test might be more instructive than one that is wholly reliant on a chemical binding of antigen and antibody.
To date, the major drawback of the DNA vaccine platform has been the suboptimal potency of the available candidate vaccines for veterinary species and humans. However, recent advances in DNA vaccine vector design and novel delivery methods, such as electroporation, have led to a substantial improvement in DNA vaccine efficacy in different animal species. The present study also demonstrated that electroporation resulted in positive antibody responses in chickens following a single-dose delivery of the DNA vaccine carrying the HA gene for the H5N1 AI virus. Compared to chickens in a previous study that were exposed to 500 μg of the DNA vaccine (unpublished data), chickens immunized with 10 μg of the DNA vaccine by electroporation (Tables 1 to 3) developed a superior antibody response, indicating that electroporation increased the potency of the DNA vaccine by over 50-fold. Electroporation transiently increases the permeability of cell membranes, resulting in an enhanced uptake of DNA vaccines and an increased and prolonged expression of the DNA vaccine transgene. The events at the local site of vaccine deposition are complemented by a low-level inflammation resulting from the killing of some of the cells affected by the electrical field; this results in the influx of antigen-presenting cells to the injection site (35). In addition, there is new evidence that injected free plasmid DNA finds its way into lymphoid organs where it is associated with antigen-presenting cells within 24 h of injection and a subsequent accumulation of antigen-specific CD4 T cells within a few days (36). Collectively, these events lead to the generation of a balanced immune response characterized by both potent and sustained humoral and cell-mediated immunity (37, 38).
It is not clear why one of the 17 birds given a high-dose vaccine by electroporation failed to produce a positive antibody response. Our recent unpublished observations indicate that a minority of birds that were immunized with DNA vaccine, but failed to mount an antibody response, developed potent cytokine responses (interleukin 4 and gamma interferon) and were protected from virus challenge. However, in this study, we did not challenge the antibody-negative (high-dose electroporated) bird, which leaves unanswered the question of whether the bird would have been protected. Here, the use of a single-vaccine delivery by electroporation to induce a protective antibody response is significant, given that other studies required two or more electroporation procedures (1). From this study and those of others (13, 33), we can infer that electroporation clearly leads to an enhanced efficacy of DNA vaccines in some veterinary species and allays the previously expressed concerns about the potency of the DNA vaccine platform in veterinary species. Of note, we were able to induce similar levels of protective antibody responses using two intramuscular injections of ≥100 μg of the DNA vaccine without electroporation, 3 weeks apart (data not shown), which suggests that DNA vaccination remains a promising option for poultry even if electroporation equipment is unavailable or its use is impractical. An ideal method of delivery will be one where mass vaccination can be done effectively and in a relatively short time. If optimally formulated DNA vaccines could be delivered using needle-free automated equipment (21, 39), this could facilitate the use of the DNA vaccine platform to confer herd immunity in the face of an AI epidemic/epizootic or panzootic. We are currently investigating an in ovo vaccination procedure that is quick and obviates the need for specialized equipment to handle live birds, but which may be ideally suited for AI-endemic areas of the world where routine vaccination is practical and advisable.
ACKNOWLEDGMENTS
We acknowledge the contributions of the following staff at the different locations where the study was conducted: J. Robert Duncan (retired), Cyril Lutze-Wallace, Hilary Kelly, Andrée Ann Dupras, Qigao Fu, Leith Kealey, Peter Neave, Sandra McIntosh, Diane Ayres (deceased), Brian Cathcart, Jim Algire, Doug Gowenluck, Gerry Dixon, Akther Farid, and Dave Deering (CFIA Ottawa Laboratory); Tim Salo, Helen Kehler, Marsha Leith, Margaret Forbes, Kevin Tierney, Sandra Radons, and Melanie VanderLoop (CFIA National Centre for Foreign Animal Diseases, Winnipeg); Ami Patel (Public Health Agency of Canada, Winnipeg); and Zoe Lawman (VIDO, Saskatoon).
Funding was provided by the CFIA through the Partnership Research Strategy and by the Canadian Poultry Research Council in collaboration with the Agriculture and AgriFood Canada's Growing Forward Initiative.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Babiuk S, Baca-Estrada ME, Foldvari M, Storms M, Rabussay D, Widera GL, Babiuk LA. 2002. Electroporation improves the efficacy of DNA vaccines in large animals. Vaccine 20:3399–3408 [DOI] [PubMed] [Google Scholar]
- 2. Swayne DE. 2006. Principles for vaccine protection in chickens and domestic waterfowl against avian influenza: emphasis on Asian H5N1 high pathogenicity avian influenza. Ann. N. Y. Acad. Sci. 1081:174–181 [DOI] [PubMed] [Google Scholar]
- 3. Robinson HL, Hunt LA, Webster RG. 1993. Protection against a lethal influenza virus challenge by immunization with a hemagglutinin-expressing plasmid DNA. Vaccine 11:957–960 [DOI] [PubMed] [Google Scholar]
- 4. Webster RG, Kawaoka Y, Taylor J, Weinberg R, Paoletti E. 1991. Efficacy of nucleoprotein and haemagglutinin antigens expressed in fowlpox virus as vaccine for influenza in chickens. Vaccine 9:303–308 [DOI] [PubMed] [Google Scholar]
- 5. Steensels M, Van Borm S, Lambrecht B, De Vriese J, Le Gros FX, Bublot M, van den Berg T. 2007. Efficacy of an inactivated and a fowlpox-vectored vaccine in Muscovy ducks against an Asian H5N1 highly pathogenic avian influenza viral challenge. Avian Dis. 51(1 Suppl):325–331 [DOI] [PubMed] [Google Scholar]
- 6. Webster RG, Webby RJ, Hoffmann E, Rodenberg J, Kumar M, Chu HJ, Seiler P, Krauss S, Songserm T. 2006. The immunogenicity and efficacy against H5N1 challenge of reverse genetics-derived H5N3 influenza vaccine in ducks and chickens. Virology 351:303–311 [DOI] [PubMed] [Google Scholar]
- 7. Tumpey TM, Kapczynski DR, Swayne DE. 2004. Comparative susceptibility of chickens and turkeys to avian influenza A H72N virus infection and protective efficacy of a commercial avian influenza H7N2 virus vaccine. Avian Dis. 48:167–176 [DOI] [PubMed] [Google Scholar]
- 8. Chambers TM, Kawaoka Y, Webster RG. 1988. Protection of chickens from lethal influenza infection by vaccine-expressed hemagglutinin. Virology 167:414–421 [PubMed] [Google Scholar]
- 9. Swayne DE, Kapczynski D. 2008. Strategies and challenges of eliciting immunity against avian influenza virus in birds. Immunol. Rev. 225:314–331 [DOI] [PubMed] [Google Scholar]
- 10. Treanor J, Wright PF. 2003. Immune correlates of protection against influenza in the human challenge model. Dev. Biol. (Basel) 115:97–104 [PubMed] [Google Scholar]
- 11. Kumar M, Chu HJ, Rodenberg J, Krauss S, Webster RG. 2007. Association of serologic and protective responses of avian influenza vaccines in chickens. Avian Dis. 51(1 Suppl):481–483 [DOI] [PubMed] [Google Scholar]
- 12. Yang M, Clavijo A, Graham J, Salo T, Hole K, Berhane Y. 2009. Production and diagnostic application of monoclonal antibodies against influenza virus H5. J. Virol. Methods 162:194–202 [DOI] [PubMed] [Google Scholar]
- 13. van Drunen Littel-van den Hurk S, Lawman Z, Wilson D, Luxembourg A, Ellefsen B, van den Hurk JV, Hannaman D. 2010. Electroporation enhances immune responses and protection induced by a bovine viral diarrhea DNA vaccine in newborn calves with maternal antibodies. Vaccine 28:6445–6454 [DOI] [PubMed] [Google Scholar]
- 14. Fynan EF, Webster RG, Fuller DH, Haynes JR, Santoro JC, Robinson HL. 1993. DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc. Natl. Acad. Sci. U. S. A. 90:11478–11482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suarez DL, Schultz-Cherry S. 2000. The effect of eukaryotic expression vectors and adjuvants on DNA vaccines in chickens using an avian influenza model. Avian Dis. 44:861–868 [PubMed] [Google Scholar]
- 16. Bachy M, Boudet F, Bureau M, Girerd-Chambaz Y, Wils P, Scherman D, Meric C. 2001. Electric pulses increase the immunogenicity of an influenza DNA vaccine injected intramuscularly in the mouse. Vaccine 19:1688–1693 [DOI] [PubMed] [Google Scholar]
- 17. Selby M, Goldbeck C, Pertile T, Walsh R, Ulmer J. 2000. Enhancement of DNA vaccine potency by electroporation in vivo. J. Biotechnol. 83:147–152 [DOI] [PubMed] [Google Scholar]
- 18. Kutzler MA, Weiner DB. 2008. DNA vaccines: ready for prime time? Nat. Rev. Genet. 9:776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patel A, Tran K, Gray M, Li Y, Ao Z, Yao X, Kobasa D, Kobinger GP. 2009. Evaluation of conserved and variable influenza antigens for immunization against different isolates of H5N1 viruses. Vaccine 27:3083–3089 [DOI] [PubMed] [Google Scholar]
- 20. World Organisation for Animal Health 2011. Avian influenza. Manual of diagnostic tests and vaccines for terrestrial animals. Chapter 2.3.4. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf Accessed 18 March 2012
- 21. Reference deleted.
- 22.Ogunremi O, Benjamin J. 2010. Development and field evaluation of a new serological test for Taenia saginata cysticercosis. Vet. Parasitol. 169:93–101 [DOI] [PubMed] [Google Scholar]
- 23. Greiner M. 1995. Two-graph receiver operating characteristic (TG-ROC): a Microsoft-EXCEL template for the selection of cut-off values in diagnostic tests. J. Immunol. Methods 185:145–146 [DOI] [PubMed] [Google Scholar]
- 24. Webster RG, Kawaoka Y, Bean WJ, Beard CW, Brugh M. 1985. Chemotherapy and vaccination: a possible strategy for the control of highly virulent influenza virus. J. Virol. 55:173–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. 2002. Development of a real-time reverse transcription PCR assay for type A influenza virus and avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaverin NV, Rudneva IA, Ilyushina NA, Varich NL, Lipatov AS, Smirnov YA, Govorkova EA, Gitelman AK, Lvov DK, Webster RG. 2002. Structure of antigenic sites on the haemagglutinin molecule of H5 avian influenza virus and phenotypic variation of escape mutants. J. Gen. Virol. 83:2497–2505 [DOI] [PubMed] [Google Scholar]
- 27. Philpott M, Hioe C, Sheerar M, Hinshaw VS. 1990. Hemagglutinin mutations related to attenuation and altered cell tropism of a virulent avian influenza A virus. J. Virol. 64:2941–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Philpott M, Easterday BC, Hinshaw VS. 1989. , Neutralizing epitopes of the H5 hemagglutinin from a virulent avian influenza virus and their relationship to pathogenicity. J. Virol. 63:3453–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Domenech J, Dauphin G, Rushton J, McGrane J, Lubroth J, Tripodi A, Gilbert J, Sims LD. 2009. Experiences with vaccination in countries endemically infected with highly pathogenic avian influenza: the Food and Agriculture Organization perspective. Rev. Sci. Tech. 28:293–305 [DOI] [PubMed] [Google Scholar]
- 30. Capua I, Cattoli G, Marangon S, Bortolotti L, Ortali G. 2002. Strategies for the control of avian influenza in Italy. Vet. Rec. 150:223. [PubMed] [Google Scholar]
- 31. Halvorson DA. 2009. Prevention and management of avian influenza outbreaks: experiences from the United States of America. Rev. Sci. Tech. 28:359–369 [DOI] [PubMed] [Google Scholar]
- 32. Halvorson DA, Frame DD, Friendshuh AJ, Shaw DP. 1998. Outbreaks of low pathogenicity avian influenza in U.S.A., p 36–46 In Swayne DE, Slemons RD. (ed), Proceedings of the Fourth International Symposium of Avian Influenza, the University of Georgia, Athens, GA [Google Scholar]
- 33. McCapes RH, Bankowski RA. 1985. Use of avian influenza vaccines in California turkey breeders—medical rationale, p 31–35 In Easterday BC. (ed), Proceedings of the Second International Symposium on Avian Influenza, the University of Georgia, Athens, GA [Google Scholar]
- 34. Price RJ. 1982. Commercial avian influenza vaccines, p 178–179 In Bankowski RA. (ed), Proceedings of the First Avian Influenza Symposium, 1981. Carter Composition Corp., Richmond, VA [Google Scholar]
- 35. van Drunen Littel-Van Den Hurk S, Hannaman D. 2010. Electroporation of DNA immunization: clinical application. Expert Rev. Vaccines 9:503–517 [DOI] [PubMed] [Google Scholar]
- 36. Rush CM, Mitchell TJ, Garside P. 2010. A detailed characterisation of the distribution and presentation of DNA vaccine encoded antigen. Vaccine 28:1620–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Provinciali M, Barucca A, Pierpaoli E, Orlando F, Pierpaoli S, Smorlesi A. 2011. In vivo electroporation restores the low effectiveness of DNA vaccination against HER-2/neu in aging. Cancer Immunol. Immunother. 61:363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roos AK, Eriksson F, Timmons JA, Gerhardt J, Nyman U, Gudmundsdotter L, Bråve A, Wahren B, Pisa P. 2009. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS One 4:e7226 doi:10.1371/journal.pone.0007226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ogunremi O, Pasick J, Berhane M. Needle-free delivery of an inactivated H5N3 avian influenza vaccine elicits potent antibody responses in chickens. Can. J. Vet. Res., in press [PMC free article] [PubMed] [Google Scholar]