Abstract
Understanding the pathogenesis of an infectious disease is critical for developing new methods to prevent infection and diagnose or cure disease. Adherence of microorganisms to host tissue is a prerequisite for tissue invasion and infection. Fungal cell wall adhesins involved in adherence to host tissue or abiotic medical devices are critical for colonization leading to invasion and damage of host tissue. Here, with a main focus on pathogenic Candida species, we summarize recent progress made in the field of adhesins in human fungal pathogens and underscore the importance of these proteins in establishment of fungal diseases.
INTRODUCTION
Pathogenic fungi are an important cause of superficial mucosal and disseminated infections in humans. In the nosocomial setting, invasive fungal infections, which are difficult to diagnose, are increasingly common and cause high morbidity and mortality. Those most frequently occurring are infections caused by Candida spp. (candidiasis), but many other species, including Cryptococcus neoformans, Aspergillus fumigatus, and dimorphic fungi causing endemic mycoses (e.g., Histoplasma capsulatum, Coccidioides immitis, Blastomyces dermatitidis, and Paracoccidioides brasiliensis), are medically important (1). Candida albicans is the most frequent cause of candidiasis, but in recent years non-albicans species have caused significant disease. Candida glabrata, for example, in some studies has been encountered in 20 to 24% of the human bloodstream infections that were caused by Candida (2, 3).
One of the striking characteristics of Candida spp. and other pathogenic fungi is their ability to adhere tightly to different surfaces, including the human skin, and to endothelial and epithelial mucosal host tissues. Adhesion is considered an important first step in the establishment of fungal infections. Candida spp. also stick to inert abiotic surfaces such as intravascular and urinary catheters, prosthetic cardiac valves, and denture prostheses (4, 5). In addition, interaction between Candida, a normal inhabitant of the human microflora, and other host microbes as well as between different Candida cells (“flocculation”) occurs. Altogether, this may result in the formation of large surface-attached multi- or monospecies communities, designated biofilms. This form of Candida growth is a significant medical problem due to reduced susceptibility to antifungal substances of cells inside biofilms (6). Both C. albicans and C. glabrata owe their success as a pathogen, in part, to a large repertoire of adhesins present on the cell surface (7–9). Fungal adhesins have been recognized as major virulence factors that contribute to pathogenesis of these organisms (10–14). The main focus of this review is on the biosynthesis, structure, and function of adhesins reported in pathogenic Candida spp. However, adhesion in additional pathogenic fungi is starting to be addressed, and, where data already exist, adhesins from other fungal pathogens are also discussed. Comparisons are made with studies in the model yeast Saccharomyces cerevisiae, which contains proteins that are involved in flocculation and agglutination in contrast to the adhesins in pathogenic fungi that are involved in binding to host tissues or abiotic medical devices.
ADHESINS ARE OUTER-SURFACE COMPONENTS OF THE FUNGAL CELL WALL
The cell wall of a fungal cell is responsible for its shape and provides a number of essential functions, including protection against environmental stresses. An extensive literature exists on cell wall structure and biosynthesis of baker's yeast, C. albicans, and some other species (15–20). Approximately 60 to 70% of the total cell wall mass in Candida spp. is accounted for by the carbohydrates β-1,3- and β-1,6-glucan and chitin. In addition, the cell walls of many fungi, including Candida spp., contain a diversity of glycoproteins. In Candida, on average about 80 to 90% of the cell wall protein mass are mannose residues added by N-glycosylation, O-glycosylation, and/or glycosylphosphatidylinositol (GPI) anchoring. The majority of the cell wall proteins are GPI proteins that are covalently bound to β-1,6-glucan via a remnant of their GPI anchor. These proteins are mostly present in the outer part of the cell wall, and among them are several proteins that govern primary host-pathogen interactions, such as superoxide dismutases, aspartyl proteases, phospholipases, and adhesins.
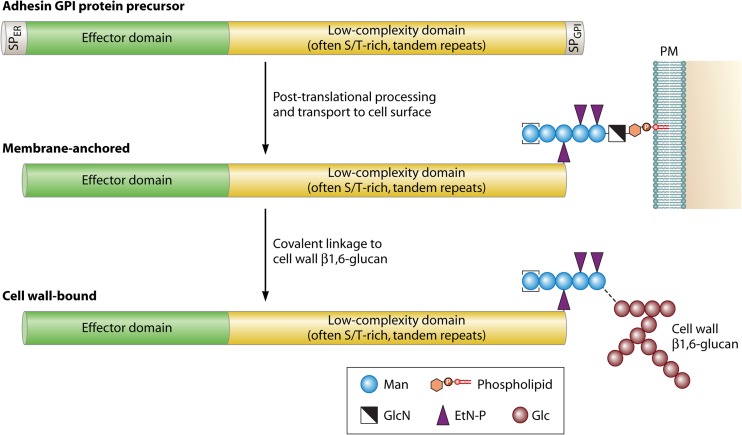
Most known fungal adhesins are GPI-modified wall proteins. The primary structure of GPI protein precursors includes conserved features, which therefore can be used to identify putative adhesins by bioinformatic means. At their N terminus, they have a signal peptide for entry into the endoplasmic reticulum (ER), and at their C-terminal end they have a peptide for anchoring to a preformed GPI lipid in the membrane of the ER (Fig. 1). Mature GPI proteins lack transmembrane domains. Most known mature adhesins are large proteins (usually >800 amino acids [aa]) with a modular structure; their N-terminal domain has a high complexity and mediates specific protein-protein, protein-sugar, or other protein-ligand interactions. These are believed to be largely responsible for the specific interactions with their substrates, e.g., host cell surface proteins or carbohydrates (14, 21, 22). It is followed by a variable domain of low complexity that often is rich in serine/threonine (Ser/Thr) and usually contains tandem repeats (TRs). The repeat regions are subject to significant intraspecies length polymorphisms due to slippage and/or recombination events during DNA replication (14, 23–25), which leads to removal or addition of repeat units. Longer repeat regions can confer greater adherence, while shorter repeat regions may result in decreased adhesion, possibly because the N-terminal effector domain remains buried in the cell wall (14).
Fig 1.
Generic structure and posttranslational processing steps leading to cell wall incorporation of fungal adhesins. Abundant protein N- and O-glycosylation, the latter especially taking place in the low-complexity domain, is not depicted for simplicity reasons. EtN-P, ethanolamine phosphate; Glc, glucose; GlcN, glucosamine; Man, mannose; PM, plasma membrane; PI, phosphatidylinositol.
Originally, binding specificities of adhesins were studied mostly by two complementary approaches. The first approach to assess gene function is to generate knockout mutant strains and study their phenotypes. However, fungal adhesin genes are often members of multigene families. Therefore, this approach is often hampered by functional redundancy as well as by compensatory mechanisms leading to upregulation of other adhesion genes whose products have similar or at least partially overlapping functions. Moreover, many adhesin genes show only low levels of expression under the experimental conditions that are frequently used in the laboratory. To overcome these problems, a successful alternative approach is to heterologously express full or partial adhesins on the cell surface of S. cerevisiae for gain-of-function studies. S. cerevisiae strains are available which lack a functional copy of the gene encoding flocculin-regulating transcription factor Flo8, rendering them very poorly adherent to most substrates. This approach thus allows analysis of adherence to potential binding ligands mediated by the gene of interest (22, 26). For instance, heterologous expression studies in S. cerevisiae showed that adherence of C. glabrata to epithelial and endothelial cells is mediated, at least in part, by the proteins encoded by the EPA gene family (7, 22, 27, 28). Recently, fungal adhesin research has moved toward more structural studies using nanotechnology in which X-ray crystallography, nuclear magnetic resonance (NMR), and atomic force microscopy (AFM) are used to obtain detailed information with respect to the structure and ligand-binding specificities of adhesins in C. albicans, C. glabrata, and S. cerevisiae (termed flocculins in the last-named organism) (29–33). Clearly, these high-resolution approaches largely improve our understanding of how fungal adhesins modulate adhesion, aggregation, biofilm formation, and host-immune responses.
CANDIDA ALBICANS ADHESINS
Until now, research mainly has addressed the three gene families ALS, HWP, and IFF/HYR as adhesins of C. albicans (listed in Table 1). They all conform to the domain organization described above and outlined in Fig. 1. Additionally, a recent bioinformatics approach (“FungalRV”) identified a plethora of proteins which have not previously been implicated in adhesion (81) but share at least some sequence and feature similarities with known adhesins. The value of this approach still needs to be validated experimentally. For example, one of the FungalRV top hits in C. albicans, Pga13, could not be confirmed to represent an adhesin in a recent study (82).
Table 1.
Adhesins in C. albicans
| Protein name(s)a | Structural propertiesb | Substrate and/or functional propertiesc | Reference(s) |
|---|---|---|---|
| Als familyd | 21 | ||
| Als1 | 1,260 aa; contains amyloid-forming sequences | Endothelial and epithelial cells; fibronectin-, laminin-, and fucose-containing glycans; abiotic surfaces such as glass and plastics; cell-cell interaction; biofilm induced; contributes to biofilm formation and pathogenesis | 32, 34–36, 37, 38, 39–41, 42 |
| Als2 | ∼2,531 aa (Als2 in CGD is incomplete; length inferred from comparison to Als2 in strain 1161 (GenBank accession no. AAC64236) | Endothelial cells; abiotic surfaces such as glass and plastics; contributes to biofilm formation and pathogenesis | 35, 36, 37 |
| Als3 | 1,155 aa; contains amyloid-forming sequences | Endothelial and epithelial cells; fibronectin, laminin, saliva-coated particles, and type IV collagen; abiotic surfaces such as glass and plastics; Streptococcus gordonii and Staphylococcus aureus; cell-cell interaction; transferrin receptor in iron acquisition; invasin; induction of C. albicans endocytosis; host cell damage; contributes to biofilm formation and pathogenesis; hypha specific | 26, 35, 43, 37, 41, 44–47, 48, 49 |
| Als4 | 2,100 aa | Endothelial cells; abiotic surfaces such as glass and plastics; functional overlap with Als2 | 36, 37 |
| Als5 | 1,347 aa; contains amyloid-forming sequences | Extracellular matrix proteins; abiotic surfaces such as such as glass and plastics; deletion mutant more adherent to endothelial and epithelial cells; aggregation; contributes to biofilm formation and pathogenesis | 50, 43, 37, 51, 52–54 |
| Als6 | 1,366 aa | Gelatin; abiotic surfaces such as glass and plastics; deletion mutant more adherent to endothelial and epithelial cells | 43, 37, 54 |
| Als7 | 1,568 aa; copy numbers of TR units are highly variable | Abiotic surfaces such as glass and plastics; deletion mutant more adherent to endothelial and epithelial cells | 43, 37, 54, 55 |
| Als9 | 1,890 aa; allelic diversity; N-terminal ligand binding (Als9-2) | Endothelial cells (N-terminal Als9-2); laminin; abiotic surfaces such as glass and plastics | 32, 37, 54 |
| Hwp1 familye | 41, 56 | ||
| Hwp1 | 634 aa; glutamine-rich N-terminal domain serves as host transglutaminase substrate; putative site for proteolytic processing; 2 Hwp1 repeats | Saliva- or fibronectin-coated surfaces; buccal epithelial cells displaying keratin 13 and SRP3; polystyrene, but less binding to silicone; low attachment to Streptococcus gordonii; role in biofilm formation; hypha specific; Tup1 repressed | 26, 40, 57–60 |
| Hwp2/Pga8 | 908 aa; short N-terminal high-complexity region; putative site for proteolytic processing; 2 Hwp1 repeats | Epithelial cells; polystyrene; role in biofilm formation on silicone; expressed in hyphae | 61, 57, 62, 63 |
| Rbt1 | 721/714 aa; N-terminal high-complexity region; putative sites for proteolytic processing; propeptide found in growth medium; 2 Hwp1 repeats | Serum; hypha induced; Tup1 repressed; required for full virulence | 57, 64, 63, 65 |
| Eap1/Pga47 | 653/1,121 aa; alleles differ in number of 6-aa repeats; short N-terminal high-complexity region; putative site for proteolytic processing; 2 Hwp1 repeats | Epithelial cells; Streptococcus gordonii; polystyrene; role in biofilm formation, filamentation, and mating | 26, 66, 67–70 |
| Ywp1/Pga24 | 533 aa; N-terminal high-complexity region; 2 sites for proteolytic processing; propeptide found in growth medium; Sap9 cleaved; 2 Hwp1 repeats | Mutant shows increased adhesion and biofilm formation; expressed in yeast cells | 71, 56 |
| Iff/Hyr familyf | |||
| Hyr1 | 919 aa | Mediates resistance to neutrophil killing; anti-Hyr1 AB is immunoprotective; hypha specific; Bcr1 dependent | 40, 72, 73, 74 |
| Rbr3/Iff1 | 1,562 aa; 10 Iff/Hyr repeats; 2 putative sites for proteolytic processing | Upregulated at low pH; expression is repressed by Rim101 and activated by Nrg1 | 75, 76 |
| Hyr3/Iff2 | 1,249 aa; 4 Iff/Hyr repeats; putative proteolytic-processing site | 75 | |
| Iff3 | 941 aa; 2 Iff/Hyr repeats; putative site for proteolytic processing | 75 | |
| Iff4 | 1.526 aa | Epithelial cells; plastics; implicated in virulence | 77–79 |
| Iff5 | 1,308 aa; 5 Iff/Hyr repeats | 75 | |
| Iff6 | 1,086 aa; putative site for proteolytic processing | ||
| Hyr4/Iff7 | 1,225 aa; 3 Iff/Hyr repeats | 75 | |
| Iff8 | 714 aa | ||
| Iff9 | 940 aa; 2 Iff/Hyr repeats | 75 | |
| Flo9/Iff10 | 1,244 aa | ||
| Iff11 | 511 aa; no GPI anchor peptide; secreted protein | Required for normal cell wall structure and virulence | 80 |
Protein names are from the Candida Genome Database (CGD) (http://www.candidagenome.org/).
All listed proteins contain signal peptides for ER entry; all except Iff11 contain C-terminal signals for GPI anchoring.
Only the most relevant phenotypes are listed.
Als family proteins share similar domain structures with N-terminal effector domains that are 55 to 90% identical across the whole family, central domains with tandem repeats (TR), and variable Ser/Thr-rich C-terminal domains.
Hwp1 family proteins have in common the presence of one or more Hwp1 repeats.
Iff/Hyr family proteins share a protein structure with a conserved putative N-terminal effector domain followed by a low-complexity C-terminal domain. The latter may contain a variable number of Iff/Hyr repeats.
Als family.
The Als family consists of eight large cell surface glycoproteins with a high degree of sequence similarity (Als1 to Als7 and Als9). The N-terminal parts of mature Als proteins comprise tandem immunoglobulin (Ig)-like domains for mammalian protein interaction, followed by a threonine-rich conserved β-sheet amyloid-forming “T region” and variable numbers of tandem repeats (TRs) (21). TRs seem to interact with hydrophobic surfaces and to facilitate aggregation (83, 84). A highly glycosylated serine- and threonine-rich spacer region in the C-terminal part extends the covalently attached proteins from the fungal surface into the environment and allows the ligand-binding domains to swivel and interact. Additional data on ALS family composition and gene organization and the difficulties in the assembly of the ALS genes in the C. albicans genome project are found in an excellent review by Hoyer and colleagues (21).
Various disease models revealed host site-dependent expression of ALS genes indicating protein-specific functions. The contributions of Als1, Als2, and Als5 to pathogenesis were confirmed by mouse and reconstituted human oral epithelium (RHE) infection models (34–36). Gene deletions, heterologous expression, and blocking experiments confirmed adhesive functions for, e.g., the family members Als1, Als2, Als3, and Als4 (36–38, 43, 50, 85). Recently, the structure and ligand-binding properties of the N-terminal domain of Als1 and the protein encoded by the second allele of ALS9 (NT-Als9-2) were resolved using NMR and X-ray crystallography (32). Salgado and coworkers showed that NT-Als9-2 is capable of binding flexible C termini of peptides in extended conformations (32). This is consistent with results from earlier studies showing that Als proteins bind to a number of structurally unrelated proteins and peptides from randomly generated sequences (51). In addition, the N-terminal part of Als1 protein specifically binds fucose-containing glycans (39). Adhesion through Als proteins can be activated and increased dramatically by amyloid nanodomain formation (see section below). Furthermore, Als1, Als2, and Als3 appear to be important for biofilm formation, and complementary roles in this process promoting monospecies biofilm formation were observed for Als1, Als3, and Hwp1 (36, 40, 41). Recently, single-molecule AFM uncovered the finding that, during the yeast-to-hypha transition, an increase in the distribution and adhesion of Als proteins is observed, accompanied by dramatically increased surface hydrophobicity and the unfolding and extension of mannosylated Als proteins (29). Earlier work has shown a link between cell surface hydrophobicity and outer-chain N-mannosylation status (86). It was postulated that β-1,2-oligomannosides present in the acid-soluble phosphomannan part of N-glycans might form a tight, inflexible helix that has a hydrophobic face. Consistent with this idea, consensus N-glycosylation sites are abundantly present in fungal adhesins, for instance, in repeat regions of Als proteins.
A high degree of allelic variability, especially in TR domains, is present in the ALS family (21), and variations in tandem repeat copies of ALS3 were shown to modulate protein function and adhesion (87). Beside a role in adhesion, effects on the fungal cell size were noticed for Als1 (21) and on host cell damage and cytokine induction for the hypha-specific Als3 (44). Als3 was also found to mediate C. albicans adherence to Staphylococcus aureus and supported mixed-species biofilm formation with Streptococcus gordonii (26, 45). Furthermore, the multifunctional Als3 protein acts as an invasin by inducing endocytosis into host cells (46) and enables iron acquisition by binding transferrin (47).
Future research will provide mechanistic insight into the different functions and binding specificities of the Als proteins and into whether this knowledge can be exploited to improve anti-Candida therapy, e.g., by immunization against the effector domains of Als proteins or by targeted inhibition of amyloid formation (88).
Amyloid formation.
Amyloids are fibrous protein structures present on the microbial cell surface. Recently, amyloids have been identified in C. albicans Als adhesins (66, 89). Amyloid formation in C. albicans has been analyzed primarily for the Als5 protein. Amyloid-forming sequences within the Als5 primary amino acid sequence were identified using the β-aggregation prediction software TANGO (90). This software predicts the potential of regions within a protein sequence to form β-strand-rich aggregates based on inter- and intramolecular interaction energies (90). Sequences predicted to form amyloids are usually found within the T region of Als adhesins. The ability of peptides corresponding to putative amyloid sequences to form β-aggregates and amyloid fibrils also has been demonstrated experimentally in vitro using transmission electron microscopy (TEM) and Congo red and thioflavin binding (66, 89). Threonine, isoleucine, and valine residues present within the T regions of C. albicans Als proteins are reported to have a significant role in amyloid formation (66, 89). Similar clusters of threonine, isoleucine, and valine residues are widely present in fungal adhesins and are also present in C. glabrata adhesins, though additional experimental evidence is needed to establish the role of these residues in aggregation and amyloid formation in C. glabrata. In addition to Als adhesins, functional amyloid formation has been reported for peptide fragments of the C. albicans adhesin Eap1 and the Flo1 and Flo11 flocculins of S. cerevisiae (66).
Recent elegant work using AFM demonstrated that amyloid interactions provide cohesive strength to C. albicans Als proteins. Possibly, initial protein binding through the Ig-like domains and/or weak hydrophobic binding through the TR domains is followed by amyloid formation, thereby strengthening cell adhesion (84). Using AFM, it was demonstrated that long-lived protein interactions are enabled by Als amyloids that can function as a molecular zipper. Formation and propagation of Als5 adhesion nanodomains on the cell surface were observed in response to mechanical stimuli, which probably causes the T region to partially unfold and expose the amyloid-forming sequence (91). The formation of amyloid clusters could thus explain why Als proteins exhibit weak binding to many ligands but mediate strong adherence. These data provided evidence that Als-mediated adhesion largely depends on conformational modifications of existing adhesins rather than or in addition to signal transduction and expression of new adhesin molecules (92).
Hwp family.
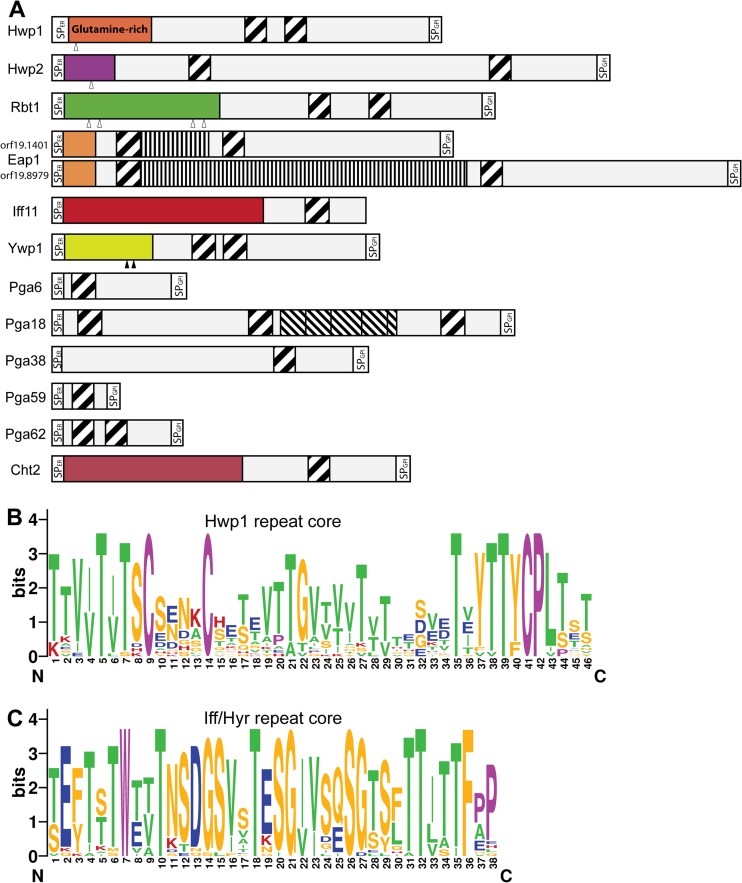
Although their N-terminal effector domains do not have sequence similarities, Hwp1, Hwp2 (“hyphal wall protein”), and Rbt1 (“repressed by Tup1”) are considered to be part of a single family because they share a highly conserved 42-aa repeat unit (Fig. 2 and Table 2) (61, 101). Expression of HWP1, HWP2, and RBT1 is hypha specific and also upregulated during mating of opaque cells (57). Their corresponding proteins are required for adhesion to host cell surface proteins, biofilm formation, cell-cell aggregation, and mating (41, 57). Within this family, Hwp1 is unique in the way that its N terminus is highly enriched in glutamine residues. These are substrates for human host transglutaminase enzymes, which covalently cross-link Hwp1 to extracellular matrix (ECM) proteins of epithelial host cells (58–60).
Fig 2.
The C. albicans Hwp1 and Iff/Hyr families contain family-specific repeat sequences. (A) Diagram showing modular structures of the protein precursors of the Hwp1 family. Included are all 12 C. albicans SC5314 proteins (genome assembly 21) containing at least one copy of the pattern T[ILV][ST]XCX(4)CX(16,20)TX[VYF][TV]T[YF]CP[ILV] (PROSITE format), which are indicated as diagonally striped boxes. N-terminal high-complexity domains of the mature proteins, believed to comprise effector domains, are presented in different colors because of their lack of sequence similarity. The two EAP1 alleles in strain SC5314 differ in length at a region that predominantly encodes repeats of a “PATEST” pattern (indicated by vertically striped boxes). A region with imperfect 41-to-50-aa serine-rich repeats in Pga18 is shown as the boxes with the thinner diagonal stripes. Signal peptides (SP) for ER entry and GPI anchoring are indicated. Putative and experimentally validated proteolytic Kex2 cleavage sites are depicted by open and black triangles, respectively. (B and C) Sequence logos (created at http://weblogo.berkeley.edu/) of the Hwp1 group (B) and of the Iff/Hyr family repeats (75) (C). Amino acid color codes for both panels are as follows: purple, conserved tryptophan in the Iff group, cysteines, and prolines; green, amyloid forming (TLVIA); red, positively charged (KRH); blue, negatively charged (DE); orange, all other amino acids. Alignments used for building the logos have been communicated to Pfam (http://pfam.janelia.org/) for creation of Pfam hidden Markov models (HMM) entries. All Iff/Hyr repeats from reference 75 match the pattern WX(2)TX (7)TX(2)G[IV](2).
Table 2.
Adhesins in non-Candida human pathogenic fungi
| Fungus | Adhesin (accession no.) | Structural propertiesa | Substrate and/or functional properties | Reference(s) |
|---|---|---|---|---|
| Aspergillus fumigatus | RodA (EAL91643.1) | 159 aa, GPI? | Hydrophobin; adhesion to collagen and bovine serum albumin | 93, 94 |
| CalA (EAL92612.1) | 177 aa | Binding to laminin and murine lung cells | 95 | |
| CspA (XP_754717.1) | 430 aa, repeats, GPI | Conidial adhesion to ECM of human alveolar epithelial cells; expression during conidial germination | 96 | |
| Cryptococcus neoformans | Cfl1 (AFR92926.1) | 309 aa | Cell-cell adhesion (flocculation); biofilm formation; hypha specific | 97 |
| Coccidioides immitis | SOWgp (AAL09436.1) | 422 aa, repeats, GPI | Binding to laminin, fibronectin, and collagen IV; role in virulence; spherule outer wall protein | 98 |
| Blastomyces dermatitidis | BAD1/WI-1 (AAA91036.1) | 1,146 aa, repeats, epidermal growth factor-like cysteine-rich consensus sequence at C terminus | Adhesion to yeast chitin and macrophages; modulation of TNF-α production by phagocytes | 99, 100 |
All proteins have signal peptides for secretion. GPI, glycosylphosphatidylinositol-anchoring signal peptide. The GPI-anchor signal prediction in RodA is controversial as it includes one of the eight cysteines, which are believed to be crucial for correct folding of this amphipathic protein.
Since the adhesin Eap1 also features the conserved 42-aa domain, it can be included in this family. In fact, seven other GPI proteins, namely, Pga6, Pga18, Pga38, Pga59, Pga62, Ywp1/Pga24, and Cht2, and Iff11, a secreted protein of the Iff family (see below), contain similar sequences (Fig. 2A and B) (102). The 42-aa-long repeat domain coincides with several amyloid-forming patches, which are thought to facilitate multimerization of adhesins (66). Moreover, the repeat unit features three conserved cysteine residues. These might be involved in the formation of intermolecular disulfide bridges between proteins, which potentially promote protein aggregation or reinforcement of cell wall integrity. Although Pga59 and Pga62 are upregulated during biofilm formation (103) and play a minor role in morphogenesis (102), their deletion mutants show no reduction in adhesion or biofilm-forming capacity (102). The repeat unit is also somewhat reminiscent of domains that have been shown to bind cell wall polymers (104), which might explain why the chitinase Cht2 has a similar domain. Thus, alternatively, these 42-aa domains might also be involved in interacting with cell wall polysaccharides and consequently contribute to cell-to-cell interactions.
EAP1 is expressed in both yeast and hyphal cells and is differentially regulated during yeast phenotypic switching. It is mainly specific to the white phase and contributes not only to biofilm formation and general epithelial cell and polystyrene adhesion (67–69) but especially to cell-cell binding in response to α-pheromone (69). The different domains of Eap1 mediate adhesion to different epitopes when expressed in S. cerevisiae (70): the N-terminal domain (Fig. 2, orange box) facilitates agar invasiveness, cell-cell contacts, and adhesion to mammalian cells, and the Ser/Thr-rich repeat regions (Fig. 2A) contribute to polystyrene adhesion (70). Similar to Als3, Eap1 and Hwp1, but not Rbt1, can facilitate binding to Streptococcus gordonii, a colonizing bacterium of the oral cavity. S. gordonii features proteins with similar amyloid sequences on its surface, but binding to Als3 and Eap1 is independent of amyloid formation (26), indicating that this binding is mediated through the N-terminal effector domain rather than the repeat units.
Strains deleted for YWP1 are hyperadherent to several surfaces and show increased biofilm formation. The Ywp1 protein is therefore considered to be involved in the release of yeast cells from surfaces (71, 105), countering the action of adhesins. The protein is processed by regulatory proteases of the Kex2 and Sap9/Sap10 group to generate 11-to-12-kDa propeptides which stay noncovalently but tightly attached to the mature protein part (105). Interestingly, the Sap9 deletion mutant is also hyperadherent (106), and a peptide immediately downstream of the Ywp1 propeptide was released from purified yeast sap9Δ sap10Δ cell walls with recombinant Sap9 (56), underlining the possible biological relevance of proteolytic activation for this protein. Similarly, Hwp1, Rbt1, and Hwp2 also contain predicted protease recognition sites within their effector domains (58, 107) (Fig. 2A); thus, it can be speculated that regulatory proteolytic events also play a role in these proteins. A fragment of the potential propeptide of Rbt1, between the second and third putative recognition sites, was found in culture supernatants (64).
Iff/Hyr family.
The 12 proteins encoded by the IFF/HYR gene family (“IFF” standing for “IPF family F” and “HYR” for “hyphally upregulated protein”) have a high degree of sequence similarity in their N-terminal effector domains (Table 1). Six of them, Iff1/Rbr3, Iff2/Hyr3, Iff3, Iff5, Iff7/Hyr4, and Iff9, also share 41-to-51-aa-long Iff-specific tandem repeats (75) (Fig. 2C). The family includes the hypha-specific proteins Hyr1 and Iff11 (74, 80). The latter represents a secretory protein that differs from other family members by lacking a GPI anchor. Deletion of IFF11 leads to significant alterations in the cell wall, suggesting that the protein has a role in cell wall organization and/or enzymatic function (80). How these functions relate to other IFF/HYR genes on the molecular level is unknown, as studies have looked at these genes only at the phenotypical level (77, 78). The molecular substrates of the Iff/Hyr family proteins have not yet been identified, but they are clearly of clinical relevance. Overexpression of IFF4 in C. albicans increased adherence to plastic and epithelial cells (79). In animal models, both overexpression and underexpression of IFF4 resulted in a reduction of virulence, indicating that a specific expression level is required for maximal virulence (108). Hyr1 has been implicated in resistance to neutrophil killing (72), and an anti-Hyr1 antibody (AB) induced immunity to disseminated candidiasis in mice (73).
ADHESINS IN NON-ALBICANS CTG-CLADE CANDIDA SPECIES
Despite several phenotypic descriptions of adhesion, adhesins have not yet been investigated deeply at the molecular level in CTG-clade species other than C. albicans. Genomic data indicate that the adhesin families described for C. albicans also exist in other species. The presence of ALS-like genes has been confirmed in Candida dubliniensis, Candida tropicalis, Candida parapsilosis, Candida lusitaniae, and Candida guilliermondii (12, 101, 108), and the IFF/HYR gene family is also widely present in CTG-clade Candida spp. (101, 109). Homologs of Rbt1 are present in C. dubliniensis, C. parapsilosis, C. tropicalis, and Candida orthopsilosis. NCBI-BLAST searches with the small N-terminal effector domains of Eap1 and Hwp2 revealed homologs only in the closely related species C. dubliniensis. A clear Hwp1 homolog is also present in C. dubliniensis, whereas a protein with only a short stretch of high sequence identity to Hwp1 is found in C. tropicalis. Of note, adhesins that fit the Als or Rbt1 definition are not present in non-CTG clade species such as C. glabrata or S. cerevisiae.
Comparative genomic analysis of the ALS and IFF/HYR gene families of the different CTG-clade species showed high genetic variability, with apparent gene losses and multiplications that have occurred during evolution (75, 101, 110). For instance, C. dubliniensis lacks an ortholog of the hyphally regulated HYR1 gene in C. albicans that was lost during evolution (110). Also, the IFF/HYR family shows duplications on three chromosomes in C. parapsilosis (101) but not in C. orthopsilosis (111). This is intriguing, as C. parapsilosis strains have the highest adhesion capacities of all clinically relevant Candida species and are mostly found growing in catheter-associated biofilms, pointing to the potentially high clinical importance of this gene family.
The ALS family is expanded to at least 13 members in C. tropicalis (101). For C. albicans and C. dubliniensis, there is a broad similarity between their ALS families. However, phylogenetic analysis showed that recombination between different ALS genes has altered some sequences since speciation (110). Furthermore, ALS5 and the multifunctional and hypha-specific ALS3 are not present in C. dubliniensis, which instead has a duplicated copy of ALS2. This high genetic variability and the recombination events occurring in fungal adhesin genes seem related to the frequent presence of intragenic tandem repeats. It should be noted, however, that adhesin gene families are often located in subtelomeric regions, which may significantly contribute to the expansion of some of these adhesin gene families.
CANDIDA GLABRATA ADHESINS
Analysis of the genome of the C. glabrata strain CBS138/ATCC 2001 revealed an exceptionally large number (66) of sequences specifying GPI-modified adhesin-like wall proteins (8, 9, 112; see also Fig. 3). All these proteins have the modular structure that is typical for adhesins: an effector domain followed by a low-complexity region that is often spiked with internal tandem repeats, termed “megasatellites,” in C. glabrata (113). Based on phylogenic analysis of their putative N-terminal ligand-binding regions, this large family can be divided into seven subfamilies (8). Only two proteins (CAGL0J05159g and CAGL0L09911g) fall outside these subfamilies, as their N-terminal domains are unrelated to those of any of the other putative adhesins (Fig. 3). The largest subfamily is the EPA family, with 17 members in CBS138. Remarkably, earlier studies reported an even larger number of EPA genes (23) in strain BG2 (114, 115). For instance, orthologs of EPA4 and EPA5, present in BG2 (116), are lacking in strain CBS138. Furthermore, none of the orthologous EPA genes in either strain are 100% identical. Relevant in this respect is the fact that the tandem repeat regions, which usually are present in fungal adhesin-encoding genes, frequently show strain-dependent size reductions or expansions occurring during DNA replication (23–25, 96). All together, these intraspecies gene variations underline the extremely large genetic plasticity with respect to fungal adhesin genes, including the EPA family, in C. glabrata.
Fig 3.
Genomic organization of putative adhesin-encoding genes in the sequenced C. glabrata strain CBS138, modified from reference 8. Chromosomes and open reading frames (ORFs) are numbered following Génolevures's systematic ORF numbering, which is also used in the Candida Genome Database. ORF sizes are to scale, but distances between ORFs are not. Many of the gene sequences, when translated, give rise to frameshifts, probably mostly due to sequencing and/or annotation errors or the presence of intronic sequences. Some unannotated ORF fragments (no ORF number), identified by BLASTX, are connected to incomplete ORFs. Colors indicate seven subfamilies, sharing homology in the N-terminal putative ligand-binding parts. Numbers of genes in each group are indicated. N-terminal domains of CAGL0L09911g and CAGL0J05170g (white) are unrelated to the other adhesins. For CAGL0E00187g (group IV, pink), only the GPI anchor peptide containing the C-terminal part was identified; its classification is therefore based on BLASTP analysis of this region. Numbers of nonadhesin ORFs separating adhesin-like ORFs and telomeres and distances of terminal adhesin-like genes to the end of the obtained telomeric DNA sequences are indicated. Numbers of the megasatellite signatures [VILF][VI][ST]H[IVS][TI][TGI] (“VVSHITT”) and SFFIT are specified only for ORFs whose protein sequences are complete in the databases. Arrows indicate directions of transcription.
Epa family.
The best-studied proteins in the Epa family are Epa1, Epa6, and Epa7. All three proteins mediate adherence to human epithelial and endothelial cells (7, 22, 27). Epa1 has also been reported to govern binding to innate immune cells (117). Remarkably, deletion of just EPA1 reduces adherence to human epithelial cells in vitro to background levels, probably because other EPA genes are transcribed only at low levels when grown under laboratory conditions (116, 118). This repression of EPA gene transcription is explained, in part, by the fact that most of the EPA genes are located within subtelomeric regions (Fig. 3) where they are subject to chromatin-based gene silencing (116, 118, 119). In fact, about two-thirds of the 66 putative adhesin genes in C. glabrata are located within these subtelomeric regions (8) and in all subtelomeric areas of strain CBS138 at least one putative adhesin gene is present (Fig. 3).
The ligand-binding domains of Epa proteins are called PA14 (“anthrax protective antigen”) domains and have lectin properties. Studies using carbochips (glycan arrays) have shown that Epa1, Epa6, and Epa7 bind to oligosaccharides that contain terminal galactose residues, such as those occurring in mucin-type O-glycans (22, 31). This is consistent with the idea that these proteins enable the fungus to bind to glycoproteins on the host cell surface. S. cerevisiae flocculins (Flo proteins) contain N-terminal domains that exhibit weak similarity to the PA14 domains in C. glabrata (23). Structural analysis of the S. cerevisiae Flo family member Flo5 showed that this protein binds to high-mannose oligosaccharides (33), giving this glycoprotein self-binding properties. Veelders et al. (33) described in detail that the crystal structure of the Flo5 effector domain, complexed to cognate ligands, revealed a beta-sandwich core that utilizes a unique DcisD calcium-binding motif for carbohydrate binding. Given the high abundance of high-mannose oligosaccharides in yeast cell walls, this confers to baker's yeast the self-aggregating properties that lead to the formation of flocs and are exploited widely in the beer- and wine-making industries. Recently, the crystal structure of Epa1 complexed to cognate disaccharide ligands, including Galβ1–3Glc and the T antigen (Galβ1–3GalNAc), has been solved, showing a similar lectin fold and DcisD calcium-binding motif (30, 31). Further studies, using point mutants of Epa6 and Epa7, or using Epa1 variants with modified binding sites that correspond to Epa2, Epa3, and Epa6, showed that substrate specificity is governed by two inner loops, CBL1 and CBL2, involved in calcium binding as well as by three outer loops, L1, L2, and L3 (22, 31). The CBL2 loop was previously also shown to determine Flo/NewFlo specificity (mannose binding versus mannose and glucose binding) in S. cerevisiae (120).
Pwp family.
Of the other putative adhesins in C. glabrata, the N-terminal effector domains of one subfamily, with seven members, also show similarity to the PA14 domains of the Epa and Flo families. These seven C. glabrata proteins were termed Pwp1 to Pwp7 (8). It is conceivable that Pwp proteins also are lectins with a role in aggregation, for instance, during biofilm formation, or host binding. In line with this, Pwp7 has been shown in vitro to play a role in adherence to human endothelial cells (112). However, solid conclusions about the functionality of the Pwp family await further functional and structural studies to elucidate their ligand-binding properties.
Other putative adhesins.
The N-terminal domains of the remaining 42 putative C. glabrata adhesins have no clear homology with those of Epa or Pwp proteins, and, with the exception of Aed1, which, like Pwp7, mediates adherence to human endothelial cells (112), their functions and ligands are unknown.
In studies where the covalently bound cell wall proteome was studied using tandem mass spectrometry, Epa3 and Epa6 were identified as well as Awp1 to Awp6, six of the non-Epa adhesin-like wall proteins (8, 121). The above-mentioned Aed1 (Awp5) is one of these proteins. Awp1 to Awp6 are members of four different adhesin subfamilies. Awp2 and Awp4 belong to a subfamily of putative adhesins with weak similarity to the C. albicans Hyr/Iff family as well as to the haze-protective mannoproteins Hpf1 and Hpf1′ in S. cerevisiae (122). Awp1 belongs to the same subfamily as Awp3. Except for Awp1, these (putative) adhesins were found by mass spectrometric analysis in isolated cell walls from strain CBS138, which was originally a feces isolate. So far, Awp1 has been detected only in a different clinical isolate (ATCC 90876, isolated from blood) grown to the stationary phase. Expression studies using a quantitative PCR (qPCR) approach showed that the corresponding genes of most of the identified wall adhesins are significantly upregulated during biofilm development (121). Interestingly, mass spectrometric analysis identified Awp6 only in cell walls under biofilm-forming conditions. Thus, expression and wall incorporation of the adhesins seem to be dependent on the genetic strain background and growth conditions. Furthermore, the analysis supports the hypothesis that C. glabrata contains a large repertoire of tightly regulated functionally diverse adhesins to enable rapid colonization of different host tissues under a variety of host-defined conditions.
Many of the C. glabrata adhesin-like proteins also contain tandem repeats, such as repeated sequences containing VSHITT or SFFIT signatures, in the region downstream of the effector domain (Fig. 3). The VSHITT repeat of about 46 amino acids is found in about half of the putative adhesins across the different subgroups, including Awp2 and Awp4 and the two unrelated adhesin-like proteins (8). Therefore, although the N-terminal effector domains of proteins in different subfamilies may not show obvious similarities, these proteins do have structural (and probably functional) relationships. Interestingly, the presence of (multiple) VSHITT or SFFIT megasatellites seems specific for adhesins in C. glabrata as it does not occur in any other protein currently present in the NCBI protein database (123). Elucidation of the exact functions of the repeat regions remains largely elusive. It is generally believed that expansion of glycosylated repeat regions improves the surface exposure and ligand-binding propensities of the N-terminal domains of adhesins (25). On the other hand, a role in cellular aggregation through amyloid-forming properties has also been suggested (66).
ASPERGILLUS FUMIGATUS ADHESINS
A. fumigatus is the main causative agent of invasive aspergillosis (IA), usually a fatal infection in patients with AIDS, solid-organ transplants, or chronic pulmonary diseases, including cystic fibrosis and allogeneic bone marrow transplants, and in patients with various hematological malignancies.
One of the best-characterized proteins required for its role in adherence to host cells in A. fumigatus is RodA (Table 2). The RodA protein is expressed primarily on the surface of conidia and is required to form the surface rodlet layer (93). RodA extracted from A. fumigatus conidia is immunologically inert, thus preventing immune recognition of fungal spores (94). Disruption of this gene decreased adherence to collagen and bovine serum albumin but not to pneumocytes, fibrinogen, or laminin (93). Although RodA is reported to be a GPI protein (94), it is not a classical wall adhesin. In fact, GPI modification of RodA would seem in conflict with the amphipathic fold of this hydrophobin, which relies on the formation of four disulfide bridges between eight conserved cysteine residues (124), the last one of which is located in the presumed GPI-anchoring signal peptide.
The A. fumigatus adhesin CalA was identified by employing a bioinformatic approach (95) and is predicted to be a secreted protein (Table 2). Recombinant CalA was shown to bind to laminin and mouse lung cells. Binding of anti-CalA antibodies to A. fumigatus conidia suggested the presence of CalA on the conidial surface.
More recently, the GPI-modified wall protein CspA (“cell surface protein A”) was identified as a putative adhesin with internal repeats (Table 2), of which the number per gene is strain dependent (96). The cspA-encoded cell wall protein is unmasked during conidial germination and is surface expressed during hyphal growth. Deletion of CspA, when combined with other cell wall proteins such as Ecm33 and Gel2, resulted in reduced conidial adherence to extracellular matrix (ECM) from human alveolar cells. Strains lacking CspA displayed cell wall weakening, whereas overexpression of CspA resulted in increased resistance to cell wall-degradative enzymes, suggesting that its main function is in cell wall integrity.
CRYPTOCOCCUS NEOFORMANS ADHESINS
C. neoformans is an environmental fungus that is found usually in soil. Much of the insight gained about its pathogenesis has come from studies focusing on capsule and melanin production—two of the most prominent virulence factors of this fungus. Surprisingly, very little is known about adhesins from C. neoformans. Recently, a secretory protein, named Cfl1 (cell flocculin 1), was reported to be an adhesin (Table 2) (97). The C. neoformans Cfl1-encoding gene was found to be specifically expressed during the hyphal growth phase, and the protein was documented to regulate morphogenesis, cell-cell adhesion, biofilm formation, and virulence (97). Paradoxically, morphotype transition in C. neoformans is typically observed during mating, but pheromone signaling components play no or minimal direct roles in virulence. Interestingly, Wang et al. have demonstrated that filamentation in C. neoformans can occur independently of pheromone signaling and mating (97). Nevertheless, elucidation of the precise function of Cfl1 requires further investigation.
ADHESINS IN FUNGI CAUSING ENDEMIC MYCOSES AND RARE FUNGAL DISEASES
The causative agents of endemic mycoses are mostly dimorphic fungi that usually grow in a filamentous (mold) form at 25°C, while at 37°C they convert to yeast growth. From this group, only a few cell wall proteins mediating adherence in C. immitis and B. dermatitidis have been characterized (Table 2). Recombinant C. immitis adhesin SOWgp (“spherule outer wall glycoprotein”) was shown to bind to mammalian ECM proteins (laminin, fibronectin, and collagen IV) (98). Deletion of SOWgp resulted in partial loss of binding of C. immitis spherules (multinucleate round cells in the parasitic cycle of this fungus) to ECM and attenuation of virulence in a murine model of coccidioidomycosis.
In the case of B. dermatitidis, BAD1/WI-1 (“Blastomyces adhesin”) has been identified as a yeast-phase-specific protein involved in adhesion. Studies performed with gene disruptants showed that BAD1 is critical for binding of B. dermatitidis yeast cells to host cells and for pathogenesis in a murine model of pulmonary infection (99). One of the major mechanisms by which the secretory protein BAD1 influences B. dermatitidis is downregulation of expression of proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α) (100).
In Fusarium oxysporum, an opportunistic filamentous fungus that has the unique ability to infect both plant and mammalian hosts, four putative GPI-modified adhesins were identified in silico. However, none of these were identified during mass spectrometric analysis of hyphal walls under adhesion-inducing conditions (125).
SUMMARY
The fungal cell wall and its constituents are of interest primarily due to its importance as a potential target for antifungal therapy and its role in pathogenesis. In recent years, tremendous progress has been made in identification and characterization of adhesins, largely by virtue of genome sequencing of many pathogenic fungi and development of novel genetic and molecular tools. Adhesins are shown to be important for disease establishment and progression by helping in colonization. This correlation has been demonstrated by a number of studies, the most important of which are the construction of deletion mutants and the demonstration of their reduced adherence and virulence. The existence of large families of adhesin proteins, e.g., the Als family in C. albicans and the Epa family in C. glabrata, with both overlapping and specific functions, conformational activation, differential gene expression patterns, and allelic variability, provides a versatile toolbox to these fungi. This may foster external reactions, for example, to enable coordinated adhesion to various tissues in specific host niches. However, there are still significant gaps in our knowledge about how fungal pathogens colonize and persist in the host. We anticipate that future studies focused on functional characterization of novel putative adhesins will not only provide new insights into their role in pathogenesis but will also help define their role related to the host niche where the organisms are found and thrive. Moreover, mechanistic insights into the mode of action of adhesins obtained using nanotechnology-based approaches may provide valuable clues to their potential to serve as future diagnostic markers or to improve antifungal therapies.
ACKNOWLEDGMENTS
P.W.J.D.G. is supported by an INCRECyT fellowship from Parque Científico y Tecnológico de Albacete. A.D.D.B. is the recipient of a postdoctoral fellowship from the junta de comunidades de Castilla—La Mancha (JCCM). M.W. was supported by the Deutsche Forschungsgemeinschaft (DFG) (WE 3537/1-2). N.C. is supported by PHRI startup funds.
Footnotes
Published ahead of print 8 February 2013
REFERENCES
- 1. Perlroth J, Choi B, Spellberg B. 2007. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 45:321–346 [DOI] [PubMed] [Google Scholar]
- 2. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borg-von Zepelin M, Kunz L, Ruchel R, Reichard U, Weig M, Gross U. 2007. Epidemiology and antifungal susceptibilities of Candida spp. to six antifungal agents: results from a surveillance study on fungaemia in Germany from July 2004 to August 2005. J. Antimicrob. Chemother. 60:424–428 [DOI] [PubMed] [Google Scholar]
- 4. Busscher HJ, Rinastiti M, Siswomihardjo W, van der Mei HC. 2010. Biofilm formation on dental restorative and implant materials. J. Dent. Res. 89:657–665 [DOI] [PubMed] [Google Scholar]
- 5. Ramage G, Martinez JP, Lopez-Ribot JL. 2006. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 6:979–986 [DOI] [PubMed] [Google Scholar]
- 6. Ramage G, Culshaw S, Jones B, Williams C. 2010. Are we any closer to beating the biofilm: novel methods of biofilm control. Curr. Opin. Infect. Dis. 23:560–566 [DOI] [PubMed] [Google Scholar]
- 7. Cormack BP, Ghori N, Falkow S. 1999. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285:578–582 [DOI] [PubMed] [Google Scholar]
- 8. de Groot PWJ, Kraneveld EA, Yin QY, Dekker HL, Groß U, Crielaard W, De Koster CG, Klis FM, Weig M. 2008. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 7:1951–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weig M, Jansch L, Gross U, De Koster CG, Klis FM, De Groot PWJ. 2004. Systematic identification in silico of covalently bound cell wall proteins and analysis of protein-polysaccharide linkages of the human pathogen Candida glabrata. Microbiology 150:3129–3144 [DOI] [PubMed] [Google Scholar]
- 10. Bernhardt J, Herman D, Sheridan M, Calderone R. 2001. Adherence and invasion studies of Candida albicans strains, using in vitro models of esophageal candidiasis. J. Infect. Dis. 184:1170–1175 [DOI] [PubMed] [Google Scholar]
- 11. Calderone RA, Fonzi WA. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327–335 [DOI] [PubMed] [Google Scholar]
- 12. Hoyer LL. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176–180 [DOI] [PubMed] [Google Scholar]
- 13. Sundstrom P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461–469 [DOI] [PubMed] [Google Scholar]
- 14. Verstrepen KJ, Klis FM. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60:5–15 [DOI] [PubMed] [Google Scholar]
- 15. Chauhan N, Li D, Singh P, Calderone R, Kruppa M. 2002. The cell wall of Candida spp., p 159–175 Calderone RA. (ed), Candida and candidiasis. ASM Press, Washington, DC [Google Scholar]
- 16. Klis FM, De Groot P, Hellingwerf K. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39( Suppl 1): 1–8 [PubMed] [Google Scholar]
- 17. Klis FM, Ram AFJ, De Groot PWJ. 2007. A molecular and genomic view of the fungal cell wall, p 95–117 Howard RJ, Gow NAR. (ed), The mycota, vol 8 Springer-Verlag, Berlin, Germany [Google Scholar]
- 18. Lesage G, Bussey H. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orlean P. 2012. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192:775–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Latgé JP. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66:279–290 [DOI] [PubMed] [Google Scholar]
- 21. Hoyer LL, Green CB, Oh SH, Zhao X. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 46:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zupancic ML, Frieman M, Smith D, Alvarez RA, Cummings RD, Cormack BP. 2008. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 68:547–559 [DOI] [PubMed] [Google Scholar]
- 23. Goossens K, Willaert R. 2010. Flocculation protein structure and cell-cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol. Lett. 32:1571–1585 [DOI] [PubMed] [Google Scholar]
- 24. MacCallum DM, Castillo L, Nather K, Munro CA, Brown AJ, Gow NAR, Odds FC. 2009. Property differences among the four major Candida albicans strain clades. Eukaryot. Cell 8:373–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verstrepen KJ, Fink GR. 2009. Genetic and epigenetic mechanisms underlying cell-surface variability in protozoa and fungi. Annu. Rev. Genet. 43:1–24 [DOI] [PubMed] [Google Scholar]
- 26. Nobbs AH, Vickerman MM, Jenkinson HF. 2010. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot. Cell 9:1622–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Domergue R, Castaño I, De Las Peñas A, Zupancic M, Lockatell V, Hebel JR, Johnson D, Cormack BP. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308:866–870 [DOI] [PubMed] [Google Scholar]
- 28. Frieman MB, McCaffery JM, Cormack BP. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a β1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46:479–492 [DOI] [PubMed] [Google Scholar]
- 29. Beaussart A, Alsteens D, El-Kirat-Chatel S, Lipke PN, Kucharíková S, Van Dijck P, Dufrêne YF. 2012. Single-molecule imaging and functional analysis of Als adhesins and mannans during Candida albicans morphogenesis. ACS Nano 6:10950–10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ielasi FS, Decanniere K, Willaert RG. 2012. The epithelial adhesin 1 (Epa1p) from the human-pathogenic yeast Candida glabrata: structural and functional study of the carbohydrate-binding domain. Acta Crystallogr. D Biol. Crystallogr. 68:210–217 [DOI] [PubMed] [Google Scholar]
- 31. Maestre-Reyna M, Diderrich R, Veelders MS, Eulenburg G, Kalugin V, Bruckner S, Keller P, Rupp S, Mösch HU, Essen LO. 2012. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc. Natl. Acad. Sci. U. S. A. 109:16864–16869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salgado PS, Yan R, Taylor JD, Burchell L, Jones R, Hoyer LL, Matthews SJ, Simpson PJ, Cota E. 2011. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 108:15775–15779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Veelders M, Brückner S, Ott D, Unverzagt C, Mösch HU, Essen LO. 2010. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. U. S. A. 107:22511–22516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alberti-Segui C, Morales AJ, Xing H, Kessler MM, Willins DA, Weinstock KG, Cottarel G, Fechtel K, Rogers B. 2004. Identification of potential cell-surface proteins in Candida albicans and investigation of the role of a putative cell-surface glycosidase in adhesion and virulence. Yeast 21:285–302 [DOI] [PubMed] [Google Scholar]
- 35. Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJ, Hoyer LL. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415–2428 [DOI] [PubMed] [Google Scholar]
- 36. Zhao X, Oh SH, Yeater KM, Hoyer LL. 2005. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology 151:1619–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aoki W, Kitahara N, Miura N, Morisaka H, Kuroda K, Ueda M. 2012. Profiling of adhesive properties of the agglutinin-like sequence (ALS) protein family, a virulent attribute of Candida albicans. FEMS Immunol. Med. Microbiol. 65:121–124 [DOI] [PubMed] [Google Scholar]
- 38. Fu Y, Rieg G, Fonzi WA, Belanger PH, Edwards JE, Filler SG. 1998. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect. Immun. 66:1783–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Donohue DS, Ielasi FS, Goossens KV, Willaert RG. 2011. The N-terminal part of Als1 protein from Candida albicans specifically binds fucose-containing glycans. Mol. Microbiol. 80:1667–1679 [DOI] [PubMed] [Google Scholar]
- 40. Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63 doi:10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18:1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525 doi:10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, Ibrahim AS, Filler SG, Zhang M, Waring AJ, Edwards JE., Jr 2004. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279:30480–30489 [DOI] [PubMed] [Google Scholar]
- 44. Murciano C, Moyes DL, Runglall M, Tobouti P, Islam A, Hoyer LL, Naglik JR. 2012. Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PLoS One 7:e33362 doi:10.1371/journal.pone.0033362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peters BM, Ovchinnikova ES, Krom BP, Schlecht LM, Zhou H, Hoyer LL, Busscher HJ, van der Mei HC, Jabra-Rizk MA, Shirtliff ME. 2012. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 158( Pt 12): 2975–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y, Filler SG. 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 10:168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 4:e1000217 doi:10.1371/journal.ppat.1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE, Jr, Filler SG. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64 doi:10.1371/journal.pbio.0050064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 78:4644–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gaur NK, Klotz SA. 1997. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 65:5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Klotz SA, Gaur NK, Lake DF, Chan V, Rauceo J, Lipke PN. 2004. Degenerate peptide recognition by Candida albicans adhesins Als5p and Als1p. Infect. Immun. 72:2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrêne YF, Lipke PN. 2011. A role for amyloid in cell aggregation and biofilm formation. PLoS One 6:e17632 doi:10.1371/journal.pone.0017632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rauceo JM, De Armond R, Otoo H, Kahn PC, Klotz SA, Gaur NK, Lipke PN. 2006. Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot. Cell 5:1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao X, Oh SH, Hoyer LL. 2007. Deletion of ALS5, ALS6 or ALS7 increases adhesion of Candida albicans to human vascular endothelial and buccal epithelial cells. Med. Mycol. 45:429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang N, Harrex AL, Holland BR, Fenton LE, Cannon RD, Schmid J. 2003. Sixty alleles of the ALS7 open reading frame in Candida albicans: ALS7 is a hypermutable contingency locus. Genome Res. 13:2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schild L, Heyken A, De Groot PWJ, Hiller E, Mock M, De Koster C, Horn U, Rupp S, Hube B. 2011. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot. Cell 10:98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ene IV, Bennett RJ. 2009. Hwp1 and related adhesins contribute to both mating and biofilm formation in Candida albicans. Eukaryot. Cell 8:1909–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Staab JF, Bradway SD, Fidel PL, Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538 [DOI] [PubMed] [Google Scholar]
- 59. Staab JF, Bahn YS, Tai CH, Cook PF, Sundstrom P. 2004. Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J. Biol. Chem. 279:40737–40747 [DOI] [PubMed] [Google Scholar]
- 60. Ponniah G, Rollenhagen C, Bahn YS, Staab JF, Sundstrom P. 2007. State of differentiation defines buccal epithelial cell affinity for cross-linking to Candida albicans Hwp1. J. Oral Pathol. Med. 36:456–467 [DOI] [PubMed] [Google Scholar]
- 61. Hayek P, Dib L, Yazbeck P, Beyrouthy B, Khalaf RA. 2010. Characterization of Hwp2, a Candida albicans putative GPI-anchored cell wall protein necessary for invasive growth. Microbiol. Res. 165:250–258 [DOI] [PubMed] [Google Scholar]
- 62. Younes S, Bahnan W, Dimassi HI, Khalaf RA. 2011. The Candida albicans Hwp2 is necessary for proper adhesion, biofilm formation and oxidative stress tolerance. Microbiol. Res. 166:430–436 [DOI] [PubMed] [Google Scholar]
- 63. Sohn K, Urban C, Brunner H, Rupp S. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89–102 [DOI] [PubMed] [Google Scholar]
- 64. Sorgo AG, Heilmann CJ, Dekker HL, Bekker M, Brul S, De Koster CG, De Koning LJ, Klis FM. 2011. Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot. Cell 10:1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Braun BR, Head WS, Wang MX, Johnson AD. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, Henry R, Litewka A, O'Meally S, Otoo HN, Khalaf RA, Dranginis AM, Gaur NK, Klotz SA, Rauceo JM, Jue CK, Lipke PN. 2010. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot. Cell 9:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li F, Palecek SP. 2003. EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot. Cell 2:1266–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li F, Svarovsky MJ, Karlsson AJ, Wagner JP, Marchillo K, Oshel P, Andes D, Palecek SP. 2007. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot. Cell 6:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sahni N, Yi S, Daniels KJ, Srikantha T, Pujol C, Soll DR. 2009. Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog. 5:e1000601 doi:10.1371/journal.ppat.1000601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li F, Palecek SP. 2008. Distinct domains of the Candida albicans adhesin Eap1p mediate cell-cell and cell-substrate interactions. Microbiology 154:1193–1203 [DOI] [PubMed] [Google Scholar]
- 71. Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. 2005. Yeast wall protein 1 of Candida albicans. Microbiology 151:1631–1644 [DOI] [PubMed] [Google Scholar]
- 72. Luo G, Ibrahim AS, Spellberg B, Nobile CJ, Mitchell AP, Fu Y. 2010. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J. Infect. Dis. 201:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Luo G, Ibrahim AS, French SW, Edwards JE, Jr, Fu Y. 2011. Active and passive immunization with rHyr1p-N protects mice against hematogenously disseminated candidiasis. PLoS One 6:e25909 doi:10.1371/journal.pone.0025909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bailey DA, Feldmann PJF, Bovey M, Gow NAR, Brown AJP. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Boisramé A, Cornu A, Da Costa G, Richard ML. 2011. Unexpected role for a serine/threonine-rich domain in the Candida albicans Iff protein family. Eukaryot. Cell 10:1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lotz H, Sohn K, Brunner H, Muhlschlegel FA, Rupp S. 2004. RBR1, a novel pH-regulated cell wall gene of Candida albicans, is repressed by RIM101 and activated by NRG1. Eukaryot. Cell 3:776–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kempf M, Apaire-Marchais V, Saulnier P, Licznar P, Lefrançois C, Robert R, Cottin J. 2007. Disruption of Candida albicans IFF4 gene involves modifications of the cell electrical surface properties. Colloids Surf. B Biointerfaces 58:250–255 [DOI] [PubMed] [Google Scholar]
- 78. Kempf M, Cottin J, Licznar P, Lefrancois C, Robert R, Apaire-Marchais V. 2009. Disruption of the GPI protein-encoding gene IFF4 of Candida albicans results in decreased adherence and virulence. Mycopathologia 168:73–77 [DOI] [PubMed] [Google Scholar]
- 79. Fu Y, Luo G, Spellberg BJ, Edwards JE, Jr, Ibrahim AS. 2008. Gene overexpression/suppression analysis of candidate virulence factors of Candida albicans. Eukaryot. Cell 7:483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bates S, De la Rosa JM, MacCallum DM, Brown AJP, Gow NAR, Odds FC. 2007. Candida albicans Iff11, a secreted protein required for cell wall structure and virulence. Infect. Immun. 75:2922–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chaudhuri R, Ansari FA, Raghunandanan MV, Ramachandran S. 2011. FungalRV: adhesin prediction and immunoinformatics portal for human fungal pathogens. BMC Genomics 12:192 doi:10.1186/1471-2164-12-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gelis S, De Groot PWJ, Castillo L, Moragues MD, Sentandreu R, Gómez MM, Valentín E. 2012. Pga13 in Candida albicans is localized in the cell wall and influences cell surface properties, morphogenesis and virulence. Fungal Genet. Biol. 49:322–331 [DOI] [PubMed] [Google Scholar]
- 83. Frank AT, Ramsook CB, Otoo HN, Tan C, Soybelman G, Rauceo JM, Gaur NK, Klotz SA, Lipke PN. 2010. Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot. Cell 9:405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrêne YF. 2012. Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 20:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gaur NK, Klotz SA, Henderson RL. 1999. Overexpression of the Candida albicans ALA1 gene in Saccharomyces cerevisiae results in aggregation following attachment of yeast cells to extracellular matrix proteins, adherence properties similar to those of Candida albicans. Infect. Immun. 67:6040–6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Masuoka J, Hazen KC. 1999. Differences in the acid-labile component of Candida albicans mannan from hydrophobic and hydrophilic yeast cells. Glycobiology 9:1281–1286 [DOI] [PubMed] [Google Scholar]
- 87. Oh SH, Cheng G, Nuessen JA, Jajko R, Yeater KM, Zhao X, Pujol C, Soll DR, Hoyer LL. 2005. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 151:673–681 [DOI] [PubMed] [Google Scholar]
- 88. Ibrahim AS, Spellberg BJ, Avanesian V, Fu Y, Edwards JE., Jr 2006. The anti-Candida vaccine based on the recombinant N-terminal domain of Als1p is broadly active against disseminated candidiasis. Infect. Immun. 74:3039–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Otoo HN, Lee KG, Qiu W, Lipke PN. 2008. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot. Cell 7:776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. 2004. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22:1302–1306 [DOI] [PubMed] [Google Scholar]
- 91. Alsteens D, Garcia MC, Lipke PN, Dufrêne YF. 2010. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. U. S. A. 107:20744–20749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Alsteens D, Ramsook CB, Lipke PN, Dufrêne YF. 2012. Unzipping a functional microbial amyloid. ACS Nano 6:7703–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Thau N, Monod M, Crestani B, Rolland C, Tronchin G, Latgé JP, Paris S. 1994. rodletless mutants of Aspergillus fumigatus. Infect. Immun. 62:4380–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, Romani L, Latgé JP. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121 [DOI] [PubMed] [Google Scholar]
- 95. Upadhyay SK, Mahajan L, Ramjee S, Singh Y, Basir SF, Madan T. 2009. Identification and characterization of a laminin-binding protein of Aspergillus fumigatus: extracellular thaumatin domain protein (AfCalAp). J. Med. Microbiol. 58:714–722 [DOI] [PubMed] [Google Scholar]
- 96. Levdansky E, Kashi O, Sharon H, Shadkchan Y, Osherov N. 2010. The Aspergillus fumigatus cspA gene encoding a repeat-rich cell wall protein is important for normal conidial cell wall architecture and interaction with host cells. Eukaryot. Cell 9:1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang L, Zhai B, Lin X. 2012. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog. 8:e1002765 doi:10.1371/journal.ppat.1002765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hung CY, Yu JJ, Seshan KR, Reichard U, Cole GT. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Finkel-Jimenez B, Wuthrich M, Brandhorst T, Klein BS. 2001. The WI-1 adhesin blocks phagocyte TNF-α production, imparting pathogenicity on Blastomyces dermatitidis. J. Immunol. 166:2665–2673 [DOI] [PubMed] [Google Scholar]
- 100. Brandhorst T, Wüthrich M, Finkel-Jimenez B, Klein B. 2003. A C-terminal EGF-like domain governs BAD1 localization to the yeast surface and fungal adherence to phagocytes, but is dispensable in immune modulation and pathogenicity of Blastomyces dermatitidis. Mol. Microbiol. 48:53–65 [DOI] [PubMed] [Google Scholar]
- 101. Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJP, Brunke S, Costanzo MC, Fitzpatrick DA, De Groot PWJ, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KAT, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MPH, Sudbery PE, Srikantha T, Zeng QD, Berman J, Berriman M, Heitman J, Gow NAR, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Moreno-Ruiz E, Ortu G, De Groot PWJ, Cottier F, Loussert C, Prévost M- C, De Koster C, Klis FM, Goyard S, d'Enfert C. 2009. The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiology 155:2004–2020 [DOI] [PubMed] [Google Scholar]
- 103. García-Sánchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d'Enfert C. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 3:536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Granger BL. 2012. Insight into the antiadhesive effect of yeast wall protein 1 of Candida albicans. Eukaryot. Cell 11:795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Albrecht A, Felk A, Pichova I, Naglik J, Schaller M, De Groot P, MacCallum D, Odds FC, Schäfer W, Klis F, Monod M, Hube B. 2006. Glycosylphosphatidylinositol-anchored proteinases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688–694 [DOI] [PubMed] [Google Scholar]
- 107. Bader O, Krauke Y, Hube B. 2008. Processing of predicted substrates of fungal Kex2 proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris. BMC Microbiol. 8:116 doi:10.1186/1471-2180-8-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhu W, Filler SG. 2010. Interactions of Candida albicans with epithelial cells. Cell Microbiol. 12:273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Richard ML, Plaine A. 2007. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot. Cell 6:119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jackson AP, Gamble JA, Yeomans T, Moran GP, Saunders D, Harris D, Aslett M, Barrell JF, Butler G, Citiulo F, Coleman DC, de Groot PWJ, Goodwin TJ, Quail MA, McQuillan J, Munro CA, Pain A, Poulter RT, Rajandream MA, Renauld H, Spiering MJ, Tivey A, Gow NAR, Barrell B, Sullivan DJ, Berriman M. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 19:2231–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Riccombeni A, Vidanes G, Proux-Wera E, Wolfe KH, Butler G. 2012. Sequence and analysis of the genome of the pathogenic yeast Candida orthopsilosis. PLoS One 7:e35750 doi:10.1371/journal.pone.0035750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Desai C, Mavrianos J, Chauhan N. 2011. Candida glabrata Pwp7p and Aed1p are required for adherence to human endothelial cells. FEMS Yeast Res. 11:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Thierry A, Dujon B, Richard GF. 2010. Megasatellites: a new class of large tandem repeats discovered in the pathogenic yeast Candida glabrata. Cell. Mol. Life Sci. 67:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kaur R, Domergue R, Zupancic ML, Cormack BP. 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8:378–384 [DOI] [PubMed] [Google Scholar]
- 115. Zupancic ML, Cormack BP. 2007. Candida cell wall proteins at the host-pathogen interface, p 327–348 d'Enfert C, Hube B. (ed), Candida comparative and functional genomics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 116. De Las Peñas A, Pan SJ, Castano I, Alder J, Cregg R, Cormack BP. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17:2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kuhn DM, Vyas VK. 2012. The Candida glabrata adhesin Epa1p causes adhesion, phagocytosis, and cytokine secretion by innate immune cells. FEMS Yeast Res. 12:398–414 [DOI] [PubMed] [Google Scholar]
- 118. Castaño I, Pan SJ, Zupancic M, Hennequin C, Dujon B, Cormack BP. 2005. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 55:1246–1258 [DOI] [PubMed] [Google Scholar]
- 119. Iraqui I, Garcia-Sanchez S, Aubert S, Dromer F, Ghigo J-M, d'Enfert C, Janbon G. 2005. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol. Microbiol. 55:1259–1271 [DOI] [PubMed] [Google Scholar]
- 120. Kobayashi O, Hayashi N, Kuroki R, Sone H. 1998. Region of Flo1 proteins responsible for sugar recognition. J. Bacteriol. 180:6503–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kraneveld EA, De Soet JJ, Deng DM, Dekker HL, De Koster CG, Klis FM, Crielaard W, De Groot PWJ. 2011. Identification and differential gene expression of adhesin-like wall proteins in Candida glabrata biofilms. Mycopathologia 172:415–427 [DOI] [PubMed] [Google Scholar]
- 122. Brown SL, Stockdale VJ, Pettolino F, Pocock KF, de Barros Lopes M, Williams PJ, Bacic A, Fincher GB, Hoj PB, Waters EJ. 2007. Reducing haziness in white wine by overexpression of Saccharomyces cerevisiae genes YOL155c and YDR055w. Appl. Microbiol. Biotechnol. 73:1363–1376 [DOI] [PubMed] [Google Scholar]
- 123. de Groot PWJ, Brandt BW. 2012. ProFASTA: a pipeline web server for fungal protein scanning with integration of cell surface prediction software. Fungal Genet. Biol. 49:173–179 [DOI] [PubMed] [Google Scholar]
- 124. Wösten HA. 2001. Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55:625–646 [DOI] [PubMed] [Google Scholar]
- 125. Prados-Rosales R, Luque-Garcia JL, Martínez-López R, Gil C, Di Pietro A. 2009. The Fusarium oxysporum cell wall proteome under adhesion-inducing conditions. Proteomics 9:4755–4769 [DOI] [PubMed] [Google Scholar]