Abstract
The SPFH protein superfamily is assumed to occur universally in eukaryotes, but information from protozoa is scarce. In the Paramecium genome, we found only Stomatins, 20 paralogs grouped in 8 families, STO1 to STO8. According to cDNA analysis, all are expressed, and molecular modeling shows the typical SPFH domain structure for all subgroups. For further analysis we used family-specific sequences for fluorescence and immunogold labeling, gene silencing, and functional tests. With all family members tested, we found a patchy localization at/near the cell surface and on vesicles. The Sto1p and Sto4p families are also associated with the contractile vacuole complex. Sto4p also makes puncta on some food vacuoles and is abundant on vesicles recycling from the release site of spent food vacuoles to the site of nascent food vacuole formation. Silencing of the STO1 family reduces mechanosensitivity (ciliary reversal upon touching an obstacle), thus suggesting relevance for positioning of mechanosensitive channels in the plasmalemma. Silencing of STO4 members increases pulsation frequency of the contractile vacuole complex and reduces phagocytotic activity of Paramecium cells. In summary, Sto1p and Sto4p members seem to be involved in positioning specific superficial and intracellular microdomain-based membrane components whose functions may depend on mechanosensation (extracellular stimuli and internal osmotic pressure).
INTRODUCTION
Stomatin is a member of the SPFH (Stomatin-Prohibitin-Flotillin/Reggie-bacterial HflC/K) superfamily, with orthologs in bacteria (1), nematodes (2), and mammals (3), where this protein was first detected (4). Like all SPFH proteins, Stomatin contains an extended SPFH domain, also called the Prohibitin or Stomatin domain, of considerable conservation (5). SPFH proteins, including Stomatin (5–9), generally can undergo homo- or heterooligomerization and become attached to the cytosolic side of the plasma membrane and membranes of intracellular vesicles. This includes proteins for perception of signals, such as mechanical and osmotic forces in structurally and functionally defined microdomains (10–13). Palmitoylation may play a role in membrane attachment (10, 13), but insertion into the cytoplasmic part of the lipid bilayer by a hook-like structure formed by an amino-terminal hydrophobic stretch is also assumed (10). Such microdomains are one aspect of so-called rafts currently discussed (14).
We first looked for sequences of the SPFH families in the Paramecium tetraurelia database (ParameciumDB; http://paramecium.cgm.cnrs-gif.fr). The SPFH superfamily members are of a molecular size of ∼30 to 50 kDa; for instance, Stomatin is usually 31 kDa, and some Stomatin-like proteins (Sto-LPs) can be >40 kDa in size (15). The most prominent subfamily, called Reggie because it is upregulated during neuronal regeneration (for a review, see reference 11), is also called Flotillin because it floats in the detergent-resistant membrane (DRM) fraction during ultracentrifugation (16). In situ they form microdomains of ∼100 nm, according to correlated light and electron microscopy (LM and EM) analysis (17, 18). While the term DRM is merely operational, we prefer the term microdomain, particularly since Reggie/Flotillin-based protein assemblies are understood as signaling platforms (19) and as a basis for ongoing membrane recycling and membrane surface control (11, 12, 20). Our present data suggest that, in the ciliate Paramecium, Stomatin is involved in such processes.
Prohibitin serves to structure the inner membrane of mitochondria (21), as has also been shown for some Sto-LPs (22). Stomatins are established regulators of mechanosensitive ion channels. A seminal observation was the dependency of touch sensation in Caenorhabditis elegans on MEC-2 (2), which is very similar to human erythrocyte membrane band 7.2 (4). In C. elegans, MEC-2 transmits mechanical stimuli by interaction with both the mechanosensitive channel protein Degenerin and microtubules (2). Homologous channels, also linked to a Stomatin ortholog and to the cytoskeleton, have been found in mammals (3, 23, 24). Thus, Stomatins and Sto-LPs appear to be universally distributed (8, 9, 25), and mechanosensation via microdomain protein assemblies may be one of the essential functions of Stomatin (13).
While this exemplifies the broad distribution of Stomatin and Sto-LPs, knowledge about SPFH proteins in general and of Stomatins in particular in protozoa is remarkably scarce, even though mechanosensation is one of the basic capabilities of unicellular organisms, including ciliates (26). For instance, in the ciliated protozoan Paramecium, electrophysiology has allowed in-depth insight into mechanoperception (27–29).
To identify SPFH proteins in Paramecium, we looked for sequence homologies in the databases of many organisms (10, 13, 25, 30). Our search, concentrating on Stomatin domain sequences, has profited from the P. tetraurelia database (ParameciumDB), which recently became available (31). (Note that in ciliates, Stomatin must not be confused with the kairomone “Stomatin.”) We thus could retrieve partial sequences of Stomatin genes, but other members of the SPFH superfamily could not be found even after extensive search.
Functionally and structurally, Paramecium cells have several advantages specifically for analyzing Stomatins. They have a highly regular structure, which facilitates localization at the LM and EM level (32–34). They also possess well-established intracellular trafficking pathways (35). In mammalian cells, SPFH proteins play a crucial role in vesicle trafficking, based not only on Reggie/Flotillin (11, 20) but also on Stomatin. In essence, we expected Stomatins at defined places of the Paramecium cell, because they may occur specifically at sites with mechanosensitivity and at sites participating in vesicle recycling based on the following basic background information.
First, Paramecium can change its swimming behavior in a mechanosensitive way, as summarized by Machemer (27–29) and specified in more detail below in Discussion. They reverse the ciliary beat direction (ciliary reversal) when hitting an obstacle during helical forward swimming (avoidance reaction) (29). Second, a Paramecium cell possesses two contractile vacuole complexes (CVCs) composed of a membrane labyrinth (spongiome), radial canals, and a vacuole. CVCs expel an excess of water by exocytosis in periodic intervals (36) when the tensile forces on the contractile vacuole membrane reach a certain level (37). Third, in Paramecium, structural and functional changes of phago(lyso)somes (food vacuoles) during transit through the cell (cyclosis) are quite evident (35) and may also require adjustment of the osmotic tension of the organelle, as shown with mammalian macrophages (38). Fourth, several avenues of membrane recycling are known, the most obvious being shuttling between the preformed site of spent phagosome release (cytoproct) and the site of nascent phagosome formation (cytopharynx) at the bottom of the cytostome (35).
In the present work, we cloned the Paramecium Stomatin (STO) genes and determined their expression and their domain structure, including molecular modeling. We also prepared antibodies (Abs) for LM and EM immunolocalization. Functional tests included gene silencing by homology-dependent posttranscriptional downregulation by feeding an appropriate Escherichia coli strain transfected with portions of the respective genes (39). This was complemented by behavioral tests, i.e., ciliary beat analysis and probing of the pulsation of the CVC and of phagocytosis performance. We thus envisaged the effects on mechanosensation by the organelles where we could localize Stomatins.
In P. tetraurelia, we found 3 family clusters of Stomatins (STO1 to STO3, STO4 to STO6, and STO7 to STO8), with a total of 20 members. This expansion is due to several whole-genome duplications (31) that have generated paralogs of high similarity, leading to gene families (40). We have prepared Abs against sequences which may be reasonably assumed to recognize several family members, because the antigenic peptide selected has sequence identity (Sto1p family) or similarity (Sto4p and Sto8p family) to more than one family member. Similarly, genomic sequences used for posttranscriptional gene silencing by feeding transformed bacteria will, in most cases, silence some but not all family paralogs (including ohnologs) when they possess >85% nucleotide identity (39), thus restricting the possibility of completely silencing individual Stomatin families.
LM and EM immunostaining labeled small domains near the cell surface, parts of the CVC, some vesicles around phagosomes (food vacuoles), and small intracellular vesicles, notably recycling vesicles dedicated to phagosome formation. Accordingly, silencing affected functions of these organelles, including mechanosensitive responses of the cell, contractile vacuole activity, and phagosome activity.
MATERIALS AND METHODS
Cell cultures.
The wild-type strain of P. tetraurelia used was strain 7S, which is derived from stock strain 51S. Cells were grown at 25°C in a medium of dried lettuce monoxenically inoculated with Enterobacter aerogenes as a food organism and supplemented with sitosterol. The ND7 mutant was used as a negative control. For further details, see Kissmehl et al. (41) and Ladenburger et al. (42).
Genomic analysis.
The Paramecium genome database (http://paramecium.cgm.cnrs-gif.fr) (31, 40) was searched with SPFH protein homologues from different species using BLASTP and BLASTN. Positive hits were further analyzed by performing forward and reverse BLAST searches at the NCBI database. Conserved motif searching was performed using the BLAST Conserved Domain Database (CDD) (43).
RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed essentially as described previously (42) to determine the positions of introns in expressed sequences. In brief, total RNA was isolated from log-phase cultures of strain 7S according to the TRIzol protocol (Life Technologies, Carlsbad, CA). RT-PCR was performed in a programmable T3 thermocycler (Biometra, Göttingen, Germany) using a standard oligo(dT) primer and PowerScript reverse transcriptase (Clontech Laboratories, Mountain View, CA) for first-strand synthesis. The subsequent PCR was performed with primers specific for the respective ohnologs under the following conditions: denaturation step (95°C for 30 s), annealing step (55°C for 60 s), and elongation step (72°C for 60 s), for a total of 35 cycles. A final elongation step of 10 min was added. PCR products were cloned into the plasmid pCR2.1 by using the TOPO-TA cloning kit (Invitrogen, Karlsruhe, Germany) according to the manufacturer's suggestions. Primer sequences used for RT-PCR are listed in Table S1 in the supplemental material.
Gene silencing.
Gene silencing by feeding was performed as described by Galvani and Sperling (39). In brief, complete cDNA sequences of Stomatin genes were cloned into the vector pPD-X using the HindII and XhoI restriction enzyme sites. Positive clones were transfected into E. coli HT115 cells and plated onto LB agar plates containing tetracycline (5 μg/ml) and ampicillin (50 μg/ml). Cultures were grown to an optical density at 600 nm of ∼0.4 and induced with isopropyl-ß-d-thiogalactopyranoside (IPTG; 125 μg/ml). After a 4-h induction at 37°C, the bacteria were pelleted and resuspended in lettuce medium plus 125 μg/ml IPTG and ampicillin (50 μg/ml) to a final optical density of 0.25. Single starved Paramecium cells were transferred into 100 μl of feeding solution and incubated overnight in a humid chamber. The next morning, single cells were transferred into fresh feeding solution. The Paramecium cells were tested on the following 2 days (72 and 96 h after treatment). Efficiency of the silencing method was routinely controlled by silencing ND7, a gene relevant for exocytosis, which can be easily tested (42). In addition, silenced cells, after immunostaining with anti-tubulin and anti-Stomatin Abs, were scanned and quantitatively evaluated.
Antibody production.
Abs against subgroup-specific peptides were produced by Biotrend (Köln, Germany) according to their standard protocols. The peptide sequences used were the following: Sto1a,b,c, Cys-Ala-Pro-Arg-Tyr-Pro-Arg-Glu-Glu-Leu-Pro-Glu-Asn-Gln-Glu; Sto4b, Cys-Val-Gln-Asp-Ser-Ile-Glu-Ala-Tyr-Leu-Asp-Lys-Ser-Thr-Glu; and Sto8b, Cys-Gln-Ile-Ser-Glu-Thr-Leu-Lys-Glu-Gln-Ser-Lys-Lys-Arg-Val.
Peptides were conjugated to keyhole limpet hemocyanin prior to immunization and affinity purified with the peptides used for immunization.
Immunoblotting.
Paramecium cells were prepared essentially as described before (42). In brief, cells were lysed with lysis buffer and proteins were separated under reducing conditions in SDS-PAGE. The proteins were blotted onto nitrocellulose membranes according to standard procedures and blocked with 1% bovine serum albumin (BSA) in TBST (10 mM Tris, 150 mM NaCl, 0.02% Tween 20 at room temperature). The blots were incubated with primary Abs against human Stomatin (Santa Cruz Biotechnology) for 1 h at room temperature (dilution, 1:1,000) and incubated with the respective peroxidase-conjugated secondary Ab for 1 h at room temperature. For testing of the antisera, blots were blocked with 3% BSA in phosphate-buffered saline (PBS), 0.05% Tween 20 for 1 h. Affinity-purified sera were diluted 1:1,000 in 1% PBS, 0.05% Tween 20 and applied overnight at 4°C. Secondary Ab (goat anti-rabbit horseradish peroxidase [HRP]) was diluted 1:10,000 and applied for 1 h at room temperature. Detection was performed with the SuperSignal chemiluminescence kit (Pierce, Rockford, IL).
Immunofluorescence localization.
For immunolight microscopy, cells were prepared basically as described previously (42) and exposed to affinity-purified rabbit anti-Stomatin Abs (diluted 1:100) and Cy3- or Alexa-488-conjugated goat anti-rabbit Abs (diluted 1:1,000 in PBS plus 1% BSA). For detection of a polyclonal goat anti-human Stomatin Ab (clone M14; Santa Cruz Biotechnology, Santa Cruz, CA), diluted 1:400, we applied Cy3- or Alexa 488-conjugated donkey anti-goat Abs, diluted 1:1,000 in PBS, plus 1% BSA. For better identification of surface structures, cells were also double stained with anti-α-tubulin Abs (clone DM1A; Sigma, Taufkirchen, Germany). Secondary Abs for the detection of anti-α-tubulin-Ab were Alexa 488-conjugated goat anti-mouse IgG (Life Technologies, Darmstadt, Germany) and Cy5-conjugated goat anti-mouse IgG (Jackson Immuno Research, Hamburg, Germany).
Samples were analyzed either by conventional LM or by confocal laser-scanning microscopy (LSM). Images acquired with LSM 510 software were processed with Photoshop software (Adobe Systems, San Jose, CA).
For quantification of silencing efficiency, the fluorescence intensity of representative membrane regions (10 by 3 μm; sagittal sections) of 5 representative cells per group were determined using the LSM image browser (Zeiss, Göttingen, Germany), and the background intensity was subtracted.
Immunogold electron microscopy.
The immunogold EM method used was as indicated by Ladenburger and Plattner (44). Briefly, polyclonal Abs were detected by a protein A-gold conjugate, 5 nm in diameter, on cells fixed with formaldehyde (8%) and glutaraldehyde (0.1%) after LR Gold embedding and UV polymerization at −35°C.
Functional tests.
Mechanosensory reactions have been tested with cells suspended in Dryl's solution (2 mM sodium citrate, 1 mM Na2HPO4, 1 mM NaH2PO4, and 1.5 mM CaCl2, adjusted to pH 6.9). They were transferred onto a silicon plate with engraved channels of a diameter of 80 μm. These plates were manufactured by the mechanical workshop of the Max Planck Institute, Tübingen, Germany, and are normally used to study axon guidance with isolated neurons (45). When these were partially filled with a cell suspension and covered with a coverslip, cells could not turn sideways. Cells were observed for 1 min, and backward/forward movements as well as the frequency of rotations were counted when they approached the only obstacle, i.e., the meniscus. The depolarization reaction was tested independently by transferring high K+ medium to single cells (20 mM KCl in the presence of 1 mM CaCl2).
For functional tests of phagocytosis, cells were exposed for 10 min to Indian ink in a solution of 1:30 in Dryl's solution and fixed immediately with 8% formaldehyde (freshly depolymerized from paraformaldehyde [PFA]) in PBS to stop the phagocytic process and count the number of newly formed food vacuoles by light microscopy.
Endocytosis via coated pits was tested as indicated by Ramoino et al. (46). Briefly, phagocytosis was inhibited by trifluoperazine (2.5 μg/ml; Sigma-Aldrich, St. Louis, MO), and internalization of endocytic vesicles from the cell surface was followed with wheat germ agglutinin labeled with AlexaFluo-488 (Sigma-Aldrich, St. Louis, MO) for fluorescence analysis in a Zeiss Axiovert 100TV.
Phylogenetic analysis.
All sequences were aligned using the ClustalW algorithm. Phylogenetic trees were constructed (neighborhood-joining algorithm as well as Gonnet and PAM matrices) using the following programs: DAMBE (47) and MEGA (48).
Hydropathy analysis and molecular modeling.
Hydropathy plots were constructed using the algorithm of Kyte and Doolittle (49) and the GREASE program (http://fasta.bioch.virginia.edu/fasta_www2/fasta_www.cgi?rm=misc1).
Molecular modeling was performed using the Swiss model server at http://swissmodel.expasy.org/workspace/index.php.
RESULTS
Identification and characterization of Stomatins in Paramecium.
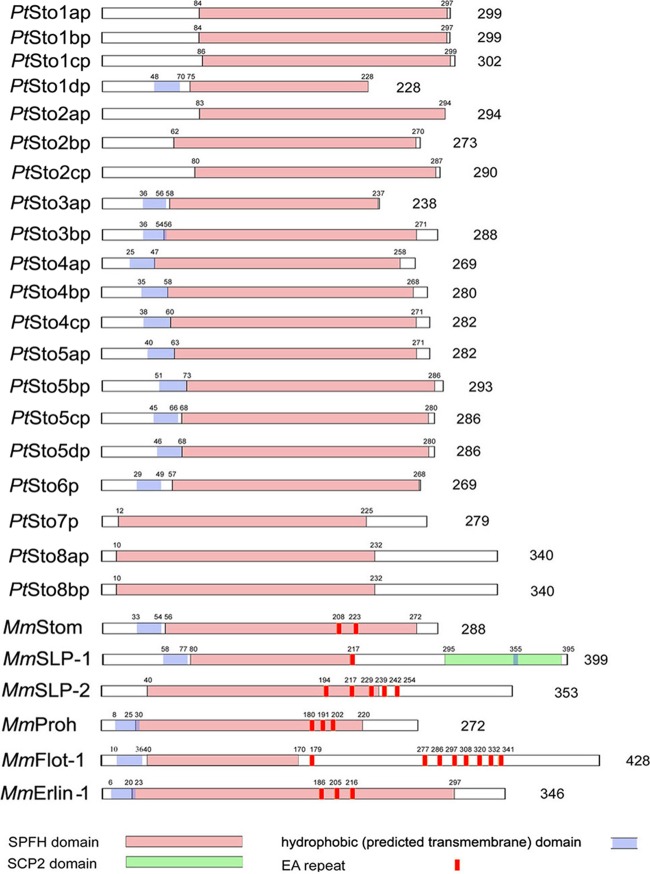
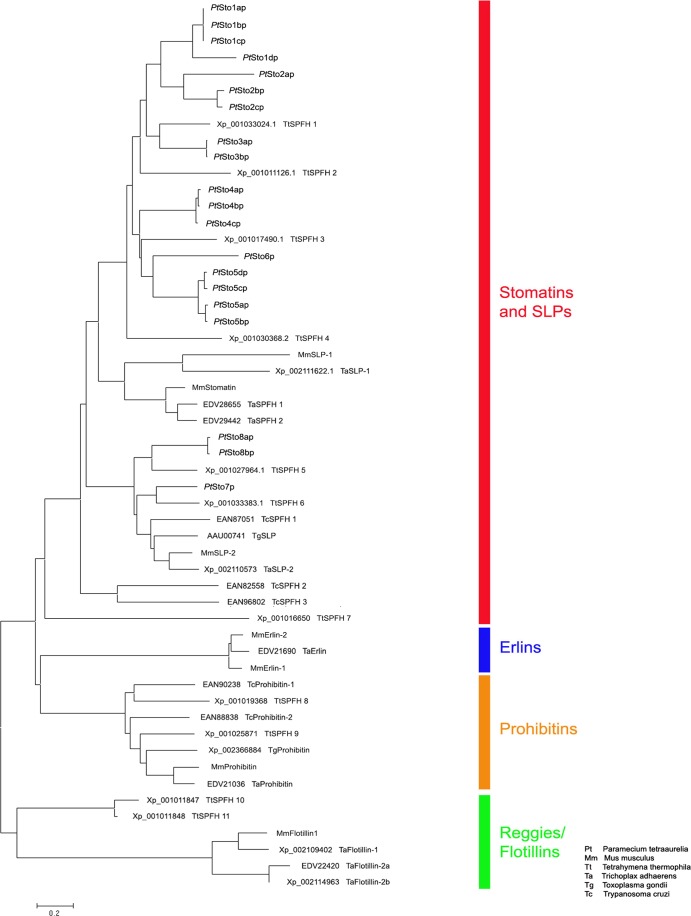
As is characteristic of SPFH superfamily members (1, 10, 25, 30), the SPFH domain (i.e., Stomatin domain) clearly occurs in the sequences of Stomatin paralogs identified in ParameciumDB (Fig. 1). Careful analysis of the P. tetraurelia genome, based on CDD analysis (43) and on primers presented in Table S1 in the supplemental material, revealed 20 putative proteins containing an SPFH domain (cd02106). Phylogenetic analysis revealed that all of them belong to the family of Stomatin proteins (Fig. 2), which is corroborated by the fact that their SPFH domain is most closely related to the Stomatin-like SPFH domain subfamily (cd03403; E values of <10−54; E values for the Prohibitin-like SPFH domain smart00244 were >10−34). Figure 1 and Fig. S1 in the supplemental material briefly characterize the genes we can assign to Stomatin in P. tetraurelia, namely, STO1a to STO1d, STO2a to STO2c, STO3a and STO3b, STO4a to STO4c, STO5a to STO5c, STO6, STO7, STO8a, and STO8b, including 7 pairs with closely related ohnologs, as frequently occurs in these cells due to a rather recent whole-genome duplication (31, 40). Dichotomy is particularly evident with STO2b and STO2c, STO3a and STO3b, STO4a and STO4b, STO5a and STO5b, STO5c and STO5d, and STO8a and STO8b (Fig. 2).
Fig 1.
Domain structure of P. tetraurelia Stomatins. Schematic presentation of P. tetraurelia Stomatin domain architecture and those of various mammalian SPFH proteins. SPFH and SCP-2 domains, hydrophobic domains (called a transmembrane domain by some), and EA repeats (absent from P. tetraurelia Sto [PtSto]) are indicated. Sequences used are the following: GSPATP00013742001 (PtSto1ap), GSPATP00018485001 (PtSto1bp), GSPATP00027276001 (PtSto1cp), GSPATP00034033001 (PtSto1dp), GSPATP00024614001 (PtSto2ap), GSPAT00010097001 (PtSto2bp), GSPATP00004121001 (PtSto2cp), GSPATP00018907001 (PtSto3ap), GSPATP00024437001 (PtSto3bp), GSPATP00030747001 (PtSto4ap), GSPATP00021746001 (PtSto4bp), GSPATP00026272001 (PtSto4cp), GSPATP00033325001 (PtSto5ap), GSPATP00022578001 (PtSto5bp), GSPATP00012426001 (PtSto5cp), GSPATP00016343001 (PtSto5dp), GSPATP00015155001 (PtSto6p), GSPATP00020490001 (PtSto7p), GSPATP00020414001 (PtSto8ap), GSPATP00023643001 (PtSto8bp), AAH10703.1 (Homo sapiens Stom [HsStom]), AAH37074.1 (Mus musculus SLP-1 [MmSLP-1]), NP_075720.1 (MmSLP-2), NP_002625.1 (HsProh), AAD40192.1 (HsFlot-1), and NP_663477 (MmErlin1). All Paramecium sequence accession numbers are from ParameciumDB (http://paramecium.cgm.cnrs-gif.fr/page/index).
Fig 2.
Phylogenetic tree of Stomatins. Protein sequences were aligned by the ClustalW method, and the tree was constructed using the Gonnet matrix and the neighbor-joining algorithm. Note that all Paramecium sequences belong to the Stomatin family.
Some of the families are related not only by the recent whole-genome duplication but also by the intermediate whole-genome duplication (e.g., the STO5 family is the result of two successive whole-genome duplications with no gene loss, as is the STO4 family, with one loss). This close relationship is reflected by the considerable similarity of the primary protein structure of the SPFH domain of these ohnolog pairs, from 88.6% (Sto3ap-Sto3bp) to 98.5% (Sto4ap-Sto4bp) (see Table S2 in the supplemental material). Only 6 Sto forms are singletons. Two of the singletons (Sto1cp and Sto4cp) are very similar to a pair (Sto1ap-Sto1bp and Sto4ap-Sto4bp, respectively). Considering criteria based on sufficient difference of the isoforms, we may have to deal with 8 to 10 structurally and possibly functionally diversified Stomatin protein isoforms in a Paramecium cell.
According to RT-PCR analysis (data not shown), all 20 members seem to be expressed. Sequencing revealed 2 annotation errors in the database: in STO5a and STO6, automatic annotation seems to have misplaced exon-intron boundaries, resulting in unusually large introns (see Fig. S1B in the supplemental material).
The apparent strong expansion of the Stomatin family and the reduction of other SPFH families seems to be a common feature of ciliates, as the close relative Tetrahymena thermophila as well as the more distant Ichthyophthirius multifiliis also show this feature, albeit to a lesser degree: there are 20 Stomatins in P. tetraurelia and 7 Stomatin, 2 Prohibitin, and 2 Reggie/Flotillin genes in T. thermophila and I. Multifiliis (note that these organisms, in contrast to Paramecium, do contain Reggie/Flotillin genes). The aberrant length of several T. thermophila Stomatin genes, e.g., T. thermophila SPFH-2 (TtSPFH-2) (see Fig. S1C in the supplemental material), might be an annotation artifact, as these sequences were not experimentally verified. It may be identified as a Stomatin, just like TtSPFH-1 and TtSPFH-3 to TtSPFH-7, whereas TtSPFH-8 and TtSPFH-9 present themselves as Prohibitin genes and TtSPFH-10 and TtSPFH-11 as Reggie/Flotillin genes in our analysis.
Other unicellular eukaryotes, like Toxoplasma (1 Stomatin, 1 unusually large Stomatin, and 1 Prohibitin gene) or Trypanosoma (2 Prohibitin, 1 Stomatin, and 2 unusually large “Stomatin” genes) do not show this Stomatin expansion. Monosiga (1 Stomatin, 2 Prohibitin, 1 Erlin, and 2 Reggie/Flotillin genes) and Trichoplax (2 Stomatin, 1 Stomatin-LP, 1 Prohibitin, 3 Reggie/Flotillin, and 1 Erlin gene) almost show the full diversity of SPFH proteins that can be seen in mammals (e.g., in mouse there is 1 Stomatin, 3 Stomatin-like protein, 2 Erlin, 2 Reggie/Flotillin, and 2 Prohibitin genes). This is in line with the recent finding that many genes play a role during evolution only until multicellularity (50). Most members of the SPFH superfamily have been implicated in the formation of membrane microdomains, which may recruit multiprotein complexes for signaling (10, 19) and trafficking (11, 12, 20). Thus, SPFH proteins have acquired increased importance during evolution, and Stomatin appears as an early type which is absent only from yeast (51).
How do Paramecium Stomatins compare to mammalian Stomatins? In brief, mammalian Stomatins are characterized by a stretch of highly charged, hydrophilic amino acids at their very N terminus (amino acids [aa] 1 to 24), followed by a hydrophobic sequence of 29 aa that most likely is associated with the membrane (52). In P. tetraurelia Stomatins, however, the charged hydrophilic N terminus can be identified only in members of the STO1 to STO6 families, whereas the hydrophobic region is consistently present only in the STO4 to STO6 families and a few members of other families (STO1d, STO3a, and STO3b). With Stomatins in higher eukaryotes, a small, hydrophobic sequence at the C terminus (aa 264 to 272) has been shown to be crucial for oligomerization (52). These features can also be identified in Stomatin of C. elegans (see Fig. S2 in the supplemental material). No direct homolog of the hydrophobic oligomerization sequence at the carboxy terminus could be identified in Paramecium, although other, short hydrophobic stretches at roughly corresponding regions were present in the STO4 to STO6 family members as well as in most members of the STO1 to STO3 families (see Fig. S2). In addition, mammalian Stomatin has been shown to be palmitoylated at Cys-29 and, to a lesser degree, at Cys-86 (7). In other SPFH proteins, like the Reggie/Flotillin proteins, these lipid modifications have been shown to be crucial for membrane attachment (53). When analyzing the P. tetraurelia Stomatins for the presence of hypothetical palmitoylation sites, only the STO4 to STO6 family and most members of the STO1 family (Sto1ap, Sto1bp, and Sto1cp) consistently show these recognition sites and interestingly also do contain a hydrophobic region. Exceptions are Sto1dp, Sto3ap, and Sto3b, which contain a hydrophobic domain but no recognizable palmitoylation site. No hypothetical myristoylation sites could be detected.
We performed molecular modeling of the SPFH core domain of the Sto1ap, Sto4ap, and Sto8bp genes using the structure of the SPFH domain of the PH1511 gene of Pyrococcus horikoshii (54) as the template. All Stomatin families thus could be modeled (Fig. 3). As the similarity between the Paramecium Stomatins and the template was lower than that of murine Stomatin (31.3% for Sto1ap, 33.1% for Sto4ap, 35.1% for Sto8bp, and 47.3% for murine Stomatin), the resulting z scores (a measure for model accuracy) of the models of Sto1ap (−3.46) and Stp4ap (−2.72) also were lower than that of murine Stomatin (−2.26). Surprisingly, the z score of the Sto8bp model was higher (−2.07).
Fig 3.
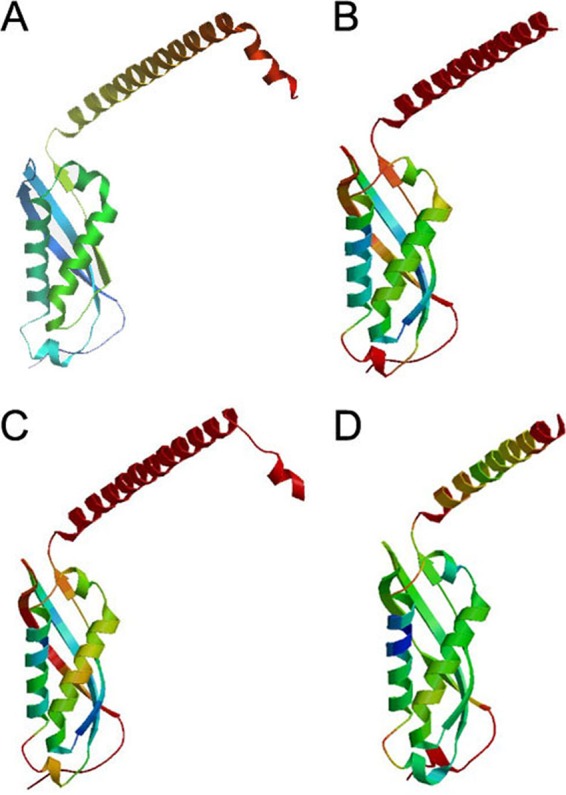
Molecular modeling of P. tetraurelia Stomatin. The SPFH domain of the only H. sapiens Stomatin (A) and representative members of each Stomatin family cluster of P. tetraurelia (B to D) were modeled with the SPFH domain of P. horikoshii (54) as a template. Although all family cluster members (PtSto1ap, z score = −3.46 [B]; PtSto4ap, z score = −2.72 [C]; PtSto8bp, z score = −2.07 [D]) could be modeled with a z score comparable to that of human Stomatin (z score = −2.26 [A]), subgroup 3 differed insofar as the homology region was slightly smaller, resulting in a smaller model region.
All models revealed the typical α/β-fold domain that has already been described for the PH1511 and PH0470 genes of P. horikoshii and for the murine Flotillin-2/Reggie-1 gene (5). This α/β-fold domain consists of 2 large and 2 small alpha helices (at positions 154 to 159, 164 to 181, 186 to 188, and 192 to 205 for Sto1ap; 117 to 122, 127 to 144, 149 to 151, and 155 to 172 for Sto4ap; and 81 to 86, 91 to 108, 113 to 115, and 119 to 132 for Sto8bp) and 4 antiparallel beta strands (at positions 124 to 128, 131 to 135, 139 to 152, and 211 to 223 for Sto1ap; 87 to 91, 94 to 98, 102 to 115, and 174 to 186 for Sto4ap; and 51 to 55, 58 to 62, 66 to 79, and 138 to 150 for Sto8bp). C terminal to this core domain is a predicted α-helical coiled-coil domain necessary for oligomerization (5, 55). Such predicted coiled-coil domains are present at the respective positions in all three family clusters (although some members of cluster 1, like Sto1dp and Sto3ap, seem to be truncated at their C terminus and, therefore, lack this feature).
Localization of Stomatins.
The peptides used for Ab production are highlighted in the full protein sequence presented in Fig. S3 in the supplemental material. Sequences used for Ab production from Sto1ap are shared by the three family members 1a, 1b, and 1c. With Sto4p, isoform 4c deviates by one and isoform 4a by two amino acids. With Sto8p, subtype 8a differs by two residues from Sto8bp. Any other paralogs differ by >3 residues. Therefore, the Abs produced against Sto1p will recognize Sto1a, -b, and -c, those against Sto4ap may also recognize Sto4cp and possibly Sto4ap, and those against Sto8bp may also bind to Sto8ap. Western blotting with the sera revealed bands in the range of 34 kDa (see Fig. S4D, lane 1, Sto1ap, 1bp, 1cp) and 38 kDa (lane 3, Sto8ap, bp) for the monomers and several bands in the range of 60 to 70 kDa, which most likely represent homo- and/or heterodimers. Interestingly, no monomers could be detected with the anti-Sto4ap, -Sto4bp, -Sto4cp serum (see Fig. S4D, lane 2).
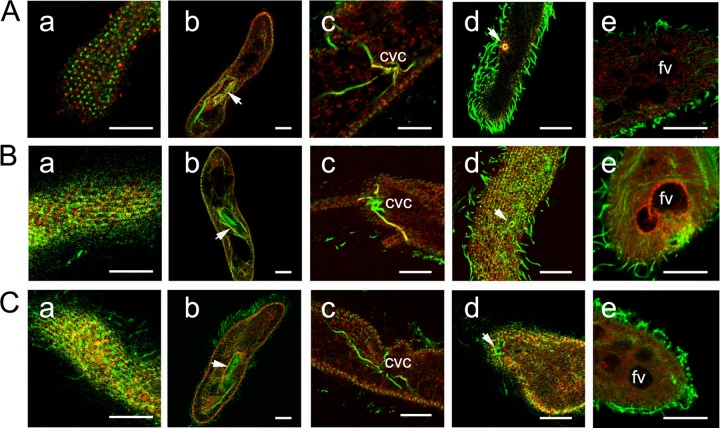
Results obtained from immunofluorescence and EM analysis are presented in Fig. 4 to 7 and are summarized in Table 1. In Paramecium, we clearly find a punctate localization of all family members investigated on the somatic (nonciliary) cell membrane over its entire extension, but no anterior-posterior gradient was recognized. Compared to other regions of the somatic membrane, all three families consistently show a somewhat stronger staining in the region of the oral cavity, which is known as a region with intense vesicle trafficking (35).
Fig 4.
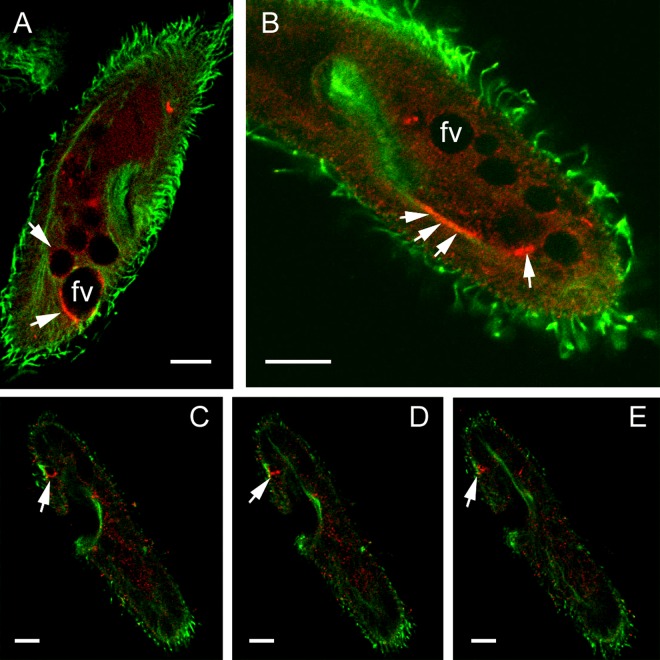
Immunofluorescence staining using Abs against peptides derived from Sto1p, Sto4p, and Sto8p. Cells were fixed and permeabilized to achieve optimal conditions for the immunoreaction as indicated in Materials and Methods. Cells then were labeled by affinity-purified Abs against peptides specific for the different Stomatin families, recognizing Sto1a,b,cp (A), Sto4a,b,c (B), and Sto8a,bp (C, red), respectively, as explained in the text. Counterstaining was performed with a mouse monoclonal anti-α-tubulin Ab (clone DM1A; green; Sigma) and with the secondary Abs specified in Materials and Methods. Note the patchy arrangement of Stomatin at the cell surface outside cilia (column a) with stronger staining in the region of the oral cavity (column b), as well as the occurrence of some Stomatin in granular form inside cells (column c). The contractile vacuole complex (column c) as well as the porus (column d) are stained only by Abs against Sto1a,b,cp and Sto4a,b,cp, whereas anti-Sto4a,b,cp stains some only phagocytic vacuoles (column e). fv, food vacuole. Scale bars, 10 μm.
Fig 7.
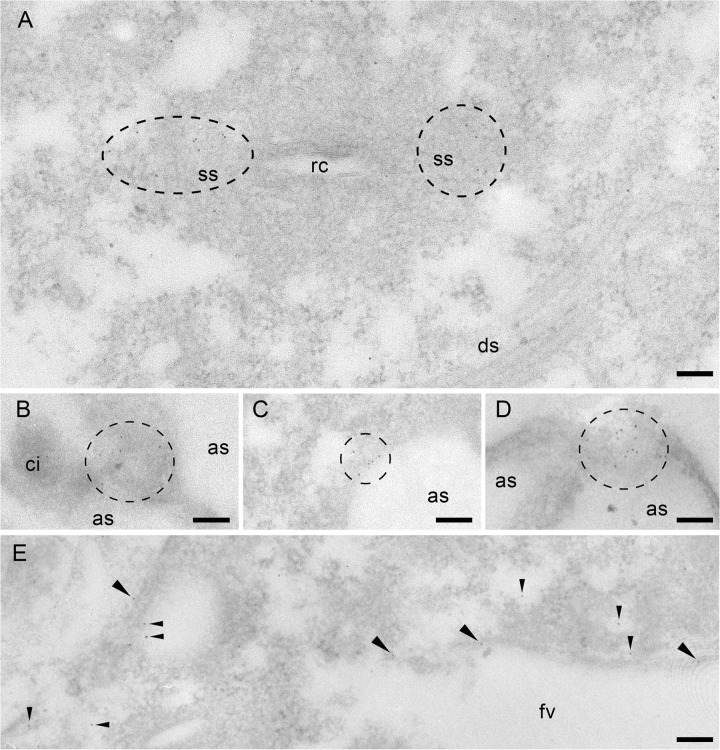
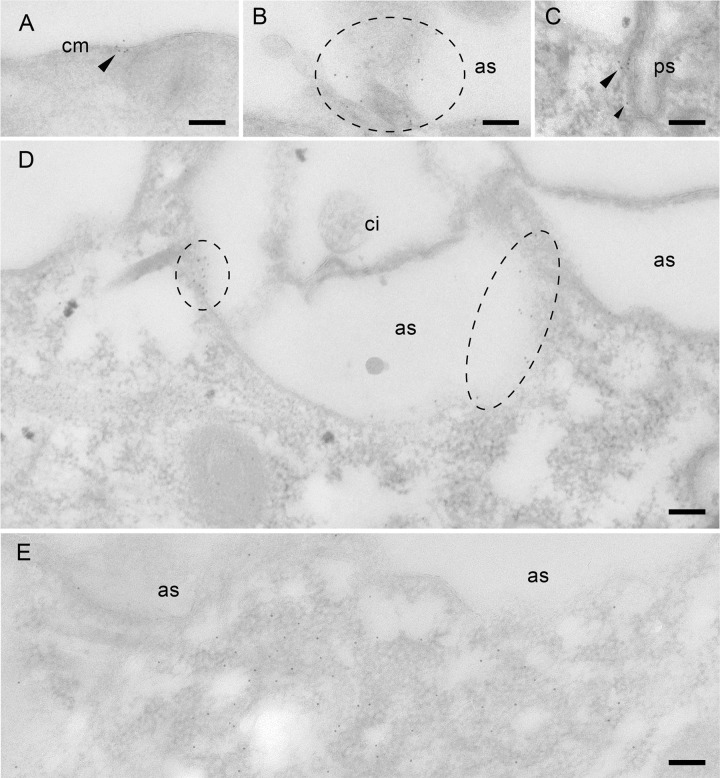
Immunogold EM analysis using anti-Sto4p (A to C and E) and anti-Sto8p (D) Abs. (A) Gold label is scattered in the CVC between the membrane labyrinth of the smooth spongiome (ss) close to the radial canals (rc) but not in the decorated spongiome (ds). With both Abs, labeling in microdomains occurs close to the cell membrane. (B to D) Label close to or at alveolar sacs (as) is particularly evident where cells are cut tangentially. Note the absence of label on a cilium (ci). (E) Label is enriched on a population of small vesicles close to the food vacuole and on the vacuole membrane itself. For further details, see Materials and Methods. Scale bars, 0.1 μm.
Table 1.
Summary of the subcellular distribution of PtSto subfamilies
| Location | Presencea of: |
||
|---|---|---|---|
| Sto1p | Sto4p | Sto8p | |
| Cell surface | ++ | ++ | ++ |
| Oral cavity | ± | ± | ± |
| CVC | ++/± (including pores) | ± (including pores) | − |
| Food vacuoles | − | ++ (only some) | − |
| Cytoproct and recycling vesicles | − | ++/± | − |
Fixed and permeabilized samples were labeled either by affinity-purified Abs against peptides specific for Sto1a,b,cp (Sto1p), Sto4a,b,cp (Sto4p), or Sto8a,bp (Sto8p). −, No staining; ++, most intense staining; ±, variable results.
Cilia were not labeled with any of the Abs applied. Many puncta, presumably small vesicles, throughout the cell were labeled with all Abs. Sto4p and, to a lesser extent, Sto1p are associated with proximal parts (radial canals) of the contractile vacuole system, whereas its preformed site for exocytosis, the porus, frequently showed staining for both families. Staining of the cytoproct and of vesicles associated with the postoral fiber system is restricted to the Sto4p family. The latter are bundles of microtubules connecting the cytoproct with the cytostome, where new phagosomes are formed (35). Most striking was the dotted labeling of some of the food vacuoles selectively with anti-Sto4p Abs.
Immunofluorescence staining was also controlled with preimmune sera, with negative results. We also applied Abs against human Stomatin as an established Stomatin reference. This also resulted in punctate staining at the cell boundary and inside cells. In Western blots, bands of comparable molecular sizes were stained (see Fig. S4 in the supplemental material). We found no mitochondrial labeling, in agreement with EM analyses (data not shown). From sequence and three-dimensional (3D) analysis we are confident that we have identified only genuine Stomatins in Paramecium. We made no lipid extractions (see Introduction), because this may not be a correlate to microdomains seen in situ, since Stomatins are partially independent of DRMs (7, 55).
Immunogold EM analysis, as presented in Fig. 6 and 7, are compatible with the LM data. The background was negligible and practically absent from sections incubated under identical conditions with the protein A-gold conjugate only. We regularly made the following observations. Abs against Sto1p (Fig. 6A and B), Sto4p (Fig. 7B), and Sto8p (Fig. 7D) label microdomains ∼0.1 to ∼0.5 μm in size at the cell membrane; the size observed depends on the local orientation of the section. Figure 5E documents labeling of cortical vesicles which cannot be functionally defined. In no case were cilia labeled (Fig. 6D and B). Also in agreement with fluorescence labeling, with anti-Sto4p Abs we see gold labeling of parts of the CVC, e.g., in regions adjacent to the radial canals (Fig. 7A) and on the contractile vacuole membrane (data now shown). Both of these structures are reported to be mechanosensitive to pressure even after isolation from the cell (56). In addition, we see some labeling of the food vacuole membrane and of adjacent vesicles (Fig. 7E). Although EM-gold analysis has much lower sensitivity, as is generally known, the higher resolution reflects the LM situation shown in Fig. 5.
Fig 6.
Immunogold EM analysis using Abs against Sto1a,b,cp. Label occurs as a small patch (microdomain) on a cross-sectioned cell membrane (cm, arrowhead) (A) and is more extended on a tangential section (encircled) close to underlying alveolar sacs (as) (B). (C) Outlines of a clathrin-coated parasomal sac (ps) are labeled. (D) Gold label on the membranes of two subcortical Ca2+ stores (as) but not on a cilia (ci). (E) Label is diffusely associated with a population of small vesicles which, as seen from the neighboring of alveolar sacs, are located below the cell surface. For further details, see Materials and Methods. Scale bars, 0.1 μm.
Fig 5.
Immunofluorescence staining of vesicles along postoral fibers and at cytoproct. Fixed and permeabilized cells were labeled by affinity-purified Abs against peptides specific for Sto4a,b,cp (red) and counterstained with a mouse monoclonal anti-tubulin Ab (clone DM1A; green; Sigma). (A) Note the patchy staining at or close to some phagocytic vacuoles. Occasionally, vesicles at or close to the postoral fibers were stained by Abs against subfamily 4 (B), as were vesicles at the cytoproct (C, D, and E; 3 consecutive confocal sections are shown). For further details, see the text. Scale bars, 10 μm.
Two most unexpected results from immunogold EM analysis are the following. (i) The membranes of some parasomal sacs are stained for Sto1p (Fig. 6C), whereas the occurrence of larger microdomains on the cell surface obscures discrimination of such details at the LM level. (ii) Unexpectedly, Sto1p- and Sto4p-labeled microdomains were also present on the membrane of alveolar sacs, specifically on the side facing the interior of the cell (Fig. 6D and 7C).
Functional analyses.
As SPFH family members have been implicated in mechanoperception in C. elegans (2) and, more recently, in mammalian neurons (23, 24), we investigated mechanosensation by transferring Paramecium cells silenced for the respective subfamily members into small, partially medium-filled silicone channels with a diameter of 80 nm and observed their behavior when they reached the meniscus. This is perceived as a mechanical obstacle. As the cells are 100 μm long and, thus, cannot turn around, untreated or mock-silenced cells show a ciliary reversal reaction when reaching the meniscus, followed by a short period of reverse swimming and rapid longitudinal rotations until another ciliary reversal occurs and the cells again swim forward until they reach the meniscus again. Interestingly, cells silenced for the STO1 family show a reduced number of ciliary reversals as well as slowed-down backward rotation series compared to control cells (Fig. 8). Some silenced cells stayed immobile at the meniscus, whereas all control cells performed ciliary reversal. Due to high similarity of the gene sequences of STO1a to those of STO1b (88%) and STO1c (86%), all of these ohnologs can be assumed to be silenced. Nonsilenced cells and aliquots mock silenced with the empty vector showed normal swimming behavior in this narrow channel only partly filled with medium. According to Fig. 8B, backward-forward movements per min, when hitting the meniscus, are considerably different in normal and silenced cells (28.0 ± 2.6 and 20.4 ± 1.97, respectively; P = 0.027), as are cell rotations (2.77 ± 0.38 and 1.00 ± 0.35, respectively; P = 0.002). This reduced mechanosensitivity was not due to defects in depolarization, as the depolarization reactions of silenced cells (depolarization by adding high K+ medium) were indistinguishable from those of control cells (data not shown).
Fig 8.
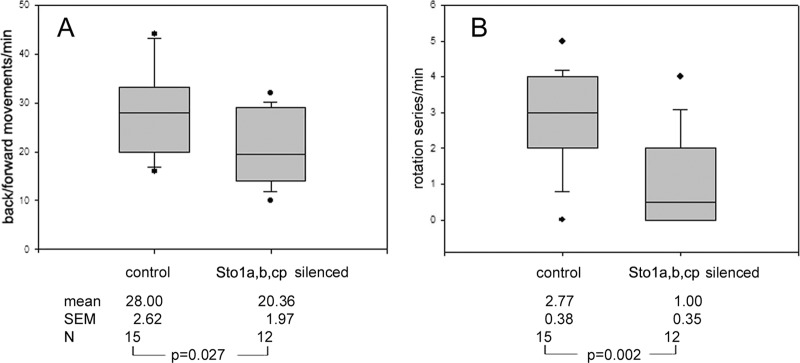
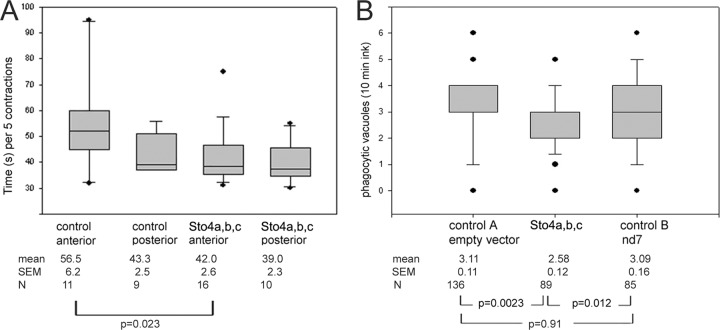
Ciliary reversal upon meeting an obstacle is impaired in cells silenced for STO1a. The entire cDNA sequence was used for silencing by feeding, as outlined in Materials and Methods. Due to high similarity of the gene sequences of STO1a to those of STO1b (88%) and STO1c (86%), all of these ohnologs can be assumed to be silenced (see the text). Silenced cells and aliquots mock silenced with the empty vector were observed when swimming through a narrow channel only partly filled with medium. Backward-forward movements per min, when hitting the meniscus, were counted (28.0 ± 2.6 for silenced cells and 20.4 ± 1.97 for mock-silenced cells; P = 0.027) (A), as were cell rotations (2.77 ± 0.38 for silenced cells and 1.00 ± 0.35 for mock-silenced cells; P = 0.002) (B). Both values are reduced significantly in silenced cells. Values are from Student's t test, and deviations are given as means ± SEM. N indicates the number of cells analyzed.
An example of the extent of Stomatin silencing achieved with the feeding of transformed bacteria (see Materials and Methods) is presented in Fig. S5 in the supplemental material, where tubulin staining is fully maintained, whereas Stomatin staining is reduced. Cosilencing of different genes has been reported only with genes of >85% nucleotide identity (39). Although most ohnologs show higher levels of identity (e.g., STO1a and STO1b, 88%; STO1a and STO1c, 86%), some are slightly below 85% identity (e.g., STO4a and STO4b, 82%) but have internal stretches with higher identity which would theoretically allow silencing. In pilot experiments we also checked the silencing efficiency by quantitative fluorescent measurements of stained membrane regions in silenced and mock-silenced cells (see Fig. S5). These measurements resulted in a reduction of fluorescence intensity from 77.8 to 16.6 relative units after silencing of STO1a, reflecting a silencing efficiency of about 80%. Whether the remaining 20% reflect incomplete silencing of all ohnologs detected by the Ab (STO1a, STO1b, and STO1c) or whether one of the family members was not silenced at all remains open. A positive control of silencing efficiency was the silencing of the exocytosis-relevant ND7 gene, which also routinely showed a similar degree of silencing by the method applied, i.e., feeding of transformed bacteria (42).
Other processes where mechanosensation is involved include the pumping process of the contractile vacuole complex. We determined separately the frequency of the anterior and of the posterior contractile vacuoles, respectively, in cells silenced for the different subfamily members. As seen in Fig. 9A, contraction frequency of the anterior contractile vacuole, but not the posterior contractile vacuole, is decreased in cells silenced for Sto4a,b,c compared to control cells, which exhibit an ∼75% higher activity. This correlates with the different sensitivity of the two CVCs to Ca2+-mobilizing second messengers (57).
Fig 9.
Increase in pumping frequency and of phagocytosis in cells silenced for STO4a. The entire cDNA sequence was used for silencing by feeding, as outlined in Materials and Methods. Due to the high similarity of the gene sequence of STO4a to that of STO4c (89%), this ohnolog may also be silenced, whereas the probability is lower for STO4b (82%). Silenced cells and aliquots mock silenced with the empty vector were analyzed. (A) The pumping frequency of the anterior and posterior contractile vacuoles was analyzed. A significant increase in pumping velocity of the anterior, but not of the posterior, contractile vacuole in cells silenced for Sto4a,b,cp was observed; the control value is ∼75% higher than that after silencing. (B) Silenced and mock-silenced cells were exposed to Indian ink for 10 min and fixed immediately thereafter. Silenced cells contained a significantly lower number of food vacuoles (phago[lyso]somes) than controls. The statistical evaluation is depicted in Fig. 8.
Furthermore, as we could see a strong Sto4a,b,c staining of vesicles at or close to the phagocytic vacuoles, we also investigated phagocytosis in silenced cells. Figure 9B clearly shows a significant decrease of phagocytic vacuole formation by ∼20% in cells silenced for STO4a, STO4b, and STO4c compared to control cells.
Since we detected immunogold staining on parasomal sacs, where constitutive endocytosis via coated pits takes place, we also investigated the uptake of wheat germ agglutinin-Alexa 488 in cells silenced for STO1a, STO1b, and STO1c and in control cells. However, no difference could be detected (data not shown).
DISCUSSION
Since SPFH proteins in general and Stomatin in particular are widely expressed across species, from bacteria to humans (9, 10, 13, 25, 51), they were expected to exist in ciliated protozoa as well. In fact, we now can show, for the first time, the existence of Stomatins in P. tetraurelia and suggest their functional involvement in mechanosensation. Thus, we also expected that SPFH-based microdomains would occur in these cells as a defined variation of a more general concept (14). No other SPFH proteins were detected.
Basic characteristics of Paramecium Stomatins.
Based on ParameciumDB mining, SPFH sequences have been identified which all can be attributed to Stomatin. Stomatins all contain a rather conservative Stomatin domain but have no EA (or similar) repeats in the carboxy-terminal half, as occur in Reggie/Flotillin genes (58). This is also the case with Stomatins of Paramecium (Fig. 1; also see Fig. S3 in the supplemental material). 3D structure modeling revealed considerable similarity of most family members to prokaryotic and mammalian Stomatin (Fig. 3). The SPFH core domain is considered relevant for membrane attachment, whereas the coiled-coil domains are important for oligomerization (5, 59); in addition, a small domain close to the C terminus may be important for oligomerization (52). Stomatin paralogs analyzed so far match several of these criteria (e.g., structure of the SPFH core domain and predicted coiled-coil domain, with the exception of the small hydrophobic sequence at the C terminus, which is lacking). This correlates with the formation of microdomains near the Paramecium cell surface, as evidenced at the LM (Fig. 4 and 5) and EM levels (Fig. 6 and 7). Thus, P. tetraurelia Stomatins might exert a microdomain scaffolding function, as shown for Reggie/Flotillin proteins (11, 12, 19, 60) and envisaged for mammalian Stomatin (61).
How Stomatin members are attached to membranes remains undetermined. For other organisms, preferred assumptions are partial insertion in the lipid bilayer by a hook-like hydrophobic stretch and a palmitoylation site, both in the amino-terminal part (10). As described above, we found such a clear hydrophobic stretch in only a few isoforms. Also, any fatty acylation remains open, as does the relevance of any kind of oligomerization, since these parameters have not been tested yet. Nevertheless, we see that Stomatin isoforms seem to be arranged in microdomains. Their sizes seen in immuno-EM are quite variable, but this is known to depend on the accessibility of the epitopes on the surface of ultrathin sections. The size ranges between 0.1 and 1 μm. (Consider that coalescence from smaller areas is also known to occur with, e.g., Reggie/Flotillin [62].)
Stomatin in lower eukaryotes.
Information on SPFH proteins in protozoa is very scarce, mostly anecdotal, and not concise. Even for Dictyostelium discoideum, a generally well-studied protozoan (unrelated to Paramecium), knowledge about Stomatin is restricted. A search with a mammalian Reggie/Flotillin sequence in Dictyostelium reveals a homolog most closely related to mammalian Stomatin (63). Alternatively, the vacuole-bound protein vacuolin was described as being more closely related to Reggie/Flotillin (59). In the parasitic flagellate Leishmania, Stomatin-like proteins (Sto-LPs) have been found, but functional evidence is scarce (64). The species closest to Paramecium endowed with a Stomatin ortholog is the malaria parasite, Plasmodium falciparum, which, together with ciliates, belongs to the phylum Alveolata. In Plasmodium this protein is exported to form microdomains in the membrane of the parasitophorous vacuole (65) and, thus, again gives little insight into the function specifically within the protozoan cell. In conclusion, protozoa belong the kingdom with the least information about SPFH proteins in general and about Stomatin in particular.
Topological and functional correlation of Stomatins on the cell surface.
After finding Stomatin-related sequences in ParameciumDB, we started our analysis by assuming that Stomatin-related effects would be easily amenable to detailed analysis in this unicellular organism because of some specific properties. These cells are capable of clear-cut behavioral responses, as documented by a plethora of electrophysiological research. They possess a highly regular structural design and established vesicle-trafficking avenues. Thus, it is possible to pinpoint different functions, particularly in conjunction with gene silencing. However, we have to consider the number of rather similar paralogs/ohnologs, particularly those retained from the last whole-genome duplication (40). The Abs designed against specific subfamily members may bind to additional paralogs. Also, from silencing of selected subfamily members, one cannot expect total inhibition effects, since close similarity may allow for some complementation by nonsilenced paralogs.
Irrespective of such restrictions, we found that a differential localization of Stomatin family members and silencing specifically affected the organelles to which the respective Stomatin subfamilies have been localized. This includes membranes with established or presumed mechanosensitivity. Aquaporins also occur in ParameciumDB and, thus, could mediate osmotic equilibration. However, the precise type of mechanosensitive channels in Paramecium remains to be specified on a molecular level. Different Stomatins may associate with different channels.
Electrophysiology has established that mechanosensitive Ca2+ influx channels are enriched in the anterior somatic (nonciliary) membrane (66) and, thus, are activated upon collision with an obstacle or experimentally by touching of the anterior end (67, 68). The depolarizing receptor potential generated by Ca2+ influx and K+ efflux triggers an action potential by activation of voltage-gated/depolarization-dependent Ca2+ influx channels located exclusively in the ciliary membrane (66, 69), as reviewed by Machemer (27–29).
STO1 silencing clearly reduces the ciliary reversal response to mechanical stimulation when a cell hits an obstacle, e.g., the meniscus of a small droplet in which it swims (Fig. 8). Instead of performing ciliary reversal accompanied by ongoing rotations, STO1-silenced cells may just stop or perform a significantly weaker reaction. In separate experiments, we showed by chemical depolarization that signal transduction for ciliary reversal is unimpaired. From this we suggest that the small patches of Sto1p on the cell membrane serve the same purpose as the Stomatin ortholog and Sto-LPs, Unc-1 and Unc-24, in C. elegans, i.e., assembly of mechanosensitive channels in microdomains. This now appears to be an old principle maintained in evolution. In mouse sensory neurons, Sto-LP3 (or SLP3), a homolog to C. elegans MEC-2 (3), in connection with the acid-sensitive ion channel (ASIC), serves mechanoperception (70). In human primary sensory neurons, it has also been shown that downregulation of Stomatin does not affect electrical activity (23). Similarly, we find that chemical depolarization can induce normal ciliary reversal reactions in Paramecium after Stomatin downregulation (data not shown).
In Paramecium, many small cytoplasmic vesicles are fluorescently stained with Abs against the different Stomatin families, but their nature is not fully clear. In pilot experiments we have excluded lipid bodies and mitochondria (data not shown) which, in higher eukaryotes, may harbor SPFH-type proteins (10). Association of Stomatin with endosomes and lysosomes could not be selectively ascertained. This is interesting considering the association of Sto4p with clathrin-coated pits (below) and of Stomatin with lysosomes isolated by Triton WR1339 accumulation from mammalian cells (71). (Note, however, that such lysosomes contain a substantial amount of residual autophagic components [72]). EM-gold analysis reveals label on some cortical vesicles and around some food vacuoles, possibly with acidic compartments, thus indicating involvement of Stomatin in vesicle trafficking. Specifically, Sto4p is clearly associated with vesicles recycling from the cytoproct to the cytopharynx, where new food vacuoles form. Overall, this suggests that Stomatin is engaged in delivery of specific membrane cargo to certain sites of the cell, just as was found with Reggie/Flotillin in mammalian cells (11), including neurons (12).
Unexpected labeling.
Among the larger organelles stained with anti-Stomatin Abs are some which are subject to osmotic tension, as outlined above. A novelty is the association of Sto1p and Sto4p with the membrane of subplasmalemmal Ca2+ stores, the alveolar sacs (Fig. 6 and 7) whose biogenesis occurs by selective vesicle flow (73). Some less-understood trafficking phenomena are known to be regulated by Stomatin (7, 74). As alveolar sacs look very different in the EM, compact and flat or swollen, depending on the osmotic conditions during fixation, they may also be engaged in osmotic regulation, an aspect to be settled by further work.
Particularly unexpected is the labeling of clathrin-coated pits (parasomal sacs), the regularly arranged sites of constantly ongoing endocytosis, by anti-Sto4p Abs (Fig. 6C). This is in clear contrast to what we found for Reggie/Flotillin in mammalian cells (18). Remarkably, these sites also contain one type from of a family of primeval types of Ca2+ channels recently identified (44). It is known from mammalian cells that cation channels serve cell volume regulation (75). In addition, in Paramecium the CVC contributes to cell volume regulation depending on osmotic conditions (76). The context of these phenomena remains to be established.
Since the mechanosensitive channels that normally underlay the ciliary reversal reaction in Paramecium are anteriorly enriched, with a gradient fading toward the middle of the cell (66), one would expect that Sto1p would display the same arrangement, but this is not the case. This contradiction may be resolved if one assumes, in analogy to Reggie/Flotillin in mammalian cells (11, 12), a general involvement of Stomatin in the dynamically controlled delivery, local positioning, and internalization of different membrane proteins (receptors, ion channels, etc.) to and from the cell membrane, respectively. It may well be that in Paramecium, hyperpolarization-sensitive channels, which have the opposite distribution pattern (69), would also be delivered to the cell surface in a similar pathway. Altogether, Stomatin-based membrane cargo delivery could also be the explanation for the occurrence of Stomatin labeling on many intracellular, including cortical, vesicles.
Stomatin has been localized to the basis of flagella of Chlamydomonas reinhardtii (77) and to the entire length of cilia of airway epithelia (78). Functional genomic analysis of C. elegans also indicates Stomatin as a ciliary component (79). The implication of this wide coincidence is regulation of beat activity by Stomatin. In contrast, in Paramecium we find no Stomatin associated with cilia, in agreement with the localization of mechanosensitive channels on the extraciliary cell membrane (66, 69), where we exclusively detect Stomatin microdomains.
Possible function of Stomatin in the contractile vacuole complex.
Water is sequestered by the CVC by the osmotic gradient generated by an electrogenic proton pump (H+-ATPase), which is located in the terminal branches (decorated spongiome) of the organelle (80, 81) and is involved in osmolyte exchange (82). This complex organelle is a most important regulator of cell volume in the course of osmotic stress (76). Within the organelle the mechanical stress rises during diastole, which finally induces exocytosis of contents and collapse of the vacuole and of the ampullae (which connect the vacuole with the radial canals emanating from the vacuole) during systole (36). Labeling with Abs against Sto1p and/or Sto4p is concentrated precisely at these strategic sites, including the radial canals. These and the vacuole all are mechanosensitive. In fact, isolated CVC fragments can undergo periodic expansion and collapse (56) which clearly requires a mechanosensor in the CVC membrane. In Chlamydomonas, type TRP5 mechanosensitive channels are associated with the CVC (77). Interestingly, only the anterior, but not the posterior, contractile vacuole in a cell is sensitive to Stomatin silencing (Fig. 9). Remarkably, such a difference is also found in the sensitivity to Ca2+-mobilizing metabolites (57). The CVC porus (exocytosis site) is also labeled with Abs against Sto1p (Fig. 4Ad) and, though less intensely, Sto4p (Fig. 4Bd), as well as with Abs against specific Ca2+ channels recently identified (44). All this suggests a method of Stomatin-mediated mechanosensitive Ca2+ mobilization for CVC discharge. With isolated CVCs, internal tension does not suffice to induce fluid expulsion, since this requires a porus for exocytotic membrane fusion (56).
Topological and functional correlation of Stomatins on food vacuoles.
In Paramecium, phagosomes also fuse with lysosomes (35), which themselves are also osmotically sensitive in mammalian cells (83). In higher eukaryotic systems the osmotic sensitivity of components of the endolysosomal system (83, 84), including phagosomes (38), is well known. Moreover, association of Reggie2/Flotillin1 with macrophage phagosome membranes has been reported (85). Stomatin may serve as a scaffold for the assembly of unknown mechanosensors in this case, and actin coating may serve to protect the organelle membrane. In fact, mechanical stress as an inducer of local actin polymerization is well established, at least for the cell surface (86, 87), and actin covers phagosomes in Paramecium (88).
Other aspects of association with the cytoskeleton.
The Stomatin homolog MEC-2 of C. elegans is reported to be associated with microtubules (89, 90), whereas for mammalian Stomatin, band 7.2, a connection to actin filaments is reported (7, 91). In Aspergillus, StoA associated with microtubules contributes to vesicle delivery for polar growth (92). Both Stomatin (93) and actin (94, 95) are established components around mammalian phagosomes. Association with actin recalls strict coincidence with Reggie/Flotillin in mammalian neuronal and epithelial cells, as shown by double immunogold EM analysis (96). The divergent reports on interaction with either one of the two cytoskeletal elements may not really be contradictory, as both usually interact with each other (97).
Although interaction with cytoskeletal elements may represent a general principle for SPFH protein function, it is unknown whether this association would be direct or indirect. Nevertheless, interaction with any of the two cytoskeletal elements should be warranted for Paramecium Stomatin forms. Concerning Stomatin associated with the cell surface (Fig. 6 and 7), we know that the subplasmalemmal space, though very narrow, contains actin (41). This is also true of the surface of some stages of the food vacuoles (88), in agreement with their decoration by Stomatin in the current paper, although we do not yet know whether this occurs in equivalent cyclosis stages. Of particular interest would be any correlation with acidification which starts a short time after pinch-off (35). The question arises from the association of Sto-LP3 (SLP3) with the ASIC for mechanoperception in mouse sensory neurons (70).
On the CVC, none of the numerous actin isoforms could be detected (88), but the organelle is known to be supported by microtubules over its entire extension (36). Interestingly, previous work found that acetylation of α-tubulin stabilizes microtubules in C. elegans, and this increases mechanosensation (98). The authors noted that the responsible α-tubulin acyltransferase also occurs in Paramecium. This would comply with the old observation that in Paramecium, Abs against acetylated α-tubulin recognize microtubule subpopulations around the oral cavity and over the entire extension of the CVC (99). To sum up, in agreement with work with metazoans, cytoskeletal elements are available in the Paramecium cell at the sites where we localized Stomatin forms and where silencing has affected function.
Conclusions.
In Paramecium, Stomatins are involved in mechanoperception, including not only the ciliary reversal reaction (in the absence of Stomatin in cilia) but also probably the osmoregulatory (CVC) and the phago(lyso)somal system. Locally the isoforms from different Stomatin families may in part overlap, though there are preferences in the localization and function, as summarized in Table 1. In particular, Sto4p contributes to recycling between spent and nascent phagosomes. Apart from this, many questions arise from our present work, not only concerning the mode of oligomerization (homo- and/or heteromeric) but also membrane attachment and intracellular pathways. Beyond that, some sites of Stomatin localization are without precedent for SPFH proteins, such as their occurrence on cortical Ca2+ stores and clathrin-coated pits in the cell membrane.
ACKNOWLEDGMENTS
We thank Alexander Bledowski for competent technical assistance in the immunogold EM localization series, Marianne Wiechers for blot analyses, and Linda Sperling (CNRS Gif-sur-Yvette) for the first hints on Stomatin in Paramecium.
We thank the Deutsche Forschungsgemeinschaft for financial support.
Footnotes
Published ahead of print 2 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00324-12.
REFERENCES
- 1. Hinderhofer M, Walker CA, Friemel A, Stuermer CAO, Möller HM, Reuter A. 2009. Evolution of prokaryotic SPFH proteins. BMC Evol. Biol. 9:10 doi:10.1186/1471-2148-9-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang M, Gu G, Ferguson EL, Chalfie M. 1995. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature 378:292–295 [DOI] [PubMed] [Google Scholar]
- 3. Mannsfeldt AG, Carroll P, Stucky CL, Lewin GR. 1999. Stomatin, a MEC-2 like protein, is expressed by mammalian sensory neurons. Mol. Cell Neurosci. 13:391–404 [DOI] [PubMed] [Google Scholar]
- 4. Hiebl-Dirschmied CM, Adolf GR, Prohaska R. 1991. Isolation and partial characterization of the human erythrocyte band 7 integral membrane protein. Biochim. Biophys. Acta 1065:195–202 [DOI] [PubMed] [Google Scholar]
- 5. Kuwahara Y, Unzai S, Nagata T, Hiroaki Y, Yokoyama H, Matsui I, Ikegami T, Fujiyoshi Y, Hiroaki H. 2009. Unusual thermal disassembly of the SPFH domain oligomer from Pyrococcus horikoshii. Biophys. J. 97:2034–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Snyers L, Umlauf E, Prohaska R. 1998. Oligomeric nature of the integral membrane protein stomatin. J. Biol. Chem. 273:17221–17226 [DOI] [PubMed] [Google Scholar]
- 7. Snyers L, Umlauf E, Prohaska R. 1999. Association of stomatin with lipid-protein complexes in the plasma membrane and the endocytic compartment. Eur. J. Cell Biol. 78:802–812 [DOI] [PubMed] [Google Scholar]
- 8. Salzer U, Prohaska R. 2001. Stomatin, flotillin-1, and flotillin-2 are major integral proteins of erythrocyte lipid rafts. Blood 97:1141–1143 [DOI] [PubMed] [Google Scholar]
- 9. Salzer U, Mairhofer M, Prohaska R. 2007. Stomatin: a new paradigm of membrane organization emerges. Dyn. Cell Biol. 1:20–33. [Google Scholar]
- 10. Browman DT, Hoegg MB, Robbins SM. 2007. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 17:394–402 [DOI] [PubMed] [Google Scholar]
- 11. Stuermer CAO. 2010. The reggie/flotillin connection to growth. Trends Cell Biol. 20:6–13 [DOI] [PubMed] [Google Scholar]
- 12. Stuermer CAO. 2011. Reggie/flotillin and the targeted delivery of cargo. J. Neurochem. 116:708–713 [DOI] [PubMed] [Google Scholar]
- 13. Lapatsina L, Brand J, Poole K, Daumke O, Lewin GR. 2012. Stomatin-domain proteins. Eur. J. Cell Biol. 91:240–245 [DOI] [PubMed] [Google Scholar]
- 14. Simons K, Gerl MJ. 2010. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11:688–699 [DOI] [PubMed] [Google Scholar]
- 15. Edqvist J, Blomqvist K. 2006. Fusion and fission, the evolution of sterol carrier protein-2. J. Mol. Evol. 62:292–306 [DOI] [PubMed] [Google Scholar]
- 16. Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. 1997. Flotillin and epideremal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 272:13793–13802 [DOI] [PubMed] [Google Scholar]
- 17. Lang DM, Lommel S, Jung M, Ankerhold R, Petrausch B, Laessing U, Wiechers MF, Plattner H, Stürmer CAO. 1998. Identification of reggie-1 and reggie-2 as plasma membrane-associated proteins which cocluster with activated GPI-anchored cell adhesion molecules in non-caveolar micropatches in neurons. J. Neurobiol. 37:502–523 [DOI] [PubMed] [Google Scholar]
- 18. Langhorst MF, Reuter A, Jäger Wippich FAFM, Luxenhofer G, Plattner H, Stürmer CA. 2008. Trafficking of the microdomain scaffolding protein reggie-1/flotillin-2. Eur. J. Cell Biol. 87:211–226 [DOI] [PubMed] [Google Scholar]
- 19. Stuermer CAO, Plattner H. 2005. The “lipid raft” microdomain proteins reggie-1 and reggie-2 (flotillins) are scaffolds for protein interaction and signalling. Biochem. Soc. Symp. 72:109–118 [DOI] [PubMed] [Google Scholar]
- 20. Solis GP, Schrock Y, Hülsbusch N, Wiechers M, Plattner H, Stuermer CAO. 2012. Reggie/flotillins regulate E-cadherin-mediated contact formation by affecting EGFR trafficking. Mol. Biol. Cell 23:1812–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tatsuta T, Model K, Langer T. 2005. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16:248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Christie DA, Lemke CD, Elias IM, Chau LA, Kirchhof MG, Li B, Ball EH, Dunn SD, Hatch GM, Madrenas J. 2011. Stomatin-like protein 2 binds cardiolipin and regulates mitochondrial biogenesis and function. Mol. Cell. Biol. 31:3845–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martinez-Salgado C, Benckendorff AG, Chiang LY, Wang R, Milenkovic N, Wetzel C, Hu J, Stucky CL, Parra MG, Mohandas N, Lewin GR. 2007. Stomatin and sensory neuron mechanotransduction. J. Neurophysiol. 98:3802–3808 [DOI] [PubMed] [Google Scholar]
- 24. Wetzel C, Hu J, Riethmacher D, Benckendorff A, Harder L, Eilers A, Moshourab R, Kozlenkov A, Labuz D, Caspani O, Erdmann B, Machelska H, Heppenstall PA, Lewin GR. 2007. A stomatin-domain protein essential for touch sensation in the mouse. Nature 445:206–209 [DOI] [PubMed] [Google Scholar]
- 25. Morrow IC, Parton RG. 2005. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic 6:725–740 [DOI] [PubMed] [Google Scholar]
- 26. Kung C, Martinac B, Sukharev S. 2010. Mechanosensitive channels in microbes. Annu. Rev. Microbiol. 64:313–329 [DOI] [PubMed] [Google Scholar]
- 27. Machemer H. 1988. Electrophysiology, p 185–215 In Görtz HD. (ed), Paramecium. Springer-Verlag, Berlin, Germany [Google Scholar]
- 28. Machemer H. 1988. Motor control of cilia, p 216–235 In Görtz HD. (ed), Paramecium. Springer-Verlag, Berlin, Germany [Google Scholar]
- 29. Machemer H. 1989. Cellular behaviour modulated by ions: electrophysiological implications. J. Protozool. 36:463–487 [Google Scholar]
- 30. Tavernarakis N, Driscoll M, Kyrpides NC. 1999. The SPFH domain: implicated in regulating targeted protein runover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 24:425–427 [DOI] [PubMed] [Google Scholar]
- 31. Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Câmara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouël A, Lepère G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Bétermier M, Weissenbach J, Scarpelli C, Schächter V, Sperling L, Meyer E, Cohen J, Wincker P. 2006. Global trends of whole genome duplications revealed by the genome sequence of the ciliate Paramecium tetraurelia. Nature 444:171–178 [DOI] [PubMed] [Google Scholar]
- 32. Allen RD. 1988. Cytology, p 4–40 In Görtz HD. (ed), Paramecium. Springer-Verlag, Berlin, Germany [Google Scholar]
- 33. Plattner H. 2002. My favorite cell–Paramecium. Bioessays 24:649–658 [DOI] [PubMed] [Google Scholar]
- 34. Plattner H. 2010. Membrane trafficking in protozoa: SNARE proteins, H+-ATPase, actin, and other key players in ciliates. Int. Rev. Cell Mol. Biol. 280:79–184 [DOI] [PubMed] [Google Scholar]
- 35. Allen RD, Fok AK. 2000. Membrane trafficking and processing in Paramecium. Int. Rev. Cytol. 198:277–317 [DOI] [PubMed] [Google Scholar]
- 36. Allen RD, Naitoh Y. 2002. Osmoregulation and contractile vacuoles. Int. Rev. Cytol. 215:351–394 [DOI] [PubMed] [Google Scholar]
- 37. Tani T, Allen RD, Naitoh Y. 2001. Cellular membranes that undergo cyclic changes in tension: direct measurement of force generation by an in vitro contractile vacuole of Paramecium multimicronucleatum. J. Cell Sci. 114:785–795 [DOI] [PubMed] [Google Scholar]
- 38. Beningo KA, Wang Y-L. 2002. Fc-receptor-mediated phagocytosis is regulated by mechanical properties of the target. J. Cell Sci. 115:849–856 [DOI] [PubMed] [Google Scholar]
- 39. Galvani A, Sperling L. 2002. RNA interference by feeding in Paramecium. Trends Genet. 18:11–12 [DOI] [PubMed] [Google Scholar]
- 40. Arnaiz O, Cain S, Cohen J, Sperling L. 2007. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 35:D439–D444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kissmehl R, Sehring IM, Wagner E, Plattner H. 2004. Immunolocalization of actin in Paramecium cells. J. Histochem. Cytochem. 52:1543–1559 [DOI] [PubMed] [Google Scholar]
- 42. Ladenburger EM, Sehring IM, Korn I, Plattner H. 2009. Novel types of Ca2+ release channels participate in the secretory cycle of Paramecium cells. Mol. Cell. Biol. 29:3605–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2009. CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ladenburger EM, Plattner H. 2011. Calcium-release channels in Paramecium. Genomic expansion, differential positioning and partial transcriptional elimination. PLoS One 6( 11): e27111 doi:10.1371/journal.pone.0027111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vielmetter J, Stolze B, Bonhoeffer F, Stuermer CAO. 1990. In vitro assay to test differential substrate affinities of growing axons and migratory cells. Exp. Brain Res. 81:283–287 [DOI] [PubMed] [Google Scholar]
- 46. Ramoino P, Fronte P, Fato M, Beltrame F, Diaspro A. 2002. Mapping cholesteryl ester analogue uptake and intracellular flow in Paramecium by confocal fluorescence microscopy. J. Microsc. 208:167–176 [DOI] [PubMed] [Google Scholar]
- 47. Xia X, Xie Z. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371–373 [DOI] [PubMed] [Google Scholar]
- 48. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 50. King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of meatazoans. Nature 451:783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Green JB, Young J. 2008. Slipins: ancient origin, duplication and diversification of the stomatin protein family. BMC Evol. Biol. 8:44 doi:10.1186/1471-2148-8-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Umlauf E, Mairhofer M, Prohaska R. 2006. Characterization of the stomatin domain involved in homo-oligomerization and lipid raft association. J. Biol. Chem. 281:23349–23356 [DOI] [PubMed] [Google Scholar]
- 53. Neumann-Giesen C, Falkenbach B, Beicht P, Claasen S, Lüers G, Stuermer CA, Herzog V, Tikkanen R. 2004. Membrane and raft association of reggie-1/flotillin-2: role of myristoylation, palmitoylation and oligomerization and induction of filopodia by overexpression. Biochem. J. 378:509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yokoyama H, Fujii S, Matsui I. 2008. Crystal structure of a core domain of stomatin from Pyrococcus horikoshii illustrates a novel trimeric and coiled-coil fold. J. Mol. Biol. 376:868–878 [DOI] [PubMed] [Google Scholar]
- 55. Mairhofer M, Steiner M, Mosgoeller W, Prohaska R, Salzer U. 2002. Stomatin is a major lipid-raft component of platelet α granules. Blood 100:897–904 [DOI] [PubMed] [Google Scholar]
- 56. Tani T, Allen RD, Naitoh Y. 2000. Periodic tension development in the membrane of the in vitro contractile vacuole of Paramecium multimicronucleatum: modification by bisection, fusion and suction. J. Exp. Biol. 203:239–251 [DOI] [PubMed] [Google Scholar]
- 57. Plattner H, Sehring IM, Mohamed IK, Miranda K, De Souza W, Billington R, Genazzani A, Ladenburger EM. 2012. Calcium signaling in closely related protozoan groups (Alveolata): non-parasitic ciliates (Paramecium, Tetrahymena) vs. parasitic Apicomplexa (Plasmodium, Toxoplasma). Cell Calcium 51:351–382 [DOI] [PubMed] [Google Scholar]
- 58. Solis GP, Hoegg M, Munderloh C, Schrock Y, Malaga-Trillo E, Rivera-Milla E, Stürmer CA. 2007. Reggie/flotillin proteins are organized into stable tetramers in membrane microdomains. Biochem. J. 403:313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wienke D, Drengk A, Schmauch C, Jenne N, Maniak M. 2006. Vacuolin, a flotillin/reggie-related protein from Dictyostelium oligomerizes for endosome association. Eur. J. Cell Biol. 85:991–1000 [DOI] [PubMed] [Google Scholar]
- 60. Langhorst MF, Reuter A, Stuermer CAO. 2005. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell. Mol. Life Sci. 62:2228–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mairhofer M, Steiner M, Salzer U, Prohaska R. 2009. Stomatin-like protein-1 interacts with stomatin and is targeted to late endosomes. J. Biol. Chem. 284:29218–29229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rajendran L, Masilamani M, Solomon S, Tikkanen R, Stuermer CAO, Plattner H, Illges H. 2003. Asymmetric localization of flotillins/reggies in preassembled platforms confers inherent polarity to hematopoietic cells. Proc. Natl. Acad. Sci. U. S. A. 100:8241–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eichinger L, Pachebat JA, Glöckner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, Bankier AT, Lehmann R, Hamlin N, Davies R, Gaudet P, Fey P, Pilcher K, Chen G, Saunders D, Sodergren E, Davis P, Kerhornou A, Nie X, Hall N, Anjard C, Hemphill L, Bason N, Farbrother P, Desany B, Just E, Morio T, Rost R, Churcher C, Cooper J, Haydock S, van Driessche N, Cronin A, Goodhead I, Muzny D, Mourier T, Pain A, Lu M, Harper D, Lindsay R, Hauser H, James K, Quiles M, Madan Babu M, Saito T, Buchrieser C, Wardroper A, Felder M, Thangavelu M, Johnson D, Knights A, Loulseged H, Mungall K, Oliver K, Price C, Quail MA, Urushihara H, Hernandez J, Rabbinowitsch E, Steffen D, Sanders M, Ma J, Kohara Y, Sharp S, Simmonds M, Spiegler S, Tivey A, Sugano S, White B, Walker D, Woodward J, Winckler T, Tanaka Y, Shaulsky G, Schleicher M, Weinstock G, Rosenthal A, Cox EC, Chisholm RL, Gibbs R, Loomis WF, Platzer M, Kay RR, Williams J, Dear PH, Noegel AA, Barrell B, Kuspa A. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Diniz MC, Costa MP, Pacheco ACL, Kamimura MT, Silva SC, Carneiro LDG, Sousa AP, Soares CE, Souza CS, de Oliveira DM. 2009. Actin-interacting and flagellar proteins in Leishmania spp.: bioinformatics predictions to functional assignments in phagosome formation. Genet. Mol. Biol. 32:652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hiller NL, Akompong T, Morrow JS, Holder AA, Haldar K. 2003. Identification of a stomatin orthologue in vacuoles induced in human erythrocytes in malaria parasites. A role for microbial raft proteins in apicomplexan vacuole biogenesis. J. Biol. Chem. 278:48413–48421 [DOI] [PubMed] [Google Scholar]
- 66. Ogura A, Machemer H. 1980. Distribution of mechanoreceptor channels in the Paramecium surface membrane. J. Comp. Physiol. A 135:233–242 [Google Scholar]
- 67. Naitoh Y, Eckert R. 1969. Ionic mechanisms controlling behavioral responses of Paramecium to mechanical stimulation. Science 164:963–965 [DOI] [PubMed] [Google Scholar]
- 68. Satow Y, Murphy AD, Kung C. 1983. The ionic basis of the depolarizing mechanoreceptor potential of Paramecium tetraurelia. J. Exp. Biol. 103:253–264 [Google Scholar]
- 69. Machemer H, Ogura A. 1979. Ionic conductances of membranes in ciliated and deciliated Paramecium. J. Physiol. 296:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Price MP, Thompson RJ, Eshcol JO, Wemmie JA, Benson CJ. 2004. Stomatin modulates gating of acid-sensing ion channels. J. Biol. Chem. 279:53886–53891 [DOI] [PubMed] [Google Scholar]
- 71. Bagshaw RD, Mahuran DJ, Callahan JW. 2005. A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of the organelle. Mol. Cell Proteomics 4:133–143 [DOI] [PubMed] [Google Scholar]
- 72. Plattner H, Henning R, Brauser B. 1975. Formation of Triton WR 1339 filled rat liver lysosomes. II. Involvement of autophagy and of pre-existing lysosomes. Exp. Cell Res. 94:377–391 [DOI] [PubMed] [Google Scholar]
- 73. Kissmehl R, Froissard M, Plattner H, Momayezi M, Cohen J. 2002. NSF regulates membrane traffic along multiple pathways in Paramecium. J. Cell Sci. 115:3935–3946 [DOI] [PubMed] [Google Scholar]
- 74. Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. 2004. Association of stomatin with lipid bodies. J. Biol. Chem. 279:23699–23709 [DOI] [PubMed] [Google Scholar]
- 75. Hua SZ, Gottlieb PA, Heo J, Sachs F. 2010. A mechanosensitive ion channel regulating cell volume. Am. J. Physiol. Cell Physiol. 298:C1424–C1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Iwamoto M, Sugino K, Allen RD, Naitoh Y. 2005. Cell volume control in Paramecium: factors that activate the control mechanisms. J. Exp. Biol. 208:523–537 [DOI] [PubMed] [Google Scholar]
- 77. Fujiu Nakayama K, Y, Iida H, Sokabe M, Yoshimura K. 2011. Mechanoperception in motile flagella of Chlamydomonas. Nat. Cell Biol. 13:630–632 [DOI] [PubMed] [Google Scholar]
- 78. Fricke B, Stewart GW, Treharne KJ, Mehta A, Knöpfle G, Friedrichs N, Müller KM, von Düring M. 2003. Stomatin immunoreactivity in ciliated cells of the human airway epithelium. Anat. Embryol. 207:1–7 [DOI] [PubMed] [Google Scholar]
- 79. Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, McKay SJ, Huang P, Swoboda P, Jones SJ, Marra MA, Baillie DL, Moerman DG, Shaham S, Leroux MR. 2005. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15:935–941 [DOI] [PubMed] [Google Scholar]
- 80. Fok AK, Aihara MS, Ishida M, Nolta KV, Steck TL, Allen RD. 1995. The pegs on the decorated tubules of the contractile vacuole complex of Paramecium are proton pumps. J. Cell Sci. 108:3163–3170 [DOI] [PubMed] [Google Scholar]
- 81. Wassmer T, Sehring IM, Kissmehl R, Plattner H. 2009. The V-ATPase in Paramecium: functional specialization by multiple gene isoforms. Pflugers Arch. Eur. J. Physiol. 457:599–607 [DOI] [PubMed] [Google Scholar]
- 82. Stock C, Grθnlien HK, Allen RD, Naitoh Y. 2002. Osmoregulation in Paramecium: in situ ion gradients permit water to cascade through the cytosol to the contractile vacuole. J. Cell Sci. 115:2339–2348 [DOI] [PubMed] [Google Scholar]
- 83. Burster T, Beck A, Poeschel S, θren, Baechle AD, Reich M, Roetzschke O, Falk K, Boehm BO, Youssef S, Kalbacher H, Overkleeft H, Tolosa E, Driessen C. 2007. Interferon-γ regulates cathepsin G activity in microglia-derived lysosomes and controls the proteolytic processing of myelin basic protein in vitro. Immunology 121:82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Swanson JA. 1999. Pathways through the macrophage vacuolar compartment. Adv. Cell Mol. Biol. Membr. Org. 5:267–284 [Google Scholar]
- 85. Dermine JF, Duclos S, Garin J, St-Louis F, Rea S, Parton RG, Desjardins M. 2001. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J. Biol. Chem. 276:18507–18512 [DOI] [PubMed] [Google Scholar]
- 86. Swanson JA. 2008. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9:639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Brugués J, Maugis B, Casademunt J, Nassoy P, Amblard F, Sens P. 2010. Dynamical organization of the cytoskeletal cortex probed by micropipette aspiration. Proc. Natl. Acad. Sci. U. S. A. 107:15415–15420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sehring IM, Reiner C, Mansfeld J, Plattner H, Kissmehl R. 2007. A broad spectrum of actin paralogs in Paramecium tetraurelia cells display differential localization and function. J. Cell Sci. 120:177–190 [DOI] [PubMed] [Google Scholar]
- 89. Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. 2010. The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U. S. A. 107:21517–21522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Delmas P, Hao J, Rodat-Despoix L. 2011. Molecular mechanisms of mechanotransduction in mammalian sensory neurons. Nat. Rev. Neurosci. 12:139–153 [DOI] [PubMed] [Google Scholar]
- 91. Snyers L, Thinès-Sempoux D, Prohaska R. 1997. Colocalization of stomatin (band 7.2b) and actin microfilaments in UAC epithelial cells. Eur. J. Cell Biol. 73:281–285 [PubMed] [Google Scholar]
- 92. Takeshita N, Diallinas G, Fischer R. 2012. The role of flotillin FloA and stomatin StoA in the maintenance of apical sterol-rich membrane domains and polarity in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 83:1136–1152 [DOI] [PubMed] [Google Scholar]
- 93. Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. 2001. The phagosome proteome: insight into phagosome functions. J. Cell Biol. 152:165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kinchen JM, Ravichandran KS. 2008. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 9:781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liebl D, Griffiths G. 2009. Transient assembly of F-actin by phagosomes delays phagosome fusion with lysosomes in cargo-overloaded macrophages. J. Cell Sci. 122:2935–2945 [DOI] [PubMed] [Google Scholar]
- 96. Langhorst MF, Solis GP, Hannbeck S, Plattner H, Stuermer CAO. 2007. Linking membrane microdomains to the cytoskeleton: regulation of the lateral mobility of reggie-1/flotillin-2 by interaction with actin. FEBS Lett. 581:4697–4703 [DOI] [PubMed] [Google Scholar]
- 97. Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5:599–609 [DOI] [PubMed] [Google Scholar]
- 98. Tominaga T, Naitoh Y, Allen RD. 1999. A key function of non-planar membranes and their associated microtubular ribbons in contractile vacuole membrane dynamics is revealed by electrophysiollogically controlled fixation of Paramecium. J. Cell Sci. 112:3733–3745 [DOI] [PubMed] [Google Scholar]
- 99. Callen AM, Adoutte A, Andrew JM, Baroin-Tourancheau A, Bré MH, Ruiz PC, Clerot JC, Delgado P, Fleury A, Jeanmaire-Wolf R. 1994. Isolation and characterization of libraries of monoclonal antibodies directed against various forms of tubulin in Paramecium. Biol. Cell 81:95–119 [DOI] [PubMed] [Google Scholar]