Abstract
Botrytis cinerea causes gray mold on a great number of host plants. Infection is initiated by airborne conidia that invade the host tissue, often by penetration of intact epidermal cells. To mimic the surface properties of natural plant surfaces, conidia were incubated on apple wax-coated surfaces, resulting in rapid germination and appressorium formation. Global changes in gene expression were analyzed by microarray hybridization between conidia incubated for 0 h (dormant), 1 h (pregermination), 2.5 h (postgermination), 4 h (appressoria), and 15 h (early mycelium). Considerable changes were observed, in particular between 0 h and 1 h. Genes induced during germination were enriched in those genes encoding secreted proteins, including lytic enzymes. Comparison of wild-type and a nonpathogenic MAP kinase mutant (bmp1) revealed marked differences in germination-related gene expression, in particular related to secretory proteins. Using promoter-GFP reporter strains, we detected a strictly germination-specific expression pattern of a putative chitin deacetylase gene (cda1). In contrast, a cutinase gene (cutB) was found to be expressed only in the presence of plant lipids, in a developmentally less stringent pattern. We also identified a coregulated gene cluster possibly involved in secondary metabolite synthesis which was found to be controlled by a transcription factor also encoded in this cluster. Our data demonstrate that early conidial development in B. cinerea is accompanied by rapid shifts in gene expression that prepare the fungus for germ tube outgrowth and host cell invasion.
INTRODUCTION
Germination of spores is a fundamental step in fungal development, leading to the conversion of a dormant cell into a growing hypha. It involves the breaking of dormancy by external signals, a pregermination phase that is visibly accompanied by isotropic swelling, and then the formation of a germ tube that marks the establishment of polar growth (1, 2). Basic requirements for the initiation of germination of fungal spores, for example, of the saprotrophic fungi Neurospora crassa and Aspergillus nidulans, are usually high humidity and the availability of nutrient sources such as sugars (2). Spore germination in response to sugars has been shown in A. nidulans to be dependent on Gα subunits, cyclic AMP (cAMP), and protein kinases (3, 4). Similarly, the cAMP-protein kinase A pathway is required for spore germination in Schizosaccharomyces pombe (5) and for yeast-to-hypha transition on solid media in the pathogenic yeast Candida albicans, a process related to spore germination (6). In plant-pathogenic fungi, including Blumeria graminis (7), Phyllosticta ampelicida (8), Colletotrichum graminicola (9), and Colletotrichum gloeosporioides f. sp. aeschynomenes (1), breaking of spore dormancy is often induced by surface contact stimuli, such as surface hardness and hydrophobicity, either alone or in combination with nutrient signals. Although some signal transduction components, such as cAMP, Ras2, and Rac1, have been found to be involved in the germination process of these fungi, the regulatory mechanisms are still poorly understood (1, 9, 10). After germination on appropriate surfaces such as host cuticles, germ tube elongation occurs until perception of physical and/or chemical stimuli by the germ tube tip, resulting in the cessation of germ tube elongation and appressorium formation (7, 11, 12, 13, 14, 15). Appressoria, which are morphologically highly diverse in different plant pathogens, initiate the penetration into the plant tissue (16).
Transcriptome studies with N. crassa, Fusarium graminearum, and Ustilago maydis have been performed using spores germinated in minimal (N. crassa and U. maydis) or full (F. graminearum) media in (partly shaken) suspensions, with no permanent surface contact of the spores (17, 18, 19). Blumeria microarray-based studies with germinated conidia forming infection structures, including haustoria, on the surface and within host epidermal cells have focused mainly on transcriptomic changes related to metabolic pathways, such as lipid degradation, glycolysis, pentose phosphate pathway, and amino acid biosynthesis (20, 21). Comprehensive studies with Magnaporthe oryzae have addressed gene expression changes, after 4 to 12 h, that are mainly correlated with appressorium formation (22, 23). The study presented here focused on gene expression changes during the early stages of conidial germination and appressorium differentiation of Botrytis cinerea, in response to a combination of physical (hard surface) and chemical (sugars, plant lipids) signals.
Botrytis cinerea (teleomorph Botryotinia fuckeliana) is a major necrotrophic pathogen of a wide host range of dicotyledonous plant species, causing serious pre- and postharvest losses in the production of economically important fruits, vegetables, and ornamental flowers (24). New infections usually occur by wind-dispersed conidia, which germinate on the plant surface and invade the tissue either through wounds or by direct penetration of cuticles and cell walls of epidermal cells (25). The host tissue is rapidly killed and degraded by various mechanisms, such as secretion of cell wall-degrading enzymes and release of toxins and other necrosis-inducing factors (26). Because of the crucial role of B. cinerea conidia for epidemiology and infection, a detailed knowledge of the molecular events during the early stages of their development is of great interest not only scientifically but also for the development of new control strategies.
Germination of B. cinerea conidia can be induced either in rich media in the absence of any surface, in the presence of carbon sources on a hard surface, or without any nutrients on a hard and hydrophobic surface (27). In the presence of carbon sources, germ tube outgrowth is preceded by rapid turnover of the disaccharide trehalose, which serves as a carbohydrate reserve and as an osmoprotectant (28). Based on the germination phenotypes of mutants, two signaling pathways have been described that are involved in the control of germination induction. For normal carbon source-induced germination on hard surfaces, an intact cAMP signaling pathway, which includes Ras2, the Gα3 subunit of the heterotrimeric G protein, and adenylate cyclase, is required (27, 29). In contrast, hydrophobic-surface-induced germination is dependent on a functional mitogen-activated protein kinase (MAPK) cascade (Ste11-Ste7-BMP1) (27, 30). B. cinerea mutants lacking either of the three MAPK components, or the putative adaptor protein Ste50, did not germinate on hydrophobic surfaces in pure water (30). After germination, the germ tube elongates on the host surface to different lengths before tip growth stops and penetration is initiated. The appressorium morphology is not very distinct, being characterized by a more or less distinct terminal swelling and, usually, no septum separating it from the germ tube. Penetration into the host tissue is accompanied by rapid death of the invaded plant cells. In addition to their role in germination, the cAMP and the BMP1 MAPK signaling pathways are required for appressorium differentiation, penetration, and host colonization, similar to the situation in other plant-pathogenic fungi (31, 32). The MAPK cascade ste11, ste7, bmp1, and ste50 mutants did not form appressoria on hard surfaces, they failed to penetrate into host tissue, and they were nonpathogenic (27, 30). Other signaling elements found to be involved in germination are the small GTPase Cdc42 and phospholipase C (Plc1) (33). Taken together, these findings show that germination induction is a complex process involving several signaling pathways which is currently not fully understood.
Transcriptome profiling analyses during germination have been published for N. crassa (17), the saprotrophic chytridiomycete Blastocladiella emersonii (18), and the wheat pathogen F. graminearum (19). These studies revealed major changes of gene expression patterns starting already during the pregermination swelling stage. Genes involved in DNA replication, protein synthesis, and degradation of storage reserves were found to be activated during the early stages of the germination process.
In the present study, we have performed a transcriptome analysis of the early stages of conidial germination of B. cinerea. In order to get insight into the molecular events during the germination process as well as during early stages of the infection process, we have chosen optimized conditions for germination and subsequent appressorium formation on a hard, wax-coated surface. To obtain further evidence for genes that might be involved in the early infection process, we included the bmp1 MAP kinase mutant in the analysis.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The Botrytis cinerea laboratory strain B05.10 (wild type) and the MAP kinase deletion bmp1 mutant were used for most experiments (27). For construction of green fluorescent protein (GFP) reporter strains, B. cinerea strain B05.Hyg-3 was used (34). For maintenance and spore production, B. cinerea was cultivated on tomato malt extract agar (TMA) as described previously (16). Conidia were harvested between 9 and 12 days after inoculation from sporulating plates using a glass spatula and 10 ml of distilled water and were washed with distilled water in 15-ml centrifugation tubes by three rounds of centrifugation at 3,500 rpm (27). Freshly harvested conidia (0 h) were used for RNA extraction or resuspended in Gamborg minimal medium (3 g/liter of Gamborg B5; 10 mM KH2PO4, pH 5.5) containing 10 mM fructose. Conditions for germination on glass, polyethylene, and polystyrene surfaces were as described previously (27). Conidia were rated as germinated when the spore wall was broken by the emerging germ tube.
Analysis of pregermination conidial swelling.
For analysis of conidial swelling prior to germination, conidia were incubated in the presence or absence of 10 mM fructose, on apple wax-coated petri dishes, or on round glass coverslips (15-mm diameter; Roth, Karlsruhe, Germany). Time-lapse microscopic observations of individual conidia were made with an Axiovert Observer A1 inverted microscope (Zeiss, Jena, Germany) equipped with a Neofluar 40×/1.3 oil immersion objective. Using a Canon Power-Shot G9 camera controlled by the program RemoteCapture, pictures were taken at 5-min intervals until germ tube outgrowth was visible. Based on the measured length and width of the spore, the changes in spore volume (V) were calculated according to the formula V = 4/3 · π · a/2 · b2/4, where a is length and b is diameter of the spore. For each condition, at least three independent experiments (n ≥ 30) were performed.
Nuclear staining.
For staining of nuclei, the germination medium was removed, and the conidia covered with 50 μl of a fixing solution containing 3% formaldehyde and 10 mg/liter of Hoechst 33342, followed by incubation for 40 min at room temperature in the dark. When the germlings had developed for more than 1.5 h, they were fixed in the germination medium by heat treatment (60°C for 15 min) in a humid chamber, followed by 30 min of incubation in a solution containing 5 mg/liter of Hoechst 33342. Stained germlings were evaluated with a fluorescence microscope, and pictures were taken at different focal planes for subsequent counting of the nuclei. For each time point, a minimum of 50 spores was analyzed in three independent experiments.
Preparation of coated surfaces for germination.
Apple wax was prepared by soaking ripe apples (cv. Golden Delicious) three times for 10 s in chloroform. The resulting solution was washed out sequentially with equal volumes of 0.1 N HCl, 0.1 N NaOH, and several changes of distilled water. The chloroform was removed under vacuum in a rotary evaporator at 30°C and dried under a stream of nitrogen gas. The wax was dissolved in hexane at a concentration of 2 g/liter and stored as stock solution at −20°C. Petri dishes (9-cm diameter) were coated by spreading the dissolved wax over the surfaces at a concentration of 10 μg/cm2 and subsequently air dried. For preparation of cutin hydrolysate, peeled skins of 20 to 25 ripe apples (cv. Golden Delicious) were prepared, treated with pectinolytic (Fructozym P; Erbslöh, Germany) and cellulolytic (Rohament CT; Röhm Enzymtechnologie, Germany) enzymes, and hydrolyzed as described previously (35). The cutin hydrolysate was dissolved in hexane and used at 10 μg/cm2 for surface coatings and induction tests. For coatings with trihydroxypalmitic acid (THPA; Sigma), 2-g/liter stock solutions in methanol were prepared.
For germination assays, conidia were inoculated in 30-μl droplets containing Gamborg B5 basal salt mixture with 10 mM fructose onto apple wax-coated polystyrene surfaces, if not indicated otherwise below, and incubated at 20°C in the dark.
Analysis of GFP fluorescence.
For fluorescence microscopy, an Axio Observer A1 (Zeiss, Jena, Germany; Semrock GFP-A basic filter set; excitation wavelength, 469 nm; emission wavelength, 525 nm) was used. For quantification of fluorescence levels of cutB-GFP germlings, ImageJ 1.43u software (National Institutes of Health, Bethesda, MD) was used. Pictures were taken with the same exposure time for all treatments, taking care to avoid overexposure. Fluorescence of the germlings (marked as regions of interest) was quantified, and background levels were subtracted. The obtained values were classified as five levels of intensity, as follows: 0, <10; 1, 10 to 69; 2, 70 to 129; 3, 130 to 190; and 4, >190 (the units are pixel values from 0 [black] to 255 [white]).
DNA manipulations.
The cda1-GFP and cutB-GFP promoter reporter strains were constructed by using a plasmid encoding a B. cinerea codon-optimized GFP (Bc-GFP1) and a hygromycin resistance cassette for selection (36). For expression of Bc-gfp1 under the control of either cda1 or cutB promoter, fragments covering 1,313 bp (cda1) and 1,281 bp (cutB) upstream of the start codons were amplified (primers are listed in Table S1 in the supplemental material) and used to replace the constitutive oliC promoter fragment. The plasmids were linearized with KpnI and used to transform B. cinerea B05.Hyg-3 (34).
Construction of the rum1 mutant was performed by amplification of the entire coding region of rum1 with an additional 365 bp of the promoter and 350 bp of the terminator region using the primers rum1-KO (see Table S1 in the supplemental material). The fragment was further cloned into the vector pLOB1 (16) via SacI and ApaI restriction sites. Natural restriction sites for EcoRV (168 bp downstream of the start codon) and for EcoRI (157 bp upstream of the stop codon) in the coding region of rum1 were used for replacing the coding region by a hygromcin resistance cassette. Genotypic verification of rum1 deletion strains was performed by quantitative reverse transcription-PCR (qRT-PCR) analysis of rum1 expression using the primers P104380fw/rev and PCR analysis using the primer pair rum1-screen (see Table S1 and Fig. S2 in the supplemental material).
RNA isolation and qRT-PCR.
For germination on 9-cm-diameter wax-coated petri dishes under standard conditions, 2 × 106 conidia were suspended in 22.5 ml of Gamborg fructose medium and transferred into the dishes (9 cm). After incubation at 20°C in the dark for various times, the medium was poured off, and the germlings were removed from the dishes using a cell scraper. Germlings were centrifuged for 5 min at 4°C and 4,000 rpm, washed once with ice-cold water, and ground in liquid nitrogen to a fine powder. RNA was isolated with the RNeasy plant minikit (Qiagen, Hilden, Germany).
For quantitative RT-PCR analysis, 1 μg of RNA of each sample was reverse transcribed into cDNA with oligo(dT) primers using a Verso cDNA kit (Thermo Fisher Scientific, Surrey, United Kingdom). PCR was performed using a MyIQ real-time PCR cycler (Bio-Rad, Munich, Germany). Expression of the genes was calculated by the 2−ΔΔCT method (37). For normalization of expression levels, several genes were tested for constant expression during germination (see Table S4 in the supplemental material). Based on the results, ubq1 and actA were subsequently used for normalization. Data are shown as normalized fold expression relative to the expression in ungerminated conidia. Means of three biological replicates, with two technical replicates each, are shown. The primers used in this study are listed in Table S1 in the supplemental material.
Microarray expression analysis.
Ten micrograms of total RNA was converted into cDNA using a SuperScript II cDNA conversion kit (Invitrogen, Carlsbad, CA). Labeling of double-stranded cDNA with Cy3 nonamers and microarray hybridization were performed by Nimblegen (Roche NimbleGen, Inc.). For hybridization, Nimblegene 4-plex arrays, containing 4 × 72,000 arrays per slide, were used. Three 60-mer oligonucleotides per gene were immobilized for a total number of 20,885 predicted genes of the two sequenced B. cinerea strains B05.10 and T4, with 3,189 T4-specific genes and 1,776 B05.10-specific genes (designated BC1G_XXXX) (38). Genes annotated according to strain T4 (http://urgi.versailles.inra.fr/gb2/gbrowse/BOTRYTIS_T4_pub/) are referred to as in the abbreviated form PXXXXXX, instead of BofuT4_PXXXXXX.1, throughout this article. Of these total genes, predicted genes encoding small coding sequences (<100 amino acids [aa]) and lacking expressed sequence tag (EST) support and any functional or topological domain were excluded, which reduced the number of genes evaluated in our study to 15,738. After hybridization of the arrays with the Cy3-labeled cDNA probes, they were scanned with a Genepix 4000b dual-laser scanner (Axon Instruments, Foster City, CA). Hybridization signals from all data sets, comprising three independent experiments, were normalized together, using the quantile function, and averaged for each gene (39). Thresholds of gene expression were determined by referring the hybridization signals to those of 9,557 random probes. Genes were considered expressed when their signal was 29 or higher, within a dynamic range of 20 to 216. This threshold was selected because for all time points, it was close to the 99th percentile of random probes hybridization signals (see Fig. S1 in the supplemental material). According to this classification, 12,091 (76.8%) of the supported genes were found be expressed at one or more time points. For analysis of time-dependent expression changes, genes showing an expression signal of at least 29 at one of the time points investigated were included in the analysis. Genes showing at least 2-fold, significant (P < 0.05, Student's t test) changes in expression at a given time point were considered differentially expressed. Groups of coregulated genes with selected expression profiles across the five time points were generated using ArrayStar software, version 3.0.1 (DNASTAR Inc., Madison, WI). Genes lacking statistical support (P < 0.05) for the chosen expression profile were discarded.
Gene annotation and categorization.
Functional annotation of B. cinerea proteins was performed as described previously (40), with the following modifications. For functional categorization, all expressed genes (12,092) were first analyzed according to their different FunCat categories (41). In addition, some genes with specific IntroPro entry were included. The following functional categories were generated: cell cycle and DNA processing (FunCat, 10); transcription (FunCat, 11); protein synthesis and processing (FunCat, 12 and 14; Interpro, 009053); amino acid metabolism (FunCat, 01_01); lipid, fatty acid, and isoprenoid metabolism (FunCat, 01_06 and 02_25); C-compound and carbohydrate metabolism (FunCat, 01_05 and 16_13); secondary metabolism (FunCat, 01_20; InterPro, 013968); cell wall (FunCat, 42_01); energy and respiration (FunCat, 02); cellular communication and signal transduction (FunCat, 30, 34, and 36; InterPro, 011009, 000182, 000198, 000195, and 000008); cellular transport (FunCat, 20; InterPro, 011701); and stress and detoxification (FunCat, 32). Prediction of secreted proteins was done using SignalP3.0, excluding proteins containing further transmembrane domains (TMHMM) or an organellar location (TargetP). Genes with CAZyme (carbohydrate-active enzyme) entry were obtained by automated functional annotation as described previously (40).
Microarray data accession number.
The entire microarray data set described in this article is available at the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds?term = GSE43569) under accession number GSE43569.
RESULTS
Pregermination swelling, kinetics of germination, and nuclear division during germination.
Germination is a multistep process that starts with initiation of metabolism and the breaking of dormancy. Swelling, i.e., increase in spore volume before germination, is the first cytological evidence of activation of dormant spores in many fungi (42). We checked whether this also holds true for B. cinerea. When B. cinerea conidia were incubated in the presence of 10 mM fructose on glass slides, pregermination swelling was observed to start after about 1 h and continued until germ tube emergence. Swelling was dependent on active conidia, it did not occur in heat-killed conidia, and it was not observed in conidia incubated at high density, which inhibits germination. Surprisingly, no swelling was observed if conidia were incubated on a hydrophobic surface in water, in the absence of nutrients, although these conidia germinated (Fig. 1). Therefore, in B. cinerea conidia, pregermination swelling depends on the availability of nutrients during germination.
Fig 1.
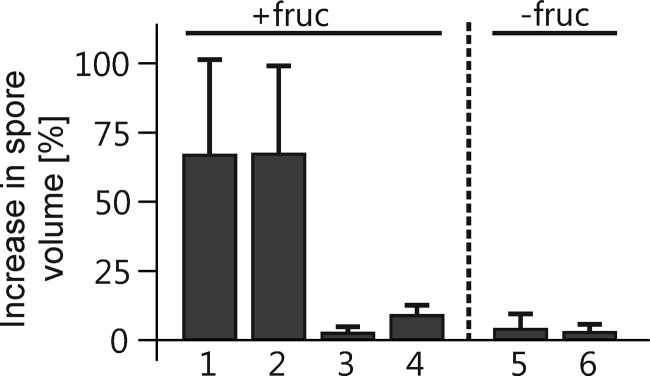
Pregermination swelling of B. cinerea wild-type conidia in minimal medium with or without 10 mM fructose. Bars 1, 3, 4, and 5, apple wax-coated glass slides; bars 2 and 6, untreated glass slides. For bar 3, heat-killed conidia (15 min at 65°C) were used. For bar 4, a high conidium concentration (107/ml) was used. For the other bars, 105/ml conidia were used. Swelling of each spore until germination was determined by subtracting the calculated volume of each spore immediately after inoculation from the volume at the moment of germination. Under conditions that did not lead to spore germination (bars 3 and 4), swelling of the spores was measured after 5 h of incubation. Values are the means of three independent experiments (n ≥ 30 spores each).
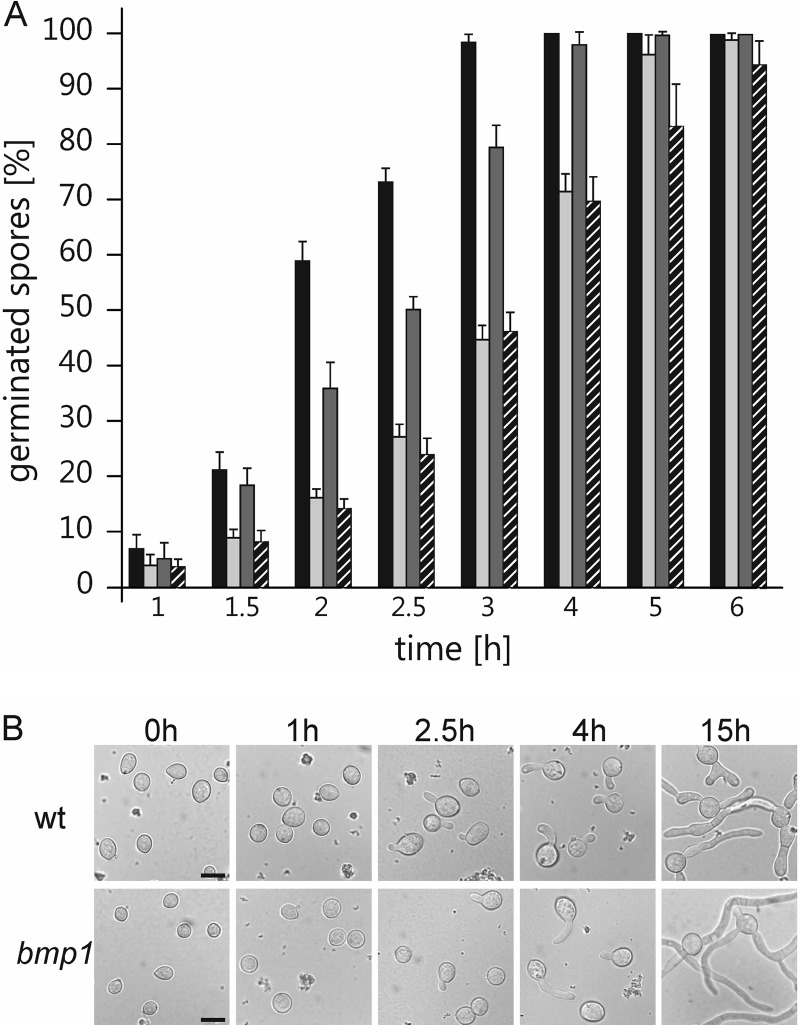
In order to achieve optimal germination conditions, similar to those on natural plant surfaces, we compared germination of B05.10 wild-type conidia on different artificial surfaces. For all experiments, conidia suspended in minimal medium containing 10 mM fructose were used. Compared to polystyrene and glass surfaces, surfaces generated by apple wax-coated petri dishes induced the most rapid germination of conidia. On wax surfaces, germination was completed already after 3 h, while on pure plastic and glass surfaces, complete germination was not observed until 4 to 5 h of incubation (Fig. 2A). For the transcriptome analyses shown below, we therefore used wax-coated surfaces. Compared to that of the wild type, germination of the MAP kinase bmp1 mutant on wax-coated surfaces was delayed by approximately 1 h (Fig. 2).
Fig 2.
Germination kinetics of B. cinerea conidia. (A) Germination kinetics of B05.10 wild-type and bmp1 mutant conidia on different surfaces in minimal medium containing 10 mM fructose as follows: the wild type on apple wax-coated polystyrene (black), the wild type on glass (light gray), the wild type on polypropylene (dark gray), and the bmp1 mutant on apple wax-coated polystyrene (hatched). Standard deviations of four independent experiments are shown. (B) Time course of germination of wild-type and bmp1 mutant conidia on apple wax-coated polystyrene. Scale bars, 10 μm.
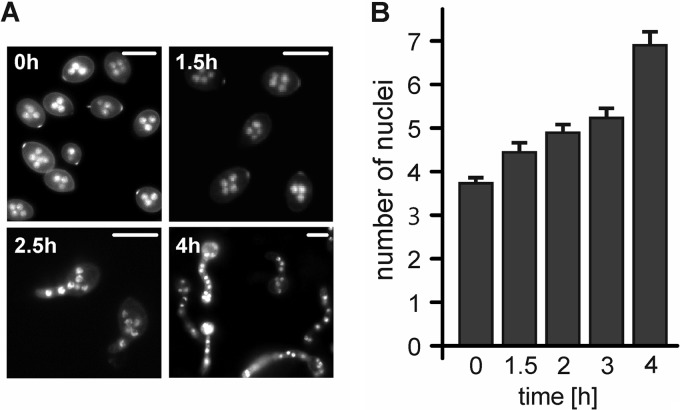
In conidia of N. crassa, A. nidulans, and F. graminearum, DNA replication and nuclear division have been found to start early during germination (43, 44, 45, 46). In B. cinerea, analysis of nuclear division is complicated by the fact that macroconidia are multinucleate. In resting conidia of B. cinerea strain B05.10, we observed between 2 and 7 nuclei (average, 3.7 ± 0.2). When germination was induced in 10 mM fructose on apple wax surfaces, a significant increase in the mean nuclear number was observed after 90 min. At this time, germination had started in approximately 20% of the conidia (Fig. 3). However, it took more than 4 h, when all conidia had germinated, until the average number of nuclei had approximately doubled. During germination, some of the nuclei migrated into the developing germ tube, but no regular pattern in this process was observed. These data indicated that the cell cycle and nuclear division start early during the germination process.
Fig 3.
Nuclear division of B. cinerea conidia during germination. (A) Hoechst 33342 staining of germinating B. cinerea conidia and increase in average number of nuclei during early germination. Scale bars, 10 μm. (B) Graph showing the average numbers of nuclei in germinating conidia incubated on apple wax-coated polystyrene in minimal medium containing 10 mM fructose. Error bars show standard errors of three independent experiments (n = 50).
Transcriptome profiling of the germination process.
In accordance with our previous biochemical studies and the cytological analyses, the following time points for isolation of RNA were used for microarray hybridization (Fig. 2B). As a reference, freshly harvested, nongerminated conidia (0 h) were used. After 1 h, degradation of trehalose had occurred (28), and conidia started to show swelling (Fig. 1). After 2.5 h, the majority of the conidia had germinated. After 4 h, most of the germ tubes had stopped elongation and formed terminal, appressorium-like thickenings. After 15 h, the germlings had resumed elongation and formed saprotrophic hyphae, with intermittent penetration attempts.
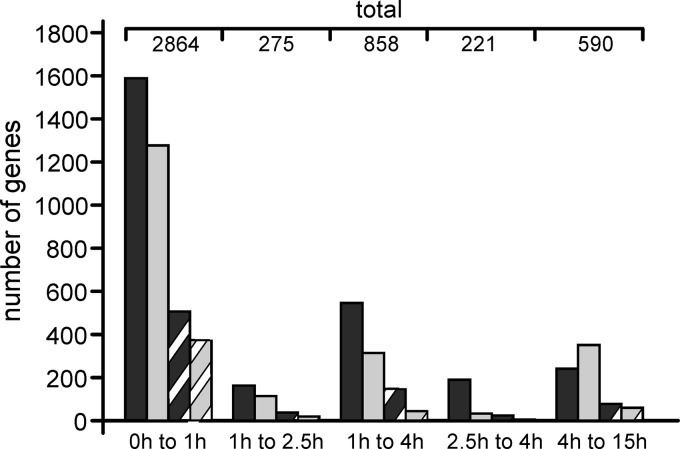
For each time point, microarray hybridizations were performed with three biological replicates of the wild-type strain, using Nimblegene whole-genome arrays representing gene models of the two sequenced B. cinerea strains B05.10 and T4 (40) (GEO database accession number GSE43569). The microarray expression data were first evaluated for stage-specific changes in global transcript abundance by performing pairwise comparisons between different time points (Fig. 4; see also Table S2 in the supplemental material). By far the largest changes were observed between resting conidia and conidia incubated for 1 h (0 h to 1 h), with a total of 2,864 genes being at least 2-fold up- or downregulated. Considerably fewer changes were observed between 1 h and later time points, when most morphological changes, including germ tube outgrowth and appressorium formation, occurred. Interestingly, gene expression changes between 1 h and 4 h were much higher than between 1 h and 2.5 h or between 2.5 h and 4 h. These data indicate that gene expression changes between the three time points largely occur in a continuous, gradual manner. In addition, they confirm the validity and comparability of the data for different time points.
Fig 4.
Changes in gene expression between different stages of wild-type conidium development. Numbers of genes showing at least 2-fold upregulation (dark gray) or downregulation (light gray) and at least 4-fold upregulation (dark hatching) or downregulation (light hatching) are shown (P < 0.05; Student's t test).
Correlation of gene expression profiles with gene function.
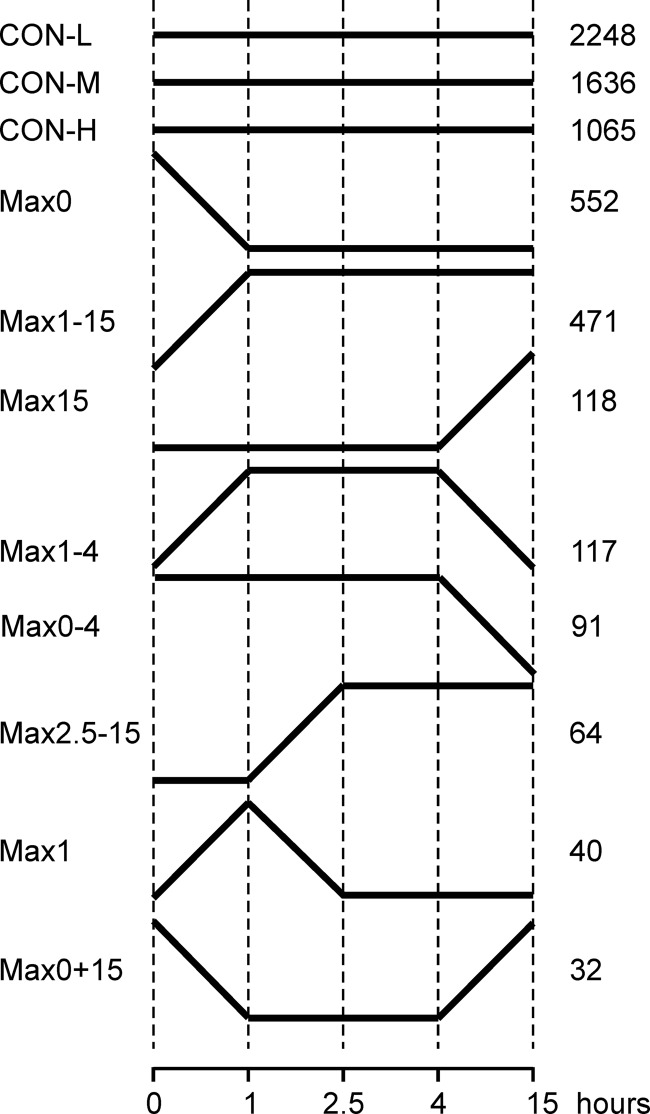
To identify genes with similar regulation during the germination process, groups of coregulated genes were generated according to the microarray data (Fig. 5; see also Table S3 in the supplemental material). The large majority of these genes (4,949 genes, or 40.1% of the expressed genes) was constitutively expressed (CON groups), showing less than 2-fold variation of expression during the five developmental stages examined. Subdivision of the CON genes into high (CON-H; expression signals of >214), medium (CON-M; 211 to 214), and low (CON-L; 29 to 211) expression revealed group CON-L to be the largest, followed by CON-M and CON-H. The next two largest groups, Max0 and Max1-15, comprised genes that showed either maximal or minimal expression in nongerminated conidia, followed by constant expression levels. The sixth largest group, Max15, comprised mycelium-specific genes. The seventh group, Max1-4, is described in more detail below. Four smaller groups are shown in Fig. 5, including one with genes that are upregulated only after 1 h (Max1), at the pregermination state.
Fig 5.
Identification of gene groups with similar transcriptional profiles across the five time points of germination and germling development.
The genes that were up- or downregulated between 0 h and 1 h (Fig. 4) and the genes belonging to groups shown in Fig. 5 (except for Max0+15), were analyzed for functional categories of their encoded proteins (Fig. 6; see also Table S3 in the supplemental material). Of the total of 12,092 expressed genes, 4,558 (37.7%) could be functionally categorized. Interestingly, the expression levels of constitutively expressed (CON) genes showed a clear correlation with the proportion of genes that could be assigned to functional categories: while the CON-L group contained only 31.8% genes that could be functionally categorized, the proportions were 47.8% for CON-M and 66.9% for CON-H. Remarkably, genes upregulated after 1 h could be functionally categorized to more than 60% (0-1_up), or even 70% (Max1, Max1-4, and Max1-15), which is clearly above the average of genes (37.7%).
Fig 6.
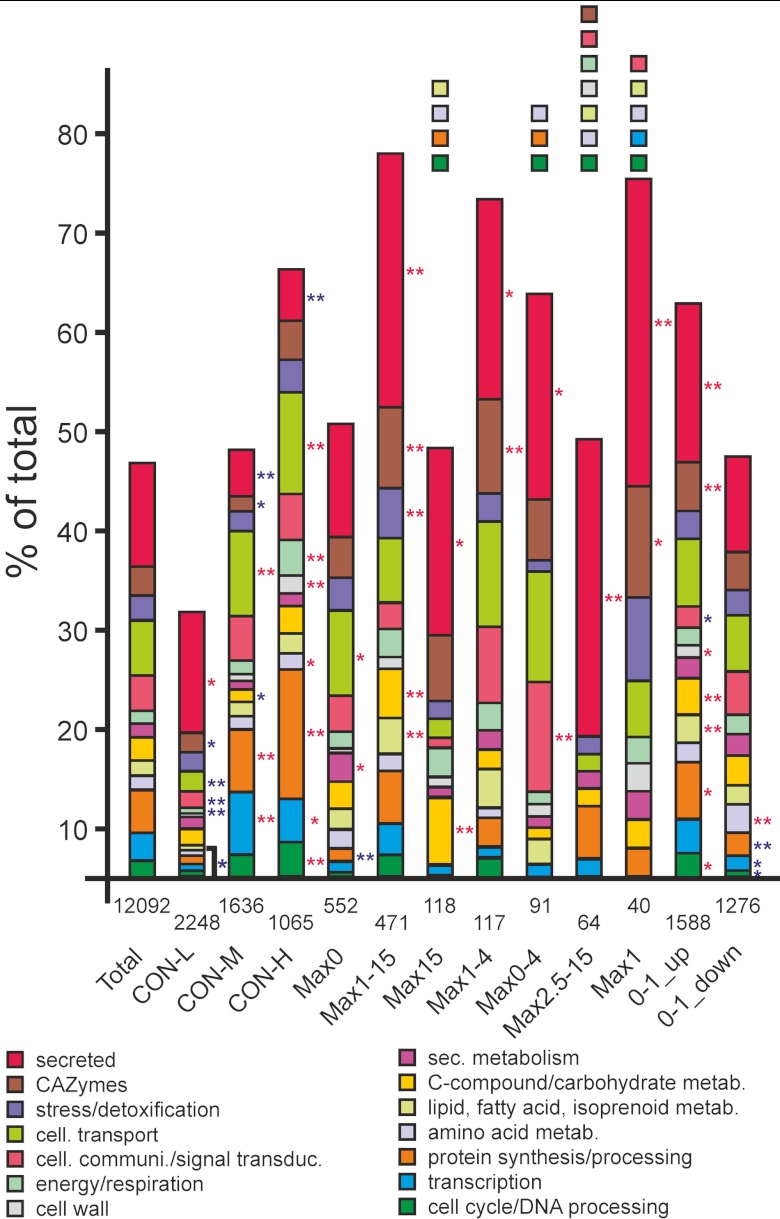
Distribution of functional categories among genes belonging to different expression groups. For each functional category, the observed percentage of genes within the group is shown. Numbers indicate the total number of genes in each group. Asterisks indicate P values showing significant differences between observed and expected frequencies for a functional category within the total number of genes in the respective group (*, P < 0.01; **, P < 0.001; Fisher test). Red asterisks indicate overrepresentation and blue asterisks underrepresentation. The colored boxes above the bars indicate functional categories that are absent.
Interesting differences of certain functional groups and their occurrence in specific expression groups were observed. In the CON-L group, an increased frequency of genes encoding secreted proteins was observed, whereas the frequency of genes belonging to cell cycle, transcription, protein synthesis, cellular transport, and other categories was significantly lower than expected. This pattern was largely inverted in groups CON-M and CON-H, which contained relatively few genes encoding secreted proteins but a strong enrichment of genes involved in cellular transport, energy and respiration (CON-H only), C-compound and carbohydrate metabolism, and protein synthesis and processing. Interestingly, all of the groups including genes induced during germination (i.e., genes activated after 1 h or 2.5 h) were enriched in genes encoding secreted proteins and carbohydrate-active enzymes (CAZymes). The only exception was the Max2.5-15 group, which did not contain any CAZyme genes.
The bmp1 mutant shows downregulation of germination-related secreted proteins.
Germination of the bmp1 mutant conidia was somewhat retarded when compared to the wild type (Fig. 2). In addition, germ tubes continue to elongate on hydrophobic surfaces and never differentiate appressoria, indicating that they are defective in signaling leading to appressorium formation (Fig. 2B) (27, 30). By comparing gene expression in wild-type and bmp1 mutant germlings, we aimed to identify genes that are involved in the appressorium differentiation process. For this, the expression profile of the bmp1 mutant after 3.5 h was compared to that of the wild type after 2.5 h, representing stages with similar degrees of germling development (Fig. 2). A remarkably high proportion of wild-type germination-specific genes (32.6% on average) were not upregulated in the bmp1 mutant (Table 1; see also Table S3 in the supplemental material). In contrast, all three constitutively expressed groups contained a very low proportion (4.0%) of BMP1-regulated genes, i.e., genes that show at least 2-fold-lower expression in the bmp1 mutant than in the wild type, in the same or a comparable stage of development. In the germination-specific groups, the induced expression of 71.4% of the genes encoding secreted proteins was found to be dependent on the presence of BMP1. A similar picture was observed for the germination-induced groups, Max1-15 and Max2.5-15, which also showed a high proportion of BMP1-regulated genes, in particular those encoding secreted proteins (Table 1). Although these comparisons are based on a single time point of development for the bmp1 mutant, they suggest that the BMP1 MAP kinase plays an important role in early gene activation in germinating conidia, especially genes encoding secreted proteins.
Table 1.
Expression of genes encoding secreted proteins in the wild type versus the bmp1 mutant
| Category | Total | No. of genes encoding secreted proteins (% of total wild type) | Total no. of bmp1 low genesa (% of total wild type) | No. of bmp1 low genesa encoding secreted proteins (% of wild type) |
|---|---|---|---|---|
| Max1-2.5 | 13 | 5 (38.5) | 7 (53.8) | 4 (80.0) |
| Max2.5 + Max2.5-4 | 5 | 2 (40.0) | 3 (60.0) | 2 (100.0) |
| Max1-4 | 117 | 21 (17.9) | 34 (29.1) | 14 (66.7)*** |
| Total germination specificb | 135 | 28 (20.7) | 44 (32.6) | 20 (71.4)*** |
| Max1-15 | 471 | 108 (22.9) | 102 (21.7) | 43 (39.8)*** |
| Max2.5-15 | 64 | 17 (26.6) | 39 (60.9) | 14 (82.4) |
| Total germination inducedc | 670 | 153 (22.8) | 185 (27.6) | 77 (50.3)*** |
| Max0 | 552 | 57 (10.3) | 27 (4.9) | 0 (0.0) |
| Max15 | 118 | 20 (16.9) | 12 (10.2) | 2 (10.0) |
| CON | 4,949 | 361 (7.3) | 199 (4.0) | 19 (5.3) |
Genes showing at least 2-fold-lower expression (P < 0.05) in the bmp1 mutant (3.5 h) compared to the wild type (2.5 h). Significant differences (P < 0.001; Student's t test) between observed and expected frequencies are indicated (***).
Comprising the Max1-2.5, Max2.5, Max2.5-4, and Max1-4 groups.
Comprising all germination-specific groups and groups Max1-15 and Max2.5-15.
Confirmation of microarray data by qRT-PCR.
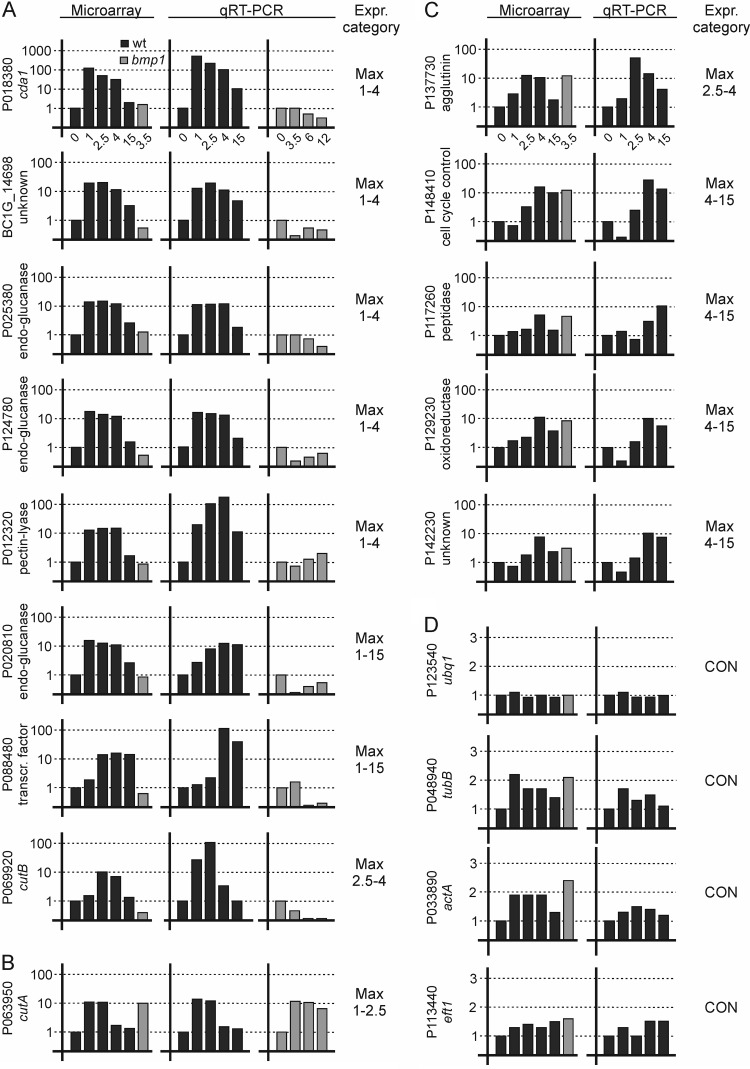
Quantitative reverse transcription-PCR (qRT-PCR) was used to confirm the results of the microarray hybridizations and to obtain more detailed expression profiles for selected genes. For this, we first tested ubq1 (ubiquitin), actA (actin), tubB (β-tubulin), eft1 (translation elongation factor 1α) and rps24 (ribosomal protein S24) for their suitability as constitutively expressed reference genes during the germination process. For most of these genes, both microarray and qRT-PCR data indicated a somewhat lower expression in resting spores (0 h) than in later stages of development (Fig. 7D; see also Table S4 in the supplemental material). An exception was ubq1, which was found to be most stably expressed. We therefore used ubq1 and actA for normalization in the qRT-PCR analyses. In Fig. 7A to C and in Table S4 in the supplemental material, the time course of transcript levels is shown for 14 genes, which were selected because of their distinct, germination-induced expression profiles based on microarray data. For these genes, the microarray data were largely confirmed by qRT-PCR. Ten of these genes encode proteins with predicted signal peptides for secretion. Eight genes that showed weak expression in the bmp1 mutant after 3.5 h in the microarray experiment were confirmed to be downregulated in the bmp1 mutant at all time points analyzed (Fig. 7A).
Fig 7.
Time course of expression of selected genes in the wild type (black bars) and in the bmp1 mutant (gray bars). (A) Germination-induced genes, downregulated during germination in the bmp1 mutant. (B) Germination-induced cutA gene, similar expression in the wild type and the bmp1 mutant. (C) Late germination induced genes, similar expression in the wild type and the bmp1 mutant. (D) Constitutive (CON-H) genes.
Expression analysis of cda1, a germination-specific gene.
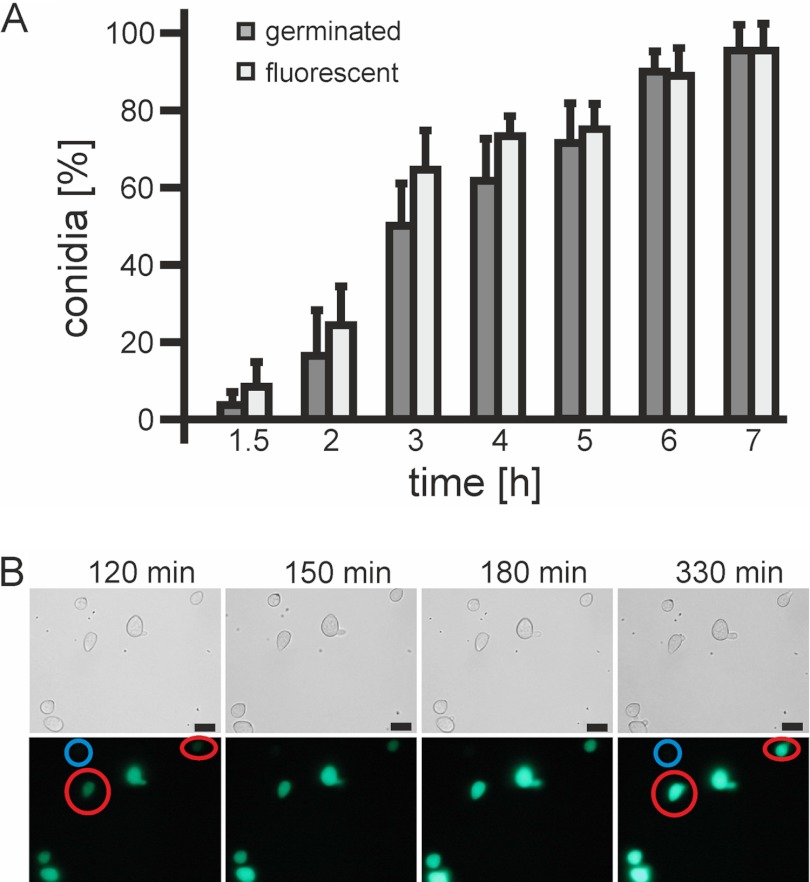
cda1 was identified as the most rapidly and strongly induced gene during germination (Fig. 8; see also Table S4 in the supplemental material). The predicted Cda1 protein contains 641 amino acids, including a signal peptide, and shows 49% identity (61% similarity) to the chitin deacetylase of Colletotrichum lindemuthianum, ClCDA, a member of the carbohydrate esterase 4 superfamily (47). Expression levels of cda1 in the wild type were maximal after 1 h and thereafter decreased to still high levels at 2.5 and 4 h and to much lower levels at 15 h. In contrast, cda1 expression was very low in the bmp1 mutant (Fig. 8; see also Table S4). To monitor the induction of cda1 in vivo, a cda1-GFP promoter fusion was constructed and transformed into B. cinerea strain B05.Hyg-3 (34). Expression of GFP in conidia was strictly correlated with germination (Fig. 8A). Starting from weak background levels, a strong increase in fluorescence occurred before germ tube outgrowth in all spores (Fig. 8B). Induction of cda1 was independent of the germination conditions and occurred in a similar manner on hard surfaces and in suspensions, in either minimal or full media (data not shown).
Fig 8.
Germination-specific expression of cda1 revealed by a B. cinerea cda1-GFP promoter reporter strain. Conidia were germinated on a polypropylene surface in minimal medium with 10 mM fructose. (A) Time course of germination and appearance of GFP fluorescence. The means of three experiments are shown, with standard deviations (n > 150). (B) Time-lapse microscopy of germinating conidia during a 3.5-h observation period. Conidia that started to show fluorescence before germ tube outgrowth are circled in red. A nonfluorescent spore that did not germinate during the observation period is circled in blue.
A cutinase-encoding gene (cutB) requires plant lipids for germination-specific expression.
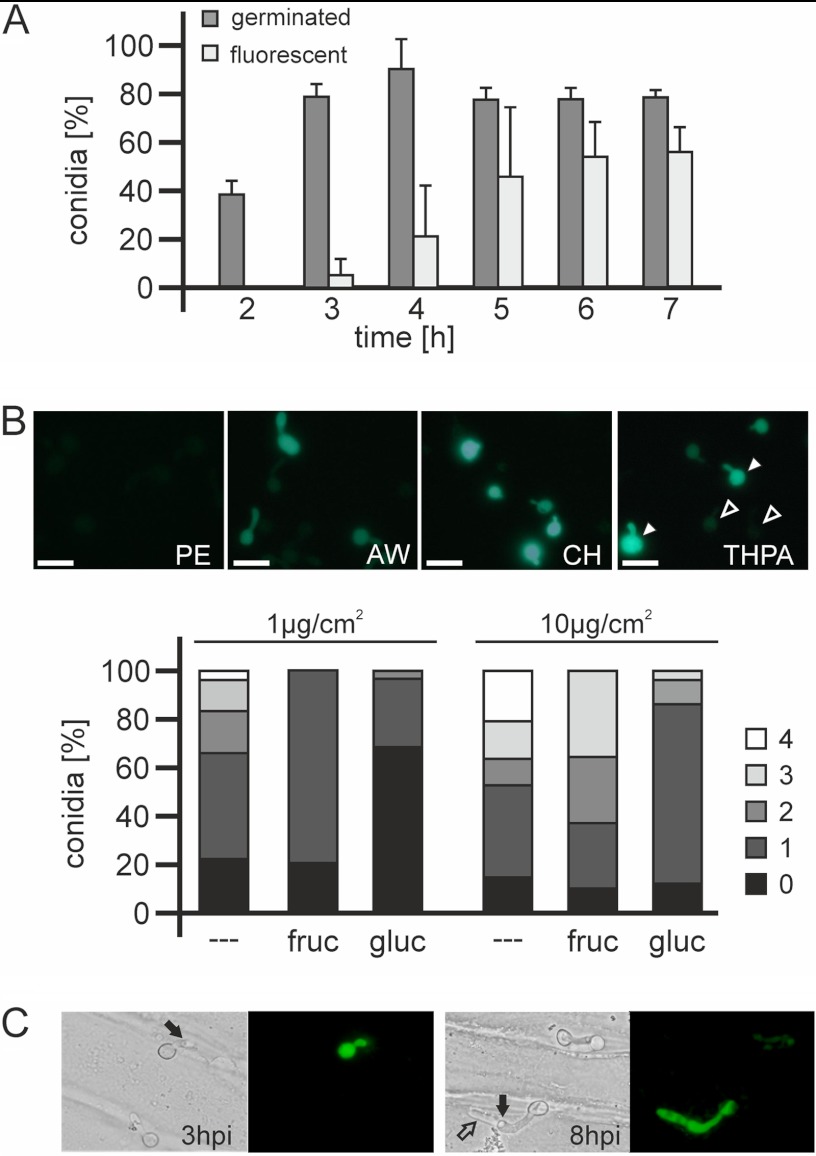
Based on microarray expression data, several genes encoding putative cutinases were found to be induced during germination (Fig. 9; see also Table S4 in the supplemental material; other data not shown). Previously, cutA has been described to be induced early after germination on the host plant surface (48). Based on the microarray data, cutA showed maximal expression between 1 h and 2.5 h, while expression of another cutinase gene, called cutB, showed a peak of expression between 2.5 h and 4 h (Fig. 7; see also Table S4). cutB encodes a 206-amino-acid protein with 34% identity to Fusarium solani Cut1 and containing a predicted 18-amino-acid leader peptide. Expression of cutB was analyzed using a B. cinerea cutB-GFP promoter reporter strain. GFP fluorescence was very weak in conidia germinated on artificial surfaces such as polypropylene (Fig. 9B). In contrast, induction of cutB was observed on surfaces covered with plant lipids. While moderate fluorescence appeared in germlings on glass surfaces coated with apple wax, strong fluorescence could be observed if the surfaces were coated with cutin hydrolysate or the cutin monomer THPA (Fig. 9B). Interestingly, individual conidia responded differently to the inducing surfaces. While some spores showed bright fluorescence after 3.5 h, other spores in a similar developmental stage remained weakly fluorescent or nonfluorescent, or fluorescence started to increase later (Fig. 9A and B; other data not shown). This behavior was independent of the concentrations of inducing plant lipids applied to the glass slides (data not shown), indicating that it was due to an intrinsic cell-to-cell variability of the cutB promoter. In the presence of sugars, cutB expression was reduced, but not abolished, by carbon catabolite repression. Fructose was observed to have a weaker suppressive effect than that of glucose, and at higher concentrations of the inducer THPA, repression by the sugars was less effective (Fig. 9B). To verify the physiological relevance of cutB induction, cutB-GFP conidia were germinated on heat-killed onion epidermal cell layers. Many germlings showed bright GFP fluorescence covering the spore, the germ tube, and the infection hyphae, indicating that cutB is similarly induced on the host cuticle (Fig. 9C). Again, cutB expression was not uniformly distributed, because some of the germlings failed to induce the GFP reporter, although penetration occurred.
Fig 9.
Analysis of cutB expression using a B. cinerea cutB-GFP promoter reporter strain. (A) Correlation of germination (dark gray) and increased GFP expression (light gray) of conidia incubated in the presence of 10 mM fructose and THPA. Values are the means of three experiments, with standard deviations (n = 90). (B) The top shows induction of GFP fluorescence in the presence of 10 mM fructose on polyethylene foil (PE) or on glass slides coated with 10 μg/cm2 of either apple wax (AW), cutin hydrolysate (CH), or THPA (3.5 h). On the THPA-coated surface, two germlings each showing strong (solid arrowheads) and very low (open arrowheads) fluorescence are highlighted. The lower part shows partial suppression of cutB promoter-driven GFP expression by 10 mM fructose (fruc) or glucose (gluc) (4 h). Fluorescence intensity levels are scored as described in Materials and Methods. Values for each condition are the means of two experiments (n = 50). (C) GFP fluorescence of conidia during germination and penetration on heat-killed onion epidermis cells. Solid arrows indicate penetration sites, and the open arrow indicates intracellular infection hyphae. Scale bars, 20 μm.
A putative secondary metabolite gene cluster showing coordinate regulation during germination.
Among the genes showing peak expression at 4 h postinoculation (hpi), we identified eight adjacent genes that could be involved in the synthesis of secondary metabolites (see Fig. S2 in the supplemental material). The putative cluster contained genes encoding a fatty acid elongase, two cytochrome 450 monooxygenases, a transferase, a thioesterase, a small secreted protein of unknown function, a major facilitator superfamily (MFS) transporter, and a transcription factor of the Zn2Cys6 zinc cluster family. When the expression of these genes was reexamined by qRT-PCR, we found three genes on one side of the cluster to be rather stably expressed during germination. In contrast, four of the five other genes, except for the transcription factor-encoding gene (called rum1, for regulator of unknown metabolite 1), were strongly downregulated after 1 h and 2.5 h, followed by a steep increase after 4 h. In contrast to the microarray data, qRT-PCR did not reveal a clear decrease in expression of these genes between 4 and 15 h (data not shown). In order to clarify the role of rum1 in the regulation of the gene cluster, the gene was deleted in B. cinerea strain B05.10. In the rum1 mutant, expression of most the cluster genes was moderately reduced in conidia (0 h) compared to that in the wild type, while a strong downregulation was observed for the four adjacent genes only (see Fig. S2).These data narrow down the gene cluster to five genes, including rum1. Rum1 seems to be required for maintaining high levels of expression of the cluster genes. A preliminary phenotypic analysis of the rum1 mutant did not reveal any differences concerning germination, appressorium formation, or pathogenicity compared to the B05.10 wild-type strain (data not shown). The role of the hypothetical secondary metabolite cluster therefore remains unclear.
DISCUSSION
A transcriptome analysis of the germination process of Botrytis cinerea conidia has been performed under conditions that mimic the situation on the host plant cuticle. On apple wax-coated surfaces and a minimal medium containing fructose, rapid and rather synchronous germination of conidia within 3 h was achieved. The majority of germinated conidia stopped germ tube elongation and formed appressorium-like terminal thickenings within 4 to 5 h, indicating the initiation of penetration. Germination on apple wax was more rapid than on artificial hydrophobic surfaces. The germination-accelerating activity of apple wax might be due to an additional chemical stimulus. Chemical components of host cuticles have been found to affect germination and differentiation of several plant-pathogenic fungi (7, 13, 14, 15). In the powdery mildew Blumeria graminis f. sp. hordei, conidial germination and differentiation were found to be triggered by components of the leaf surface wax, in particular long-chain aldehydes (11).
In many fungi, spore germination has been observed to be preceded by swelling (2). Using video microscopy, we observed that pregermination swelling of B. cinerea conidia occurred only in the presence of carbohydrate nutrients, while in the absence of nutrients, hydrophobic-surface-induced germination was not accompanied by swelling of the conidia (Fig. 1). This indicates a profound difference in the developmental programs leading to germination either in the presence or in the absence of exogenous nutrients. The major difference in signaling is the predominant role of cAMP for carbon source-induced germination and the essential role of the BMP1 MAP kinase cascade for hydrophobicity-induced germination (27, 28, 30). Carbon source-induced germination, but not hydrophobicity-induced germination in the absence of nutrients, is accompanied by trehalose degradation (28). We therefore speculate that the stronger mobilization of nutrient reserves might be responsible for conidial swelling. In our study, germination was accompanied by an increase in the number of nuclei (Fig. 3). However, we did not observe a synchronization of germination or appressorium formation and nuclear division, as has been reported for M. oryzae or C. gloeosporioides (49, 50, 51). Previously, synchronous division of individual nuclei in germinating B. cinerea conidia has been observed, using a B. cinerea strain with GFP-expressing nuclei (33). An explanation for this discrepancy from our result is difficult, but it might be related to the different surfaces used, namely, untreated glass slides in the work of Kokkelink et al. (33) and apple wax-coated surfaces in our study, resulting in germination kinetics that differed by more than 1 h (Fig. 2).
For the transcriptome studies, resting conidia (0 h) and conidia incubated for 1, 2.5, 4, and 15 h on apple wax-coated surfaces in the presence of 10 mM fructose were used. Before RNA isolation, conidia were washed, and the germlings were scraped off the plates and washed once in ice-cold water. While we cannot exclude that these procedures led to changes in the mRNA composition, the differential expression data obtained were found to be quite robust and did not seem to be due to artifacts. The Nimblegene array represented 15,738 gene models of the two sequenced B. cinerea strains, T4 and B0510, that were supported by the size of the open reading frames, the presence of functional or topological domains, or expression data. After normalization and threshold subtraction, 12,092 (76.8%) genes were classified as being expressed during at least one of the five time points analyzed. However, this number is only a rough estimation, because it was difficult to define the threshold that distinguished expression signals from background noise. The threshold of >29 was selected because it was beyond the 99th percentile values of the random probes for all time points except one. Extensive changes of expression, equaling 23.7% of the expressed genes, were observed during the first hour of incubation, prior to germ tube outgrowth. In contrast, during the time period when most of the morphological changes occurred, between 1 h and 4 h, much lower changes of gene expression were observed (Fig. 4). In F. graminearum, pregermination changes in gene expression (from 0 to 2 h) have been reported to be even larger, with 3,421 (66.5%) of 5,146 expressed genes, also exceeding gene expression changes during later development (19). In M. oryzae conidia, 2,154 (approximately 21%) of all genes were found to be up- or downregulated after complete germination under non-appressorium-inducing conditions after 7 h (22). Although these data are not completely comparable to each other, they confirm data in the literature on a rapid mRNA turnover in germinating conidia (2, 42). In B. cinerea and other fungi, the first biochemical evidence for the breaking of dormancy and initiation of the germination program is the breakdown of trehalose, detectable after 30 min and almost completed after 1 h in the presence of fructose on a glass surface (28). It remains to be shown when transcriptional changes start to occur after perception of the germination stimuli. From the expressed genes, 11 major groups of coregulated genes were extracted. As expected, the majority of the genes (4,949, or 40.9% of the expressed genes) belonged to the three constitutive groups (CON-L, CON-M, and CON-H) (Fig. 5). Furthermore, many genes showed preferential expression in the dormant stage (Max0; 552 genes) and in the mycelium stage (Max15; 471 genes). As already indicated by the large changes observed between 0 and 1 h, a considerable number of germination-induced genes with specific expression patterns (including Max1, Max1-4, Max2.5-15, and Max1-15; all together, 692) were identified. Overall, the gene expression patterns determined by the microarrays were confirmed by qRT-PCR analysis, although some quantitative differences were observed between the two methods (Fig. 7; see also Table S4 in the supplemental material).
The groups revealed interesting differences in the distribution of functional categories. By far the lowest proportion of genes which could be assigned to functional categories (31.8%) were found in the CON-L group. This illustrates a particular lack of knowledge about the function of fungal genes that are weakly expressed during all growth stages. In contrast, the high proportion of genes with functional evidence (66.9%) in the CON-H group was correlated with a prevalence of well-known genes involved in major functions of primary metabolism, including cell cycle and DNA processing, transcription, protein synthesis and processing, and respiration. Consistent with the analysis in N. crassa and F. graminearum, functional categories of cell cycle, DNA processing, and protein synthesis were also found to be overrepresented in B. cinerea conidia among the genes induced between 0 and 1 h (17, 19). Genes induced 1 h after germination could be functionally categorized to more than 60% (0-1_up), or even more than 70% (Max1, Max1-4, and Max1-15). Furthermore, all of these categories were enriched in genes encoding secreted proteins (20.7% of the total number of expressed genes), whereas a lower frequency of genes encoding secreted proteins were observed to be expressed in dormant spores (10.3%). This peak of secretory activity during early germination has not been reported for N. crassa or Fusarium oxysporum (17, 19). In M. oryzae, genes encoding secreted proteins have been found to be induced mainly during appressorium formation. Some of these proteins, such as the cutinase Cut2, the hydrophobin MPG1, and the SnodprotI homolog MSPI, have been shown to be involved in virulence (22).
A B. cinerea early secretome, derived from conidia germinated for 16 h in a minimal medium enriched with low-molecular-weight host plant compounds, has been published recently (52). Of the 105 secreted proteins identified, 14 are encoded by the germination-specific and 21 by the germination-induced genes identified in our study. In addition, 35 of the proteins corresponded to constitutively expressed genes. Overall, these data document a significant overlap of the proteomic and our transcriptomic data. Five proteins in the secretome have been described as virulence factors, namely, the endopolygalacturonases PG1 and PG2 (53, 54), the pectin methylesterase PME1 (55, 56), the endo-β-1,4-xylanase Xyn11A (57), and the cerato-platanin family protein SPL1 (58). They were found in the Max1-15 (PG1, PG2, and PME1), Max1 (Xyn11A), and Max15 (SPL1) groups. Eleven of the secreted proteins, including PG1 and PG2, were encoded by genes that are downregulated in the bmp1 mutant, and 9 of them belonged to the germination-induced gene groups. The bmp1 mutant also showed a clear downregulation of the PG3-encoding gene (P094200) which is expressed Max1-15 in the wild type. While PG3 has not been described as a virulence factor, previous expression data revealed a massive induction of PG3 in infected apples (59).
In M. oryzae, 357 genes (22), or even 1,026 genes (23), have been found to be differentially expressed during appressorium initiation or maturation. This large number can be explained by the fact that M. oryzae appressoria are morphologically highly distinct and specialized structures. In contrast, B. cinerea appressoria are morphologically much less differentiated and usually not separated from the germ tube by a septum. Comparison of the genes induced during germination and appressorium formation between B. cinerea and M. oryzae did not reveal clear similarities (23). For example, of the 31 genes involved in acetyl coenzyme A (acetyl-CoA) utilization or production, 13 were found in M. oryzae to be upregulated and six downregulated during appressorium formation. In contrast, only three of these genes were germination-specifically expressed in B. cinerea, and only one of them was an ortholog of an appressorium-specific gene in M. oryzae. Similarly, 17 out of 22 genes involved in fatty acid oxidation and glyoxylate cycle were specifically induced in appressoria of M. oryzae, but none of their orthologs were induced during B. cinerea germination. This suggests that appressorium formation in M. oryzae differs from that in B. cinerea significantly also at the molecular level. It also indicates that fatty acid mobilization and the glyoxylate cycle, which are essential for providing the high concentrations of glycerol in the M. oryzae appressoria that are required for high turgor pressure-mediated penetration, do not play a similarly important role in B. cinerea. Nevertheless, a common feature of both fungi was the high frequency of genes encoding secreted proteins, including CAZymes, which were found to be upregulated both in M. oryzae appressoria and in B. cinerea germlings.
Comparison of gene expression in germlings of the wild type and bmp1 mutant revealed a high proportion of genes that were induced during germination in the wild type and that appeared to be dependent for their induction on the presence of the MAP kinase BMP1 (Table 1). Even more strikingly, 50.3% of the germination-induced genes encoding secreted proteins were downregulated in the bmp1 mutant. In contrast, only 5.3% of CON genes encoding secreted proteins were BMP1 regulated. However, these data are based on microarray analysis of only a single time point (3.5 h) for the bmp1 mutant. Nevertheless, for eight genes, dependence on BMP1 was confirmed by a qRT-PCR time course analysis with the wild-type and the bmp1 mutant (Fig. 7A; see also Table S2 in the supplemental material). These results indicate a significantly lower induction of secretory activity during germination in the bmp1 mutant. In M. oryzae, the BMP1-orthologous MAP kinase PMK1 is also essential for appressorium differentiation and infection. Transcriptome studies have revealed a total of 481 genes that are PMK1 regulated. Of these genes, 238 and 174 are induced during early and late appressorium development, respectively (23). However, only 13 orthologs of these genes were found to be downregulated in the B. cinerea bmp1 mutant, and only 5 of these were upregulated during germination and appressorium differentiation. In the M. oryzae genome, a total of 206 genes encode CAZymes. Seventy-two (35%) of these genes were significantly upregulated in appressoria, but only 9 (12.5%) of these genes were downregulated in the pmk1 mutant. Of the 316 genes encoding CAZymes in B. cinerea, 48 (15%) were upregulated during germination, and 30 (62.5%) of these were downregulated in the bmp1 mutant. Overall, this comparison revealed a rather small overlap between the two fungi of the genes that are upregulated during early stages of infection, and differences in their regulation by the BMP1/PMK1 MAP kinase pathway. The most rapidly and strongly induced gene during germination, cda1, showed more than 700-fold-increased expression within the first hour of spore incubation. Analysis of a cda1-gfp promoter reporter strain confirmed that the appearance of GFP fluorescence was strictly correlated with germination, reaching strong levels before germ tube appearance. Germination-specific expression of cda1 was found to be independent of the germination conditions and strongly reduced in the bmp1 mutant. cda1 is a member of a gene family encoding secreted proteins similar to chitin deacetylases and chitin binding proteins in other fungi (47). In the rust fungus Uromyces fabae, differentiation-specific induction of chitin deacetylase activity has been speculated to be involved in protecting the fungal hyphae from degradation by plant chitinases (60). Functional analysis of cda1 is difficult, because it is a member of a family of three cda genes, including cda1, cda2 (P079220/BC1G_03291), and cda3 (P005177/BC1G_06509), which possibly have redundant functions. While cda2 shows a rather stable expression during development, both cda1 and cda3 showed maximum expression at 1 to 4 h (see Table S2 in the supplemental material; other data not shown).
Because germination was induced on apple wax-coated surfaces, cutinase genes that require the presence of host plant lipids were observed in the microarrays among the germination-specific genes. cutA has previously been shown to be induced in B. cinerea mycelium in the presence of cutin monomers and repressed by glucose (48, 61, 62). We found that cutA and cutB were induced during germination with different kinetics (Fig. 7). With a cutB promoter reporter strain, GFP fluorescence could be observed in germinating spores only on surfaces covered with plant lipids such as apple wax or cutin monomers. Similar to cutA, cutB was subject to catabolite repression. The degree of repression by glucose was stronger than that by fructose and was reduced with increasing concentrations of the inducing lipid (THPA) (Fig. 9B). In contrast to the highly synchronized expression observed for cda1, expression of cutB was found to be variable in different germlings. The delayed induction of cutB in some germlings, sometimes even after penetration, indicates that perception of the inducing plant lipids occurs with variable efficiency and kinetics, which is in striking contrast to the strictly controlled induction of cda1 during early germination. In addition to cutA and cutB, the B. cinerea genome encodes several more cutinase genes which show different patterns of expression (reference 48 and unpublished data). The redundancy of cutinases makes it difficult to assess the role of their activity for host cell wall penetration by the analysis of mutants (61).
A limitation of our approach to search for genes that are induced during germination is the fact that it does not identify genes with different expression that might be crucial for the germination process, including regulatory genes. Nevertheless, we identified a BMP1-regulated transcription factor gene (P088480; BC1G_06861) with germination-induced expression (Max2.5-15) (see Table S2 in the supplemental material). Targeted knockout mutagenesis, however, did not reveal any obvious phenotype concerning germination, penetration, or infection (data not shown). Another candidate for functional analysis was the zinc cluster transcription factor rum1, which is part of a coregulated cluster of five genes that appears to be involved in secondary metabolite synthesis (see Fig. S2). Next to this cluster, three more genes are located that might perform a similar function but were only weakly regulated by Rum1. B. cinerea produces two types of secondary metabolites which have been reported to be virulence factors, namely, sesquiterpenoids, including botrydial, and polyketides, including botcinic acid (63, 64). Although the rum1 deletion mutant showed a strongly reduced expression of the cluster genes, it did not reveal any defects concerning germination, penetration, or infectivity. A metabolite profiling of the rum1 mutant might provide an idea about the role of this gene cluster.
In summary, our data demonstrate that early conidial development in B. cinerea is accompanied by rapid shifts in gene expression that likely orchestrate germ tube outgrowth, appressorium differentiation, and host cell invasion. For some of the germination-induced genes, a role in infection has already been demonstrated, while for the other genes, this remains to be analyzed. Besides their functional characterization, it will be a challenge to achieve a more comprehensive description of the regulatory processes that are involved in germination and early infection of B. cinerea.
ACKNOWLEDGMENTS
We thank Marc-Henri Lebrun, J. Amselem, and A. Simon for making the Botrytis whole-genome arrays accessible to us. Furthermore, we are grateful to Nathalie Müller for help with characterization of rum1 deletion strains.
This study was supported by the German Science Foundation (DFG project travel grant for E. Silva (HA1486/9-1), DFG project HA1486/5-1, the Conicyt CCTE/PBF16 Program, FONDECYT 3120001 (postdoctoral fellowship), and Project UNAB (DI-23-12/R).
Footnotes
Published ahead of print 15 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00295-12.
REFERENCES
- 1. Barhoom S, Sharon A. 2004. cAMP regulation of pathogenic and saprophytic fungal spore germination. Fungal Genet. Biol. 41:317–326 [DOI] [PubMed] [Google Scholar]
- 2. d'Enfert C. 1997. Fungal spore germination: insights from the molecular genetics of Aspergillus nidulans and Neurospora crassa. Fungal Genet. Biol. 21:163–172 [Google Scholar]
- 3. Fillinger S, Chaveroche MK, Shimizu K, Keller N, d'Enfert C. 2002. cAMP and ras signaling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001–1016 [DOI] [PubMed] [Google Scholar]
- 4. Lafon A, Han KH, Seo JA, Yu JH, d'Enfert C. 2006. G protein and cAMP-mediated signaling in aspergilli: a genomic perspective. Fungal Genet. Biol. 43:490–502 [DOI] [PubMed] [Google Scholar]
- 5. Hatanaka M, Shimoda C. 2001. The cyclic AMP/PKA signal pathway is required for initiation of spore germination in Schizosaccharomyces pombe. Yeast 18:207–217 [DOI] [PubMed] [Google Scholar]
- 6. Maidan MM, Thevelein JM, van Dijck P. 2005. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem. Soc. Trans. 33:291–293 [DOI] [PubMed] [Google Scholar]
- 7. Carver TLW, Ingerson SM. 1987. Responses of Erysiphe graminis germlings to contact with artificial and host surfaces. Physiol. Mol. Plant Pathol. 30:359–372 [Google Scholar]
- 8. Kuo K, Hoch HC. 1996. Germination of Phyllosticta ampelicida pycnidiospores: prerequisite of adhesion to the substratum and the relationship of substratum wettability. Fungal Genet. Biol. 20:18–29 [DOI] [PubMed] [Google Scholar]
- 9. Chaky J, Anderson K, Moss M, Vaillancourt L. 2001. Surface hydrophobicity and surface rigidity induce spore germination in Colletotrichum graminicola. Phytopathology 91:558–564 [DOI] [PubMed] [Google Scholar]
- 10. Nesher I, Minz A, Kokkelink L, Tudzynski P, Sharon A. 2011. Regulation of pathogenic spore germination by CgRac1 in the fungal plant pathogen Colletotrichum gloeosporioides. Eukaryot. Cell 10:1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hansjakob A, Bischof S, Bringmann G, Riederer M, Hildebrandt U. 2010. Very-long-chain aldehydes promote in vitro prepenetration processes of Blumeria graminis in a dose- and chain length-dependent manner. New Phytol. 188:1039–1054 [DOI] [PubMed] [Google Scholar]
- 12. Mendoza-Mendoza A, Berndt P, Djamei A, Weise C, Linne U, Maraheil M, Vranes M, Kämper J, Kahmann R. 2009. Physical-chemical plant-derived signals induce differentiation in Ustilago maydis. Mol. Microbiol. 71:895–911 [DOI] [PubMed] [Google Scholar]
- 13. Podila GK, Rogers LM, Kolattukudy PE. 1993. Chemical signals from avocado surface wax trigger germination and appressorium formation in Colletotrichum gloeosporioides. Plant Physiol. 103:267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reisige K, Gorzelanny C, Daniels U, Moerschbacher BM. 2006. The C28 aldehyde octacosanal is a morphogenetically active component involved in host plant recognition and infection structure differentiation in the wheat stem rust fungus. Physiol. Mol. Plant Pathol. 68:33–40 [Google Scholar]
- 15. Tsuba M, Katagiri C, Takeuchi Y, Taada Y, Zamoka N. 2002. Chemical factors of the leaf surface involved in morphogenesis of Blumeria graminis. Physiol. Mol. Plant Pathol. 60:51–57 [Google Scholar]
- 16. Mendgen K, Hahn M, Deising H. 1996. Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annu. Rev. Phytopathol. 34:367–386 [DOI] [PubMed] [Google Scholar]
- 17. Kasuga T, Townsend JP, Tian C, Gilbert LB, Mannhaupt G, Taylor JW, Glass NL. 2005. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 33:6469–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salem-Izacc SM, Koide T, Vencio RZ, Gomes SL. 2009. Global gene expression analysis during germination in the chytridiomycete Blastocladiella emersonii. Eukaryot. Cell 8:170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seong KY, Zhao X, Xu JR, Güldener U, Kistler HC. 2008. Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 45:389–399 [DOI] [PubMed] [Google Scholar]
- 20. Both M, Csukai M, Stumpf MP, Spanu PD. 2005. Gene expression profiles of Blumeria graminis indicate dynamic changes to primary metabolism during development of an obligate biotrophic pathogen. Plant Cell 17:2107–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Both M, Eckert SE, Csukai M, Müller E, Dimopoulos G, Spanu PD. 2005. Transcript profiles of Blumeria graminis development during infection reveal a cluster of genes that are potential virulence determinants. Mol. Plant Microbe Interact. 18:125–133 [DOI] [PubMed] [Google Scholar]
- 22. Oh Y, Donofrio N, Pan H, Coughlan S, Brown DE, Meng S, Mitchell T, Dean RA. 2008. Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol. 9:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soanes DM, Chakrabarti A, Paszkiewicz KH, Talbot NJ. 2012. Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 8( 2): e1002514 doi:10.1371/journal.ppat.1002514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williamson B, Tudzynski B, Tudzynski P, van Kan JAL. 2007. Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8:561–580 [DOI] [PubMed] [Google Scholar]
- 25. Holz G, Coertze S, Williamson B. 2004. The ecology of Botrytis on plant surfaces, p 9–27 In Elad Y, Williamson B, Tudzynski P, Delen N. (ed), Botrytis: biology, pathology and control. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 26. van Kan JAL. 2006. Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11:249. [DOI] [PubMed] [Google Scholar]
- 27. Doehlemann G, Berndt P, Hahn M. 2006. Different signalling pathways involving a Gα protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59:821–835 [DOI] [PubMed] [Google Scholar]
- 28. Doehlemann G, Berndt P, Hahn M. 2006. Trehalose metabolism is important for heat stress tolerance and spore germination of Botrytis cinerea. Microbiology 152:2625–2634 [DOI] [PubMed] [Google Scholar]
- 29. Schumacher J, Kokkelink L, Huesmann C, Jimenez-Teja D, Collado IG, Barakat R, Tudzynski P, Tudzynski B. 2008. The cAMP-dependent signaling pathway and its role in conidial germination, growth, and virulence of the gray mold Botrytis cinerea. Mol. Plant Microbe Interact. 11:1443–1459 [DOI] [PubMed] [Google Scholar]
- 30. Schamber A, Leroch M, Diwo J, Mendgen K, Hahn M. 2010. The role of MAP kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea. Mol. Plant Pathol. 11:105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. D'Souza CA, Heitman J. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349–364 [DOI] [PubMed] [Google Scholar]
- 32. Zhao X, Mehrabi R, Xu JR. 2007. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 6:1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kokkelink L, Minz A, Al-Masri M, Giesbert S, Barakat R, Sharon A, Tudzynski P. 2011. The small GTPase BcCdc42 affects nuclear division, germination and virulence of the gray mold fungus Botrytis cinerea. Fungal Genet. Biol. 48:1012–1019 [DOI] [PubMed] [Google Scholar]
- 34. Noda J, Brito N, Espino JJ, Gonzalez C. 2007. Methodological improvements in the expression of foreign genes and in gene replacement in the phytopathogenic fungus Botrytis cinerea. Mol. Plant Pathol. 8:811–816 [DOI] [PubMed] [Google Scholar]
- 35. Walton TJ, Kolattukudy PE. 1972. Determination of the structure of cutin monomers by a novel depolymerization procedure and combined gas chromatography and mass spectrometry. Biochemistry 11:1886–1896 [DOI] [PubMed] [Google Scholar]
- 36. Leroch M, Mernke D, Koppenhoefer D, Schneider P, Mosbach A, Doehlemann G, Hahn M. 2011. Living colors in the gray mold pathogen Botrytis cinerea: codon-optimized genes encoding green fluorescent protein and mCherry, which exhibit bright fluorescence. Appl. Environ. Microbiol. 77:2887–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 38. Silva E, Valdés J, Holmes D, Valenzuela PDT. 2006. Generation and analysis of expressed sequence tags from Botrytis cinerea. Biol. Res. 39:367–376 [DOI] [PubMed] [Google Scholar]
- 39. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 40. Amselem J, Cuomo CA, van Kan JA, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier JM, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun MH, Dickman M. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7( 8): e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Güldener U, Mannhaupt G, Münsterkötter M, Mewes HW. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32:5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Osherov N, May GS. 2001. The molecular mechanisms of conidial germination. FEMS Microbiol. Lett. 199:153–160 [DOI] [PubMed] [Google Scholar]
- 43. Bergen LG, Morris NR. 1983. Kinetics of the nuclear division cycle of Aspergillus nidulans. J. Bacteriol. 156:155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dayton JS, Means AR. 1996. Ca(2+)/calmodulin-dependent kinase is essential for both growth and nuclear division in Aspergillus nidulans. Mol. Biol. Cell 7:1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harris SD, Morrell JL, Hamer JE. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmidt JC, Brody S. 1976. Biochemical genetics of Neurospora crassa conidial germination. Bacteriol. Rev. 40:1–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blair DE, Hekmat O, Schüttelkopf AW, Shresta B, Tokuyasu K, Withers SG, van Aalten DM. 2006. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry 45:9416–9426 [DOI] [PubMed] [Google Scholar]
- 48. van Kan JAL, van't Klooster JW, Wagemakers CA, Dees DC, van der Vlugt-Bergmanns CJ. 1997. Cutinase A of Botrytis cinerea is expressed, but not essential, during penetration of gerbera and tomato. Mol. Plant Microbe Interact. 10:30–38 [DOI] [PubMed] [Google Scholar]
- 49. Nesher I, Barhoom S, Sharon A. 2008. Cell cycle and cell death are not necessary for appressorium formation and plant infection in the fungal plant pathogen Colletotrichum gloeosporioides. BMC Biol. 6:9 doi:10.1186/1741-7007-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saunders DGO, Dagdas YF, Talbot NJ. 2010. Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Plant Cell 22:2417–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. 2006. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312:580–583 [DOI] [PubMed] [Google Scholar]
- 52. Espino JJ, Gutierrez-Sanchez G, Brito N, Shah P, Orlando R, Gonzalez C. 2010. The Botrytis cinerea early secretome. Proteomics 10:3020–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. ten Have A, Mulder W, Visser J, van Kan JA. 1998. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact. 11:1009–1016 [DOI] [PubMed] [Google Scholar]
- 54. ten Have A, Breuil WO, Wubben JP, Visser J, van Kan JA. 2001. Botrytis cinerea endopolygalacturonase genes are differentially expressed in various plant tissues. Fungal Genet. Biol. 33:97–105 [DOI] [PubMed] [Google Scholar]
- 55. Kars I, Krooshof GH, Wagemakers L, Joosten R, Benen JA, van Kan JA. 2005. Necrotizing activity of five Botrytis cinerea endopolygalacuronases produced in Pichia pastoris. Plant J. 43:213–225 [DOI] [PubMed] [Google Scholar]
- 56. Valette-Collet O, Cimerman A, Reignault P, Levis C, Boccara M. 2003. Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol. Plant Microbe Interact. 16:360–367 [DOI] [PubMed] [Google Scholar]
- 57. Brito N, Espino JJ, Gonzalez C. 2006. The endo-β-1,4-xylanase Xyn11A is required for virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 19:25–32 [DOI] [PubMed] [Google Scholar]
- 58. Frías M, Gonzalez C, Brito N. 2011. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192:483–495 [DOI] [PubMed] [Google Scholar]
- 59. Wubben JP, Mulder W, ten Have A, van Kan JA, Visser J. 1999. Cloning and partial characterization of endopolygalacturonase genes from Botrytis cinerea. Appl. Environ. Microbiol. 65:1596–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deising H, Siegrist J. 1995. Chitin deacetylase activity of the rust Uromyces viciae-fabae is controlled by fungal morphogenesis. FEMS Microbiol. Lett. 127:207–212 [Google Scholar]
- 61. Reis H, Pfiffi S, Hahn M. 2005. Molecular and functional characterization of a secreted lipase from Botrytis cinerea. Mol. Plant Pathol. 6:257–267 [DOI] [PubMed] [Google Scholar]
- 62. van der Vlugt-Bergmans CJ, Wagemakers CA, van Kan JAL. 1997. Cloning and expression of the cutinase A gene of Botrytis cinerea. Mol. Plant Microbe Interact. 10:21–29 [DOI] [PubMed] [Google Scholar]
- 63. Collado IG, Sanchez AJ, Hanson JR. 2007. Fungal terpene metabolites: biosynthetic relationships and the control of the phytopathogenic fungus Botrytis cinerea. Nat. Prod. Rep. 24:674–686 [DOI] [PubMed] [Google Scholar]
- 64. Dalmais B, Schumacher J, Moraga J, Le Pêcheur P, Tudzynski B, Collado IG, Viaud M. 2011. The Botrytis cinerea phytotoxin botcininc acid requires polyketide synthases for production and has a redundant role in virulence with botrytidial. Mol. Plant Pathol. 12:564–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Espino JJ, Brito N, Noda J, Gonzalez C. 2005. Botrytis cinerea endo-β-1,4-glucanase Cel5A is expressed during infection but is not required for pathogenesis. Physiol. Mol. Plant Pathol. 66:213–221 [Google Scholar]
- 66. Master ER, Zheng Y, Storms R, Tsang A, Powlowski J. 2008. A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: recombinant expression, purification and characterization. Biochem. J. 411:161–170 [DOI] [PubMed] [Google Scholar]