Abstract
Ure2p, normally a regulator of nitrogen catabolism in Saccharomyces cerevisiae, can be a prion (infectious protein) by forming a folded in-register parallel amyloid called [URE3]. Using S. cerevisiae as a test bed, we previously showed that Ure2p of Candida albicans (CaUre2p) can also form a prion, but that Ure2p of C. glabrata (CgUre2p) cannot. Here, we constructed C. glabrata strains to test whether CgUre2p can form a prion in its native environment. We find that while CaUre2p can form a [URE3] in C. glabrata, CgUre2p cannot, although the latter has a prion domain sequence more similar to that of ScUre2p than that of CaUre2p. This supports the notion that prion formation is not a conserved property of Ure2p but is a pathology arising sporadically. We find that some [URE3albicans] variants are restricted in their transmissibility to certain recipient strains. In addition, we show that the C. glabrata HO can induce switching of the C. glabrata mating type locus.
INTRODUCTION
Ure2p of the yeast Saccharomyces cerevisiae (ScUre2p) is involved in nitrogen catabolite repression, the ability to differentiate between the presence of good and poor nitrogen sources in the growth environment (1, 2). On rare occasions Ure2p, normally a soluble dimer, forms amyloid filaments, polymers of Ure2p. Once formed, Ure2p amyloid is maintained in the cell and is transferred from mother to daughter and between cells during incomplete matings (cytoductions) (3–6). Thus, Ure2p can misfold, resulting in the formation of a prion—an infectious protein—named [URE3]. Proteins with homology to S. cerevisiae Ure2p are found in many ascomycete yeasts, but few have been functionally characterized.
ScUre2p has an N-terminal Q/N-rich domain that is important for the stability of the protein in vivo (7), but it comprises the part that converts to amyloid in the prion form of the protein (5, 8). Because this N-terminal domain is sufficient for prion propagation or for infection in the complete absence of the remainder of the molecule (4, 6, 9), it is called the prion domain. The C-terminal domain is sufficient, if overexpressed, for the nitrogen regulation function and is homologous to glutathione-S-transferases. In the normal form, the Ure2p prion domain is unstructured (10), and in forming amyloid it becomes a folded in-register parallel β-sheet (5, 11).
Many of the Ure2p homologs in ascomycete yeasts have an asparagine/glutamine-rich N-terminal domain and a glutathione S-transferase-like C-terminal domain (12, 13). Recently, we showed that the Ure2p homologs of the human-pathogenic yeasts Candida albicans and Candida glabrata can functionally replace ScUre2p and in their native environment regulate a similar set of genes. In addition, we showed that CaUre2p can form a prion in S. cerevisiae with a similarly folded in-register parallel β-sheet architecture but that CgUre2p cannot (14, 15).
Because of the tools available for S. cerevisiae, few tests of non-S. cerevisiae prion candidates have been made in their native hosts. Sup35p is a subunit of the translation termination factor, and ScSup35p rarely can form a prion called [PSI+] (3). Sup35p of the ascomycete yeasts Pichia methanolica, Candida albicans, and Kluyveromyces lactis have been shown to be capable of prion formation, but in each case, S. cerevisiae was used as the host (16–19). Nakayashiki et al. have presented evidence that K. lactis Sup35p forms [PSI+] in K. lactis itself as well as in S. cerevisiae (19). In contrast, Ure2p of K. lactis does not form [URE3] in K. lactis (20) or in S. cerevisiae (13). S. paradoxus strains for the study of [URE3] have been developed (21), and in agreement with work in the S. cerevisiae host (12), S. paradoxus Ure2p can propagate [URE3] in its own environment (14). In the only three cases examined, results in S. cerevisiae are consistent with those in the native host, but the myriad of chaperones and other factors affecting prion propagation (reviewed in reference 22) suggest that there could be differences between the native host and S. cerevisiae.
Here, we constructed C. glabrata strains with the Ure2p-regulated DUR3 promoter controlling ADE2 and showed that CaUre2p can form a [URE3] prion in such cells, but that CgUre2p cannot, even though it is in its native environment. This supports our view that the prion-forming ability of Ure2p is not conserved but rather occurs sporadically among yeast species.
MATERIALS AND METHODS
C. glabrata strains for assaying Ure2p function.
We set up a Ure2p activity system in C. glabrata based on a similar system developed by Schlumpberger et al. (23). C. glabrata does not contain a DAL5 gene, which is often used in S. cerevisiae to study nitrogen regulation and to monitor the presence of the [URE3] prion. However, C. glabrata Ure2p (CgUre2p) represses CgDUR3 transcription (14), so the CgDUR3 promoter was fused to CgADE2 in C. glabrata to measure Ure2p activity. Yeasts were transformed by the Li acetate method (24), and media were as previously described (14).
The parental strain was BG88b [MATa ura3::G418 his3::ura3(5FOA)] (25). CgADE2 was replaced with FRT-HIS3-FRT, producing strain HCg8, and then CgHIS3 was deleted by recombination, making strain HCg12 [MATa ura3::G418 his3::ura3(5FOA) ade2::FRT]. Specifically, the CgADE2 5′ untranslated region (UTR) (nucleotides [nt] −510 to +20 relative to the start ATG of ADE2) was amplified from BG88b using primers HE776 and HE782. The CgADE2 3′ UTR (nt 1714 to 2152 relative to the start ATG; the ADE2 open reading frame [ORF] ends at nt 1713) was amplified from BG88b using primers HE780 and HE781. CgHIS3 flanked by FRT sites was amplified from strain 37A (26) using primers HE783 and HE779. CgHIS3 starts 514 bp upstream of the start codon and terminates at the stop codon. This HIS3 fragment is flanked by FRT sites (GAAGTTCCTATACTTTCTAGAGAATAGGAACTTC). Flanking this PCR product are nt 3 to 20 and nt 1714 to 1737 of ADE2 (ADE2 stops at 1713). The 3 PCR products were fused using oligonucleotides HE776 and HE781, and the resulting fusion product was transformed into BG88b, selecting for His+ colonies. His+ colonies unable to grow without adenine were identified.
CgHIS3 was deleted from the HCg8 ADE2 locus by expression of the Flp recombinase using plasmid pRD16 (27), forming strain HCg12. HCg12 could not grow on medium without adenine and formed red colonies on medium containing 30 mg/liter adenine. Thus, the ade2 mutant phenotype in C. glabrata is similar to that in S. cerevisiae, suggesting that CgADE2 is useful as a marker in C. glabrata to assay for Ure2p activity.
CgDUR3 (CAGL0K03157g), the homologue of S. cerevisiae YHL016c, is a plasma membrane transporter for both urea and polyamines (28, 29). DUR3 expression is derepressed more than 100-fold upon deletion of URE2 in C. glabrata. CgGAP1 (CAGL0L03267g), the homologue of S. cerevisiae (ScYKR039w) encoding the general amino acid permease, is the only other gene derepressed 100-fold, but CgGAP1 has a higher basal expression level than CgDUR3 (14).
HCg16.
For the strain HCg16 [MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT-HIS3-FRT] (the single colon refers to a promoter-ORF fusion) the CgDUR3 5′ UTR (nt −685 to −1 of DUR3) was amplified from BG88b with a 5′ NotI site and a 3′ PstI site using oligonucleotides HE784 and HE788. The CgADE2 ORF was amplified from BG88b with a 5′ PstI site and a 3′ HindIII site using oligonucleotides HE786 and HE787. CgHIS3 flanked by FRT sites and containing a 5′ HindIII site and a 3′ SalI site was amplified from strain 37A using oligonucleotides HE789 and HE790. The CgDUR3 3′ UTR (nt 2153 to 2728 of DUR3; DUR3 ends at nt 3175) was amplified from BG88b with a 5′ SalI site and a 3′ ApaI site using oligonucleotides HE791 and HE792. PCR products were cloned into pBC KS+ (Agilent Technologies) in the following order: DUR3 5′ UTR→ADE2 ORF→FRT-HIS3-FRT→DUR3 3′ UTR, creating pH1104. All PCR products were checked by sequencing. The cassette was released from pH1104 by digestion with NotI and XhoI (which cleaves in the 3′ UTR of DUR3 at nt 2696) and transformed into BG88b, selecting for cells capable of growing without histidine, producing HCg16.
HCg18.
For the HCg18 [MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT] strain, the CgHIS3 marker was removed from HCg16 through Flp-mediated recombination using plasmid pRD16 (as described for HCg12).
HCg19 and HCg20.
For the HCg19 and HCg20 [MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT-HIS3-FRT] strains, the CgADE2 ORF was replaced with FRT-HIS3-FRT as described for HCg12. Transformants were selected on plates lacking histidine and containing either arginine (1 g/liter) or urea (10 mM) as a nitrogen source (arginine is converted to urea and ornithine). Although the DUR3 urea permease is disrupted, HCg18 can still grow on plates containing urea as the sole nitrogen source, although very slowly. Growth on medium containing urea as the sole nitrogen source should activate the DUR3 promoter and result in the production of adenine.
HCg23 and HCg25.
For HCg23 and HCg25 [MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT], the HIS3 marker was removed from HCg19 (creating HCg23) and from HCg20 (creating HCg25) through Flp-mediated recombination using plasmid pRD16.
Deleting CgURE2 to create HCg27.
For HCg27 [MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT ure2::FRT-HIS3-FRT], the CgURE2 5′ UTR (nt −501 to −1 of URE2) was amplified from BG88b with a 5′ NotI site and a 3′ BamHI site using oligonucleotides HE815 and HE816. CgHIS3 flanked by FRT sites and containing a 5′ BamHI site and a 3′ PstI site was amplified from strain 37A using oligonucleotides HE819 and HE820. The CgURE2 3′ UTR (nt 1074 to 1565 of URE2; the URE2 ORF ends at nt 1068) was amplified from BG88b with a 5′ XhoI site and a 3′ ApaI site using oligonucleotides HE821 and HE822. The PCR products were cloned into pBC KS+ (Agilent Technologies) in the following order: URE2 5′ UTR→FRT-HIS3-FRT→URE2 3′ UTR, creating pH1111. All PCR products were checked by sequencing. The cassette was released from pH1111 by digestion with NotI and ApaI and transformed into HCg23, selecting for cells capable of growing without histidine.
HCg29.
For HCg29 [MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT ure2::FRT], the HIS3 marker was removed from HCg27 through Flp-mediated recombination using plasmid pRD16.
Replacing the CgURE2 ORF with the CaURE2 ORF.
For HCg30 [MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT Cgure2:CaURE2–FRT-HIS3-FRT], the CaURE2 ORF from the Darlington strain (30) was amplified with a 5′ BamHI site and a 3′ PstI site using oligonucleotides HE817 and HE818. This CaURE2 PCR product was cloned between the CaURE2 5′ UTR and FRT-HIS3 in pH1111 (see HCg27), creating pH1113. The cassette was released by digestion with NotI and ApaI and transformed into HCg29, selecting for cells capable of growing without histidine.
HCg32.
For HCg32 [MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT ure2:URE2(C. albicans)-FRT], the HIS3 marker was removed from HCg30 through Flp-mediated recombination using plasmid pRD16.
C. glabrata expression plasmids.
The centromeric S. cerevisiae plasmid pRS316 (31) is capable of replicating in C. glabrata (32, 33) with a copy number of 10 to 30. The ADH1 promoter cassette was amplified from pVT103 (34) (using oligonucleotides HE66 and HE67) and cloned in the PvuII window of pRS316, replacing the multiple cloning region. In the resulting plasmid, pH130, the ADH1 and URA3 promoters are facing each other. By site-directed mutagenesis using oligonucleotide HE127, a PstI site was removed from URA3, resulting in pH393.
No inducible promoter system is available in C. glabrata. The TPI1 constitutive strong promoter (nt −959 to −1 of TPI1; CAGL0H08327g, triosephosphate isomerase) was amplified from C. glabrata CBS138 genomic DNA using oligonucleotides HE892 and HE893. The ScADH1 promoter from pH393 was replaced with the CgTPI1 promoter using flanking NheI and BamHI sites, resulting in pH1232. In order to check if pH1232 could be used as an expression vector in C. glabrata, green fluorescent protein (GFP) was inserted behind the promoter as a BamHI-XhoI fragment from pH199 (35), resulting in plasmid pH1251. Transformation of this plasmid into C. glabrata BG88b resulted in very bright GFP fluorescence in all of the cells, while the vector pRS316 showed only very faint background fluorescence. Thus, pH1232 can be used as an expression plasmid in C. glabrata.
CgURE2 was amplified from BG88b with primers HE297 and HE299, introducing a BamHI site upstream of the start codon and an XhoI site downstream of the stop codon, and inserted as a BamHI/XhoI fragment into expression vector pH1232, resulting in pH1259. CaURE2 was transferred as a BamHI/XhoI fragment from pH563 (12) into pH1232, resulting in pH1258. CgURE2 lacking the 5′ prion domain (starting at nt 292; M98) was amplified with primers HE941 and HE299 and cloned as a BamHI/XhoI fragment into pH1232, resulting in pH1274. CaURE2 lacking the 5′ prion domain (starting at nt 262; Q88) was amplified with primers HE942 and HE188 and cloned as a BamHI/XhoI fragment into pH1232, resulting in pH1272.
[URE3] induction constructs.
The centromeric S. cerevisiae plasmid pRS313 (31) containing HIS3 was modified by site-directed mutagenesis to remove 2 BglII sites (oligonucleotides HE123 and HE124), 1 PstI site (oligonucleotide HE126), 1 NheI site (oligonucleotide HE125), and 2 HindIII sites (oligonucleotides HE122 and HE125), resulting in pH339. An ADH1 promoter cassette was amplified from pVT103 (34) using oligonucleotides HE66 and HE67 and cloned in the PvuII window of pH339, replacing the multiple cloning region. In the resulting plasmid, pH403, the ADH1 and HIS3 promoters are facing each other.
The NheI/BamHI-bordered ADH1 promoter in pH403 was replaced with the CgTPI1 promoter from pH1232, creating pH1286. The CgTPI1 promoter and a fragment of CgURE2 containing the first 100 amino acids was transferred as an NheI/XhoI fragment into pH403, creating pH1287. The CgTPI1 promoter and a fragment of CaURE2 containing the first 89 amino acids was transferred as an NheI/XhoI fragment into pH403, creating pH1288.
Switching C. glabrata mating type loci.
The HO ORF (CAGL0G05423) was amplified from C. glabrata BG88b using oligonucleotides HE894 and HE895 and cloned as a BamHI/XhoI fragment into pH1232, resulting in pH1254. pH1254 was transformed into HCg25, and transformants capable of growing without uracil were streaked as single colonies on YPAD (14). Nearly all of the YPAD colonies had lost plasmid pH1254. The MAT locus was scored using two sets of primers described by Butler et al. (36), as well as BG88b (MATa) and CBS138 (MATα) as controls. Among 20 clones in which HO had been expressed using pH1254, four isolates were identified that had become MATα. The same procedure, starting with strain HCg32, produced 7 MATα clones of 20 examined. Thus, CgHO has the ability to change the mating type locus. HCg25 MATα was named HCg58.
The 5′ UTR of CgLEU2 (CAGL0H03795g; nt −399 to +17 of LEU2) was amplified from BG88b using oligonucleotides HE831 and HE833 with a 5′ BamHI site and a 3′ PstI site. CgHIS3, flanked by FRT sites and containing a 5′ BamHI site and a 3′ PstI site, was amplified from strain 37A using oligonucleotides HE819 and HE820 and cloned into pBC KS+, creating pH1107. The 3′ UTR of CgLEU2 (nt 1099 to 1511 of LEU2 with the LEU2 ORF terminating at nt 1098) was amplified with oligonucleotides HE834 and HE832 with a 5′ XhoI site and a 3′ ApaI site. PCR products were cloned into pBS KS+ (Agilent Technologies) in the order LEU2 5′ UTR→FRT-HIS3-FRT→LEU2 3′ UTR, creating pH1285. The cassette was released from pH1285 by digestion with BamHI and ApaI and transformed into HCg58, selecting for cells capable of growing without histidine, producing the leu2 C. glabrata strain HCg62.
Transfection of [URE3albicans] from C. glabrata extracts into S. cerevisiae cells.
Six Ade+ C. glabrata isolates expressing CaUre2p (HCg32+pH1258+pH1288) were grown in SD medium to saturation, diluted into 50 ml YPAD, and grown for 2 to 3 doublings. As a positive control, S. cerevisiae YHE1161 (BY302 [URE3] (MATa his3 leu2 trp1 CaURE2 PDAL5:ADE2 PDAL5:CAN1 kar1 [14]) was used, which propagates [URE3] based on CaUre2p (15). Cells were collected by centrifugation and washed 2 times with water, resuspended in 600 μl of water, and disrupted in a BioSpec Mini-Beadbeater-8. Lysates were cleared by 5 min of centrifugation at 4°C and sonicated with 3 cycles in a Branson sonifier 250 equipped with a microtip (45 s, 20% duty cycle, output 4).
S. cerevisiae BY302 (14) was used as the recipient for the protein extract. Cells were grown to early log phase (optical density at 600 nm [OD600] of 0.8) in YPAD, washed once with water and 2 times with ST buffer (1 M sorbitol, 10 mM Tris-HCl, pH7.4), and resuspended in 5 ml ST buffer. Cells were protoplasted by adding 100 U lyticase (L2524; Sigma) and incubating them for 40 min at 30°C. Protoplasts were collected by centrifugation at 250 × g, washed twice in STC buffer (1 M sorbitol, 10 mM Tris-HCl, pH 7.4, 10 mM CaCl2), and resuspended in 1 ml STC buffer. Protoplasts (100 μl) were mixed with 1 μl single-stranded DNA (ssDNA; 10 μg/μl; SPB1136; Open Biosystems), 4.6 μg LEU2 plasmid pRS425 (37), and 9 μl sonicated protein extract. The mixture was incubated for 10 min at room temperature. PTC buffer (900 μl; 20% [wt/vol] polyethylene glycol 8000, 10 mM Tris-HCl, pH 7.4, 10 mM CaCl2) was added, and incubation was continued at room temperature for 20 min. Protoplasts were collected by centrifugation at 400 × g, resuspended in 200 μl SOS buffer (1 M sorbitol, 7 mM CaCl2, 1/3 yeast extract-peptone-dextrose [YPD]), allowed to recover for 30 min at 30°C, mixed with 10 ml CS+A5 medium (1 M sorbitol, leucine dropout medium containing 5 mg/liter adenine) at 50°C, and spread on plates containing 20 ml of the same medium. Plates were placed in a 30°C incubator for 6 days.
Cytoductions.
A cytoduction recipient isogenic to BY302 was constructed with the opposite mating type and different markers. TRP1 was amplified from S. cerevisiae S288c using oligonucleotides HE899 and HE900, and BY302 cells that had become Ade+ after receiving protein extracts of HCg32 expressing CaURE2 and CaURE2N were restored to Trp+ by transformation with the PCR product. The mating type of BY302 was changed from MATa to MATα by transient overexpression of HO from plasmid pJH298 (CEN4 LEU2 GAL10::HO; kindly provided by James Haber), resulting in YHE1355. HIS3 was amplified from S288c using oligonucleotides HE950 and HE951 and used to make YHE1355 His+, resulting in YHE1362. Subsequent growth on YPAD with ethidium bromide resulted in the [rho0] strain YHE1364. TRP1 amplified from S. cerevisiae as described above also was used to transform YHE1160, YHE1161, YHE1162, and YHE1170 to the corresponding Trp+ strains YHE1345, YHE1346, YHE1347, and YHE1348, respectively.
RESULTS
Ure2p activity in C. glabrata.
Candida glabrata strains HCg25 (CgURE2), HCg32 (CaURE2; genomic replacement), and HCg29 (ure2Δ) have the Ure2p-regulated DUR3 promoter fused to ADE2, making growth on medium without adenine a measure of Ure2p activity (Table 1). We tested nitrogen sources for Ure2p activation (making cells Ade−) (Fig. 1). The ure2Δ strain HCg29 grew equally well on all nitrogen sources, producing single colonies within 2 days, except on media containing urea, where the absence of the Dur3 urea permease slowed growth.
Table 1.
Yeast strains and plasmids
| Strain | Description | Source or reference |
|---|---|---|
| Saccharomyces cerevisiae | ||
| BY302 | MATa his3 leu2 trp1 CaURE2 PDAL5:ADE2 PDAL5:CAN1 kar1 | 14 |
| YHE1160 | BY302 [URE3albicans1160] | 14 |
| YHE1161 | BY302 [URE3albicans1161] | 14 |
| YHE1162 | BY302 [URE3albicans1162] | 14 |
| YHE1170 | BY302 [URE3albicans1170] | 14 |
| YHE1181 | MATα ura2 leu2 kar1 CaURE2 PDAL5:ADE2 PDAL5:CAN1 [rho0] | This work |
| YHE1364 | BY302 changed to MATα HIS3 [rho0] | This work |
| Candida glabrata | ||
| BG88b | MATa ura3::G418 his3::ura3(5FOA) | 25 |
| HCg8 | MATa ura3::G418 his3::ura3(5FOA) ade2::FRT-HIS3-FRT | This work |
| 37A | Clinical isolate | 26 |
| HCg12 | MATa ura3::G418 his3::ura3(5FOA) ade2::FRT | This work |
| HCg16 | MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT-HIS3-FRT | This work |
| HCg18 | MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT | This work |
| HCg19, HCg20 | ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT-HIS3-FRT | This work |
| HCg23, HCg25 | MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT | This work |
| HCg27 | MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT ure2::FRT-HIS3-FRT | This work |
| HCg29 | MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT ure2::FRT | This work |
| HCg30 | MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT ure2:CaURE2–FRT-HIS3-FRT | This work |
| HCg32 | MATa ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT ure2:CaURE2-FRT | This work |
| HCg62 | MATα ura3::G418 his3::ura3(5FOA) dur3:ADE2-FRT ade2::FRT leu2::FRT-HIS3-FRT | This work |
| Plasmids | ||
| pRS316 | CEN ARS URA3 Ar | 31 |
| pVT103 | ADH1 promoter with multiple cloning site | 34 |
| pH1232 | pRS316 with ADH1 promoter, MCS from VT103 CEN ARS URA3 Ar ADH1 promoter | This work |
| pH1259 | pH1232 with CgURE2 | This work |
| pH1258 | pH1232 with CaURE2 | This work |
| pH1274 | pH1232 with CgURE2NΔ (lacks aa 1–97) | This work |
| pH1272 | pH1232 with CaURE2NΔ (lacks aa 1–87) | This work |
| pH1286 | CEN ARS HIS3 CgTPI1 promoter | This work |
| pH1287 | CEN ARS HIS3 CgTPI1 promoter CgURE2N (aa 1–100) | This work |
| pH1288 | CEN ARS HIS3 TPI1 promoter CaURE2 (aa 1–89) | This work |
| pH1254 | CEN ARS URA3 Ar CgTPI1 promoter CgHO | This work |
Fig 1.
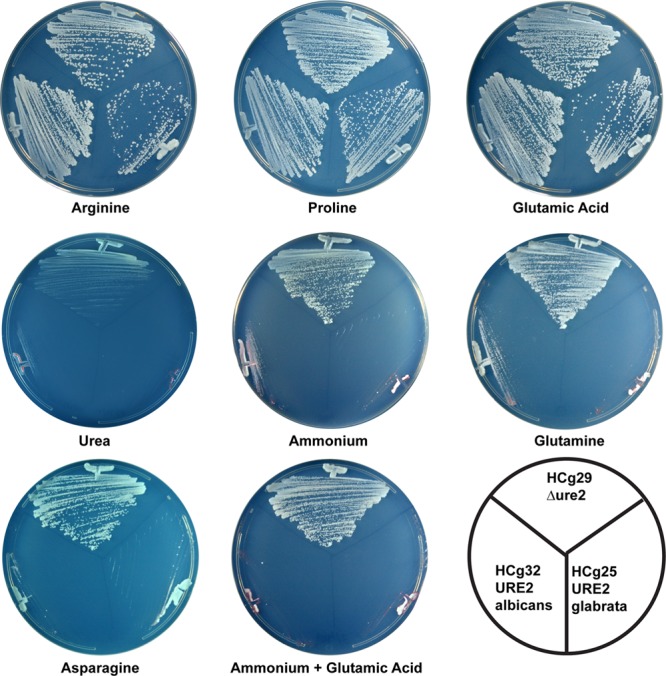
Nitrogen sources that activate Ure2p and repress DUR3:ADE2. Amino acids were tested at 1 g/liter, ammonium as present in yeast nitrogen base, and urea at 10 mM. Medium containing 1 g/liter glutamic acid was adjusted to pH 5.6. Growth was examined after 2 days at 30°C. At later times, all strains grew on all nitrogen sources.
Ammonium, glutamine, asparagine, urea, or the combination of ammonium and glutamate activated C. glabrata Ure2p, but arginine, proline, or glutamate did not, a pattern similar to that observed in S. cerevisiae. CaUre2p showed the same pattern, except that urea did not activate CaUre2p and there was a bit more growth on ammonium and glutamine medium than for CgUre2p. When plates were incubated beyond the 2 days used to assess the effects of the various nitrogen sources described above, all strains formed single colonies on all media tested. This indicates that background expression of the DUR3 promoter in C. glabrata is higher than that of the promoter routinely used in S. cerevisiae, DAL5. It is nevertheless remarkable that CaURE2 functions so well in both S. cerevisiae and C. glabrata given the limited homology between the Gln3 proteins of these three organisms and the fact that Ure2p largely acts through Gln3p in S. cerevisiae (1, 38). Homology is limited to the zinc finger domain in the middle of the protein and the last 9 amino acids (14). However, it is not known whether CgUre2p or CaUre2p acts largely through Gln3p in their native species.
Expressing CgUre2p or CaUre2p from plasmids in the ure2Δ strain HCg29 resulted in a tight Ade− phenotype on medium containing asparagine plus glutamine as nitrogen sources (1 g/liter each) for some transformants, but a substantial number of transformants were Ade+ on the same medium.
[URE3] induction in C. glabrata.
Because strains HCg25 (CgURE2) and HCg32 (CaURE2) with genomic URE2 genes grew without adenine on all media tested (including the combination of asparagine and glutamine) when incubated for more than 2 days, we combined overexpression of each full-length Ure2p from plasmids with chromosomal expression, resulting in a tight Ade− phenotype and overexpressed N-terminal fragments (known to be the prion domain in the case of CaUre2p) in attempts to induce prion formation. Essentially all double transformants were Ade− in the presence of ammonium or asparagine plus glutamine even after 5 days of incubation. This was sufficient to allow selection of rare Ade+ putative [URE3] clones (Table 2).
Table 2.
Induction of [URE3] in Candida glabrataa
| Strain | URA3 plasmid | HIS3 plasmid | No. Ade+/106 cells in expt: |
||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||
| HCg25 CgURE2 | CgURE2 (pH1259) | Vector (pH1286) | <1 | <1 | <1 | <1 | <1 |
| CgURE2 (pH1259) | CgURE2 aa 1–100 (pH1287) | <1 | <1 | <1 | <1 | <1 | |
| HCg32 URE2albicans | CaURE2 (pH1258) | Vector (pH1286) | <1 | <1 | 12 | 1 | 1 |
| CaURE2 (pH1258) | CaURE2 aa 1–89 (pH1288) | 36 | 15 | 30 | 24 | 95 | |
Cells were grown on medium without adenine, adjusted to pH 5.6, and contained both ammonium and glutamate as nitrogen sources. Colonies were counted after 5 days at 30°C. No histidine was added, as this seemed to increase the background.
Confirming [URE3] generation in C. glabrata. (i) Mating in C. glabrata.
If the C. glabrata Ade+ colonies generated in strain HCg32 (CaURE2) are indeed [URE3], the Ade+ phenotype has to be transferable. However, no mating has been observed in C. glabrata (39). Three mating type-like loci, similar to those found in S. cerevisiae, are present in C. glabrata (40). Furthermore, C. glabrata has an HO endonuclease which is essential in S. cerevisiae for mating type switching (36). No mating type switching has been observed in C. glabrata grown in culture (41), but both MATa and MATα cells are found in the same clade, suggesting the occurrence of mating type switching and mating in the wild (42).
Overexpression of CgHO changed the mating type locus of HCg25 (or HCg32) from MATa to MATα, indicating that CgHO is active. HCg25 (MATa his3) was crossed with the switched strain, HCg62 (MATα leu2), on plates containing various nitrogen sources or on plates containing acetate as a carbon source (S. cerevisiae sporulation medium). Plates were incubated at several temperatures. Single colonies emerged on plates containing glutamate as the sole nitrogen source that had been placed for 4 days at 37°C and then left at room temperature for 3 weeks. These colonies propagated when transferred to a fresh plate or to a plate containing ammonium as the nitrogen source, although the growth rate remained similarly low. However, when streaked on YPAD, all single colonies showed only the parental marker combinations. Thus, no mating was observed, indicating that C. glabrata cannot be used for prion infectivity studies.
(ii) Curing of the Ade+ phenotype.
C. glabrata grows well on YPAD with 120 mM guanidine HCl (GuHCl), but no curing of the Ade+ phenotype was observed. A few colonies showed white to red sectoring, but these had to be purified several times on YPAD with 120 mM GuHCl in order to obtain Ade− colonies. Thus, guanidine curing does not seem to work in C. glabrata. It is unknown if CgHsp104p is sensitive to guanidine inhibition. CaHsp104 protein cannot be inhibited by guanidine (43).
(iii) Protein transformation into S. cerevisiae.
When spheroplasts of S. cerevisiae are exposed to ScUre2p amyloid or to a protein extract from a [URE3] cell, the recipient cells can acquire the prion (6, 44). [URE3] based on CaUre2p but propagating in S. cerevisiae can be transferred to recipient S. cerevisiae cells by cytoduction or by exposure to a protein extract from infected cells (14, 15). If C. glabrata propagates [URE3], based on CaUre2p the prion should be transferable from a protein extract of these cells into recipient S. cerevisiae similarly expressing CaUre2p. Using the genetic tools available for S. cerevisiae, the prion then can be characterized.
Extracts of six Ade+ C. glabrata clones (HCg32+pH1258+pH1288) expressing CaUre2p were transformed into spheroplasts of S. cerevisiae BY302 (CaURE2), selecting first for clones which had been transformed with a LEU2 plasmid included in the transformation mix and then checking Leu+ clones for Ade+, a sign that the same cell had been infected with the [URE3albicans] prion (Table 3). Since prion-infected cells should be guanidine curable, BY302 cells that received protein extracts from C. glabrata HCg32 colonies 1, 2, and 6 appear to contain [URE3albicans] (Table 3). This indicates that [URE3albicans] was present in these C. glabrata clones. All of the [URE3albicans] variants are weak, as colonies turn red on adenine-limiting (YES+W) medium or on minimal medium with no adenine (SD+H+W+L).
Table 3.
Infection of S. cerevisiae with [URE3albicans] from extracts of C. glabrata strainsa
| Protein extract source | Ade+ ScBY302 of 48 tested | Ade+ clone no. | No. Ade+/color on YES after growth on: |
|
|---|---|---|---|---|
| YPAD | YPAD + 3 mM guanidine | |||
| YHE1161 | 39 | |||
| 19 | 12/white | 0/red | ||
| 20 | 12/white | 0/red | ||
| 21 | 12/white | 0/red | ||
| 22 | 12/white | 0/red | ||
| H2O | 0 | |||
| HCg32 | ||||
| 1 | 8 | |||
| 11 | 11/red | 0/red | ||
| 12 | 12/red | 0/red | ||
| 13 | 12/red | 0/red | ||
| 14 | 12/red | 0/red | ||
| 15 | 12/red | 0/red | ||
| 16 | 12/red | 0/red | ||
| 17 | 12/red | 0/red | ||
| 18 | 12/red | 0/red | ||
| 2 | 4 | |||
| 8 | 10/red | 0/red | ||
| 9 | 10/red | 0/red | ||
| 10 | 7/white | 7/white | ||
| 3 | 0 | |||
| 4 | 0 | |||
| 5 | 1 | |||
| 7 | 12/white | 12/white | ||
| 6 | 6 | |||
| 1 | 12/red | 0/red | ||
| 2 | 12/red | 0/red | ||
| 4 | 12/red | 0/red | ||
| 5 | 12/red | 0/red | ||
| 6 | 12/red | 0/red | ||
BY302 cells were transformed with the indicated extract and the LEU2 plasmid pRS425 and plated on −Leu plates containing 5 mg/liter adenine. Forty-eight transformant colonies from each plate were picked and tested for growth on SD+H+W (−Ade −Leu). The number of colonies growing on the indicated plates is shown. The indicated Ade+ clones were then streaked for single colonies on YPAD or YPAD with 3 mM guanidine, and 12 were tested from each plate for growth on −Ade medium (SD+H+W+L) and color on YES. The number of Ade+ clones and their color on YES is shown.
(iv) Cytoduction.
A suitable cytoduction recipient was constructed from BY302 by changing its mating type and markers (see Materials and Methods). Each of several S. cerevisiae strains expressing CaUre2p and transformed to guanidine-curable Ade+ by extracts of C. glabrata (CaURE2) (Ade+ clones 1, 2, and 6) could transmit [URE3albicans] by cytoplasmic mixing to the constructed recipient (Table 4).
Table 4.
Cytoduction tests of S. cerevisiae transformed with extracts of C. glabrata Ade+ clonesa
| Protein extract | S. cerevisiae BY302 Ade+ clone no. and color on YES | No. of Ade+ cytoductants/total |
|---|---|---|
| YHE1161 | 19; white | 28/28 |
| 20; white | 30/30 | |
| HCg32 | ||
| 1 | 11; red | 26/28 |
| 12; red | 18/18 | |
| 2 | 8; red | 2/44 |
| 9; red | 27/43 | |
| 6 | 1; red | 41/42 |
| 2; red | 26/26 |
The cytoduction recipient was YHE1364, isogenic with BY302, but with different markers. Cytoductants were tested for ability to grow without adenine. Numbers are expressed as no. of Ade+ cytoductants out of the total number of cytoductants assayed.
Thus, transformation of a protein extract from C. glabrata cells selected to be Ade+ into S. cerevisiae resulted in the recipient cells becoming Ade+. The Ade+ phenotype was curable by growth on YPAD containing 3 mM guanidine HCl and was transferrable by cytoduction. This indicates that the Ade+ phenotype in the recipient S. cerevisiae cells was due to the presence of [URE3albicans]. We conclude that the C. glabrata donor cells contain [URE3albicans].
Transmission variants of [URE3albicans].
Transient overexpression of CaUre2p in a S. cerevisiae strain in which ScURE2 has been replaced with CaURE2 results in an increase of cells containing [URE3albicans] (14). Each of four [URE3albicans]-containing isolates from strain BY302 (MATa CaURE2 PDAL5:ADE2 kar1) could be cured of [URE3albicans] by growth on medium containing 3 mM guanidine hydrochloride (14), and, when crossed with strain YHE1181 (MATα CaURE2 PDAL5:ADE2 [rhoo]), diploids remained capable of propagating [URE3]. However, whereas [URE3albicans] was readily cytoduced from two of the [URE3] isolates, YHE1161 and YHE1162, into strain YHE1181, no [URE3albicans] cytoductants could be identified when the other two isolates, YHE1160 and YHE1170, were used as donors (Table 5).
Table 5.
Variants of [URE3albicans] differa
| [URE3albicans] donor | [ure-o] ρ0 recipient | Cytoductants |
Diploids |
No. of Ade+ mated cytoductants | ||
|---|---|---|---|---|---|---|
| % Ade+ | n | % Ade+ | n | |||
| YHE1160 | YHE1181 | 0 | 86 | 35 | 48 | 0/86 |
| YHE1161 | YHE1181 | 100 | 88 | 72 | 47 | 86/88 |
| YHE1162 | YHE1181 | 100 | 89 | 54 | 48 | 89/89 |
| YHE1170 | YHE1181 | 0 | 91 | 38 | 38 | 0/91 |
Cells were tested for the presence of [URE3albicans] by growth on adenine dropout media. When SD medium was used, lacking adenine and only containing the amino acids needed for growth, the same result was obtained. Cytoductants were mated to strain YHE1186 (BY302 [rho0]), and the diploid phenotype was tested similarly.
Strains YHE1160 and YHE1170 were cured of [URE3albicans] by growth on guanidine, and [URE3albicans] originating in YHE1161 or YHE1162 was introduced into each by cytoduction (see Table S2 in the supplemental material). Those cytoductants could readily transfer [URE3albicans1161] or [URE3albicans1162] by cytoduction to YHE1181 (see Table S3, middle column). Thus, there is a difference in the [URE3albicans] variants between YHE1161 and YHE1162, which can transfer their prion to YHE1181, and YHE1160 and YHE1170, which cannot.
A combination of the [URE3albicans] variant and the host background must block transfer of [URE3albicans] from YHE1160 and YHE1170 to YHE1181. Note that [URE3albicans1161] originating from YHE1161 changed during the transfers between the hosts BY302 and YHE1181, so that during its second time in the latter host it no longer supported growth on adenine dropout medium. The difference in results between the two adenine-lacking media was also displayed by the [URE3albicans] variants derived from C. glabrata when introduced into S. cerevisiae BY302.
The inability to transfer [URE3albicans] present in strain YHE1160 and YHE1170 to strain YHE1181 is due in part to the prion variant, as shown above. To determine if there is also a recipient host component to this phenomenon, isogenic donor and recipient strains were created. When using recipient strains with a background isogenic to the donor, the [URE3albicans] present in YHE1160 and YHE1170 is infectious (Table 6). It is thus a combination of the prion variant and the host background that prevents transfer. In meiotic crosses, the infection potential of these [URE3] prions segregated as a multigene trait in the spores, making definition of the genes involved impractical.
Table 6.
All [URE3albicans] variants are transmitted to a recipient isogenic to donorsa
| [URE3alb] donor | Recipient | No. of cytoductants growing on indicated medium/total |
|
|---|---|---|---|
| SD+U+L | −Ade | ||
| YHE1160 TRP+ | YHE1181 | 3/26 | 1/26 |
| YHE1161 TRP+ | YHE1181 | 48/48 | 48/48 |
| YHE1162 TRP+ | YHE1181 | 48/48 | 48/48 |
| YHE1170 TRP+ | YHE1181 | 0/42 | 0/42 |
| YHE1160 TRP+ | YHE1364 | 93/93 | 6/96 |
| YHE1161 TRP+ | YHE1364 | 48/48 | 48/48 |
| YHE1162 TRP+ | YHE1364 | 48/48 | 48/48 |
| YHE1170 TRP+ | YHE1364 | 94/94 | 32/96 |
YHE1364 is isogenic to BY302, the parent of the [URE3albicans] donors. Cytoductants are listed as Ade+ colonies per number of cytoductants tested. Adenine prototrophy was assessed on minimal medium plus the other required amino acids (SD+U+L) or on complete defined medium lacking only adenine (−Ade).
DISCUSSION
Here, we show that Ure2p of C. glabrata cannot form a prion at detectable frequency even in its own environment. As a control, we show that Ure2p of C. albicans can do so. Like previous results on K. lactis Sup35p and Ure2p (19, 20) and S. paradoxus Ure2p (14), this result supports the use of S. cerevisiae as a test bed for potential prion proteins (PrP). The one noted exception is the failure to find prion formation by PrP in S. cerevisiae. It is possible that the cell surface location of the normal form, PrPC, and the formation of the prion form, PrPSc, either on the cell surface or in the endosomal system has not been properly set up in the attempts to make PrP a prion in yeast.
Our work confirms that prion-forming ability is not generally conserved. While the CgUre2p prion domain sequence is much closer to that of S. cerevisiae than is that of CaUre2p, only CaUre2p can form prions in S. cerevisiae or C. glabrata. Indeed, CgUre2p also does not form amyloid in vitro, while the C. albicans protein readily does so (15). Aigle's group has likewise found that the Ure2p of Kluyveromyces lactis is incapable of forming [URE3] in K. lactis itself (20). The fact that prion formation by Ure2p or Sup35p is not restricted to S. cerevisiae has been used as an argument that it is a conserved trait, and that prion formation therefore must have a function for yeast (and those other species) (45, 46). Our results indicate that the ability of a protein from each of two species to form prions does not imply that that ability is conserved; it may well be a sporadically occurring trait. The same point is inferred from the finding that sequence is not important for prion-forming ability by the Ure2p and Sup35p prion domains (47–49). Our results parallel the fact, long known for mammalian prion proteins, that the ability of mammalian PrP to assume the prion form varies dramatically from one species to another and even within the same species (for examples, see reference 50).
In the course of preparing strains for this study, we have verified that, through Ure2p, C. glabrata selectively activates or represses transcription of genes in response to the availability of nitrogen sources in the growth environment. We showed previously that C. glabrata Ure2p regulates a set of genes similar to that of S. cerevisiae; however, we did not show that glabrata reacts to the nitrogen state of the environment, as does S. cerevisiae. As in S. cerevisiae, ammonia, asparagine, and glutamine activate Ure2p, whereas proline does not (1, 51, 52). Glutamate derepresses some Ure2p-regulated genes but not others in S. cerevisiae (53). Some genes, notably the general amino acid permease GAP1, are transcribed when glutamate is present as the sole nitrogen source but the protein is channeled to the vacuole for degradation (54, 55). Medium containing both ammonium and glutamate was used to isolate the first [URE3] prion (56). In C. glabrata the combination of ammonium and glutamate is among the strongest repressors of DUR3 promoter activity, and it was used here to select [URE3] candidates.
In preparing isogenic strains of opposite mating type to attempt cytoductions, we showed that the S. cerevisiae HO endonuclease homologue of C. glabrata can perform mating type locus switching, as is the case in S. cerevisiae. Although we did not observe mating, and no mating has been reported yet for C. glabrata, it is unclear why mating type switching has remained functional, if not for the purpose of creating mating partners.
ACKNOWLEDGMENTS
This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
We thank Brendan Cormack (Johns Hopkins University) for generously supplying several strains and plasmids and Jim Haber (Brandeis University) for the HO plasmid.
Footnotes
Published ahead of print 8 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00015-13.
REFERENCES
- 1. Magasanik B, Kaiser CA. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1–18 [DOI] [PubMed] [Google Scholar]
- 2. Cooper TG. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26:223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wickner RB. 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science 264:566–569 [DOI] [PubMed] [Google Scholar]
- 4. Masison DC, Wickner RB. 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270:93–95 [DOI] [PubMed] [Google Scholar]
- 5. Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283:1339–1343 [DOI] [PubMed] [Google Scholar]
- 6. Brachmann A, Baxa U, Wickner RB. 2005. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24:3082–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shewmaker F, Mull L, Nakayashiki T, Masison DC, Wickner RB. 2007. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 176:1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baxa U, Taylor KL, Wall JS, Simon MN, Cheng N, Wickner RB, Steven A. 2003. Architecture of Ure2p prion filaments: the N-terminal domain forms a central core fiber. J. Biol. Chem. 278:43717–43727 [DOI] [PubMed] [Google Scholar]
- 9. Masison DC, Maddelein Wickner M-LRB. 1997. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl. Acad. Sci. U. S. A. 94:12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. 2005. Is the prion domain of soluble Ure2p unstructured? Biochemistry 44:321–328 [DOI] [PubMed] [Google Scholar]
- 11. Baxa U, Wickner RB, Steven AC, Anderson D, Marekov L, Yau Tycko W-MR. 2007. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry 46:13149–13162 [DOI] [PubMed] [Google Scholar]
- 12. Edskes HK, Wickner RB. 2002. Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full-length protein. Proc. Natl. Acad. Sci. U. S. A. 99( Suppl 4): 16384–16391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baudin-Baillieu A, Fernandez-Bellot E, Reine F, Coissac E, Cullin C. 2003. Conservation of the prion properties of Ure2p through evolution. Mol. Biol. Cell 14:3449–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edskes HK, Engel A, McCann LM, Brachmann A, Tsai Wickner H-FRB. 2011. Prion-forming ability of Ure2 of yeasts is not evolutionarily conserved. Genetics 188:81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Engel A, Shewmaker F, Edskes HK, Dyda F, Wickner RB. 2011. Amyloid of the Candida albicans Ure2p prion domain is infectious and has a parallel in-register β-sheet structure. Biochemistry 50:5971–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. 2000. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol. Microbiol. 35:865–876 [DOI] [PubMed] [Google Scholar]
- 17. Kushnirov VV, Kochneva-Pervukhova NV, Cechenova MB, Frolova NS, Ter-Avanesyan MD. 2000. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 19:324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santoso A, Chien P, Osherovich LZ, Weissman JS. 2000. Molecular basis of a yeast prion species barrier. Cell 100:277–288 [DOI] [PubMed] [Google Scholar]
- 19. Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. 2001. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol. Cell 7:1121–1130 [DOI] [PubMed] [Google Scholar]
- 20. Safadi RA, Talarek N, Jacques N, Aigle M. 2011. Yeast prions: could they be exaptations? The URE2/[URE3] system in Kluyveromyces lactis. FEMS Yeast Res. 11:151–153 [DOI] [PubMed] [Google Scholar]
- 21. Talarek N, Maillet L, Cullin C, Aigle M. 2005. The [URE3] prion is not conserved among Saccharomyces species. Genetics 171:23–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liebman SW, Chernoff YO. 2012. Prions in yeast. Genetics 191:1041–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schlumpberger M, Prusiner SB, Herskowitz I. 2001. Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol. 21:7035–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Geitz RD, Woods RA. 2002. Transformation of yeast by the Liac/SS carrier DNA/PEG method. Methods Enzymol. 350:87–96 [DOI] [PubMed] [Google Scholar]
- 25. Cormack BP, Falkow S. 1999. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151:979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyazaki H, Miyazaki Y, Gerber A, Parkinson T, Hitchcock C, Falconer DJ, Ward DJ, Marsden K, Bennett JE. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PHD1, in Candida glabrata. Antimicrob. Agents Chemother. 42:1695–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaur R, Ma B, Cormack BP. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. U. S. A. 104:7628–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. ElBerry HM, Majumdar ML, Cunningham TS, Sumrada RA, Cooper TG. 1993. Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J. Bacteriol. 175:4688–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uemura T, Kashiwagi K, Igarashi K. 2007. Polyamine uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. J. Biol. Chem. 282:7733–7741 [DOI] [PubMed] [Google Scholar]
- 30. Kakeya H, Miyazaki Y, Miyazaki H, Nyswaner K, Grimberg B, Bennett JE. 2000. Genetic analysis of azole resistance in the Darlington strain of Candida albicans. Antimicrob. Agents Chemother. 44:2985–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou P, Szczypka MS, Young R, Thiele DJ. 1994. A system for gene cloning and manipulation in the yeast Candida glabrata. Gene 142:135–140 [DOI] [PubMed] [Google Scholar]
- 33. Hanic-Joyce PJ, Joyce PB. 1998. A high-copy-number ADE2-bearing plasmid for transformation of Candida glabrata. Gene 211:395–400 [DOI] [PubMed] [Google Scholar]
- 34. Vernet T, Dignard D, Thomas DY. 1987. A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52:225–233 [DOI] [PubMed] [Google Scholar]
- 35. Edskes HK, Gray VT, Wickner RB. 1999. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. U. S. A. 96:1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe KH. 2004. Evolution of the MAT locus and its HO endonuclease in yeast species. Proc. Natl. Acad. Sci. U. S. A. 101:1632–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122 [DOI] [PubMed] [Google Scholar]
- 38. Mitchell AP, Magasanik B. 1984. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:2758–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muller H, Hennequin C, Gallaud J, Dujon B, Fairhead C. 2008. The asexual yeast Candida glabrata maintains distinct a and α haploid mating types. Eukaryot. Cell 7:848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Srikantha T, Lachke SA, Soll DR. 2003. Three mating type loci in Candida glabrata. Eukaryot. Cell 2:328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brockert PJ, Lachke SA, Srikantha T, Pujol C, Galask R, Soll DR. 2003. Phenotypic switching and mating type switching of Candida glabrata at sites of colonization. Infect. Immun. 71:7109–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brisse S, Pannier C, Angoulvant A, de Meeus T, Diancourt L, Faure O, Muller H, Peman J, Viviani MA, Grillot R, Dujon B, Fairhead C, Hennequin C. 2009. Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples. Eukaryot. Cell 8:287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zenthon JF, Ness F, Cox B, Tuite MF. 2006. The [PSI+] prion of Saccharomyces cerevisiae can be propagated by an Hsp104 orthologue from Candida albicans. Eukaryot. Cell 5:217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brachmann A, Toombs JA, Ross ED. 2006. Reporter assay systems for [URE3] detection and analysis. Methods 39:35–42 [DOI] [PubMed] [Google Scholar]
- 45. Shorter J, Lindquist S. 2005. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6:435–450 [DOI] [PubMed] [Google Scholar]
- 46. Harrison LB, Yu Z, Stajich JE, Dietrich FS, Harrison PM. 2007. Evolution of budding yeast prion-determinant sequences across diverse fungi. J. Mol. Biol. 368:273–282 [DOI] [PubMed] [Google Scholar]
- 47. Ross ED, Baxa U, Wickner RB. 2004. Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24:7206–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ross ED, Edskes HK, Terry MJ, Wickner RB. 2005. Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. U. S. A. 102:12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Toombs JA, McCarty BR, Ross ED. 2010. Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30:319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vorberg I, Groschup MH, Pfaff E, Priola SA. 2003. Multiple amino acid residues within the rabbit PrP protein inhibit formation of its abnormal isoform. J. Virol. 77:2003–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. ter Schure EG, van Riel NA, Verrips CT. 2000. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 24:67–83 [DOI] [PubMed] [Google Scholar]
- 52. Hofman-Bang J. 1999. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotech. 12:35–71 [DOI] [PubMed] [Google Scholar]
- 53. Rai R, Daugherty JR, Tate JJ, Buford TD, Cooper TG. 2004. Synergistic operation of four cis-acting elements mediate high level DAL5 transcription in Saccharomyces cerevisiae. FEMS Yeast Res. 5:29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stanbrough M, Magasanik B. 1995. Transcriptional and posttranscriptional regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacteriol. 177:94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rubio-Texeira M, Kaiser CA. 2006. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway. Mol. Biol. Cell 17:3031–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lacroute F. 1971. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 106:519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]


