Abstract
The amino acid cysteine has long been known to be toxic at elevated levels for bacteria, fungi, and humans. However, mechanisms of cysteine tolerance in microbes remain largely obscure. Here we show that the human pathogenic yeast Candida albicans excretes sulfite when confronted with increasing cysteine concentrations. Mutant construction and phenotypic analysis revealed that sulfite formation from cysteine in C. albicans relies on cysteine dioxygenase Cdg1, an enzyme with similar functions in humans. Environmental cysteine induced not only the expression of the CDG1 gene in C. albicans, but also the expression of SSU1, encoding a putative sulfite efflux pump. Accordingly, the deletion of SSU1 resulted in enhanced sensitivity of the fungal cells to both cysteine and sulfite. To study the regulation of sulfite/cysteine tolerance in more detail, we screened a C. albicans library of transcription factor mutants in the presence of sulfite. This approach and subsequent independent mutant analysis identified the zinc cluster transcription factor Zcf2 to govern sulfite/cysteine tolerance, as well as cysteine-inducible SSU1 and CDG1 gene expression. cdg1Δ and ssu1Δ mutants displayed reduced hypha formation in the presence of cysteine, indicating a possible role of the newly proposed mechanisms of cysteine tolerance and sulfite secretion in the pathogenicity of C. albicans. Moreover, cdg1Δ mutants induced delayed mortality in a mouse model of disseminated infection. Since sulfite is toxic and a potent reducing agent, its production by C. albicans suggests diverse roles during host adaptation and pathogenicity.
INTRODUCTION
Although sulfite is potentially toxic, in yeasts the molecule is also a normal intermediate during reductive sulfate assimilation. Due to its antimicrobial and antioxidative properties, sulfite is also well known for its common use as a preservative in food industries, in particular during the production of wine (1). The molecular basis of sulfite tolerance has therefore been intensively studied in fermenting yeasts and led to the identification of the sulfite efflux pump Ssu1 in Saccharomyces cerevisiae (2, 3). Overexpression of SSU1 in S. cerevisiae resulted in enhanced sulfite resistance, and the gene was shown to be positively controlled by the transcription factor Fzf1 (4–6). Beside a role in sulfite detoxification, other biological functions have been addressed for Ssu1 in S. cerevisiae, and with a pathobiological interest, in the human pathogenic yeast Candida albicans. Of particular note, transcriptional profiling in both S. cerevisiae and C. albicans during nitrosative stress identified a small set of upregulated genes, including SSU1 (7, 8). This nitric oxide-induced response was found in S. cerevisiae to depend on Fzf1, which, however, is absent from C. albicans. Instead, Cta4 was identified in C. albicans as a major nitrosative stress regulator. Nitric oxide-induced expression of SSU1 in C. albicans was to some extent dependent on this transcription factor but not sulfite resistance (9).
A sophisticated function of active sulfite secretion was hypothesized in dermatophytes. The particular ability of these specialized filamentous fungi to grow within keratinized host structures was correlated with sulfite secretion as a means to reduce the keratin stabilizing disulfide bonds (10–12). A putative role of Ssu1 in this process of sulfitolysis was supported by the observation that SSU1 is constitutively expressed at a high level in these microorganisms (13). In humans, sulfite is produced as an intermediate during oxidative cysteine catabolism, a process which is regulated by the key enzyme cysteine dioxygenase Cdo1. A homologous enzyme has recently been detected also in dermatophytes (14). Apart from potential functions in active sulfite formation, such a pathway may at the same time also represent an alternative means of cysteine detoxification, like in humans (15). Elevated concentrations of cysteine are known to be growth inhibiting not only for humans or dermatophytes, but also for a broad variety of other microorganisms (15–24). We have recently reported a functional role of Ssu1 and Cdo1 for keratin degradation and cysteine tolerance by targeted mutagenesis in the dermatophyte Arthroderma benhamiae (25).
C. albicans, a common commensal of the human gastrointestinal tract, has also long been known for the growth sensitivity to increasing cysteine levels (23, 26). Cysteine and other thiols are present in the mammalian gut at considerable concentration, where they support eminent intestinal functions, including tissue redox homeostasis, mucus fluidity and nutrient absorption (27). Therefore, mechanisms which favor cysteine tolerance in C. albicans are likely to be important during host adaptation processes.
In order to uncover mechanisms supporting cysteine and sulfite tolerance in C. albicans, the present work was started by investigating a potential role of the putative sulfite efflux pump Ssu1 in this process. It is known from prior work that C. albicans ssu1Δ mutants are sensitive to toxic sulfite, a phenotype shared with S. cerevisiae (9). Since in humans sulfite can be produced from cysteine, we found it reasonable to discover that C. albicans ssu1Δ mutants were also sensitive to cysteine and that cysteine induced the expression of the SSU1 gene. In humans, the conversion of cysteine to sulfite depends on cysteine dioxygenase Cdo1. Therefore, we were attracted by a homologue of this key enzyme in C. albicans, Cdg1. Sulfite production from cysteine in C. albicans was abolished in a C. albicans cdg1Δ knockout mutant. A transcriptional regulator governing CDG1 and SSU1 expression as well as sulfite tolerance was identified.
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains used in the present study are listed in Table S1 in the supplemental material. All C. albicans strains were routinely propagated on YPD agar plates (10 g yeast extract, 20 g of peptone, 20 g glucose, and 15 g of agar per liter) at 30°C and stored as frozen stocks in liquid YPD medium with 15% glycerol at −80°C. A library of C. albicans transcription factor mutant strains (28) was obtained via The Fungal Genetics Stock Center (University of Missouri, Kansas City, MO) (29).
Plasmid constructions.
A construct for the deletion of the SSU1 gene was generated as follows: an ApaI-XhoI fragment with SSU1 upstream sequences was cloned after amplification by PCR with the primers SSU1-1 and SSU1-2 (all of the primers are listed in Table S2 in the supplemental material). Genomic DNA from C. albicans strain SC5314 was used as a template. A SacII-SacI fragment containing SSU1 downstream sequences was obtained with the primers SSU1-3 and SSU1-4. The SSU1 upstream and downstream fragments were successively substituted for WOR1 upstream and downstream fragments in plasmid pWOR1M2 via the introduced restriction sites, respectively (30), to result in pSSU1M2, in which the SAT1 flipper is flanked by SSU1 sequences. In the same way, constructs were generated for the deletion of orf19.1867 (alias SSU12 or MAE1) with the primers SSU1L1-1/SSU1L1-2 and SSU1L1-3/SSU1L1-4, for the deletion of CDG1 with the primers CDG1-3/CDG1-4 and CDG1-5/CDG1-6, for the deletion of ZCF2 with the primers ZCF2-5/ZCF2-2 and ZCF2-3/ZCF2-4, and for the deletion of CTA4 with the primers CTA4-1/CTA4-2 and CTA4-3/CTA4-4 to yield plasmids pSSU1L1M2, pCDG1M2, pZCF2M2, and pCTA4M2, respectively. For reinsertion of the SSU1 gene at its original locus in the homozygous ssu1Δ mutants SSU1M4A/B, the SSU1 gene of strain SC5314 was amplified by PCR with primers SSU1-1 and SSU1-6. Primer SSU1-6 introduced a BamHI site directly after the stop codon. The ApaI-BamHI fragment was cloned in the ApaI-BglII-digested plasmid pSAP2KS1 (31) to result in pSSU1KS1. The SacII-SacI downstream SSU1 fragment from pSSU1M2 was cloned in pSSU1KS1 to replace the SacII-SacI downstream SAP2 fragment, resulting in plasmid pSSU1KS2. In the same manner, reinsertion of the CDG1 gene at its wild-type locus in mutants CDG1M4A/B was performed, using primer CDG1-3 and primer CDG1-7, which introduced a BglII site directly after the stop codon. The ApaI-BglII fragment was cloned in the ApaI-BglII digested plasmid pSAP2KS1, resulting in pCDG1KS1. The SacII-SacI downstream CDG1 fragment from pCDG1M2 was cloned in pCDG1KS1, resulting in plasmid pCDG1KS2. A PSSU1-GFP reporter construct was generated as follows: an ApaI-SalI fragment containing SSU1 upstream sequences was amplified from genomic DNA of strain SC5314 with the primers SSU1-1 and SSU1-7, and a PstI-SacI downstream SSU1 fragment was amplified with the primers SSU1-8 and SSU1-4. The upstream and downstream SSU1 fragments were successively cloned via the introduced restriction sites in the plasmid pOPT1G22 (32), resulting in the plasmid pSSU1G2, in which the promoter PSSU1 is fused to the GFP gene.
C. albicans transformant construction.
C. albicans was transformed by an electroporation protocol (33) with the following gel-purified, linear DNA fragments: the ApaI-SacI fragments from pSSU1M2, pSSU1L1M2, pCDG1M2, pZCF2M2, and pCTA4M2 were used to delete the genes SSU1, SSU12, CDG1, ZCF2, and CTA4 in strain SC5314 (see Fig. S1 in the supplemental material), and strains SSU1M4A/B were used for the construction of ssu1Δ ssu12Δ and ssu1Δ cdg1Δ double-knockout mutants. The ApaI-SacI fragments from pSSU1KS2 and pCDG1KS2 were used to reinsert the SSU1 and CDG1 genes in the ssu1Δ and cdg1Δ deletion mutants, respectively, and the ApaI-SacI fragment from plasmid pSSU1G2 was used to integrate the PSSU1-GFP fusion into one of the two SSU1 alleles in strain SC5314, CTA4M4A/B, and ZCF2M4A/B, respectively (see Fig. S1 and S2 in the supplemental material). Selection of transformants on nourseothricin (Werner Bioagents, Jena, Germany) and sensitive derivatives thereof was carried out by the SAT1-flipping method as described previously (34). The correct genomic integration of the constructs was confirmed by Southern hybridization.
Southern and Northern hybridization.
Genomic DNA from C. albicans strains was isolated as described before (35). DNA (15 μg) was digested with appropriate restriction enzymes, separated in 1% (wt/vol) agarose gels, ethidium bromide stained, transferred by vacuum blotting onto nylon membranes, and fixed by UV cross-linking. Southern hybridization was performed with enhanced chemiluminescence-labeled probes, using the Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare, Braunschweig, Germany) and according to the instructions of the manufacturer. Total RNA from C. albicans was isolated by the hot acidic phenol method (36), purified with the RNeasy Minikit (Qiagen), and DNase-treated on-column by use of the RNase-free DNase set (Qiagen) to remove contaminations with genomic DNA. A total of 20 μg of RNA per sample was separated on formaldehyde-containing agarose gels and blotted onto nylon membranes. For the detection of SSU1 transcripts, an SSU1-specific probe was prepared by PCR with the primers SSU1-10 and SSU1-11, using SC5314 DNA as a template, and labeled with digoxigenin-11-dUTP (Roche). In accordance with the instructions of the manufacturer, Northern hybridization was carried out in DIG-Easy Hyb buffer, followed by binding of antidigoxigenin-alkaline phosphate Fab fragments and the use of CDP-Star Ready-To-Use solution (Roche). Signals were detected on Amersham ECL-Hyperfilms (GE Healthcare).
Quantitative real-time reverse transcription-PCR (qRT-PCR).
Total RNA (100 ng) was used to carry out qRT-PCR within a one-step approach using the Brilliant III SYBR green Ultra-Fast QRT PCR master mix kit (Agilent Technologies, La Jolla, CA) (for the primers used, see Table S2 in the supplemental material). RT-PCR was performed on a Stratagene Mx3005P and the threshold cycle was determined by the instrument's MxPro software version 4.10 (Agilent Technologies). Expression was calculated by the ΔΔCT method as described previously (37). Expressions were normalized to the expression of the ACT1 gene; for all samples, three biological replicates were analyzed.
Growth and sensitivity assays.
Sulfite resistance of C. albicans strains was monitored on YPD-TA agar plates containing sulfite, prepared as described previously (13). Briefly, appropriate amounts of a freshly prepared Na2SO3 stock solution were spread by plating onto 25 ml of YPD agar containing 75 mM l-tartaric acid (pH 3.5) for a final sodium sulfite concentration as indicated in Results. Before use, the sulfite was allowed to dry and diffuse into the agar overnight at room temperature. To test for the ability of the C. albicans strains to grow in the presence of l-cysteine or l-methionine, overnight precultures grown at 30°C in YPD were diluted 1:100 in 10 ml of fresh SD medium (20 g glucose and 6.7 g of YNB with ammonium sulfate [MP Biomedicals] per liter) containing increasing concentrations of the amino acids as indicated in Results, followed by incubation at 30°C. The influence of cysteine on germ tube formation was assayed in 1 ml of Dulbecco modified Eagle medium (DMEM; 1 g of glucose/liter, without l-glutamine) cell culture medium (PAA Laboratories) at 37°C.
Detection of sulfite.
A commercially available sulfite detection kit was used for sulfite identification in C. albicans culture supernatants (R-Biopharm, Darmstadt, Germany). In brief, precultures of strains SC5314, CDG1M4A, and CDG1KS2A were grown in 50 ml of liquid YPD medium. After 24 h, the cultures were centrifuged, washed once with 25 ml of SD medium and resuspended in 50 ml of fresh SD medium. The cultures were supplemented with 10 mM cysteine and grown for 24 h at 30°C. Afterward, supernatants were harvested and used for sulfite detection according to the instructions of the manufacturer. Each culture was prepared in triplicate.
In vivo experiments.
Female BALB/c mice (Charles River Laboratories, 18 to 20 g) were housed in groups of five in individually ventilated cages and cared for in strict accordance with the principles outlined in the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (http://conventions.coe.int/Treaty/en/Treaties/Html/123.htm). Experiments were approved by the governmental ethics committee (reg. no. 03-007/07). Mice were infected via the lateral tail vein with 104 C. albicans yeast cells per g of body weight in phosphate-buffered saline. Animals were monitored at least twice daily and euthanized if moribund (defined by lethargy, inability to ambulate, and/or hypothermia). Gross pathological alterations were recorded during necropsy.
Fluorescence microscopy and flow cytometry.
Fluorescence microscopy was performed with a Zeiss LSM 510 Meta laser scanning microscope. Imaging scans were acquired with an argon laser with a wavelength of 488 nm and corresponding filter settings for green fluorescent protein (GFP) and parallel/overlay transmission images. The cells were inspected with a ×63 immersion oil objective. Flow cytometry of C. albicans wild-type and GFP reporter strains was performed using a BD FACSCanto II flow cytometer and the FACSDiva (BD Biosciences, Heidelberg, Germany) software to determine the median fluorescence values. The analyzed C. albicans samples were derived from SD medium overnight cultures which were diluted 1:100 in fresh SD medium supplemented with the indicated compounds and incubated at the conditions as specified in Results.
Statistical analysis.
Unless otherwise indicated, data were expressed as the means ± the standard deviations (SD). Differences were analyzed by the two-tailed unpaired Student t test, a P value of <0.05 was considered statistically significant. Mouse survival was analyzed by using Kaplan-Meyer curves and the log-rank test.
RESULTS
C. albicans ssu1Δ mutants are growth sensitive to cysteine.
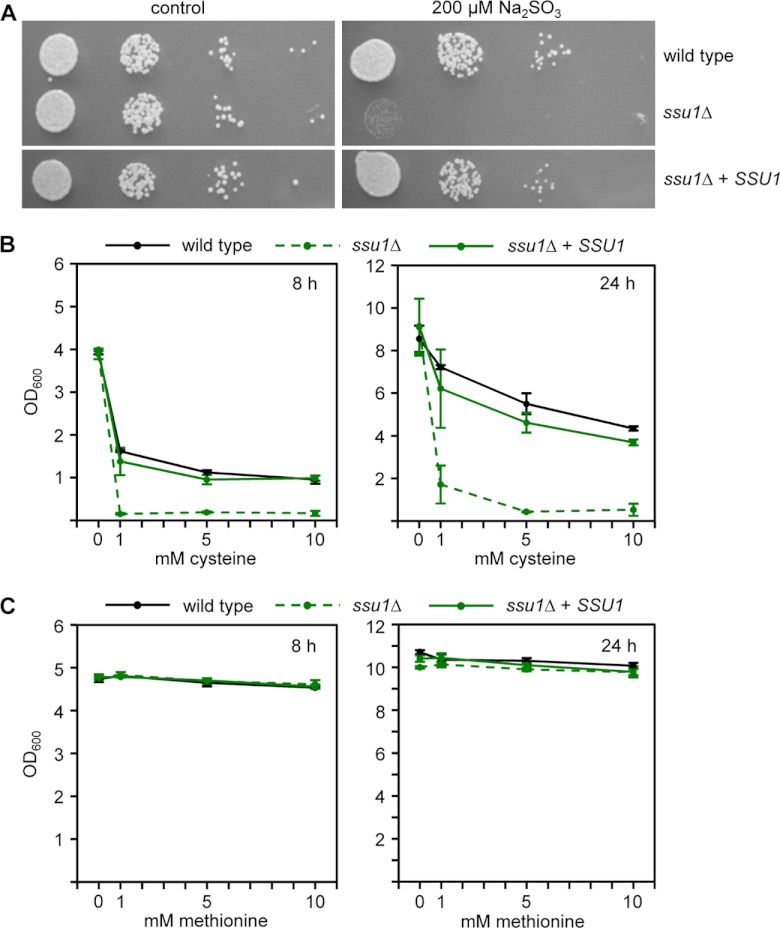
In order to test the possibility whether Ssu1 contributes to cysteine tolerance of C. albicans, specific ssu1Δ deletion mutants and reconstituted strains were constructed in the C. albicans wild-type strain SC5314 by the SAT1-flipping methodology (see Fig. S1 in the supplemental material) (34). Wild-type and transformants were assayed for their ability to grow in the presence of sulfite, because sulfite resistance has previously been reported to depend on SSU1 in both S. cerevisiae and C. albicans (6, 9). Confirming prior findings, our C. albicans ssu1Δ mutants were strongly inhibited in growth on YPD-TA agar containing elevated sulfite concentrations (Fig. 1A). In the next step, ssu1Δ mutants were tested for growth in the presence of increasing cysteine levels. In this experiment, growth was also monitored in the presence of the sulfur containing amino acid methionine. Compared to wild-type and reconstituted strains, a growth-inhibitory effect was identified for ssu1Δ mutants in the presence of cysteine (Fig. 1B) but not for methionine (Fig. 1C). In order to find out whether other genes homologous to SSU1 are present in the C. albicans genome, a BLAST search with SSU1 DNA sequences was carried out (http://www.candidagenome.org/). This approach identified the uncharacterized gene orf19.1867 (alias SSU12 or MAE1) with considerable sequence similarity to SSU1 (52.1% on the protein level). This gene was also deleted in the C. albicans wild type, as well as in the ssu1Δ mutant (Materials and Methods). In the presence of sulfite or cysteine, C. albicans ssu12Δ deletion mutants behaved like the wild type, and ssu1Δ ssu12Δ double knockout strains behaved like ssu1Δ single mutants (data not shown). These observations excluded a contribution of SSU12 to the tolerance of cysteine and/or sulfite under the tested conditions.
Fig 1.
C. albicans ssu1Δ mutants are growth sensitive to sulfite and cysteine. (A) Overnight cultures of strains SC5314 (wild type), SSU1M4A/B (ssu1Δ) and SSU1KS2A/B (ssu1Δ + SSU1) were serially 10-fold diluted, and aliquots were inoculated on YPD-TA agar plates containing 200 μM sodium sulfite. As a control, YPD-TA agar plates without sodium sulfite were used. The agar plates were incubated for 28 h at 30°C. (B and C) C. albicans overnight cultures of the same strains were diluted 1:100 in fresh SD medium containing increasing concentrations of cysteine (B) or methionine (C). The optical densities of the cultures were measured after growth for 8 h and 24 h at 30°C. The addition of cysteine to the medium significantly reduced the growth of the ssu1Δ mutant compared to wild-type and reconstituted strains. The two independently constructed mutants and complemented strains behaved identically, and the results for only one of them are shown.
Cysteine activates SSU1 gene expression.
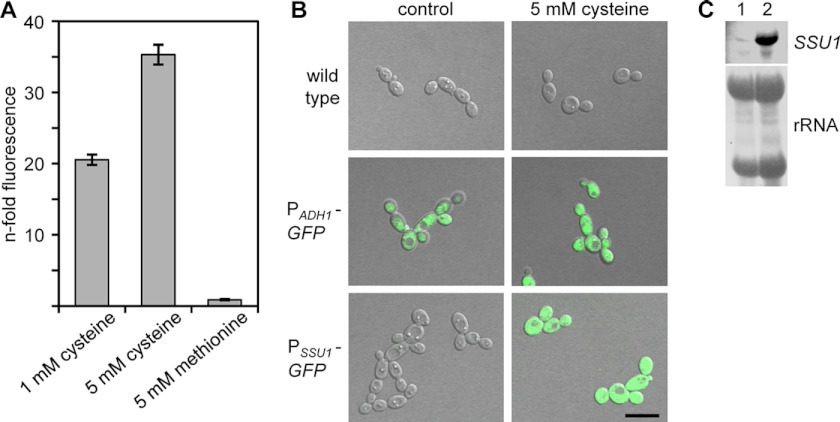
The finding that Ssu1 supports cysteine tolerance in C. albicans instigated the question whether cysteine itself acts as an inducer of SSU1 gene expression. C. albicans reporter strains were constructed which express the GFP gene under the control of the promoter PSSU1 in one of the wild-type SSU1 alleles (see Fig. S2 in the supplemental material). Overnight cultures of the reporter strains SSU1G1A/B were diluted and grown in fresh SD medium in the absence and presence of cysteine. After 6 h, the samples were inspected by flow cytometry for quantification of the GFP intensity as a measurement of gene expression (Fig. 2A). Herein, SSU1 was found to be strongly activated only during growth of the cells in the presence of cysteine, but not in the presence of methionine. Enhanced PSSU1-dependent GFP expression in the presence of cysteine was also demonstrated by fluorescence microscopy (Fig. 2B). As a positive control, GFP expression from the constitutively active C. albicans ADH1 promoter was monitored (38). Northern analysis further proved SSU1 gene induction in response to cysteine (Fig. 2C).
Fig 2.
SSU1 gene expression is upregulated in C. albicans in the presence of cysteine. (A) The fluorescence of C. albicans cells of reporter strains SSU1G1A/B was measured by flow cytometry after growth for 6 h in SD medium containing the indicated substances. The bars represent the n-fold fluorescence intensities measured in the presence versus absence of the tested compound for each strain. The two independently constructed reporter strains behaved identically, and only one of them is shown. (B) Detection of fluorescence on the level of single cells in the presence of cysteine. After 6 h of growth in SD medium in the absence or presence of 5 mM cysteine, respectively, overlay fluorescence microscopy pictures were taken from cells of the wild type, the positive control SCADH1G4A (PADH1-GFP), and the reporter strains SSU1G1A/B (PSSU1-GFP). The two independently constructed PSSU1-GFP reporter strains behaved identically, and the results for only one of them are shown (scale bar, 10 μm). (C) Detection of SSU1 mRNA by Northern hybridization analysis in response to cysteine. After 6 h of growth in SD medium in the absence (lane 1) or presence of 5 mM cysteine (lane 2), SSU1 mRNA was detected in cells of the wild-type strain SC5314 (upper panel). rRNA is shown as a loading control (lower panel).
C. albicans produces sulfite in the presence of cysteine, relying on Cdg1.
In mammals, toxic concentrations of cysteine are eliminated by the oxidative cysteine catabolism, via the formation of sulfite (15). Given our knowledge that cysteine induced the expression of the sulfite efflux pump Ssu1, we reasoned that excess of cysteine may be converted to sulfite by C. albicans as well. Using a commercially available detection kit, sulfite was found in the supernatant of C. albicans wild-type cells in the presence of cysteine but not in the absence of cysteine or in the uninoculated control media (Fig. 3A). The C. albicans genome harbors a putative homologue of cysteine dioxygenase, C. albicans Cdg1 (uncharacterized orf19.7314), and a putative cysteine degradation pathway resulting in sulfite formation has been predicted (http://www.candidagenome.org/ [39]). Notably, the CDG1 and SSU1 genes are located next to each other in the genome, in divergent orientation, the two start codons separated by an intergenic region of 2378 bp. To clarify whether sulfite production by C. albicans in the presence of cysteine depends on Cdg1, the CDG1 gene was deleted in the wild-type strain SC5314 (see Fig. S1 in the supplemental material). Wild-type, cdg1Δ mutant, and complemented strains were compared for the production of sulfite in the culture supernatant during growth in the presence of cysteine. Here, sulfite was specifically absent in the culture supernatant of the cdg1Δ mutant (Fig. 3A). This finding demonstrated a central role of cysteine dioxygenase Cdg1 for sulfite formation under the tested conditions. In contrast to the ssu1Δ mutant, cdg1Δ mutant cells displayed only a slightly reduced growth in the presence of elevated cysteine levels after 8 h and showed even enhanced growth after 24 h compared to the wild-type and reconstituted strains (Fig. 3B). This observation suggests that cysteine toxicity under these conditions largely results from the formation of sulfite, which is no longer produced by cdg1Δ mutant cells. However, the possibility remained that cysteine itself could be a substrate of Ssu1, which would allow cysteine detoxification in cdg1Δ mutants independent from sulfite formation via cysteine dioxygenase. To exclude this alternative, we deleted the CDG1 gene in the C. albicans ssu1Δ mutant background. This approach should rescue ssu1Δ mutant cells from increased cysteine sensitivity under these conditions, in case that cysteine toxicity is mediated via sulfite formation. Proving this assumption, the ssu1Δ cdg1Δ double-knockout strain displayed reduced sensitivity to cysteine, like the cdg1Δ single mutant (Fig. 3B). In order to monitor whether CDG1 gene expression in C. albicans is also upregulated in the presence of cysteine, transcription of CDG1 in the wild-type SC5314 was measured by qRT-PCR during growth in the absence or presence of cysteine (Fig. 3C). A strong upregulation of CDG1 (and SSU1) expression in response to cysteine was detected, further supporting a role of C. albicans cysteine dioxygenase in the elimination of cysteine via sulfite.
Fig 3.
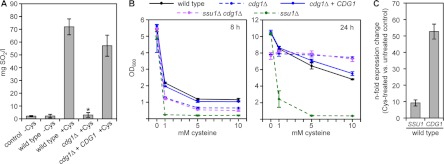
Growth of C. albicans in the presence of cysteine results in sulfite formation in culture supernatants. (A) Sulfite production was inspected in the supernatants of the wild type, mutants CDG1M4A/B (cdg1Δ), and complemented strains CDG1KS2A/B (cdg1Δ + CDG1) after 24 h of growth in SD medium containing 10 mM cysteine. Supernatants of the wild type grown for 24 h in SD medium without cysteine and uninoculated SD medium with 10 mM cysteine were used as controls. The results are means ± the SD from three independent experiments. Sulfite as SO2 was not detectable in supernatants of the cdg1Δ mutant compared to wild type and complemented strain. The asterisk (*) indicates that the detected difference was significant (P < 0.05). The independently constructed A/B mutants behaved identically, and the results for only one of them are shown. (B) Overnight precultures of wild type, SSU1M4A (ssu1Δ), CDG1M4A/B (cdg1Δ), complemented strains CDG1KS2A/B (cdg1Δ + CDG1), and ssu1Δ cdg1Δ double mutants were diluted 1:100 in fresh SD medium containing various concentrations of cysteine. The optical densities of the cultures were measured after growth for 8 h and 24 h at 30°C. The results are means ± the SD from three independent experiments. (C) qRT-PCR measurements show an upregulation of CDG1 and SSU1 gene expression levels in the wild type in the presence or absence of 5 mM cysteine after 6 h of growth in SD medium.
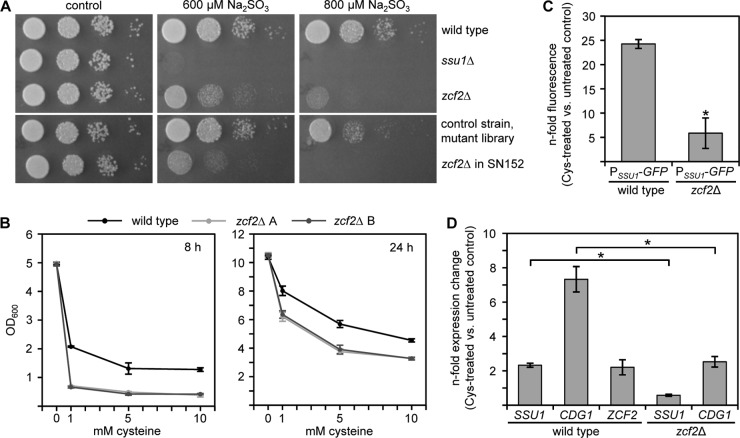
Zcf2 governs sulfite resistance, as well as cysteine-inducible CDG1 and SSU1 expression.
In order to identify a potential transcriptional regulator of sulfite tolerance in C. albicans, we screened a library of transcription factor mutants in the presence of sulfite for enhanced sulfite sensitivity. This library contained a collection of 165 genetically matched C. albicans strains, each one with the deletion of a specific transcription regulator (28). By this approach, we identified only one factor, Zcf2, the lack of which resulted in a strongly increased sensitivity to sulfite (Fig. 4A). In order to further verify the role of Zcf2 in sulfite tolerance, independent zcf2Δ deletion mutants were constructed in the C. albicans wild-type SC5314 (see Fig. S1 in the supplemental material). Phenotypic analysis of the resulting strains ZCF2M4A/B confirmed the increased sensitivity of the zcf2Δ mutant to sulfite (Fig. 4A). Inspection of the zcf2Δ mutant during growth in SD medium plus cysteine revealed that the absence of Zcf2 also resulted in increased cysteine sensitivity (Fig. 4B). Next, by using the zcf2Δ mutant, we tested whether Zcf2 is necessary for cysteine inducible SSU1 and/or CDG1 expression. To study SSU1 expression, the zcf2Δ mutant was used as a host strain for integration of the PSSU1-GFP reporter construct at the SSU1 locus (strains ZCF2SSU1G1A/B). Flow cytometry measurements discovered that SSU1 upregulation in the presence of cysteine is dependent on a functional Zcf2 (Fig. 4C). Strongly decreased SSU1 upregulation in zcf2Δ mutant cells under these conditions was confirmed by qRT-PCR (Fig. 4D). Using qRT-PCR, we also monitored CDG1 expression, showing that upregulation of the CDG1 gene upon cysteine in the zcf2Δ mutant was not as strong as in the wild type (Fig. 4D). qRT-PCR analysis in wild-type cells further revealed that expression of ZCF2 itself was also positively controlled in the presence of cysteine (Fig. 4D). In conclusion, these analyses suggest that Zcf2 supports both sulfite and cysteine tolerance; second, Zcf2 governs the cysteine inducible expression of SSU1, and at least to some extent, CDG1.
Fig 4.
Zcf2 supports sulfite and cysteine tolerance and governs cysteine-inducible SSU1/CDG1 expression. (A) Overnight cultures of wild type, SSU1M4A (ssu1Δ), and ZCF2M4A/B (zcf2Δ) were serially diluted and inoculated on YPD-TA agar plates containing sodium sulfite as indicated, or without sulfite for control. The two independently constructed zcf2Δ mutants behaved identically, and the results for only one of them are shown. The zcf2Δ mutants (zcf2Δ in SN152) and the corresponding control strain from the originally screened transcription factor mutant library were also included (lower panel). Photographs were taken after 28 h of growth at 30°C. (B) C. albicans overnight cultures of wild-type and ZCF2M4A/B (zcf2Δ A/B) strains were diluted 1:100 in fresh SD medium containing cysteine as indicated. The optical densities of the cultures were measured after growth for 8 h and 24 h at 30°C. The results are means ± the SD from three independent experiments. The addition of cysteine to the medium significantly reduced the growth of the zcf2Δ mutant compared to the wild type. (C) The fluorescence of C. albicans cells of reporter strains SSU1G1A/B (PSSU1-GFP/wild type) and ZCF2SSU1G1A/B (PSSU1-GFP/zcf2Δ) was measured by flow cytometry after growth for 6 h in SD medium containing 1 mM cysteine. The bars represent the n-fold fluorescence intensities measured in the presence versus absence of cysteine for each strain. The results are means ± the SD from three independent experiments. An asterisk (*) indicates that the detected differences were significant (P < 0.05). The two independently constructed A/B reporter strains behaved identically, and the results for only one of them are shown. (D) qRT-PCR measurements show an upregulation of CDG1, SSU1, and ZCF2 gene expression levels in the wild type after growth for 6 h in SD medium in the presence or absence of 1 mM cysteine, whereas expression levels of SSU1 and CDG1 are decreased in the zcf2Δ mutant. The results are means ± the SD from three independent experiments. An asterisk (*) indicates that the detected differences were significant (P < 0.05).
Cysteine-inducible expression of SSU1 is not relying on the nitrosative stress regulator Cta4.
Formerly, the nitric oxide-induced expression of SSU1 in C. albicans was revealed to depend to some extent on the nitrosative stress regulator Cta4 (9). Therefore, we also investigated whether Cta4 contributes to cysteine inducible SSU1 expression and/or Ssu1 mediated cysteine/sulfite tolerance. Deletion of CTA4 in the wild-type SC5314 (see Fig. S1 in the supplemental material) resulted in a slightly increased sensitivity of C. albicans to sulfite in our assay (see Fig. S3A in the supplemental material). This finding is in contrast to prior observations, where no phenotype of a cta4Δ mutant on sulfite was recorded (9). The phenotype we detected, however, was not comparable to the effect of ZCF2 deletion. The sensitivity of cta4Δ mutant cells to cysteine was not increased under the tested conditions (Fig. S3B). Also, SSU1 gene expression in the presence of cysteine was found to be induced in the cta4Δ mutant cells at a level comparable to wild-type cells, demonstrated by PSSU1-GFP reporter strains in the background of the cta4Δ mutants (see Fig. S3C in the supplemental material). qRT-PCR measurements confirmed this finding and also revealed that in the cta4Δ mutant CDG1 is still strongly induced in the presence of cysteine, albeit to a decreased extent (see Fig. S3D in the supplemental material). In summary, our results suggest that Cta4, unlike Zcf2, does not play a major role for cysteine and/or sulfite tolerance under the tested conditions.
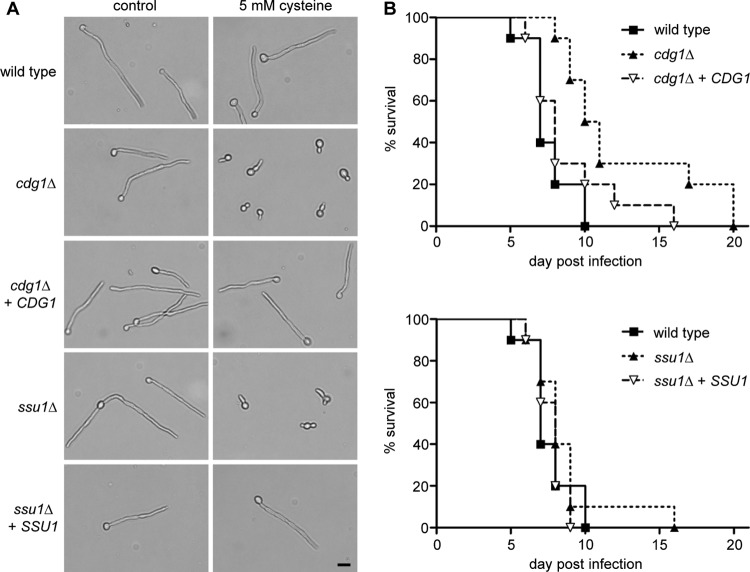
cdg1Δ mutants display reduced hyphae formation in the presence of cysteine and are attenuated in virulence.
We also tested whether C. albicans hyphae development is compromised by the lack of Cdg1 (or Ssu1) in the presence of cysteine. Cells of the wild-type, cdg1Δ and ssu1Δ mutant, and complemented strains were diluted in cell culture medium at 37°C for 6 h in the presence or absence of 5 mM cysteine. The presence of cysteine resulted in a strong repression of hyphae formation specifically in cultures of both mutants, cdg1Δ and ssu1Δ (Fig. 5A). Here, only the production of very short germ tubes was monitored. This observation stimulated the question whether Cdg1 and Ssu1 contribute to the pathogenicity of C. albicans. To test this possibility, the standard model of disseminated candidiasis in mice was applied. Immunocompetent mice were infected intravenously with the C. albicans wild type, ssu1Δ and cdg1Δ knockout mutants, and complemented strains, respectively. Mice infected with the C. albicans cdg1Δ mutant survived significantly longer than mice infected with either the corresponding complemented strain or wild-type C. albicans (Fig. 5B). We performed histological analysis of the kidneys of moribund mice. Histologically, the mutant strain showed filamentation indistinguishable from wild-type cells. We therefore conclude that the mutant did not have a gross filamentation defect in vivo, at least in the inspected tissue samples (not shown). In contrast to knockout of CDG1, deletion of SSU1 did not affect C. albicans virulence in this model.
Fig 5.
Cdg1 contributes to cysteine tolerance during hyphal development and to the pathogenicity of C. albicans. (A) Decreased hypha formation of cdg1Δ and ssu1Δ mutants in the presence of cysteine. C. albicans cells of SC5314 (wild type) and mutant strains SSU1M4A/B (ssu1Δ) and CDG1M4A/B (cdg1Δ), as well as complemented strains SSU1KS2A/B (ssu1Δ + SSU1) and CDG1KS2A/B (cdg1Δ + CDG1), were grown overnight at 30°C in SD medium and diluted 1:100 in DMEM in the absence or presence of 5 mM cysteine, followed by incubation at 37°C with gentle shaking. Microscopic photographs were taken after 6 h (scale bar, 10 μm). The two independently constructed mutants A/B and complemented strains A/B behaved identically, and the results for only one of them are shown. (B) Survival of mice infected with strain SC5314 (wild type), cdg1Δ mutant CDG1M4A, and complemented strain CDG1KS2A (cdg1Δ + CDG1), as well as ssu1Δ mutant SSU1M4A and complemented strain SSU1KS2A (ssu1Δ + SSU1) (n = 10 per group). Mortality induced by the cdg1Δ mutant is significantly delayed (log-rank test, P < 0.05 compared to the wild-type and complemented strains, respectively).
DISCUSSION
From our findings, we conclude that elevated levels of cysteine are converted by C. albicans to sulfite. Central to this process is cysteine dioxygenase Cdg1. Sulfite in turn can be detoxified by C. albicans via support of the putative sulfite efflux pump Ssu1 (Fig. 6). The pathway of sulfite production from cysteine may support the tolerance of C. albicans to high cysteine levels under certain conditions, but at the same time, sulfite secretion and resistance per se may display specific roles during host adaptation and/or pathogenicity.
Fig 6.
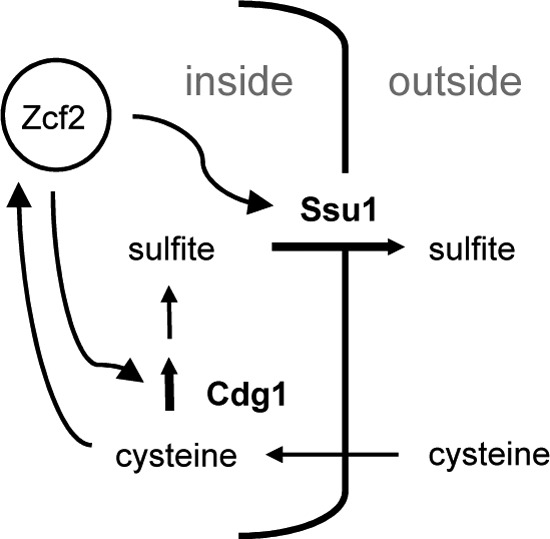
Proposed, simplified model for sulfite production by C. albicans from cysteine. Excess of environmental cysteine leads to induced expression of both cysteine dioxygenase Cdg1 and the sulfite efflux pump Ssu1, controlled by transcription factor Zcf2. Cdg1 manages a key step in the conversion of cysteine to sulfite, the export of which is supported by Ssu1.
Putative function of cysteine and sulfite tolerance in the context of C. albicans commensalism and/or infection.
The knowledge that sulfite is also a normal metabolite generated during reductive sulfate assimilation in bacteria, fungi and plants suggests mechanisms avoiding endogenous sulfite toxicity. On the other hand, active sulfite secretion may serve direct functions. A specific biological role of sulfite secretion by fungal microorganisms has been proposed for dermatophytes, i.e., acting as a reducing agent in the process of keratin degradation (12, 13). Testing this hypothesis for C. albicans, we were not able to compare the growth ability of wild-type and mutant strains in keratin media in which dermatophyte growth is assumed to rely on sulfitolysis. We found that C. albicans was unable to grow under such conditions (data not shown). Considering that sulfite is a potent reducing agent and antimicrobial preservative, its secretion and/or resistance by microbes may in general interfere with the cellular survival, especially in habitats of complex communities. This situation may also occur in the human intestinal tract, where C. albicans resides as a common commensal together with a multitude of competing microorganisms. In addition, other molecules might be related to sulfite and cysteine metabolism, e.g., hydrogen sulfide and sulfur dioxide, both of which can act as crucial gasotransmitters in humans (40). Addressing the question of whether such gases or sulfite produced by C. albicans are involved in commensalism will be of future interest. In principle, sulfite can modify any protein in which function or stability relies on discrete disulfide bridges. Such putative targets also include certain antimicrobial peptides, key effectors in the protection of mucosal surfaces against infectious microbes. As an example, recent evidence suggested that the reduction of disulfide bonds of β-defensin 1 altered the antimicrobial activity of the molecule (41).
In mammals, cysteine and methionine are normally consumed as part of dietary proteins, being transported across the intestinal epithelium. Circulating free amino acids are removed from plasma by diverse tissues, by the liver, for example, for the synthesis of glutathione (42). In addition, experiments in rats detected also cysteine secretion from intestinal mucosa to the gut lumen. This process was assumed to explain the observation that cysteine accounted for 15 to 20% of the mucosal thiol pool in rat jejunum, putatively playing an important role in intestinal function (43); the intestinal mucosa of rats was found to contain millimolar thiol concentrations (44). It can be speculated that microbial pathogens such as C. albicans, which can induce endogenous, systemic infections via penetration of gastrointestinal mucosae, benefit from specialized mechanisms of cysteine tolerance.
Role of Ssu1 and Cdg1 in the process of cysteine tolerance in C. albicans.
Intracellular cysteine levels in mammals are strictly maintained, in order to allow cysteine-dependent metabolism at concentrations below the threshold of cysteine toxicity, e.g., in rats in a range between 20 and 100 μmol/kg in the liver and between 80 and 200 μmol/liter in plasma, respectively (15). Cysteine dioxygenase has been described in mammals as a key enzyme in the process of catabolic elimination of toxic cysteine concentrations via sulfite. Homologues of cysteine dioxygenases have recently been identified in bacteria (45), in dermatophytes (14), and in the present study, in C. albicans. Interestingly, however, such sequences have not been detected in the model yeast S. cerevisiae. In mammals, sulfite is further oxidized by sulfite oxidase to nontoxic sulfate, which in turn is excreted by the urine. Sulfite oxidases are widespread also in filamentous fungi; however, homologous sequences or genes associated with the implied pathway (gene ontology GO:0008482) have not been identified in the C. albicans genome (http://www.candidagenome.org/). The latter observation further supports a role of the putative sulfite efflux pump Ssu1 for the process of cysteine conversion and sulfite elimination. This assumption was supported by our findings that the ssu1Δ mutants were sensitive to both cysteine and sulfite. Further, we showed that cysteine sensitivity of ssu1Δ mutants during yeast growth was decreased by the deletion of CDG1, i.e., by inhibition of sulfite formation. During our virulence studies in a model of disseminated infection of mice, we observed significantly delayed mortality in mice infected with the C. albicans cdg1Δ, but not the ssu1Δ mutant. Nevertheless, it remains elusive whether the decreased virulence potential of the cdg1Δ mutant is a result of altered cysteine sensitivity or the failure of sulfite generation per se via this pathway. Interestingly, in vitro inspection of the cdg1Δ mutant discovered strongly decreased hyphae formation in the presence of cysteine, whereas yeast growth was only slightly affected. Whether such phenotypic differences indicate a specific impact on the morphogenetic switch or merely reflect enhanced cysteine sensitivity of hyphae remains to be investigated. For the future, it may also be of particular interest to test additional infection models with the cdg1Δ mutant. Ideally, such experiments should elucidate the commensal state of C. albicans in the human intestinal tract and the penetration of gut mucosae.
SSU1 and CDG1 gene regulation.
Whereas SSU1 was found to be induced in C. albicans in the presence of cysteine, a global gene expression analysis in the presence of cysteine did not detect increased SSU1 expression levels in S. cerevisiae (46). In S. cerevisiae, the inducible response of SSU1 gene expression to nitric oxide stress is known to be governed by the transcriptional regulator Fzf1, which, however, has not been identified in the C. albicans genome (8). Instead, the transcription factor Cta4 was recently shown to control the activation of the nitric oxide-inducible gene YHB1 and also, to some extent, the gene SSU1 in C. albicans (9). Our findings on SSU1 expression in the cta4Δ mutant background suggest that SSU1 gene activation in response to cysteine was not relying on Cta4. Nevertheless, in contrast to others (9), we cannot fully exclude a role of Cta4 in sulfite tolerance. We found a small but detectable growth inhibition of the cta4Δ mutant upon increasing sulfite concentrations; however, this was not comparable to the effect displayed by the zcf2Δ mutant. In addition, sensitivity to cysteine was not increased in C. albicans cta4Δ mutants. Therefore, it seems likely that the activation of SSU1 upon cysteine as well as sulfite resistance are mainly under the control of a pathway different from the major nitrosative stress response in C. albicans. We have identified a candidate transcriptional regulator for this process, Zcf2, the lack of which resulted in an increased sensitivity to sulfite and cysteine. The discovered function of Zcf2 may be quite specific, since C. albicans zcf2Δ mutants did not exhibit any other growth defects either in our experiments or in any assays used for exhaustive phenotyping of this transcription factor mutant library (28). The role of Zcf2 in the control of SSU1 expression and sulfite tolerance is further supported by our observation that SSU1 expression upon cysteine was strongly decreased in the zcf2Δ mutant. In light of previous (9) and present findings on the putative regulation of SSU1 in C. albicans, the gene appears to be a target of both Cta4 and Zcf2, depending on the stimulus. Nevertheless, it should be tested in the future if Zcf2 is involved in the nitric oxide stress response, and genome-wide analyses may identify additional target genes of this regulator.
Conclusion.
Given the antimicrobial activity of cysteine and cysteine derivatives, these compounds have even been suggested as potential antimicrobial drugs. Nevertheless, the underlying molecular basis for such growth-inhibiting effects remains largely unknown. Different possible mechanisms of cysteine toxicity have been suggested in the past in diverse microbial species, including a putative interference of cysteine with the biosynthesis of branched-chain amino acids, a possible interaction with membrane bound respiratory enzymes, and the promotion of oxidative DNA damage (20, 22, 47, 48). Our finding that C. albicans can convert cysteine to sulfite adds to the knowledge on cysteine toxicity in this organism. At the same time, the fungus can detoxify sulfite by the transporter Ssu1. Therefore, it appears to be of particular interest to find out whether active sulfite secretion by such mechanisms is used by C. albicans in the human host and what the consequences of sulfite secretion are during commensalism and/or during symptomatic infection.
ACKNOWLEDGMENTS
This study was supported by Deutsche Forschungsgemeinschaft (DFG) grant STA 1147/1-1 and by the DFG funded excellence graduate school Jena School for Microbial Communication and the Hans Knoell Institute.
We thank Ronny Martin and Bettina Bardl for technical assistance and advice and Oliver R. Homann and Alexander D. Johnson for providing the C. albicans transcription factor mutant library. Sequence data for C. albicans was obtained from the Candida Genome Database (http://www.candidagenome.org/).
Footnotes
Published ahead of print 15 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00336-12.
REFERENCES
- 1. Taylor SL, Higley NA, Bush RK. 1986. Sulfites in foods: uses, analytical methods, residues, fate, exposure assessment, metabolism, toxicity, and hypersensitivity. Adv. Food Res. 30:1–76 [DOI] [PubMed] [Google Scholar]
- 2. Avram D, Bakalinsky AT. 1997. SSU1 encodes a plasma membrane protein with a central role in a network of proteins conferring sulfite tolerance in Saccharomyces cerevisiae. J. Bacteriol. 179:5971–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu X, Wightman JD, Geller BL, Avram D, Bakalinsky AT. 1994. Isolation and characterization of sulfite mutants of Saccharomyces cerevisiae. Curr. Genet. 25:488–496 [DOI] [PubMed] [Google Scholar]
- 4. Avram D, Leid M, Bakalinsky AT. 1999. Fzf1p of Saccharomyces cerevisiae is a positive regulator of SSU1 transcription and its first zinc finger region is required for DNA binding. Yeast 15:473–480 [DOI] [PubMed] [Google Scholar]
- 5. Donalies UE, Stahl U. 2002. Increasing sulphite formation in Saccharomyces cerevisiae by overexpression of MET14 and SSU1. Yeast 19:475–484 [DOI] [PubMed] [Google Scholar]
- 6. Park H, Bakalinsky AT. 2000. SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast 16:881–888 [DOI] [PubMed] [Google Scholar]
- 7. Hromatka BS, Noble SM, Johnson AD. 2005. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 16:4814–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sarver A, DeRisi J. 2005. Fzf1p regulates an inducible response to nitrosative stress in Saccharomyces cerevisiae. Mol. Biol. Cell 16:4781–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiranand W, McLeod I, Zhou H, Lynn JJ, Vega LA, Myers H, Yates JR, III, Lorenz MC, Gustin MC. 2008. CTA4 transcription factor mediates induction of nitrosative stress response in Candida albicans. Eukaryot. Cell 7:268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kunert J. 1972. Keratin decomposition by dermatophytes: evidence of the sulphitolysis of the protein. Experientia 28:1025–1026 [DOI] [PubMed] [Google Scholar]
- 11. Kunert J. 1973. The role of sulfite excretion in keratin catabolism by dermatophytes. Dermatol. Monatsschr. 159:456–457 (In German.) [PubMed] [Google Scholar]
- 12. Kunert J. 1992. Effect of reducing agents on proteolytic and keratinolytic activity of enzymes of Microsporum gypseum. Mycoses 35:343–348 [DOI] [PubMed] [Google Scholar]
- 13. Léchenne B, Reichard U, Zaugg C, Fratti M, Kunert J, Boulat O, Monod M. 2007. Sulphite efflux pumps in Aspergillus fumigatus and dermatophytes. Microbiology 153:905–913 [DOI] [PubMed] [Google Scholar]
- 14. Kasperova A, Kunert J, Horynova M, Weigl E, Sebela M, Lenobel R, Raska M. 2011. Isolation of recombinant cysteine dioxygenase protein from Trichophyton mentagrophytes. Mycoses 54:e456–462 [DOI] [PubMed] [Google Scholar]
- 15. Stipanuk MH, Dominy JE, Jr, Lee JI, Coloso RM. 2006. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J. Nutr. 136:1652S–1659S [DOI] [PubMed] [Google Scholar]
- 16. Allen EH, Hussey GG. 1971. Inhibition of the growth of Helminthosporium carbonum by l-cysteine. Can. J. Microbiol. 17:101–103 [DOI] [PubMed] [Google Scholar]
- 17. Carlsson J, Granberg GP, Nyberg GK, Edlund MB. 1979. Bactericidal effect of cysteine exposed to atmospheric oxygen. Appl. Environ. Microbiol. 37:383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Lucca AJ, Walsh TJ, Daigle DJ. 1996. N-Acetylcysteine inhibits germination of conidia and growth of Aspergillus spp. and Fusarium spp. Antimicrob. Agents Chemother. 40:1274–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galgoczy L, Kovacs L, Krizsan K, Papp T, Vagvolgyi C. 2009. Inhibitory effects of cysteine and cysteine derivatives on germination of sporangiospores and hyphal growth of different zygomycetes. Mycopathologia 168:125–134 [DOI] [PubMed] [Google Scholar]
- 20. Gomez RF, Montville T, Blais K. 1980. Toxic effect of cysteine against Salmonella typhimurium. Appl. Environ. Microbiol. 39:1081–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris CL. 1981. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J. Bacteriol. 145:1031–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kari C, Nagy Z, Kovacs P, Hernadi F. 1971. Mechanism of the growth inhibitory effect of cysteine on Escherichia coli. J. Gen. Microbiol. 68:349–356 [DOI] [PubMed] [Google Scholar]
- 23. Wain WH, Price MF, Cawson RA. 1975. A re-evaluation of the effect of cysteine on Candida albicans. Sabouraudia 13( Pt 1): 74–82 [PubMed] [Google Scholar]
- 24. Königsbauer H. 1951. Über die Wirkung schwefelhaltiger Aminosäuren auf das Wachstum verschiedener Fadenpilze “in vitro. ” Mycopathologia 5:173–177 [Google Scholar]
- 25. Grumbt M, Monod M, Yamada T, Hertweck C, Kunert J, Staib P. 25 January 2013. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J. Invest. Dermatol. [Epub ahead of print.] doi:10.1038/jid.2013.41. [DOI] [PubMed]
- 26. Winsten S, Murray TJ. 1956. Virulence enhancement of a filamentous strain of Candida albicans after growth on media containing cysteine. J. Bacteriol. 71:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Circu ML, Aw TY. 2011. Redox biology of the intestine. Free Radic. Res. 45:1245–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783 doi:10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCluskey K, Wiest A, Plamann M. 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 35:119–126 [DOI] [PubMed] [Google Scholar]
- 30. Ramirez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhäuser J. 2008. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 4:e1000089 doi:10.1371/journal.ppat.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staib P, Lermann U, Blass-Warmuth J, Degel B, Würzner R, Monod M, Schirmeister T, Morschhäuser J. 2008. Tetracycline-inducible expression of individual secreted aspartic proteases in Candida albicans allows isoenzyme-specific inhibitor screening. Antimicrob. Agents Chemother. 52:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reuss O, Morschhäuser J. 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 60:795–812 [DOI] [PubMed] [Google Scholar]
- 33. Köhler GA, White TC, Agabian N. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 35. Millon L, Manteaux A, Reboux G, Drobacheff C, Monod M, Barale T, Michel-Briand Y. 1994. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J. Clin. Microbiol. 32:1115–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 37. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dabas N, Morschhäuser J. 2007. Control of ammonium permease expression and filamentous growth by the GATA transcription factors GLN3 and GAT1 in Candida albicans. Eukaryot. Cell 6:875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Skrzypek MS, Arnaud MB, Costanzo MC, Inglis DO, Shah P, Binkley G, Miyasato SR, Sherlock G. 2010. New tools at the Candida Genome Database: biochemical pathways and full-text literature search. Nucleic Acids Res. 38:D428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li X, Bazer FW, Gao H, Jobgen W, Johnson GA, Li P, McKnight JR, Satterfield MC, Spencer TE, Wu G. 2009. Amino acids and gaseous signaling. Amino Acids 37:65–78 [DOI] [PubMed] [Google Scholar]
- 41. Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Buchner J, Schaller M, Stange EF, Wehkamp J. 2011. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature 469:419–423 [DOI] [PubMed] [Google Scholar]
- 42. Stipanuk MH. 2004. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 24:539–577 [DOI] [PubMed] [Google Scholar]
- 43. Dahm LJ, Jones DP. 1994. Secretion of cysteine and glutathione from mucosa to lumen in rat small intestine. Am. J. Physiol. 267:G292–G300 [DOI] [PubMed] [Google Scholar]
- 44. Siegers CP, Riemann D, Thies E, Younes M. 1988. Glutathione and GSH-dependent enzymes in the gastrointestinal mucosa of the rat. Cancer Lett. 40:71–76 [DOI] [PubMed] [Google Scholar]
- 45. Dominy JE, Jr, Simmons CR, Karplus PA, Gehring AM, Stipanuk MH. 2006. Identification and characterization of bacterial cysteine dioxygenases: a new route of cysteine degradation for eubacteria. J. Bacteriol. 188:5561–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumar A, John L, Alam MM, Gupta A, Sharma G, Pillai B, Sengupta S. 2006. Homocysteine- and cysteine-mediated growth defect is not associated with induction of oxidative stress response genes in yeast. Biochem. J. 396:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cowman RA, Baron SS, Fitzgerald RJ. 1983. Cysteine toxicity for oral streptococci and effect of branched-chain amino acids. Infect. Immun. 39:1107–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park S, Imlay JA. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 185:1942–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]