Abstract
CAF-1 and HIR are highly conserved histone chaperone protein complexes that function in the assembly of nucleosomes onto chromatin. CAF-1 is characterized as having replication-coupled nucleosome activity, whereas the HIR complex can assemble nucleosomes independent of replication. Histone H3K56 acetylation, controlled by the acetyltransferase Rtt109 and deacetylase Hst3, also plays a significant role in nucleosome assembly. In this study, we generated a set of deletion mutants to genetically characterize pathway-specific and overlapping functions of CAF-1 and HIR in C. albicans. Their roles in epigenetic maintenance of cell type were examined by using the white-opaque switching system in C. albicans. We show that CAF-1 and HIR play conserved roles in UV radiation recovery, repression of histone gene expression, correct chromosome segregation, and stress responses. Unique to C. albicans, the cac2Δ/Δ mutant shows increased sensitivity to the Hst3 inhibitor nicotinamide, while the rtt109Δ/Δ cac2Δ/Δ and hir1Δ/Δ cac2Δ/Δ mutants are resistant to nicotinamide. CAF-1 plays a major role in maintaining cell types, as the cac2Δ/Δ mutant exhibited increased switching frequencies in both directions and switched at a high frequency to opaque in response to nicotinamide. Like the rtt109Δ/Δ mutant, the hir1Δ/Δ cac2Δ/Δ double mutant is defective in maintaining the opaque cell fate and blocks nicotinamide-induced opaque formation, and the defects are suppressed by ectopic expression of the master white-opaque regulator Wor1. Our data suggest an overlapping function of CAF-1 and HIR in epigenetic regulation of cell fate determination in an H3K56 acetylation-associated manner.
INTRODUCTION
Eukaryotic genomes are packaged into chromatin, an ordered yet dynamic assembly of proteins interfaced with DNA that encodes both genetic and epigenetic information. The basic repeating unit of chromatin is the nucleosome. The nucleosome contains equal molar amounts of core histones H3, H4, H2A, and H2B independent of organism and cell type (1). The nucleosome core particle has been defined at a 2.8-Å resolution, and the structure determined contains 147 bp of DNA wrapped around a core tetramer of (H3-H4)2 molecules flanked by two heterodimers of H2A-H2B (2). Chromatin-bound histone proteins have an extensive number of covalent modifications which are considered to play an effectual role in many genome-related processes (3, 4). Histone proteins are highly basic and consequently prone to promiscuous interactions and aggregation, which can negatively impact cellular health. Therefore, when not contained in chromatin, histone proteins are usually bound to committed proteins called histone chaperones that transfer histones, store them, present them to enzymes, and assemble/disassemble them onto chromatin (5, 6).
The evolutionary conserved histone chaperone complexes CAF-1 and HIR (HIRA in humans) both bind H3-H4 units, and both are characterized as containing WD-40 domain repeats (6). The CAF-1 complex is a heterotrimeric H3-H4 histone chaperone whose replication-coupled nucleosome assembly activity is conserved from Saccharomyces cerevisiae to humans (7, 8). The CAF-1 deposition activity is specifically targeted to locations of DNA synthesis by a physical interaction with the DNA polymerase clamp PCNA (9). Genetic studies in S. cerevisiae identified defects in the heritable maintenance but not formation of heterochromatin at telomeres, mating type loci, and rDNA in yeast cells lacking CAF-1-encoding genes (7, 10–13). Another evolutionarily conserved histone H3-H4 chaperone contributing to the formation of functional chromatin is the HIR complex. The HIR complex is characterized as possessing nucleosome assembly activity independent of replication and generates chromatin structures resistant to SWI/SNF remodeling (14, 15). The genes encoding the HIR complex were originally identified as factors that regulate the transcription of three of the four histone gene pairs in S. cerevisiae (16, 17). Later, it was shown that the HIR complex functions in pathways to repress cryptic and antisense transcription in both budding and fission yeast (18–20).
In addition to the CAF-1 and HIR pathway-specific functions, these two complexes also share overlapping functions in several chromatin-related processes. For example, synergistic defects in heterochromatin silencing at telomeres and HM loci were detected in S. cerevisiae strains lacking both HIR and CAF-1 (21). Furthermore, deletion of both of these factors results in increased rates of chromosome missegregation and alterations of chromatin structure at centromeres (22). A more recent study investigated the significance of HIR and CAF-1 in dynamic histone H3 exchange (23). It was found that both complexes positively affect the rate of histone H3 incorporation in a replication-independent manner. Deletion of HIR1 resulted in decreased rates of histone H3 exchange at gene-rich regions, whereas deletion of CAC1 resulted in a uniform decrease across the regions tested. Deletion of both genes resulted in a further decrease of about 25% uniformly across the genomic region tested. In accordance with this observation in yeast, a genome-wide study in HeLa cells revealed an interplay between CAF-1 and HIRA in controlling histone dynamics and genome stability (24). Specifically, when CAF-1 is absent, HIRA assembles nucleosomes during replication. While it is still challenging to study functions of both CAF-1 and HIRA in higher eukaryotes due to their essential functions (25, 26), the significance of chromatin replication during DNA synthesis and cell division in epigenome maintenance is receiving more attention (27).
White-opaque switching in Candida albicans is a unique system in a unicellular organism that allows us to study factors important for the epigenetic maintenance of specialized cell types. White-opaque switching describes a phenotypic switch between white and opaque cells, and each cell type is stable for hundreds of cell divisions under routine laboratory conditions (28). Even though white and opaque cells both grow as single-celled yeasts, they have considerable differences in gene expression profiles, mating capacity, and biofilm formation, and several environmental and stress signals modulate an otherwise low switching frequency between cell types (29, 30). White- and opaque-cell-type specification in C. albicans is under the control of interlocking transcriptional feedback loops, with Wor1 being a master regulator of the white-opaque switch (31–34). Despite the large evolutionary distance between mammals and fungi, transcriptome analysis combined with Wor1 chromatin immunoprecipitation (ChIP) chip data suggests that the Wor1 circuit shares several characteristics with the transcriptional circuits for pluripotency of mammalian embryonic stem cells (34, 35). In addition to the transcriptional loops, histone-modifying enzymes constitute another layer of regulation for the white and opaque cell types. Recently, a comprehensive deletion analysis focusing on catalytic subunits of histone-modifying enzymes identified several enzymes that can modulate frequencies of switching and discovered that the Set3/Hos2 complex is a key regulator of white-opaque switching (36). HAT Rtt109 and HDAC Hst3, enzymes that control H3K56 acetylation status in C. albicans, also play a critical role in white-opaque switching (37). This study suggests that hypoacetylation of H3K56 acetylation resulted in opaque-cell-type instability, whereas hyperacetylation of H3K56 acetylation stimulated opaque-cell formation. This modification has been shown to specifically mark nascent H3 molecules, plays a role in replication-coupled and repair-coupled nucleosome assembly, and is needed for dynamic gene induction; chromatin lacking H3K56 acetylation results in an overall decrease in histone turnover rates (38–43). Enzymatic activity of Rtt109 for H3K56 is stimulated by the histone chaperone Asf1 (44). Interestingly, Asf1 has been shown to have biochemical and genetic links to both CAF-1 and HIR nucleosome assembly (14, 42, 45).
Since CAF-1 and HIR genes are uncharacterized in C. albicans, and given the critical role H3K56 acetylation status has in C. albicans (46, 47), we decided to investigate the importance of these two factors. To identify CAF-1 and HIR pathway-specific functions as well as any overlapping functions between CAF-1 and HIR in C. albicans, we generated a parsimonious set of single- and double-deletion mutants and found conserved and novel functions for these chaperones. Deletion of the CAF-1 subunit gene CAC2 resulted in UV, thermal, and genotoxic stress sensitivities. Deletion of HIR1 resulted in an increase in histone gene expression. Combining these two gene deletions resulted in a synergistic reduction of growth and chromosome missegregation. We then utilized the white-opaque switching system to characterize contributions of these factors to cell type formation and stability. Through genetic and pharmacological tests, we discovered a novel interplay between H3K56 acetylation status, CAF-1, and HIR in white-opaque switching.
MATERIALS AND METHODS
Strains and growth conditions.
Candida albicans strains used in this study are listed in Table 1. C. albicans cells were routinely maintained on YEPD agar plates (2% dextrose, 2% Bacto-peptone, 1% yeast extract, 2% agar) at room temperature and/or at 30°C.
Table 1.
Strains used in this study
| Strain | Descriptiona | Genotype | Reference or source |
|---|---|---|---|
| JYC5 | WT | MTLa/a ura3::imm434/ura3::imm434 | 31 |
| HLY4125 | hir1Δ/Δ | MTLa/a hir1Δ::FRT/hir1Δ::FRT | This study |
| HLY4126 | cac2Δ/Δ | MTLa/a cac2Δ::FRT/cac2Δ::FRT | This study |
| HLY4127 | hir1Δ/HIR1 | MTLa/a hir1Δ::FRT/HIR1::FRT | This study |
| HLY4128 | cac2Δ/CAC2 | MTLa/a cac2Δ::FRT/CAC2::FRT | This study |
| HLY4129 | hir1Δ/Δ cac2Δ/Δ | MTLa/a hir1Δ::FRT/hir1Δ::FRT cac2Δ::FRT/cac2Δ::FRT | This study |
| HLY3997 | rtt109Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT | 37 |
| HLY4130 | rtt109Δ/Δ hir1Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT hir1Δ::FRT/hir1Δ::FRT | This study |
| HLY4131 | rtt109Δ/Δ cac2Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT cac2Δ::FRT/cac2Δ::FRT | This study |
| HLY4132 | rtt109Δ/Δ hir1Δ/HIR1 | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT hir1Δ::FRT/HIR1::FRT | This study |
| HLY4133 | rtt109Δ/Δ cac2Δ/CAC2 | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT cac2Δ::FRT/CAC2::FRT | This study |
| HLY3555 | WT | MTLa/a ura3::imm434/ura3::imm434 | 31 |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| HLY4134 | hir1Δ/Δ | MTLa/a hir1Δ::FRT/hir1Δ::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| HLY4135 | cac2Δ/Δ | MTLa/a cac2Δ::FRT/cac2Δ::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| HLY4136 | hir1Δ/HIR1 | MTLa/a hir1Δ::FRT/HIR1::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| HLY4137 | cac2Δ/CAC2 | MTLa/a cac2Δ::FRT/CAC2::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| HLY4138 | hir1Δ/Δ cac2Δ/Δ | MTLa/a hir1Δ::FRT/hir1Δ::FRT cac2Δ::FRT/cac2Δ::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-SAT1-WOR1 | ||
| HLY3998 | rtt109Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT | 37 |
| +WOR1p-GFP | WOR1/PWOR1-GFP-SAT1-WOR1 | ||
| HLY4139 | rtt109Δ/Δ hir1Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT hir1Δ::FRT/hir1Δ::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-SAT1-WOR1 | ||
| HLY4140 | rtt109Δ/Δ cac2Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT cac2Δ::FRT/cac2Δ::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-SAT1-WOR1 | ||
| HLY4141 | rtt109Δ/Δ hir1Δ/HIR1 | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT hir1Δ::FRT/HIR1::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-SAT1-WOR1 | ||
| HLY4142 | rtt109Δ/Δ cac2Δ/CAC2 | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT cac2Δ::FRT/CAC2::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-SAT1-WOR1 | ||
| HLY4055 | WT | MTLa/a ura3::imm434/ura3::imm434 | 37 |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| +MAL2-WOR1-3×HA | ADH1/adh1::PMAL2-WOR1-3XHA-SAT1 | ||
| HLY4143 | hir1Δ/Δ | MTLa/a hir1Δ::FRT/hir1Δ::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| +MAL2-WOR1-3×HA | ADH1/adh1::PMAL2-WOR1-3XHA-SAT1 | ||
| HLY4144 | cac2Δ/Δ | MTLa/a cac2Δ::FRT/cac2Δ::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| +MAL2-WOR1-3×HA | ADH1/adh1::PMAL2-WOR1-3XHA-SAT1 | ||
| HLY4145 | hir1Δ/Δ cac2Δ/Δ | MTLa/a hir1Δ::FRT/hir1Δ::FRT cac2Δ::FRT/cac2Δ::FRT | This study |
| +WOR1p-GFP | WOR1/PWOR1-GFP-URA3-WOR1 | ||
| +MAL2-WOR1-3×HA | ADH1/adh1::PMAL2-WOR1-3XHA-SAT1 | ||
| HLY4057 | rtt109Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT | 37 |
| +MAL2-WOR1-3×HA | ADH1/adh1::PMAL2-WOR1-3XHA-SAT1 | ||
| HLY4146 | rtt109Δ/Δ hir1Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT hir1Δ::FRT/hir1Δ::FRT | This study |
| +MAL2-WOR1-3×HA | ADH1/adh1::PMAL2-WOR1-3×HA-SAT1 | ||
| HLY4147 | rtt109Δ/Δ cac2Δ/Δ | MTLα/α rtt109Δ::FRT/rtt109Δ::FRT cac2Δ::FRT/cac2Δ::FRT | This sudy |
| +MAL2-WOR1-3×HA | ADH1/adh1::PMAL2-WOR1-3XHA-SAT1 | This study | |
| HLY3993 | HST3/hst3 | MTLα/α ade2/ade2 ura3::ADE2/ura3::ADE2 HST3/hst3::FRT | 37 |
| JMR114 | hog1Δ/Δ | hog1Δ::ARG4/hog1Δ::URA3 | 48 |
WT, wild type.
Plasmid and strain construction.
To generate the HIR1 deletion construct, the HIR1 upstream and downstream regions were amplified with the primer pairs 5′HRR-1/5′HRR-2 and 3′HRR-3/3′HRR-4, respectively, digested at the introduced ApaI/XhoI and NotI/SacII sites, and substituted for the CZF1-flanking sequences in pCZF1M2 (49) cloned on both sides of the SAT1 flipper cassette to generate p1073. To generate the CAC2 deletion construct, the CAC2 upstream and downstream regions were amplified with the primer pairs 5′HRR-5/5′HRR-6 and 3′HRR-7/3′HRR-8, respectively, digested at the introduced ApaI, native XhoI, and introduced SacII/SacI sites, and substituted for the CZF1-flanking sequences in pCZF1M2 (49) cloned on both sides of the SAT1 flipper cassette to generate p1074. To disrupt the endogenous copies of HIR1, p1073 was linearized using ApaI and SacII followed by two sequential rounds of transformation, selection, and recycling of the SAT1 maker as described by Reuss et al. (50). To disrupt the endogenous copies of CAC2, p1074 was linearized using ApaI and SacI, followed by sequential rounds of transformation, selection, and recycling of the SAT1 maker as described by Reuss et al. (50). To generate the HIR1 reintroduction plasmid, the entire HIR1 open reading frame (ORF) was amplified using primers HORF-1/HORF-2, digested with introduced ApaI/SalI sites, and substituted for the 5′ upstream region in p1073 to generate p1075. To generate the CAC2 reintroduction plasmid, the entire CAC2 ORF was amplified using primers CORF-1/CORF-2, digested with introduced ApaI/SalI sites, and substituted for the 5′ upstream region in p1074 to generate p1076. Reintroduction of HIR1 carried out by restriction digestion of p1075 with PshAI. Reintroduction of CAC2 was carried out by restriction digestion of p1076 with XhoI. A detailed list of primers used and further descriptions can be found in Table S1 in the supplemental material.
Transformation.
C. albicans transformation was carried out by the Tris-EDTA (TE)–lithium acetate (LiAc)–polyethylene glycol (PEG) protocol with some minor changes. Mid-log-phase cells were pelleted at 3,000 rpm, washed gently in 50 volumes of a freshly prepared TE-LiAc mix (10 mM Tris [pH 7.5], 1 mM EDTA, 100 mM lithium acetate), transferred to 1.5-ml Eppendorf tubes, and pelleted at 3,000 rpm again. Pellets were resuspended in 1 pellet volume of TE-LiAc mix, and 30 to 50 μl of gently mixed cells was added to a 1.5-ml Eppendorf tube containing a mix of freshly boiled salmon sperm DNA (carrier DNA) and transforming DNA that had been predigested with appropriate restriction enzymes. Cells and DNA material were mixed and incubated at 30°C for 30 min, and then 700 μl of freshly made TE-LiAc mix plus 40% PEG MW-3350 (Sigma-Aldrich) was used to fully and gently resuspend the cell-DNA mixture. Tubes were then incubated at 30°C overnight with rotation. Strains were then heat shocked at 42°C for 50 to 60 min, depending on the strain, and allowed to recover for 4 to 8 h in 5 ml YEPD at 30°C, also depending on the strain, before plating onto selective medium containing 200 μg/ml nourseothricin (Werner Bioagents) as described previously (50). Correct integration was checked by PCR.
RNA isolation and qPCR.
RNA from 14 ml of log-phase cells at an optical density at 600 nm (OD600) of 0.8 was purified using Qiagen RNeasy minicolumns with on-column DNase I treatment (Qiagen). Two micrograms of total RNA was used to generate cDNA using SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR (qPCR) of synthesized cDNA was performed according to the iQ SYBR green Supermix instructions (Bio-Rad) and performed with an iCycler iQ detection system (Bio-Rad). Primers specific to 3′ untranslated regions for each histone gene were used, with primer annealing and extension temperatures set at 60°C. Melting curves were used to check for single amplicon products, and a genomic dilution series was used to generate standard curves for each primer pair used. The qPCR was performed in triplicate, and data are reported as means and standard deviations.
Propidium iodide staining and fluorescence-activated cell sorting (FACS) analysis.
C. albicans cells were stained with propidium iodide for cell cycle analysis by flow cytometry as described by Susan Forsburg for Schizosaccharomyces pombe (http://flowcyt.salk.edu/protocols/ycc.html). Before analysis, cells were sonicated on a Misonix sonicator 3000 using the cup horn attachment at an output level of 3 with 1-s on/off cycling for 20 s. The distribution of fluorescence from the WOR1p-GFP reporter gene across an entire cell population was investigated. The machine used for both propidium iodide (PI) staining and GFP detection was a dual laser fluorescence-activated cell sorter (BD FACSCalibur system; Becton, Dickinson, San Jose, CA). A minimum of 10,000 cells was routinely measured. The results were analyzed and graphed with FlowJo flow cytometry analysis software (version 8.7 for Mac).
Quantitative switching assays.
White cells were maintained on YEPD 2%–agar plates before being resuspended in 1× phosphate-buffered saline (PBS), serially diluted, and plated on test plates containing one of the following: synthetic complete medium with dextrose (SCD) [2% dextrose (wt/vol), 0.15% yeast nitrogen base without amino acids or ammonium sulfate (wt/vol), 0.2 mM inositol, 0.5% ammonium sulfate (wt/vol), the appropriate amount of amino acids, 2% agar (wt/vol)]; synthetic complete medium (SC) with 2% N-acetylglucosamine (GlcNAc) (wt/vol), where GlcNAc replaced dextrose; SCD plus 5 mM or 25 mM nicotinamide (Sigma-Aldrich); or YEP with 2% maltose. Plates were maintained at room temperature and scored after 1 to 2 weeks depending on the particular strain and test condition. A total of three or four independent switching tests were done for each strain and condition.
Spot tests.
Log-phase cultures were adjusted to a final OD600 of 0.1, and a 5-fold serial dilution was prepared in a 96-well plate. Then 2.5 μl of each dilution was hand pipetted onto test and control plates using an 8-channel multipipettor. All plates contained YEPD agar with or without nicotinamide (Sigma-Aldrich), sodium chloride (Sigma-Aldrich), or hydroxyurea (Sigma-Aldrich). Plates were incubated for 2 days at 30°C or 42°C (unless indicated differently) before image capture with a Fujifilm LAS-4000 imager.
RESULTS
Characterization of single CAF-1 and HIR mutants in C. albicans.
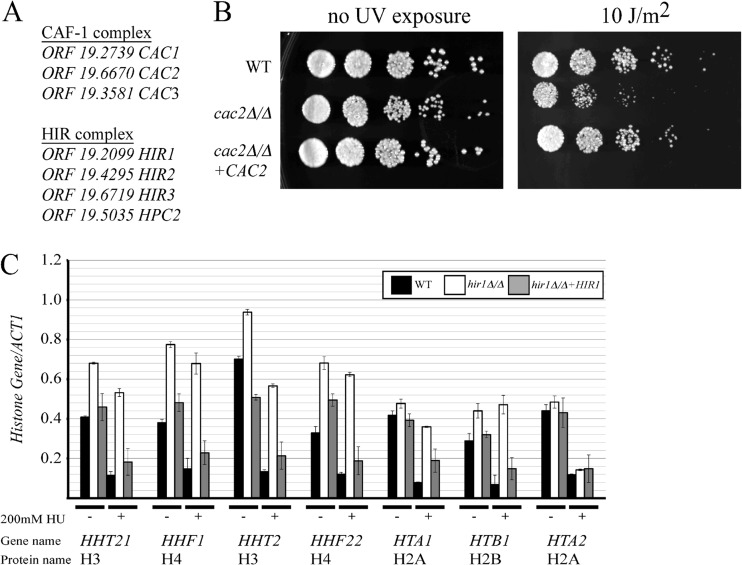
It was previously shown that deletion of any of the three CAF-1-encoding genes in S. cerevisiae (CAC1, CAC2, and CAC3) results in sensitivity to UV radiation, but no additive effects are seen when CAF-1 gene deletions are combined, indicating that deletion of any CAF-1-encoding gene is sufficient to disrupt CAF-1 function in yeast (7). The C. albicans genome encodes all three of the potential proteins of the CAF-1 complex based on sequence homology (Fig. 1A). Since deleting any one of the three genes is sufficient to disrupt CAF-1 functions, we decided to target C. albicans ORF 19.6670 (referred to here as CAC2) for gene disruption in the MTLa/a strain JYC5 by using the SAT1 flipper sequence-specific recombination system to replace the target locus with a single FLP recombination target (FRT) site after excision (50). Correct strain construction was determined by PCR. We were able to generate a homozygous CAC2 deletion strain, indicating that CAC2 is not essential in C. albicans. This strain displayed a small increase in doubling time but otherwise behaved normally under routine growth conditions. Since yeast strains with deletions of CAF-1-encoding genes are sensitive to UV radiation in S. cerevisiae (7), we decided to test if deletion of C. albicans CAC2 also resulted in sensitivity to UV radiation. Indeed, we found that deletion of CAC2 resulted in increased sensitivity to UV radiation and reintroduction of a full-length CAC2 restored this strain to wild-type levels of growth after UV radiation exposure (Fig. 1B). Therefore, ORF 19.6670 likely encodes a functional homolog of S. cerevisiae Cac2.
Fig 1.
CAF-1 and HIR have conserved functions in C. albicans. (A) List of C. albicans ORFs with proposed gene names. (B) Fivefold serial dilutions of the indicated strains on YEPD with or without exposure to UV radiation. Plates were incubated at 30°C for 24 h before image capture using a Fujifilm LAS-4000 imager. WT, wild type. (C) The indicated strains were grown to log phase at 30°C before RNA extraction, cDNA synthesis, and qPCR expression analysis. In a parallel set of cultures, the indicated strains were exposed to 200 mM hydroxyurea for 30 min before RNA extraction, cDNA synthesis, and qPCR expression analysis. The qPCR was performed in triplicate; standard deviations are shown by the error bars. cDNA was quantified using an iQ SYBR green Supermix (Bio-Rad) on an iCycler iQ detection system (Bio-Rad).
The genes encoding the HIR complex (HIR1, HIR2, HIR3, and HPC2) were first identified genetically as transcriptional repressors of three (HHT1-HHF1, HHT2-HHF2, and HTA1-HTB1) of the four histone gene pairs in S. cerevisiae (16, 17). Cells lacking HIR1 or HIR2 expressed histone genes throughout the cell cycle and failed to repress transcription of the three histone gene pairs in response to the inhibition of DNA replication (51). HTA2-HTB2 transcription is also repressed in response to hydroxyurea, but the regulation is independent of the HIR complex (16). Based on sequence homology, the C. albicans genome encodes all of the potential proteins of the HIR complex (Fig. 1A). We decided to target C. albicans ORF 19.2099 (referred to here as HIR1) for gene disruption in the MTLa/a strain JYC5 by using the SAT1 flipper system (50) and examine if histone gene expression is altered in the hir1Δ/Δ deletion strain (Fig. 1C). As in S. cerevisiae, we detected a modest increase in expression for three pairs of histone genes (HHT21-HHF1, HHT2-HHF22, and HTA1-HTB1) in the strain lacking HIR1 compared to the wild type and the HIR1 reintroduction strain. We then tested if HIR1 was necessary to rapidly repress histone gene transcription when the strain is exposed to hydroxyurea. The hir1Δ/Δ deletion strain was not able to repress the transcription of these three pairs of histone genes when exposed to hydroxyurea, in contrast to the wild type and the HIR1 reintroduction strain (Fig. 1C). The inability to repress histone gene transcription was not due to a defect in cell cycle arrest, because all strains tested arrested normally in the presence of hydroxyurea (data not shown). HTA2 expression was repressed in response to hydroxyurea in a Hir1-independent manner, as reported for S. cerevisiae (Fig. 1C). We concluded that C. albicans ORF 19.2099 encodes a functional homolog of S. cerevisiae Hir1.
Overlapping function of CAF-1 and HIR in proper chromosome segregation in C. albicans.
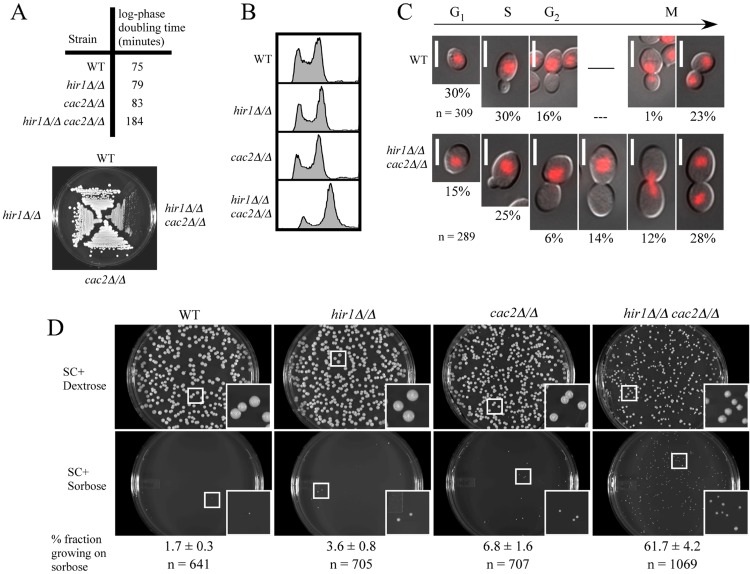
Several striking phenotypes are revealed when CAF-1 and HIR gene deletions are combined, and we became interested in seeing if any of these phenotypes were also observed in C. albicans. For example, deletion of both of these factors in S. cerevisiae results in increased rates of chromosome missegregation due to alterations of chromatin structure at centromeres (22). To investigate if CAF-1 and HIR had similar overlapping functions in C. albicans, we generated a hir1Δ/Δ cac2Δ/Δ double-deletion strain by deleting the CAC2 gene in the hir1Δ/Δ deletion background using the FLP/FRT system as described above. As observed in S. cerevisiae, we found that the hir1Δ/Δ cac2Δ/Δ mutant was not lethal, but this strain did grow significantly more slowly than either single mutant. The doubling time for the hir1Δ/Δ cac2Δ/Δ strain was more than twice that of the wild type and single mutants, and relative growth differences were obvious when cells were grown on plates for single colonies at 30°C (Fig. 2A). The growth defect was more pronounced in C. albicans than in the S. cerevisiae double mutant, because the growth defects in S. cerevisiae were observed at 37°C or 16°C (21, 22). The double mutant also showed a dramatic accumulation of 4N cells during log-phase growth by FACS analysis (Fig. 2B). This profile was not observed in wild-type cells or any of the single mutants. Both a cell cycle shift to G2/M phase and chromosomal aneuploidy could cause the observed accumulation of 4N cells. To determine if there was a defect in cell cycle progression in the hir1Δ/Δ cac2Δ/Δ strain, we analyzed bud size and nuclear localization of the propidium iodide-stained cells from the FACS analysis (Fig. 2C). As in S. cerevisiae, bud size and nuclear localization are landmarks for determining cell cycle stages in C. albicans. Based on these landmarks, we were able to determine the percentage of cells in each phase of the cell cycle and identify any abnormality in nuclear localization (Fig. 2C). We observed a decrease in the percentage of unbudded G1 cells and an increase in large-budded G2/M-phase cells. We also noticed the existence of a significant percentage (14%) of large-budded cells with the nucleus still in the mother cell in the double mutant, indicating a clear defect in chromosome segregation. This type of phenotype was not observed in wild-type cells. Conversely, fewer large-budded cells exhibited correct nuclear migration toward the daughter cells in the double mutant (6%) than in the wild-type cells (16%). Both observations indicate a defect in nuclear migration. Defects in chromosome segregation or nuclear migration are expected to cause aneuploidy. Aneuploidy in budding yeast leads to abnormally large cells (52). Consistent with the expected aneuploidy effect, the hir1Δ/Δ cac2Δ/Δ cells were observed to be larger than wild-type cells. In summary, the percentages of cells in G2/M phase were much higher for the double mutant (60%) than for the wild type (40%), consistent with a shift to 4N in the hir1Δ/Δ cac2Δ/Δ double mutant by FACS analysis. However, the percentage of 4N cells in the double mutant was higher than the observed 60% large-budded G2/M cells. This suggested that some of the 4N cells could be 4N in G1 or S phase due to the impaired chromosome separation that caused aneuploidy. These observations are consistent with what has been observed in S. cerevisiae (22).
Fig 2.
Overlapping function of CAF-1 and HIR in proper chromosome segregation in C. albicans. (A) Log-phase doubling times of strains grown in liquid YEPD medium at 30°C and relative growth of strains on streaked YEPD agar plates, photographed after 2 days at 30°C. (B) FACS profiles of propidium iodide-stained asynchronous log-phase cultures of the indicated strains. (C) Wild-type and hir1Δ/Δ cac2Δ/Δ cells used in the FACS analysis were photographed. Percentages of cells with similar morphology and DNA localization are listed below each photograph, as well as total cells examined (n). Bar, 5 μm. (D) About 200 cells of the indicated strains were plated on SC plus 2% dextrose or SC plus 2% sorbose agar and incubated at 30° for 5 days before being imaged. The fraction was calculated by dividing the number of visible colonies growing on SC plus 2% sorbose by the number of visible colonies growing on SC plus 2% dextrose. This experiment was done with two independent sets of plates, and the error associated with the fraction growing on sorbose is from these two tests. Images are from one set of plates, and the insets are at a magnification of ×3 relative to the original pictures.
To further determine if the hir1Δ/Δ cac2Δ/Δ double mutant had defects in chromosome segregation, we used a monosomy selection assay specific to C. albicans. The utilization of l-sorbose as the main carbon source in C. albicans requires monosomy of chromosome 5 (53). This quirk allows us to functionally test how faithful chromosome segregation events are, because when chromosome segregation is faithful, the ability to grow on sorbose is blocked due to chromosome 5 disomy, whereas if there are defects in chromosome segregation, then the ability to grow on sorbose is increased due to chromosome 5 monosomy. To test if our mutants had any singular or synergistic defects in proper chromosome stability, approximately 200 cells per strain were plated onto synthetic complete medium containing sorbose as the main carbon source and grown at 30°C for 6 days (Fig. 2D). Only the hir1Δ/Δ cac2Δ/Δ mutant showed significant numbers of colonies (61.7%) that grew on sorbose-containing plates, whereas very few colonies grew from wild-type (1.7%), hir1Δ/Δ (3.6%), and cac2Δ/Δ (6.8%) strains on sorbose medium. This result indicates that CAF-1 and HIR functionally overlap to maintain correct segregation of chromosome 5 in C. albicans. These findings are in accordance with previous studies in S. cerevisiae, where CAF-1 and HIR have overlapping functions in proper chromosome segregation (22), and we conclude that C. albicans CAF-1 and HIR have a similar overlapping function.
CAF-1 and HIR function in stress responses of C. albicans.
Stress response often requires an alteration of chromatin at target genes to repress or activate transcription occurring at these loci. It is thought that chromatin regulatory factors likely play a role in responding to stress because of the observation that chromatin regulatory mutants are often identified as having decreased tolerance to chemical and environmental stresses (54, 55).
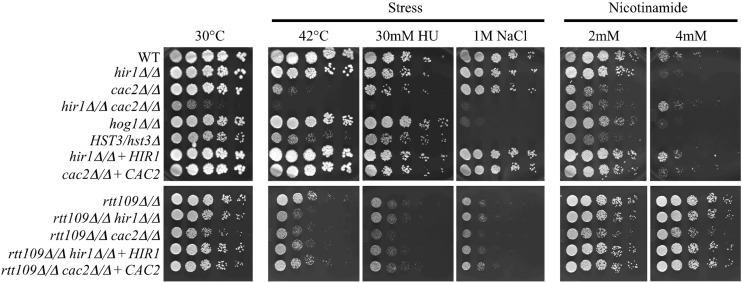
This view is further advanced by a systematic large-scale study that focused exclusively on the transcriptional dynamics of 170 target genes in response to amide stress using a combination of histone tail mutants and gene deletions in several classes of regulatory genes (56).These studies in S. cerevisiae revealed a functional role for CAF-1 and HIR in stress response, but synergistic effects were not investigated. Therefore, to determine if CAF-1 and HIR played pathway-specific or overlapping roles in stress response in C. albicans, we performed growth assays with our histone chaperone mutants in the presence of thermal (42°C), genotoxic (hydroxyurea), and osmotic (1 M NaCl) stress (Fig. 3). We found that deletion of HIR1 did not dramatically alter growth under any condition tested. However, the cac2Δ/Δ deletion strain displayed growth sensitivity under thermal and genotoxic stress but not osmotic stress. We next wanted to determine if any overlapping functions existed between CAF-1 and HIR in stress responses, so we tested the hir1Δ/Δ cac2Δ/Δ double mutant under the same stress conditions. The hir1Δ/Δ cac2Δ/Δ double mutant showed a high sensitivity to thermal, genotoxic, and osmotic stresses, as shown by limited growth. This suggested a possible synergy between CAF-1 and HIR in stress response, but the slow growth of the double mutant could have obscured greater-than-expected growth defects under these stress conditions.
Fig 3.
Growth phenotypes of chromatin regulatory mutants exposed to stresses and nicotinamide. A 5-fold serial dilution of log-phase cultures of the indicated strains. The controls were grown on YEPD agar at 30°C. The test cultures were grown on YEPD agar at 42°C and on YEPD supplemented with 30 mM hydroxyurea (HU), 1 M sodium chloride (NaCl), or 2 mM or 4 mM nicotinamide (NAM). All plates were incubated at 30°C except for the 42°C plates. All plates were imaged after 2 days using a Fujifilm LAS-4000 imager.
We then tested if the HST3/hst3 and rtt109Δ/Δ strains also displayed growth sensitivities to thermal, genotoxic, and osmotic stress (Fig. 3). It was found that these strains displayed growth sensitivities to hydroxyurea, as observed elsewhere (46, 47), and previously unreported growth sensitivities to thermal and osmotic stress in C. albicans. The osmotic sensitivities of the HST3/hst3 and rtt109Δ/Δ strains were similar to that of the hog1Δ/Δ strain. To dissect the genetic relationship between Rtt109 and the histone chaperones CAF-1 and HIR in stress response, we constructed rtt109Δ/Δ hir1Δ/Δ and rtt109Δ/Δ cac2Δ/Δ double-deletion mutants in the MTLα/α rtt109Δ/Δ strain described before (37). In general, these double-deletion mutants displayed growth phenotypes at levels comparable to that of the rtt109Δ/Δ single-deletion strain (Fig. 3), suggesting that Cac2 or Hir1 functions in the same pathway as Rtt109 in stress responses. However, they may function in additional pathways that are independent of Rtt109 in stress resistance, as deletion of HIR1 in rtt109Δ/Δ resulted in a slight increase in thermal sensitivity and the rtt109Δ/Δ cac2Δ/Δ double mutant displayed a slight increase in sensitivity to hydroxyurea. We conclude that CAF-1, HIR, and the enzymes that control H3K56 acetylation status all contribute to the stress response in C. albicans.
Growth sensitivity of chromatin regulatory mutants to nicotinamide.
Hst3 and, to some extent, Hst4 are the enzymes that remove H3K56 acetylation in S. cerevisiae (57, 58). Cells lacking Hst3p/Hst4p exhibit spontaneous DNA damage, chromosome loss, thermosensitivity, and acute sensitivity to genotoxic agents but are still viable. Unlike the case in S. cerevisiae, HST3 is an essential gene in C. albicans, and pharmacological inhibition of Hst3 with high levels of nicotinamide is toxic to C. albicans (37, 47). Several biochemical and genetic links between Rtt109-Asf1-H3K56 acetylation status and CAF-1 and HIR have been uncovered in S. cerevisiae, but less is known about the relationship between these histone chaperones, Hst3, and consequences of H3K56 hyperacetylation (14, 42, 45). Therefore, we decided to test if our histone chaperone mutants displayed any interactions with the Hst3 inhibitor nicotinamide (Fig. 3). We found that deletion of CAC2 resulted in sensitivity to nicotinamide. The level of nicotinamide sensitivity observed with the cac2Δ/Δ mutant was comparable to that with the HST3/hst3 heterozygous mutant, which is known to be haploinsufficient in response to nicotinamide (37). Deletion of HIR1 did not result in nicotinamide sensitivity. This indicates that the CAF-1 nucleosome assembly pathway normally mitigates the negative effects of H3K56 hyperacetylation in C. albicans.
We next determined if any overlapping function existed between CAF-1 and HIR in response to nicotinamide. In contrast to the nicotinamide sensitivity seen in the single cac2Δ/Δ deletion strain, the hir1Δ/Δ cac2Δ/Δ strain actually displayed a modest amount of tolerance to nicotinamide. This level of tolerance was greater than that in wild-type cells, considering that the hir1Δ/Δ cac2Δ/Δ strain displayed significant reduction in growth even in the absence of nicotinamide. Comparing the hir1Δ/Δ cac2Δ/Δ double mutant to the single rtt109Δ/Δ deletion strain, we conclude that removing the enzyme responsible for H3K56 acetylation, Rtt109, affords a significantly greater amount of tolerance to nicotinamide than inactivation of CAF-1 and HIR. Furthermore, the rtt109Δ/Δ cac2Δ/Δ double mutation completely eliminated the nicotinamide hypersensitivity seen in the single cac2Δ/Δ strain, revealing that Rtt109 and H3K56 acetylation is responsible for the nicotinamide hypersensitivity seen in the cac2Δ/Δ strain.
Taken together, our results indicate that hyperacetylation at histone H3K56 via inhibition of Hst3 by nicotinamide is toxic to C. albicans, and CAF-1 plays an important role in maintaining proper levels of H3K56 acetylation in chromatin in wild-type cells, as the cac2Δ/Δ mutant was as sensitive as the HST3/hst3 mutant to nicotinamide. Reducing levels of histone H3K56 acetylation by deleting RTT109 could rescue the sensitivity of cac2Δ/Δ cells to nicotinamide. Alternatively, blocking the histone chaperone function of HIR in cac2Δ/Δ could also alleviate the sensitivity of cac2Δ/Δ cells to nicotinamide. Thus, an interplay between functions of histone chaperones and global levels of H3K56 acetylation is important for proper cellular functions and growth.
Spontaneous and GlcNAc-induced switching profiles of CAF-1 and HIR deletions.
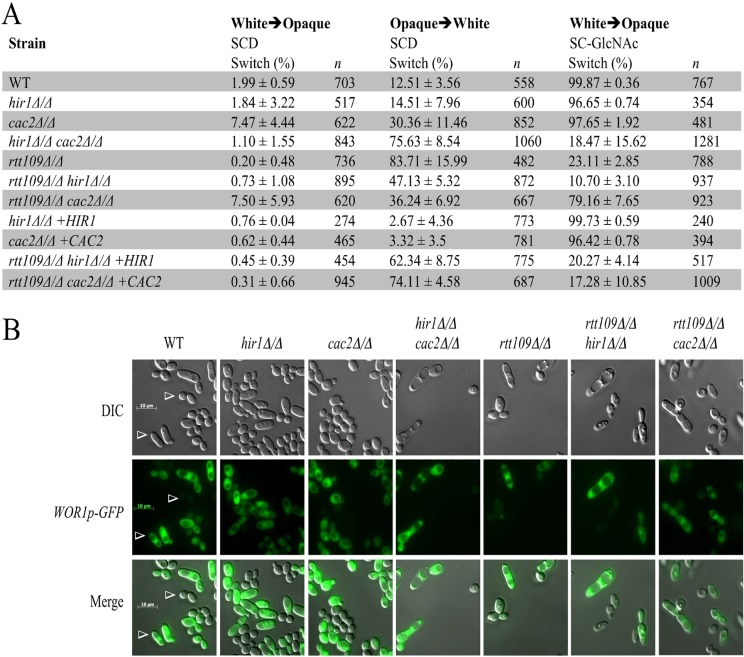
Due to the essential nature of CAF-1 and HIR in higher eukaryotes (25, 26), designing studies investigating these factors in cell type formation and maintenance is a challenge. The white-opaque switching system in C. albicans is a useful eukaryotic unicellular system for investigating the importance of CAF-1 and HIR in epigenetic regulation of cell type formation and maintenance. To this end, we first performed quantitative switching assays with three histone chaperone mutants (hir1Δ/Δ, cac2Δ/Δ, and hir1Δ/Δ cac2Δ/Δ mutants) to assess if these mutants displayed positive or negative roles in spontaneous switching and switching induced by N-acetylglucosamine (GlcNAc), an environmental molecule known to induce opaque-cell formation at high frequencies (59) (Fig. 4A). We found that deletion of CAC2 slightly increased spontaneous switching frequencies in both switching directions, whereas deletion of HIR1 resulted in frequencies that largely resembled those of the wild type. We also found that either gene was dispensable for GlcNAc-induced switching. The hir1Δ/Δ cac2Δ/Δ double-deletion strain had a spontaneous white-to-opaque switching frequency that resembled the frequency of wild-type cells but formed small sectors (data not shown). However, the double mutant spontaneously switched from opaque to white cells at a high frequency (75.6%) which was comparable to the switching frequency of the rtt109Δ/Δ deletion strain (83.7%) that we previously determined was unable to maintain the opaque cell type (37). The hir1Δ/Δ cac2Δ/Δ double-deletion strain also phenotypically copied the rtt109Δ/Δ deletion strain on GlcNAc inducing medium, because both strains showed much-reduced switching frequencies in the presence of GlcNAc (Fig. 4A). The wide-type strain switched to opaque at near 100%, whereas strains with the hir1Δ/Δ cac2Δ/Δ and the rtt109Δ/Δ deletions switched at around 18% and 23%, respectively. However, both mutants could still respond to the GlcNAc signal, as the increases in the frequency of GlcNAc-stimulated switching relative to the spontaneous switching frequency were significant. Taken together, these results suggest that the histone chaperones CAF-1 and HIR play positive overlapping roles in forming and maintaining the opaque cell type.
Fig 4.
Spontaneous and GlcNAc-induced switching phenotypes of CAF-1 and HIR mutants. (A) Quantitative switching assays were performed to determine the spontaneous and N-acetylglucosamine (GlcNAc)-induced switching frequencies for each mutant. Values are average percentages of total colonies displaying a sector or colony that switched phenotypes ± the standard deviations for the independent tests done for each strain. A total of 3 or 4 independent tests were done on different days for each strain. Switching assays were performed on synthetic complete agar plates left at room temperature for 1 to 2 weeks, depending on the strain used. (B) White- and opaque-cell phenotypes of indicated strains. The opaque cells express a WOR1p-GFP reporter gene, whereas white cells lack fluorescence (arrowheads). White and opaque cells were collected from white and opaque sectors taken from SC-GlcNAc-containing plates. Cells were mixed together and imaged. Bar, 10 μm. Images were obtained by differential interference contrast microscopy (DIC).
We next determined if any genetic interactions existed between the chaperone mutants and the rtt109Δ/Δ deletion strain in spontaneous and GlcNAc-induced switching (Fig. 4A). We found that the rtt109Δ/Δ hir1Δ/Δ double mutant resembled the rtt109Δ/Δ single mutant in spontaneous opaque-cell formation, whereas the rtt109Δ/Δ cac2Δ/Δ strain had a switching phenotype that resembled that of the cac2Δ/Δ single mutant. We also found that the rtt109Δ/Δ cac2Δ/Δ double-deletion strain could restore opaque-cell formation on GlcNAc, whereas the rtt109Δ/Δ hir1Δ/Δ double mutant had a switching frequency that resembled that of the rtt109Δ/Δ single mutant under this condition. These data suggest that removing CAF-1-mediated replication-coupled nucleosome assembly could suppress the negative effect of H3K56 hypoacetylation in both spontaneous and GlcNAc-induced opaque-cell formation. Interestingly, deletion of either CAC2 or HIR1 in the rtt109Δ/Δ deletion background resulted in a mild suppression of the abnormally high opaque-white switching frequency observed in the rtt109Δ/Δ single-deletion strain. These data reveal that removal of either CAF-1 or HIR nucleosome deposition activity could partially suppress the negative effect on opaque cell type maintenance when Rtt109 is absent. Together this mutant analysis indicates that CAF-1 is the major factor responsible for the low white-opaque switching frequency observed when Rtt109 is absent, and both histone chaperones promote opaque-white switching when Rtt109 is absent. Representative white- and opaque-cell morphology and WOR1 expression of wild-type and mutant cells are shown in Fig. 4B. All cells of the white and opaque types displayed the expected bistable fluorescence signal of the WOR1-GFP reporter gene. With regard to cellular morphologies, all strains tested displayed the expected round and elongated phenotypes of the characteristic white and opaque cell types, respectively, except for the cac2Δ/Δ strain, where the opaque cells were round rather than elongated.
Wor1 acts downstream of histone chaperone mutants.
We have previously shown that ectopic Wor1 expression can bypass the requirement for Rtt109 in opaque-cell formation (37). To dissect the relationship between the chaperones and Wor1, we transformed the maltose-inducible MAL2p-WOR1-3×HA transgene into all of our single and double mutants and tested for opaque colony formation on maltose-containing solid-agar plates (Table 2). We found that all single- and double-deletion strains carrying the WOR1 transgene were able to switch at frequencies resembling that of the wild type. This result indicates that ectopic expression of Wor1 can bypass the need for CAF-1, HIR, and Rtt109 to form opaque cells.
Table 2.
Wor1 can bypass the requirement for CAC2, HIR1, and RTT109
| Genotype | White→opaque switching on YEP-maltosea |
|
|---|---|---|
| % | n | |
| WT | ||
| Control | 0.11 ± 0.25 | 836 |
| +MAL2p-WOR1-3×HA | 99.80 ± 0.40 | 503 |
| hir1Δ/Δ | ||
| Control | 0.11 ± 0.22 | 862 |
| +MAL2p-WOR1-3×HA | 100 ± 0.00 | 580 |
| cac2Δ/Δ | ||
| Control | 1.71 ± 0.69 | 708 |
| +MAL2p-WOR1-3×HA | 99.38 ± 0.51 | 958 |
| hir1Δ/Δ cac2Δ/Δ | ||
| Control | 0.35 ± 0.40 | 858 |
| +MAL2p-WOR1-3×HA | 100.00 ± 0.00 | 736 |
| rtt109Δ/Δ | ||
| Control | 0.06 ± 0.29 | 1,272 |
| +MAL2p-WOR1-3×HA | 99.00 ± 0.15 | 1,432 |
| rtt109Δ/Δ hir1Δ/Δ | ||
| Control | 0.70 ± 0.72 | 1,032 |
| +MAL2p-WOR1-3×HA | 97.69 ± 0.72 | 1,195 |
| rtt109Δ/Δ cac2Δ/Δ | ||
| Control | 1.56 ± 0.74 | 1,079 |
| +MAL2p-WOR1-3×HA | 98.10 ± 1.32 | 1,043 |
White cells carrying a MAL2p-WOR1-3×HA transgene or control vector (GFP) were plated on YEP–2% maltose test plates and scored for opaque-cell formation after 1 to 2 weeks growth at room temperature. Values are average percentages of total colonies (n) displaying a sector that switched phenotypes ± standard deviations for 2 independent tests done on different days for each strain.
Interplay between nicotinamide inhibition of Hst3 and histone chaperone mutants in epigenetic switching.
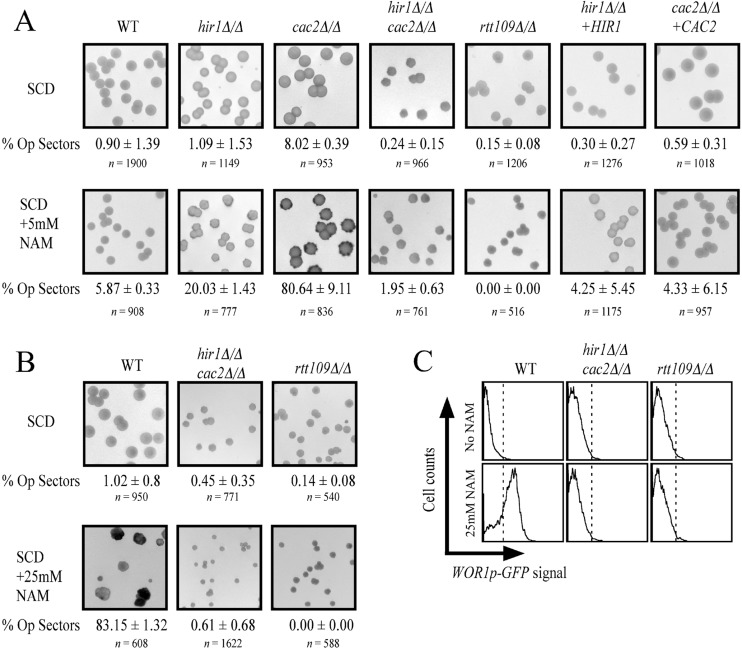
We previously found that nicotinamide induces opaque-cell formation in an RTT109-dependent manner (37). Our lab and others also discovered that nicotinamide targets the NAD-dependent histone deacetylase Hst3 in C. albicans (37, 47), and with these data together, it has become clear that hyperacetylation of H3K56 promotes opaque-cell formation and hypoacetylation makes opaque cells switch back to white at high frequency. We were interested to determine the functional relationship between the inhibition of Hst3 by nicotinamide and the nucleosome deposition chaperones CAF-1 and HIR in white-opaque switching. We grew our mutants in the presence of low nicotinamide (5 mM), where switching is mild for wild-type cells (5.87%), and found that the cac2Δ/Δ strain was sensitive to nicotinamide-induced switching, producing opaque sectors in a large majority (80.64%) of colonies scored visually by using a WOR1p-GFP reporter gene (Fig. 5A). In contrast, the hir1Δ/Δ deletion strain displayed a smaller increase in switching (∼20%). Therefore, the CAF-1-mediated nucleosome assembly pathway normally dampens nicotinamide induced switching, and the HIR complex plays a less important role. We also found that the hir1Δ/Δ cac2Δ/Δ double mutant had a low switching frequency (1.95%) on 5 mM nicotinamide, more comparable to that of wild-type cells than to that of either of the single mutants. Since the double mutant displayed some tolerance to nicotinamide (Fig. 3), we also tested it for high-nicotinamide (25 mM)-induced switching. While the wild-type strain showed 83.15% opaque sector formation, the hir1Δ/Δ cac2Δ/Δ double mutant hardly switched (0.61%) in the presence of high nicotinamide, which mimicked the low switching frequency seen in the rtt109Δ/Δ strain (Fig. 5B). Therefore, the hir1Δ/Δ cac2Δ/Δ double mutant blocked nicotinamide-induced opaque formation, phenotypically copying the rtt109Δ/Δ strain. The rtt109Δ/Δ cac2Δ/Δ strain also displayed insensitivity to nicotinamide-induced switching (data not shown). These data suggest that in the absence of CAF-1, HIR contributes to nicotinamide-induced opaque-cell formation, which is consistent with the nicotinamide growth phenotypes shown in Fig. 3.
Fig 5.
Interplay between H3K56 hyperacetylation and histone chaperone mutants in epigenetic switching. (A) White cells of the indicated strains were plated to synthetic complete medium containing 2% dextrose (SCD) and SCD supplemented with 5 mM nicotinamide and scored for opaque sector formation after 2 weeks growth at room temperature. The images shown are of representative agar plates with colonies and were taken using a Fujifilm LAS-4000 imager set to detect GFP fluorescence. The colonies are at a magnification of ×3, similar to the insets in Fig. 2D. The GFP fluorescence is seen as dark grey, whereas a lack of GFP signal displays as light grey. Correct phenotypes were also determined by correct cellular morphologies. “% Op sectors” is the percentage of total colonies displaying a sector that switched phenotypes (average ± standard deviation for independent tests done for each strain). A total of 2 or 3 independent tests were done on different days for each strain. (B) Similar switching assay, except that nicotinamide was at a concentration of 25 mM. (C) Flow cytometry detecting WOR1p-GFP fluorescence in cells taken from panel B.
DISCUSSION
CAF-1 and HIR are highly conserved histone chaperone protein complexes that function in the assembly of nucleosomes onto chromatin. CAF-1 is characterized as having replication-coupled nucleosome activity, whereas the HIR complex can assemble nucleosomes independently of replication. In this study, we used an efficient set of deletion mutants to genetically characterize single and overlapping functions of CAF-1 and HIR in C. albicans.
We confirmed that C. albicans CAF-1 and HIR play conserved roles in UV radiation recovery and repression of histone gene expression, respectively, as reported for S. cerevisiae (7, 47). These histone chaperone complexes also play overlapping roles in correct chromosome segregation in C. albicans, an observation also seen in S. cerevisiae (22). The chromosome missegregation in S. cerevisiae is due to a defect in centromeric chromatin organization, as CAF-1 and HIR proteins (Cac1, Cac2, and Hir1) colocalize with centromere-specific proteins, and cac1Δ hir1Δ cells display extracentromeric localization of the centromere-specific histone H3 variant Cse4 (22). C. albicans carries 3 to 5 kb of long unique centromeric chromatin on each of its eight chromosomes (60). These regional centromeres have unusual chromatin structures, and Cse4 incorporation onto these regions has been shown to be epigenetically regulated (60, 61). It is tempting to postulate that CAF-1 and HIR might play a similar role in controlling Cse4 localization on chromosomes in C. albicans. This might contribute to our understanding of how centromere-specific histone H3 variant Cse4 is correctly deposited to form specialized chromatin structures at centromeres in C. albicans.
We discovered a novel function of CAF-1 in alleviating the negative effect of H3K56 hyperacetylation. Unlike S. cerevisiae, nicotinamide is toxic to C. albicans. This toxicity is mediated through increasing levels of H3K56 acetylation by inhibition of Hst3, because deletion of Rtt109 results in insensitivity to nicotinamide (37, 47). In this study, we uncovered negative and positive genetic interactions between CAC2, HIR1, and H3K56 acetylation. The cac2Δ/Δ mutant is more sensitive to nicotinamide than wild-type cells, while the rtt109Δ/Δ cac2Δ/Δ and hir1Δ/Δ cac2Δ/Δ mutants are resistant to nicotinamide. Since CAF-1 and HIR function in the assembly of nucleosomes onto chromatin, one explanation of our genetic data is that high levels of H3K56 acetylation in chromatin is the major cause of nicotinamide toxicity. The fact that cac2Δ/Δ cells show increased sensitivity to nicotinamide suggests an important function for CAC2 and, by extension, that the CAF-1 complex is critical in maintaining proper levels of H3K56 acetylation in chromatin. However, it should be noted that CAF-1 subunit-specific functions are known to exist in S. cerevisiae (6), and some of the phenotypes observed here might be specific to the removal of CAC2 rather than CAF-1 disruption. Nevertheless, we found that CAC2, and most likely CAF-1, plays a critically important role in mitigating the negative effect of nicotinamide in C. albicans. Hence we have discovered a novel function of CAF-1 that might extend to other clinically important pathogenic fungi that have also displayed sensitivities to nicotinamide (47). Although in S. cerevisiae, HST3 is not essential and nicotinamide is not toxic, HST3 is essential in C. albicans, and high levels of nicotinamide are toxic to C. albicans and several other clinically important pathogenic fungi (47). Proper control of H3K56 acetylation levels in chromatin is probably important in those fungi, and CAF-1 may play a similar role in mitigating H3K56 hyperacetylation.
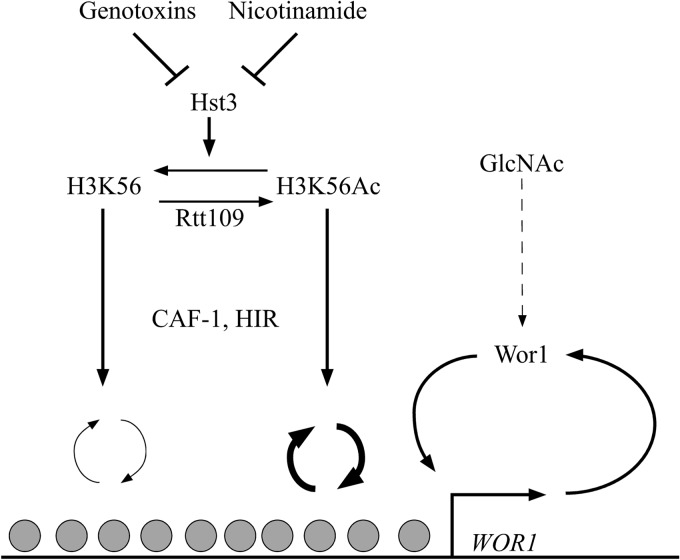
We also discovered overlapping functions of CAF-1 and HIR in epigenetic regulation of cell fate determination. Like the rtt109Δ/Δ mutant, the hir1Δ/Δ cac2Δ/Δ double mutant is defective in maintaining the opaque cell type and blocks nicotinamide-induced opaque-cell formation. Ectopic expression of Wor1 can bypass the defect of both the hir1Δ/Δ cac2Δ/Δ double mutant and rtt109Δ/Δ mutant in opaque-cell formation. These similarities suggest that the histone chaperones CAF-1 and HIR might functionally overlap in the same pathway as Rtt109 to contribute to opaque-cell-type formation and maintenance (Fig. 6). The chaperones CAF-1 and HIR function downstream of H3K56 acetylation, as deletion of CAC2 can partially suppress the defect of rtt109Δ/Δ cells in opaque-cell formation, and both rtt109Δ/Δ hir1109Δ/Δ and rtt109Δ/Δ cac2109Δ/Δ can partially stabilize the opaque cell type. Although it has not been demonstrated in C. albicans, we postulate that Rtt109 and histone chaperones control dynamic histone exchange, because a global reduction of histone H3 exchange is observed in S. cerevisiae strains lacking Rtt109 or CAF-1 and HIR. Specifically, deletion of Rtt109 and of Asf1 both slowed turnover at “hot” nucleosome regions, suggesting that H3K56 acetylation specifically enhances replacement in budding yeast (41). Similarly, it was shown that HIR plays a positive role in histone H3 exchange at gene-rich regions, whereas CAF-1 uniformly promoted histone H3 exchange across the region probed (23). Given this information, we favor the view that dynamic histone exchange might play a significant role in promoting and maintaining the opaque cell type, with the WOR1 locus being central to this process. A threshold of histone H3 exchange might be important for removing repressive chromatin marks on histone tails that are specific to white or opaque cell types. For example, several histone-modifying enzymes that add or remove histone marks, e.g., Rpd3, Hda1, Set3/Hos2, and Nat4, have already been shown to affect spontaneous switching frequencies (36). Dynamic histone exchange might also make chromatin more accessible to the transcription machinery to establish an interlocking transcriptional feedback loop to maintain cellular identity. It is known that nucleosomes serve as general transcriptional repressors and compete with transcription factors to bind DNA sequences. A recent comprehensive analysis of several genome-wide data sets showed that transcriptional activator binding sites correlate more with nucleosome binding than transcriptional repressor binding sites do (62). Binding of activators to sites usually bound by nucleosomes results in disruption, unwinding, or sliding of the corresponding nucleosomes. This permits other transcriptional activators and the transcription initiation complex to bind these regions and initiate transcription. Therefore, we postulate that histone turnover by CAF-1 and HIR is juxtaposed with transcriptional activators at WOR1 to set and maintain the cell state. We propose a convergent model of transcriptional regulation of WOR1 (Fig. 6), where promoter chromatin and transcription factors all contribute to WOR1 transcription. Genotoxins and nicotinamide downregulate Hst3, which negatively regulates H3K56Ac (37). CAF-1 and HIR function downstream of Rtt109 and assemble nucleosomes onto chromatin in an H3K56 acetylation-associated manner, thereby imparting the function of chromatin at the WOR1 locus on its transcription. The rtt109Δ/Δ mutant and the hir1Δ/Δ cac2Δ/Δ double mutant had completely blocked nicotinamide-induced opaque formation, whereas they still responded to GlcNAc induced switching, albeit not as efficiently as other strains. Therefore, nicotinamide acts through the Rtt109 and the histone chaperones in regulating white-opaque switching, while GlcNAc signals through a different pathway, possibly through regulating Wor1 activity, as suggested by Huang et al. (59). WOR1 transcription is placed downstream of all factors, since ectopic expression of WOR1 by a strong promoter can bypass all of the mutants tested. How Wor1 recruits the transcriptional machinery is currently unknown, as is the mechanism of de novo expression of WOR1.
Fig 6.
Schematic diagram of a genetic pathway for H3K56 acetylation and histone chaperones CAF-1 and HIR in epigenetic switching. H3K56 acetylation is controlled by the histone acetyltransferase Rtt109 and deacetylated by the NAD-dependent histone deacetylase Hst3. CAF-1 and HIR mediate nucleosome assembly functions downstream of Rtt109. This pathway is responsive to genotoxins and nicotinamide through inhibition of Hst3 (37) and functions in parallel to GlcNAc, which activates Wor1. Hst3 is known to actively remove H3K56 acetylation marks during chromatin maturation, whereas Rtt109 activity is on free histones before deposition (39, 40, 57, 58). Line thickness indicates relative nucleosome turnover dynamics.
We found that CAF-1 plays a major role in maintaining cell types, as the cac2Δ/Δ mutant exhibited increased frequencies of white-opaque switching in both directions. We further showed that CAF-1 interplays with H3K56 acetylation in epigenetic regulation of cell fate determination. The cac2Δ/Δ mutant switched to the opaque cell type at a high frequency under a low level of nicotinamide, demonstrating a negative interaction between the cac2Δ/Δ strain and nicotinamide. This synthetic interaction could be suppressed by deletion of RTT109 or HIR1, revealing interconnectedness between H3K56 acetylation and nucleosome assembly. Interestingly, the rtt109Δ/Δ cac1Δ/Δ mutant largely resembled the wild type in terms of phenotypic switching, revealing that the replication-coupled nucleosome assembly pathway is mostly responsible for the unstable opaque phenotype seen in the rtt109Δ/Δ strain. Altogether, our genetic data suggest that CAF-1 plays a predominant role over HIR in maintaining cell types in C. albicans. How epigenetic information is transmitted during semiconservative replication is an interesting topic (27, 63). With recent technical advances in measuring histone protein inheritance and histone partitioning in lower and higher eukaryotes, respectively (64, 65), we will have a better understanding of the mechanistic basis for epigenome maintenance. Recently, a system was developed that makes it possible to assess the effects of artificial induction of a heterochromatin domain on gene expression in vivo (66). Active gene transcription was associated with significant chromatin remodeling, with histone replacement resulting in efficient erasure of repressive marks. When the induction system was turned off, it was found that the heterochromatic domains were heritably transmitted through multiple generations. The silenced state was maintained by local recruitment of repressive chromatin marks, which induces spreading of the repressive domain, and further recruitment of additional repressive marks like DNA methylation. Together, these data indicated that dynamic competition between histone marking and turnover determines the boundaries and stability of heterochromatic domains. A combination of such temporal dynamic studies of establishment and memory of histone marks and transcription states with chromatin mutants will shed light on mechanisms of epigenetic regulation.
ACKNOWLEDGMENTS
We thank members of the Liu lab for helpful discussions.
Work in the Liu laboratory is supported by the National Institutes of Health grants R01GM/AI55155-14A and R01AI099190-01 to H.L.
Footnotes
Published ahead of print 15 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00334-12.
REFERENCES
- 1. Kornberg RD. 1977. Structure of chromatin. Annu. Rev. Biochem. 46:931–954 [DOI] [PubMed] [Google Scholar]
- 2. Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 3. Millar CB, Grunstein M. 2006. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 7:657–666 [DOI] [PubMed] [Google Scholar]
- 4. Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12:7–18 [DOI] [PubMed] [Google Scholar]
- 5. De Koning L, Corpet A, Haber JE, Almouzni G. 2007. Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14:997–1007 [DOI] [PubMed] [Google Scholar]
- 6. Eitoku M, Sato L, Senda T, Horikoshi M. 2008. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell. Mol. Life Sci. 65:414–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaufman PD, Kobayashi R, Stillman B. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11:345–357 [DOI] [PubMed] [Google Scholar]
- 8. Stillman B. 1986. Chromatin assembly during SV40 DNA replication in vitro. Cell 45:555–565 [DOI] [PubMed] [Google Scholar]
- 9. Shibahara K, Stillman B. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575–585 [DOI] [PubMed] [Google Scholar]
- 10. Enomoto S, Berman J. 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12:219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. 1997. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 11:358–370 [DOI] [PubMed] [Google Scholar]
- 12. Monson EK, de Bruin D, Zakian VA. 1997. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl. Acad. Sci. U. S. A. 94:13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith JS, Caputo E, Boeke JD. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19:3184–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, III, Kaufman PD. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15:2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL. 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19:2534–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Osley MA, Lycan D. 1987. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 7:4204–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu H, Kim UJ, Schuster T, Grunstein M. 1992. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anderson HE, Wardle J, Korkut SV, Murton HE, Lopez-Maury L, Bahler J, Whitehall SK. 2009. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol. Cell. Biol. 29:5158–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162:1557–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nourani A, Robert F, Winston F. 2006. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:1496–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaufman PD, Cohen JL, Osley MA. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sharp JA, Franco AA, Osley MA, Kaufman PD. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lopes da Rosa J, Holik J, Green EM, Rando OJ, Kaufman PD. 2011. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz David C, Pchelintsev Nikolay A, Adams Peter D, Jansen Lars ET, Almouzni G. 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44:928–941 [DOI] [PubMed] [Google Scholar]
- 25. Houlard M, Berlivet S, Probst AV, Quivy Héry J-PP, Almouzni G, Gérard M. 2006. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2:e181 doi:10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. 2002. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol. 22:2318–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13:153–167 [DOI] [PubMed] [Google Scholar]
- 28. Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lohse MB, Johnson AD. 2009. White-opaque switching in Candida albicans. Curr. Opin. Microbiol. 12:650–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soll DR. 2009. Why does Candida albicans switch? FEMS Yeast Res. 9:973–989 [DOI] [PubMed] [Google Scholar]
- 31. Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. 2006. Bistable expression of WOR1, a master regulator of white–opaque switching in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 103:12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 5:1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zordan RE, Galgoczy DJ, Johnson AD. 2006. Epigenetic properties of white–opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. U. S. A. 103:12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. 2007. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 5:e256–e256 doi:10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. 2010. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 6:e1001070–e1001070 doi:10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hnisz D, Schwarzmüller T, Kuchler K. 2009. Transcriptional loops meet chromatin: a dual-layer network controls white–opaque switching in Candida albicans. Mol. Microbiol. 74:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stevenson JS, Liu H. 2011. Regulation of white and opaque cell-type formation in Candida albicans by Rtt109 and Hst3. Mol. Microbiol. 81:1078–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen C-C, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. 2008. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134:231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Driscoll R, Hudson A, Jackson SP. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315:649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han J, Zhou H, Horazdovsky B, Zhang K, Xu Zhang R-MZ. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315:653–655 [DOI] [PubMed] [Google Scholar]
- 41. Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. 2008. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4:e1000270–e1000270 doi:10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134:244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Masumoto H, Hawke D, Kobayashi R, Verreault A. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294–298 [DOI] [PubMed] [Google Scholar]
- 44. Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, Kaufman PD, Allis CD. 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. U. S. A. 103:6988–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463–473 [DOI] [PubMed] [Google Scholar]
- 46. Lopes da Rosa J, Boyartchuk VL, Zhu LJ, Kaufman PD. 2010. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:1594–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wurtele H, Tsao S, Lepine G, Mullick A, Tremblay J, Drogaris P, Lee Thibault E-HP, Verreault A, Raymond M. 2010. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat. Med. 16:774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rauceo JM, Blankenship JR, Fanning S, Hamaker JJ, Deneault Smith J-SFJ, Nantel A, Mitchell AP. 2008. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell 19:2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramírez-Zavala B, Reuss O, Park Ohlsen Y-NK, Morschhäuser J. 2008. Environmental induction of white–opaque switching in Candida albicans. PLoS Pathog. 4:e1000089–e1000089 doi:10.1371/journal.ppat.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 51. Sherwood PW, Tsang SV, Osley MA. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. 1999. Ploidy regulation of gene expression. Science 285:251–254 [DOI] [PubMed] [Google Scholar]
- 53. Janbon G, Sherman F, Rustchenko E. 1998. Monosomy of a specific chromosome determines l-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 95:5150–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kapitzky L, Beltrao P, Berens TJ, Gassner N, Zhou C, Wuster A, Wu J, Babu MM, Elledge SJ, Toczyski D, Lokey RS, Krogan NJ. 2010. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol. Syst. Biol. 6:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sinha H, David L, Pascon RC, Clauder-Munster S, Krishnakumar S, Nguyen M, Shi G, Dean J, Davis RW, Oefner PJ, McCusker JH, Steinmetz LM. 2008. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180:1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weiner A, Chen HV, Liu CL, Rahat A, Klien A, Soares L, Gudipati M, Pfeffner J, Regev A, Buratowski S, Pleiss JA, Friedman N, Rando OJ. 2012. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 10:e1001369 doi:10.1371/journal.pbio.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. 2006. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr. Biol. 16:1280–1289 [DOI] [PubMed] [Google Scholar]
- 58. Maas NL, Miller KM, DeFazio LG, Toczyski DP. 2006. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23:109–119 [DOI] [PubMed] [Google Scholar]
- 59. Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. 2010. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806–e1000806 doi:10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. 2006. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 103:14877–14882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanyal K, Baum M, Carbon J. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. U. S. A. 101:11374–11379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Charoensawan V, Janga SC, Bulyk ML, Babu MM, Teichmann SA. 2012. DNA sequence preferences of transcriptional activators correlate more strongly than repressors with nucleosomes. Mol. Cell 47:183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaufman PD, Rando OJ. 2010. Chromatin as a potential carrier of heritable information. Curr. Opin. Cell Biol. 22:284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. 2011. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 9:e1001075 doi:10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. 2010. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328:94–98 [DOI] [PubMed] [Google Scholar]
- 66. Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. 2012. Dynamics and memory of heterochromatin in living cells. Cell 149:1447–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]