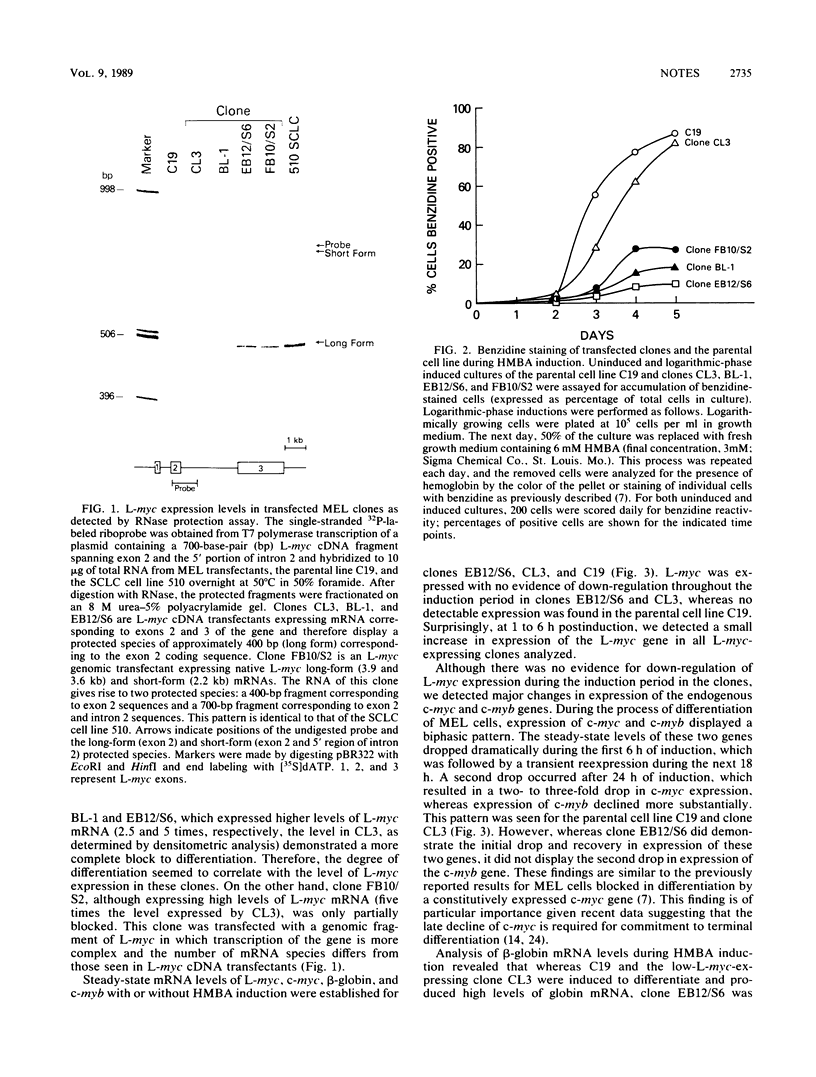
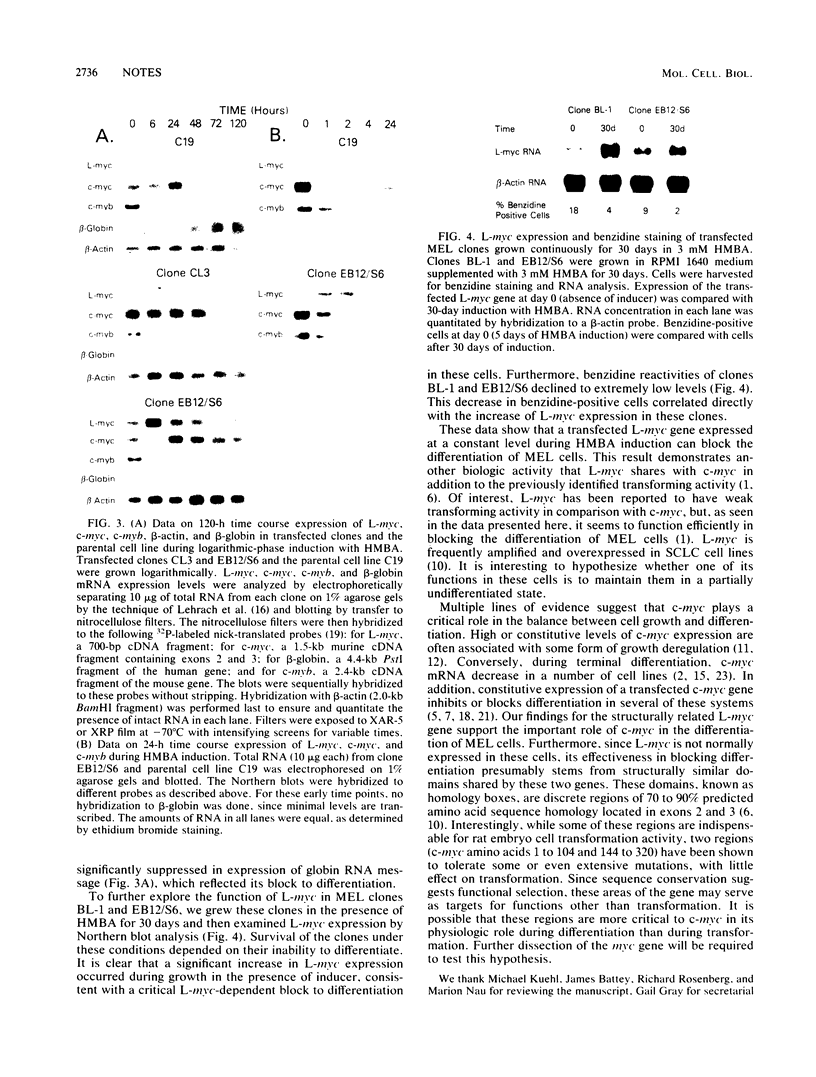
Abstract
We investigated the ability of the proto-oncogene L-myc to substitute for c-myc in blocking murine erythroleukemia differentiation. Murine erythroleukemia cells (line C19) were transfected with recombinant plasmids containing genomic and cDNA fragments of the L-myc gene driven by a Moloney murine leukemia virus long terminal repeat. Clones expressing constitutive high levels of L-myc failed to differentiate in response to the chemical inducer N,N'-hexamethylene bisacetamide (HMBA). The block to differentiation correlated with the level of L-myc expression. Furthermore, transfected clones grown in the presence of inducer for an extended period of time showed an increased level of L-myc expression. These results suggest that functional domains of the c-myc gene involved in differentiation are located in the discrete regions of homology between the c- and L-myc genes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birrer M. J., Segal S., DeGreve J. S., Kaye F., Sausville E. A., Minna J. D. L-myc cooperates with ras to transform primary rat embryo fibroblasts. Mol Cell Biol. 1988 Jun;8(6):2668–2673. doi: 10.1128/mcb.8.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Clarke M. F., Kukowska-Latallo J. F., Westin E., Smith M., Prochownik E. V. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol Cell Biol. 1988 Feb;8(2):884–892. doi: 10.1128/mcb.8.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola J. A., Cole M. D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986 Apr 24;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Hatton K. S., Tesfaye A., Yancopoulos G. D., Alt F. W. The human myc gene family: structure and activity of L-myc and an L-myc pseudogene. Genes Dev. 1987 Dec;1(10):1311–1326. doi: 10.1101/gad.1.10.1311. [DOI] [PubMed] [Google Scholar]
- Dmitrovsky E., Kuehl W. M., Hollis G. F., Kirsch I. R., Bender T. P., Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986 Aug 21;322(6081):748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- Friend C., Scher W., Holland J. G., Sato T. Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: stimulation of erythroid differentiation by dimethyl sulfoxide. Proc Natl Acad Sci U S A. 1971 Feb;68(2):378–382. doi: 10.1073/pnas.68.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Kaye F., Battey J., Nau M., Brooks B., Seifter E., De Greve J., Birrer M., Sausville E., Minna J. Structure and expression of the human L-myc gene reveal a complex pattern of alternative mRNA processing. Mol Cell Biol. 1988 Jan;8(1):186–195. doi: 10.1128/mcb.8.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keath E. J., Caimi P. G., Cole M. D. Fibroblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell. 1984 Dec;39(2 Pt 1):339–348. doi: 10.1016/0092-8674(84)90012-6. [DOI] [PubMed] [Google Scholar]
- Kelekar A., Cole M. D. Tumorigenicity of fibroblast lines expressing the adenovirus E1a, cellular p53, or normal c-myc genes. Mol Cell Biol. 1986 Jan;6(1):7–14. doi: 10.1128/mcb.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch I. R., Bertness V., Silver J., Hollis G. F. Regulated expression of the c-myb and c-myc oncogenes during erythroid differentiation. J Cell Biochem. 1986;32(1):11–21. doi: 10.1002/jcb.240320103. [DOI] [PubMed] [Google Scholar]
- Kuehl W. M., Bender T. P., Stafford J., McClinton D., Segal S., Dmitrovsky E. Expression and function of the c-myb oncogene during hematopoietic differentiation. Curr Top Microbiol Immunol. 1988;141:318–323. doi: 10.1007/978-3-642-74006-0_42. [DOI] [PubMed] [Google Scholar]
- Kume T. U., Takada S., Obinata M. Probability that the commitment of murine erythroleukemia cell differentiation is determined by the c-myc level. J Mol Biol. 1988 Aug 20;202(4):779–786. doi: 10.1016/0022-2836(88)90558-x. [DOI] [PubMed] [Google Scholar]
- Lachman H. M., Skoultchi A. I. Expression of c-myc changes during differentiation of mouse erythroleukaemia cells. Nature. 1984 Aug 16;310(5978):592–594. doi: 10.1038/310592a0. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Marks P. A., Rifkind R. A. Erythroleukemic differentiation. Annu Rev Biochem. 1978;47:419–448. doi: 10.1146/annurev.bi.47.070178.002223. [DOI] [PubMed] [Google Scholar]
- Prochownik E. V., Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. 1986 Aug 28-Sep 3Nature. 322(6082):848–850. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sarid J., Halazonetis T. D., Murphy W., Leder P. Evolutionarily conserved regions of the human c-myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci U S A. 1987 Jan;84(1):170–173. doi: 10.1073/pnas.84.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal S., Dmitrovsky E., Paez de la Cadena M. The effect of a transfected c-myc proto-oncogene on cellular differentiation. Mol Immunol. 1988 Nov;25(11):1129–1132. doi: 10.1016/0161-5890(88)90147-2. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin E. H., Gallo R. C., Arya S. K., Eva A., Souza L. M., Baluda M. A., Aaronson S. A., Wong-Staal F. Differential expression of the amv gene in human hematopoietic cells. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2194–2198. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingrove T. G., Watt R., Keng P., Macara I. G. Stabilization of myc proto-oncogene proteins during Friend murine erythroleukemia cell differentiation. J Biol Chem. 1988 Jun 25;263(18):8918–8924. [PubMed] [Google Scholar]