Abstract
Lysine acetylation of histones is one of the major epigenetic regulators of chromatin conformation and gene expression. The dynamic nature of histone acetylation is determined by the counterbalancing activity of histone acetyltransferase and histone deacetylase (HDAC) enzymes. Acetylation of histones is generally associated with open and transcriptionally active chromatin, whereas the activity of HDACs leads to histone deacetylation, condensation of chromatin, and inhibition of transcription. Aberrant silencing of tumor suppressors and other genes has been found in different types of cancer. Abnormal activity of HDACs has been implicated in tumorigenesis and therefore considerable effort has been put into the development of HDAC inhibitors as a means of modifying histone acetylation status and reexpressing aberrantly silenced tumor suppressor genes. This has led to the generation of a number of structurally diverse compounds that can effectively inhibit HDAC activity, thus altering chromatin structure in cancer cells. This unit discusses the methods and recent technological developments with respect to the studies of HDAC inhibition in cancer.
Keywords: Histone deacetylases, Histone acetyltransferases, HDAC inhibitors, Epigenetic gene silencing, Chromatin remodeling
1. Introduction
Epigenetic modifications refer to heritable and reversible changes in chromatin structure that are not due to alterations in primary DNA sequence (1, 2). The biochemical modifications that dictate epigenetic changes include methylation of cytosine residues in CpG dinucleotides and posttranslational modifications of the histone tails such as acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and ADP-ribosylation (3, 4). Among these modifications, histone acetylation is one of the major regulators of chromatin conformation and gene expression. The histone deacetylase (HDAC) family is divided into zinc-dependent (class I, IIa, IIb, and IV of which there are 11 subtype enzymes) and zinc-independent enzymes (class III, also called sirtuins), which require NAD+ for their catalytic activities (Table 1) (5). Acetylation by histone acetyltransferases (HATs) is generally associated with transcriptionally active chromatin (euchromatin) and activity of HDACs typically leads to chromatin condensation and inhibition of transcription (heterochromatin). Over the past decade, a number of HDAC inhibitors have been rationally designed and developed. These HDAC inhibitors have been examined for their ability to alter chromatin structure and reexpress aberrantly silenced genes which is associated with growth inhibition and apoptosis in cancer cells (6, 7). The field of HDAC inhibitors is moving rapidly into a new stage of development that has now started to produce success in the clinic, particularly in the field of cancer therapy. Based on their chemical structures, HDAC inhibitors are divided into four groups: hydroxamic acids, cyclic tetrapeptides, short-chain fatty acids, and benzamides (Table 2). Most of the HDAC inhibitors developed so far are nonselective reagents and among the most potent inhibitors are those that have been designed to target primarily the zinc cofactor of the enzyme active site and exhibit their effects in nano- or micro-molar levels (8, 9). However, several recent studies revealed some unique features of class IIa HDAC biochemistry and demonstrated unexpected selectivity of HDAC inhibitors presumed to be nonselective (10, 11).
Table 1.
Human histone deacetylase subunits
| Class | Subunit | Zinc-dependent | NAD+dependent |
|---|---|---|---|
| I | HDAC 1, 2, 3, 8 | Yes | No |
| IIa | HDAC 4, 5, 7, 9 | Yes | No |
| IIb | HDAC 6, 10 | Yes | No |
| III | SIRT 1, 2, 3, 4, 5, 6, 7 | No | Yes |
| IV | HDAC 11 | Yes | No |
HDAC histone deacetylase, SIRT sirtuin
Table 2.
Characteristics of some HDAC inhibitors in clinical trials
| Class | Compound | Targeted HDACs | Clinical trial stage |
|---|---|---|---|
| Hydroxamic acid | SAHA (Vorinostat) | Class I, II, IV | USFDA approved for CTCL |
| LBH-589 (Panobinostat) | Class I, II, IV | Phase III | |
| PXD-101 (Belinostat) | Class I, II, IV | Phase II | |
| ITF2357 | Class I, II | Phase I | |
| Cyclic peptide | Romidepsin (FK/228) | Class I, II | USFDA approved for CTCL |
| Short-chain fatty acid | Valproic acid | Class I HDAC 1 | Phase II |
| Phenylbutyrate | Class I | Phase I, II | |
| Benzamide | MS-275 (Entinostat) | Class I HDAC 1, 2, 3 | Phase II |
| MGC0103 | Class I, 11 | Phase II | |
CTCL cutaneous T-cell lymphoma, HDAC histone deacetylase, SAHA suberoylanilide hydroxamic acid, USFDA US Food and Drug Administration
The efficacy of HDAC inhibitors such as TSA (trichostatin A), SAHA (vorinostat), romidepsin (FK-228), LBH589 (Panobinostat), PDX101 (Belinostat), and MS-275 (Entinostat) as antitumor agents has been demonstrated in a wide range of cancer cell lines as well as in animal models (12–15). Two pharmaceutical HDAC inhibitors, SAHA and romidepsin, have already been approved by the US-FDA for the clinical treatment of cutaneous T-cell lymphoma (CTCL). A number of other promising HDAC inhibitors are currently under evaluation in advanced clinical trials. The exact mechanisms through which HDAC inhibitors mediate anticancer activity have not been fully elucidated. One model suggests that HDAC inhibitor-induced hyperacetylation of histones activates tumor-suppressor genes and represses oncogenes, thus activating intrinsic apoptotic pathways (16, 17). For example, in ER-negative breast cancer cells, inhibition of HDAC activity by specific HDAC inhibitors reactivates aberrantly silenced estrogen receptor alpha (ERα) and progesterone receptor (PR) gene expression (18–21). Pruitt et al., demonstrated that the inhibition of class III HDAC SIRT1 using a pharmacologic inhibitor, splitomicin, or siRNA reactivates epigenetically silenced SFRP1, SFRP2, E-cadherin, and CRBP1 genes in human breast and colon cancer cells despite full retention of DNA hypermethylation at promoters of reactivated genes (22). A recent study demonstrated that HDAC inhibitors induce cellular senescence through downregulation of polycomb group genes, suggesting that HDAC activity is important for self-renewal of human multipotent stem cells (MSCs) (23). In addition, a growing field of mass spectrometry-based proteomic techniques have identified several nonhistone proteins whose lysine acetylation is directly regulated by HDACs (24–26). These studies suggest that HDAC inhibitors can also affect diverse pathways in the cell and have recently been used to predict lysine acetylation motifs (27). A broader discussion of mass spectrometry techniques used in epigenetic research can be found in volume 593, Chapter 13 of this series (28).
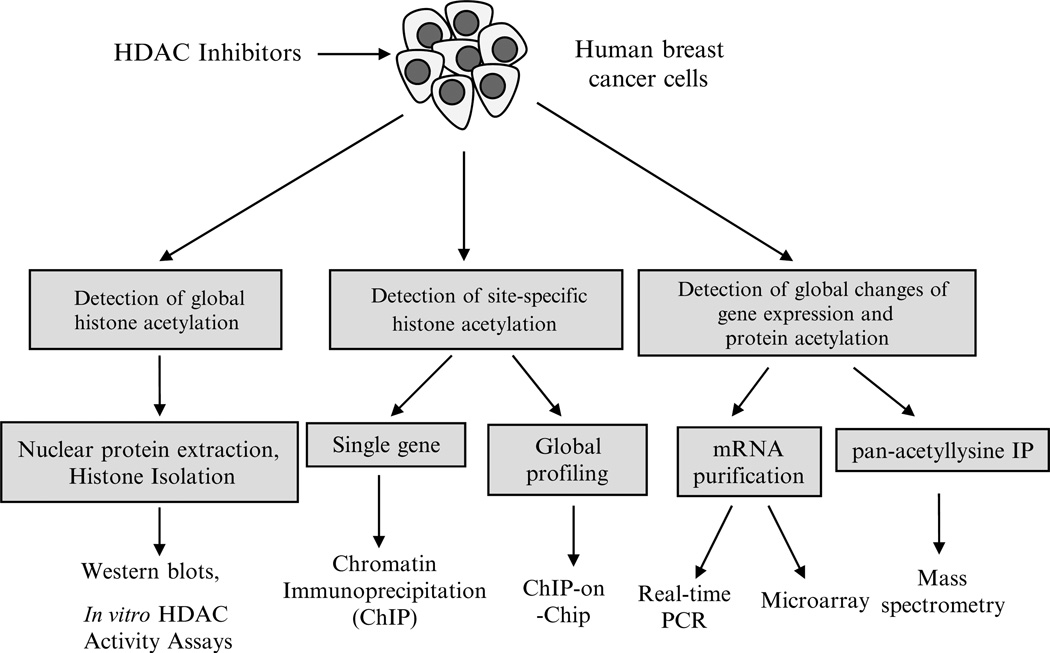
Studies investigating the effects of HDAC inhibitors on chromatin and gene transcription generally involve measuring the alterations of histone acetylation levels or expression of genes and gene products associated with acetylated histones induced by drug (Fig. 1). Although histones are often enriched prior to analysis using a classical acid extraction protocol (29–31), we find that nuclear extraction using a kit-based method described here is faster and typically sufficient. Additionally, more efficient methods for histone isolation which preserve more labile modifications such as phosphorylation have recently been developed (32).
Fig. 1.
Techniques for studying HDAC inhibition in human breast cancer. After human breast cancer cells are treated with HDAC inhibitors, immunological detection methods such as Western blot or immunochemistry can be used to determine the level of histone acetylation using specific antibodies against histone H3 or H4 or specific histone lysine residues such as AcH3K9, AcH3K27, and AcH4K20. Chromatin immunoprecipitation (ChIP) is used to determine the interaction of site-specific acetylated histones with promoters of genes of interest in breast cancer cells after HDAC inhibitor treatment. DNA sequences bound to a particular acetylated histone or nonhistone protein can be isolated by ChIP and these fragments can subsequently be hybridized to a DNA microarray (such as a tiling array). This so-called ChIP-on-chip technology allows the determination of acetylated histone binding occupancy throughout the genome of the cancer cell. Quantitative-PCR is able to precisely measure the specific gene expression changes in the presence of HDAC inhibitor treatment. Microarray-based gene expression profiling can be used to identify genes whose expression is altered by HDAC inhibitor treatment. Several mass spectrometry (MS)-based proteomic methods exist to quantitatively analyze proteins that are hyperacetylated after treatment with HDAC inhibitors as well as determine isoform specific occupancy of histone modifications.
As part of an effort to define the “histone code” of variable histone tail modifications, a significant area of research focuses on the detection of specific acetylated lysines of histones. Immunological detection (Western blots or immunochemistry) has become the method of choice to determine histone acetylation in cancer cells as a result of the growing availability of site-specific, histone family-specific, or pan-acetylation antibodies. While recent developments in mass spectrometry enable accurate quantification of isoform-specific histone modifications (33), the existing array of specific antibodies remains indispensable for the analysis of gene expression in concert with histone acetylation.
A range of methods is now available for assessing gene regulation relevant to histone acetylation in a gene-specific or genome-wide manner. The relationship between a gene of interest and site-specific histone acetylation can be analyzed by chromatin immunoprecipitation (ChIP) using antibodies (preferably monoclonal) to identify specifically modified histones bound to DNA. To globally assess genes correlated with specific histone acetylation sites, the ChIP-on-chip method (ChIP in combination with microarray) is used. This strategy has been extensively described in other recent editions of this collection (34–36). Quantitative-PCR has been widely used to quantify specific gene expression changes, and microarray expression analysis has been successfully used in our laboratory to measure the global gene expression after HDAC inhibitor treatment (37). The methods and protocols for the analysis of the cellular effects of histone deacetylase inhibition in human breast cancer cells are described below.
2. Materials
2.1. Cell Culture, HDAC Inhibitors
Phosphate-buffered saline (PBS), pH 7.2.
Dulbecco’s modified Eagle’s medium (DMEM, Mediatech) containing 5% fetal bovine serum (Mediatech) used for culture of MDA-MB-231 human breast cancer cells.
Trypsin/EDTA solution: 0.05% trypsin/0.53 mM EDTA (Mediatech).
HDAC inhibitors: SAHA (suberoylanilide hydroxamic acid, vorinostat, Cayman); TSA (7-[4-(dimethylamino) phenyl]-N-hydroxy-4,6-dimethyl-7-oxo-2,4-heptadienamide; Sigma); MS275 (Selleck Chemicals); belinostat (PXD101, Selleck Chemicals); and panobinostat (LBH589, Selleck Chemicals). These HDAC inhibitors are dissolved in 100% DMSO and stored at −20°C (see Note 1).
2.2. Histone Isolation
PBS, pH 7.2.
Lysis buffer A: 10 mM Tris–HCl, 1 mM MgCl2, 5 mM butyrate, 1% Triton X-100.
Buffer B: 0.25 M sucrose, 3 mM CaCl2, 10 mM Tris–HCl, and 5 mM butyrate.
Sulfuric acid.
Acetone.
2.3. Nuclear Protein Extraction
NE-PER® nuclear and cytoplasmic extraction reagents (Thermo Scientific/Pierce).
Protease inhibitors: benzamidine 250 mg/ml; aprotinin 2 mg/ml; leupeptin 2 mg/ml; PMSF (phenlymethlysulfonyl flouride) 0.2 M.
2.4. Western Blots and Antibodies
2.4.1. Chemiluminescence Detection
Tris–HCl SDS-PAGE precast gels, gel running and protein transfer apparatus, and PVDF membrane.
ECL plus Western blotting detection system (GE Healthcare).
2.4.2. Odyssey Quantitative Fluorescence Detection
Infrared dye 800CW goat anti-mouse secondary antibody and infrared dye 680 goat anti-rabbit secondary antibody (Li-COR).
Odyssey blocking buffer (Li-COR).
Tween 20, PBS buffer, methanol, and SDS.
The Odyssey® infrared imaging system (Li-COR).
2.4.3. Histone Antibodies
Rabbit anti-acetyl-histone H3 polyclonal IgG (Millipore), Rabbit anti-acetyl-histone H4 polyclonal IgG (Millipore), Rabbit anti-acetyl-histone H3 (Lys 9) polyclonal IgG (Millipore), Rabbit anti-acetyl-histone H3 (Lys 27) polyclonal IgG (Millipore), Rabbit anti-histone H3 polyclonal IgG (Abcam), Goat anti-rabbit immunoglobulin/HRP (DAKO).
2.5. In vitro HDAC Activity Assay
HDAC assay buffer: 50 mM Tris–Cl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2.
HDAC substrate (Calbiochem/EMD).
HDAC developer concentrate (20×) (Calbiochem/EMD).
Deacetylated standard (Calbiochem/EMD).
96-Well microplates.
Fluorimeter.
2.6. RNA Extraction, cDNA Synthesis, PCR
RNA extraction: TRIzol® reagent (Invitrogen).
First-strand cDNA synthesis: Oligo(dT)12–18, dNTP Mix (dATP, dGTP, dCTP, and dTTP), 5× first-strand buffer, DTT, RNaseOUT™, M-MLV reverse transcriptase (Invitrogen).
PCR reagent: JumpStart™ Taq ready mix (Sigma).
Real-Time PCR reagents: SYBR green or Taqman® (Applied Biosystems).
Applied Biosystems Real-Time PCR system.
2.7. Chromatin Immunoprecipitation
37% Formaldehyde.
SDS lysis buffer: 1% SDS, 10 mM EDTA pH 8.0, 50 mM Tris–HCl pH 8.1.
ChIP diluent buffer: 0.01% SDS, 1.1% Triton-X 100, 1.2 mM EDTA pH 8.0, 16.7 mM Tris–HCl pH 8.1, 167 mM NaCl.
Low salt buffer: 0.1% SDS, 1.0% Triton X-100, 2 mM EDTA pH 8.0, 20 mM Tris–HCl pH 8.1, 150 mM NaCl.
High salt buffer: 0.1% SDS, 1.0% Triton X-100, 2 mM EDTA pH 8.0, 20 mM Tris–HCl pH 8.1, 500 mM NaCl.
LiCl immune complex: 0.25 M LiCl, 1% NP40, 1% deoxycholate, 1% SDS, 1 mM EDTA pH 8.0, 10 mM Tris–HCl pH 8.1.
Elution buffer: 1% SDS, 0.1 M NaHCO3.
Protein A agarose slurry/Salmon Sperm DNA (Millipore).
Protein A agarose beads (Millipore).
Magnetic separator.
Sonifier (Branson).
3. Methods
3.1. Histone Extraction and Isolation
Histone proteins from HDAC inhibitor-treated human breast cancer cells are isolated according to previously published method (29).
Treat human breast cancer cells with HDAC inhibitors.
Harvest cells by scraping or trypsinization and centrifugation at 500 × g for 5 min and wash cells three times in ice-cold PBS.
Remove the supernatant and resuspend in 1 ml lysis buffer A with protease inhibitor and transfer to a 1.5-ml eppendorf tube on ice for 30 min.
Centrifuge for 15 min at 16,000 × g at 4°C.
Resuspend the pellet in 250 µl buffer B with protease inhibitor.
Add 11 µl 3.8N H2SO4 to make final concentration of 0.4N and leave the tube at 4°C overnight.
Centrifuge at 16,000 × g for 15 min at 4°C and transfer the supernatant to a new tube and precipitate with 10× cold acetone at −20°C.
Centrifuge 16,000 × g 15 min at 4°C and wash the pellet with cold acetone containing 0.2% H2SO4.
Dry pellet and dissolve pellet in ddH2O.
3.2. Nuclear Protein Extraction (See Note 2)
This protocol is derived from the published protocol from manufacturer (Thermo Scientific/Pierce).
Isolate approximately 20 µl packed cell volume of breast cancer cells treated with HDAC inhibitors or vehicle control by centrifugation at 500 × g for 2–3 min.
Add 200 µl of ice-cold CER I reagent with 1× protease inhibitor mixture.
Vortex vigorously for 15 s to fully resuspend the cell pellet and incubate the samples on ice for 10 min.
Add 11 µl of ice-cold CER II.
Vortex the tube for 5 s on the highest setting. Incubate tube on ice for 1 min.
Vortex the tube for 5 s and centrifuge the tube for 5 min at 16,000 × g.
Immediately transfer the supernatant (cytoplasmic extract) fraction to a clean prechilled tube and resuspend the insoluble pellet fraction containing nuclei in 100 µl of ice-cold NER with 1× protease inhibitor mixture.
Vortex for 15 s every 10 min, for a total of 40 min.
Centrifuge the tube at 16,000 × g for 10 min and transfer the supernatant (nuclear extract) fraction to a clean prechilled tube.
Store all extracts at −80°C until use.
3.3. Western Blot Analysis of Histone Acetylation
3.3.1. Chemiluminescence Western Blot
Treat cells with HDAC inhibitors.
Extract nuclear proteins or histone proteins as described above in Subheadings 3.1 and 3.2.
Dilute equal amounts of proteins in 2× SDS loading buffer, and boil at 95°C for 5 min.
Separate nuclear extract or histones on SDS-PAGE gels and transfer them onto PVDF membrane according to appropriate protocols.
Block blot in TBST with 5% nonfat dry milk for 2 h at room temperature or 4°C overnight.
Add the primary antibodies against acetylated histone proteins at a dilution of 1:2,000, and incubate the membranes with the diluted antibody at room temperature for 2 h.
Wash the membrane with TBST on a shaker with the revolution at 40–50 rpm for 3 × 10 min.
Incubate blot with secondary antibody (rabbit) at a concentration of 1:3,000 in TBST containing 5% nonfat dry milk at room temperature for 1.5 h.
Wash the blot three times for 10 min in TBST and once for 5 min in 1× TBS, and rinse with water.
Visualize the acetylated histones with the ECL kit.
3.3.2. Odyssey Western Blot (See Note 3)
Protein sample preparation, gel running and PVDF membrane transfer should be performed using standard blotting procedures as described in Subheading 3.3.1.
Place membrane in Odyssey blocking buffer (without Tween 20) for at least 1 h with gentle shaking at room temperature.
Incubate blot with primary antibodies in Odyssey blocking buffer with 0.1% Tween 20 for 2 h at room temperature or overnight at 4°C.
Rinse membrane with 1× PBST (0.2% Tween 20).
Incubate blot with infrared dye secondary antibody at a concentration of 1:5,000 in Odyssey blocking buffer with 0.1% Tween 20 and 0.01% SDS at room temperature for 60 min. Protect membrane from light during incubation.
Rinse membrane with 1× PBST (0.1% Tween 20). The membrane can be scanned wet or dry.
3.4. HDAC Activity Assay
In vitro HDAC activity assays are performed using the Calbiochem® HDAC activity assay kit according to the manufacturer’s instructions. This method is an assay system to measure HDAC activity in whole cell or nuclear extracts, immunoprecipitates, or purified recombinant human HDACs. In this unit, we describe the method of in vitro measurement of nuclear HDAC activity using the peptide substrate comprising an acetylated side chain lysine residue and a bound fluorescent group (Calbiochem/EMD Chemical).
Treat cells with HDAC inhibitors.
Extract nuclear proteins using the methods as described in Subheading 3.2.
Prepare the standard curve of deacetylation. Optimize the concentration ranges of deaceylated standard.
Add HDAC assay buffer to appropriate wells of the 96-well plate.
Add diluted cell nuclear extract or other HDAC containing samples to designated wells in triplicate.
Add HDAC substrate to each well containing nuclear extract or “no enzyme control.”
Mix thoroughly and incubate the plate at room temperature (~25°C) for 10–15 min.
Read samples in a fluorimeter at an excitation wavelength of 350–380 nm and an emission wavelength of 440–460 nm (see Note 4).
3.5. RT-PCR to Detect the Reexpression of Epigenetically Silenced Genes by HDAC Inhibitors in Human Breast Cancer Cells
Previous studies in our laboratory showed that pharmacological inhibition of histone deacetylation resulted in the expression of functional ERα mRNA and protein in ER negative human breast cancer cells (18–20). Here, we describe the method to detect the reactivation of ERα mRNA by SAHA treatment using RT-PCR in ER negative MDA-MB-231 cells. The RT-PCR primers and conditions for some other genes reactivated by HDAC inhibitors in human breast cancer cells are summarized in Table 3.
Treat ER negative human breast cancer cells (MDA-MB-231) with 1–10 µM SAHA for 24 h.
Rinse cells with ice-cold PBS and lyse cells directly by adding 1 ml of TRIZOL reagent (Invitrogen) per 10-cm2 dish and scraping with cell scraper.
Add 0.2 ml of chloroform per 1 ml of TRIzol reagent. Vortex samples for 15 s and incubate them at room temperature for 2–3 min.
Centrifuge the samples at 12,000 × g for 15 min at 4°C and transfer upper aqueous phase containing RNA into nuclease-free tubes.
Add 0.5 ml of isopropyl alcohol per 1 ml of TRIZOL reagent to precipitate the RNA and centrifuge at 12,000 × g for 10 min at 4°C.
Wash the RNA pellet with 75% ethanol, centrifuge, and dissolve RNA in 100 µl DEPC-treated water. Measure the RNA concentrations.
First strand cDNA is synthesized by mixing 3 µg total RNA with 1 µl oligo (dT)12–18 (500 µg/ml), and 1 µl of 10 mM dNTP mix (Invitrogen), then adding sterile ddH2O to 12 µl.
Heat mixture to 65°C for 5 min and quickly chill on ice.
Add 4 µl 5× first strand buffer, 2 µl 0.1 M DTT and 1 µl RNaseOUT™ Recombinant Ribonuclease Inhibitor (40 U/µl) (Invitrogen), mix contents of the tube gently and incubate at 37°C for 2 min.
Add 1 µl (200 U) of M-MLV reverse transcriptase (Invitrogen) and incubate 50 min at 37°C followed by heating at 70°C for 15 min to inactivate the reaction.
Conventional PCR was performed in cDNA samples as previously described (20) using the following primers: ERα S: GCACCCTGAAGTCTCTGGAA; AS: TGGCTAAAGTGGTGCATGAT; Actin S: ACCATGGATGATGATATCGC; AS: ACATGGCTGGGGTGTTGAAG. Amplified products are analyzed on 1.5% agarose gels.
Table 3.
RT-PCR primers and conditions for genes reactivated by HDAC inhibitors in human breast cancer cells
| Gene | Primers | Annealing temperature (°C) |
HDAC inhibitor | Reference |
|---|---|---|---|---|
| ERα | S: GCACCCTGAAGTCTCTGGAA AS: TGGCTAAAGTGGTGCATGAT |
55 | TSA | (18–21) |
| Scriptaid | ||||
| LBH | ||||
| SAHA | ||||
| PR | S: TCATTACCTCAGAAGATTTGTTTAATC AS: TGATCTATGCAGGACTAGACAA |
60 | TSA | (18–21) |
| Scriptaid | ||||
| SFRP1 | S: GGCCCATCTACCCGTGTCG AS: GATGGCCTCAGATTTCAACTCGT |
60 | Splitomicin | (22) |
| SFRP2 | S: AAGCCTGCAAAAATAAAAATGATG AS: TGTAAATGGTCTTGCTCTTGGTCT |
53 | Splitomicin | (22) |
| E-cadherin | S: CCGCCGGCGTCTGTAGGAA AS: AGGGCTCTTTGACCACCGCTCTC |
57 | Splitomicin | (22) |
| CRBP1 | S: CATCCGCACGCTGAGCACTTTTAG AS: CACGCCCCTCCTTCTCACCCTTCT |
58 | Splitomicin | (22) |
ERα estrogen receptor alpha, PR progesterone receptor, SFRP secreted frizzled-related protein, CRPB1 cellular retinol binding protein 1
3.6. Chromatin Immunoprecipitation to Analyze Changes in Regulatory Chromatin Marks by HDAC Inhibitors at the Specific Gene Promoters
ChIP is a powerful tool to study protein–DNA interaction in HDAC inhibitor treated cells. This technique can map minute-byminute changes of histone acetylation at a single promoter, or over the entire genome by using advanced ChIP on DNA microarray technology (ChIP-on-chip) (38). The protocol and reagents for ChIP used in our laboratory are described below as recommended by the manufacturer’s instructions (Millipore/Upstate).
Treat 2 × 106 human breast cancer cells with 1–10 µM HDAC inhibitors for 24 h.
Crosslink DNA and proteins by adding 37% formaldehyde directly to growth media to a final concentration of 1%, and gently shake dishes to mix, and incubate at room temperature for 10 min.
Add glycine to a final concentration of 0.125 M to quench crosslinking reactions.
Wash cells with cold PBS containing 1× protease inhibitor and scrape cells from each dish into a microcentrifuge tube.
Spin at 500 × g at 4°C for 2–5 min to pellet cells.
Resuspend cell pellet in 200 µl of SDS lysis buffer containing 1× protease inhibitor.
Sonicate cell lysate to shear DNA to ~200–1,000 bp in length (see Note 5). In between pulses, let samples sit on ice for at least 2 min.
Centrifuge samples at 14,000 × g at 4°C for 10 min and transfer 200 µl of the sonicated cell supernatant to a new microcentrifuge tube.
Dilute the sonicated cell supernatant tenfold in 1.8 ml ChIP dilution buffer with 1× protease inhibitor.
Preclear the 2-ml diluted cell supernatant with 80 µl of salmon sperm DNA/Protein A Agarose beads (50% slurry) for 30 min at 4°C with agitation.
Separate beads by brief centrifugation and transfer the supernatant to a fresh tube (if using magnetic beads, beads are separated on magnetic rack).
Remove 10 µl (1%) of the supernatant as input and save at 4°C.
Collect the supernatant by aliquoting 1 ml into fresh microfuge tubes.
Add the immunoprecipitating antibodies against acetylated H3, H4, or specific lysine residues on histone tail to the supernatant fraction and incubate overnight at 4°C with rotation. Rabbit immunoglobulin G (IgG) and H3 antibodies are used for negative and quantitative controls, respectively (see Note 6).
Add 60 µl of Protein A Agarose beads (Millipore) and mix for 1 h at 4°C with rotation.
Separate beads by brief centrifugation or magnetic rack and remove the supernatant fraction.
Wash the beads once in 1 ml of low salt immune complex wash buffer (Millipore), once in high salt immune complex wash buffer (Millipore), once in LiCl Immune complex wash buffer (Millipore), and twice in TE buffer.
Add 100 µl of elution buffer (20% SDS, 1 M NaHCO3) to each tube containing the antibody/bead complex. Mix and incubate at room temperature for 15 min. Transfer the eluate to fresh tube and wash the beads with 250 µl ChIP elution buffer. Repeat the wash and pool the eluates.
Add 20 µl 5 M NaCl to the pooled eluates and reverse cross-links at least 4 h at 65°C.
Add 1 µl of RNase A and incubate for 30 min at 37°C. Add 4 µl 0.5 M EDTA, 8 µl 1 M Tris–HCl and 1 µl Proteinase K, and incubate at 45°C for 1 h.
Extract the samples with phenol/chloroform (1:1), ethanol-precipitate DNA in the presence of 20 µg of glycogen, wash with 70% ethanol, and dissolve in 50 µl TE.
IP DNA can be further analyzed by quantitative Real-Time PCR to quantify alteration in acetylated histone marks at the promoter region of gene of interest (see Note 7). DNA immunoprecipitated by H3 antibody is used for normalization.
Acknowledgments
This work was funded by NIH SPORE grant CA88843 (to N.E.D.) and the Breast Cancer Research Foundation (to N.E.D.).
Footnotes
HDAC inhibitors should be dissolved in DMSO first for maximum solubility and then diluted in aqueous buffer of choice. Since some HDAC inhibitors, such as vorinostat, are unstable in aqueous media, we replace the drug-containing medium every day if the length of treatment time is more than 24 h.
This method has been successfully used to extract nuclear proteins and examine the expression level of nuclear histone proteins (39). The CER I reagent from cytoplasmic/nuclear protein extraction kit induces swelling of the cell leading to stress on the cellular membrane and the CER II reagent lyses the cell membrane, allowing cytoplasmic proteins to be collected. The NER reagent is then used to extract nuclear proteins from the intact nucleus. EDTA-free protease inhibitors can be used for the extraction agents. However, protease inhibitors that contain alcohols should be avoided.
By use of fluorescence-labeled antibodies rather than enzyme labels, Odyssey infrared image system quantitatively detects protein expression on the Western blot with wide linear dynamic range that cannot be achieved by conventional chemiluminescence. With two detection channels, multiple separate targets can be probed in the same experiment. Therefore, quantification accuracy is improved when the second channel is used for loading normalization. This method has been used in our recent study to quantify drug-induced changes in global histone marks (39).
This method has been successfully used with preparations of all class I and II HDACs and class III HDAC SIRT1. It is necessary to use a potent HDAC inhibitor, such as TSA, as a positive control in experiments. The exact concentration range of the deaceylated standard, substrate and inhibitors should be carefully optimized and determined.
It is important to optimize conditions for shearing crosslinked DNA to 200–1,000 bp in length. These conditions vary with different cell types, cell density, and the specific sonication equipment setting including the power output, duty cycle, and number of pulses. During the sonication, keep all the samples on ice to avoid the occurrence of protein denaturation.
It is possible that an anti-acetylated histone antibody used in ChIP will not recognize the epitope of the antigen in fixed chromatin. In such case, choose an antibody with higher affinity that has been validated as suitable for ChIP. It is important to use a negative control in every ChIP experiment (such as IgG) to detect nonspecific binding. If polyclonal antibodies are used, a control using unimmunized sera from the same species should be included.
ChIP Primers for silenced genes reactivated with HDAC inhibition in human breast cancer cells: ERα, forward: TGAACCGTCCGCAGCTCAAGATC and reverse: GTCTGACCGTAGACCTGCGCGTTG (19); SFRP1, forward: AGCCGCGTCTGGTTCTAGT and reverse: GGAGGCTGCAGGGCTG; E-cadherin, forward: TAGAGGGTCACCGCGTCTATG and reverse: GGGTGCGTGGCTGCAGCCAGG (22).
References
- 1.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 6.Marson CM. Histone deacetylase inhibitors: design, structure-activity relationships and therapeutic implications for cancer. Anticancer Agents Med Chem. 2009;9:661–692. doi: 10.2174/187152009788679976. [DOI] [PubMed] [Google Scholar]
- 7.Khan O, La Thangue NB. Drug Insight: histone deacetylase inhibitor-based therapies for cutaneous T-cell lymphomas. Nat. Clin. Pract. Oncol. 2008;5:714–726. doi: 10.1038/ncponc1238. [DOI] [PubMed] [Google Scholar]
- 8.Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv. Cancer. Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- 9.Ficner R. Novel structural insights into class I and II histone deacetylases. Curr Top. Med. Chem. 2009;9:235–240. doi: 10.2174/156802609788085304. [DOI] [PubMed] [Google Scholar]
- 10.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, McLean TH, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J. Biol. Chem. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stimson L, Wood V, Khan O, Fotheringham S, La Thangue NB. HDAC inhibitor-based therapies and haematological malignancy. Ann Oncol. 2009;20:1293–1302. doi: 10.1093/annonc/mdn792. [DOI] [PubMed] [Google Scholar]
- 13.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 14.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J. Cell. Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campas-Moya C. Romidepsin for the treatment of cutaneous T-cell lymphoma. Drugs Today (Barc) 2009;45:787–795. doi: 10.1358/dot.2009.45.11.1437052. [DOI] [PubMed] [Google Scholar]
- 16.Nebbioso A, Clarke N, Voltz E, Germain E, Ambrosino C, Bontempo P, Alvarez R, Schiavone EM, Ferrara F, Bresciani F, Weisz A, de Lera AR, Gronemeyer H, Altucci L. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat. Med. 2005;11:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 17.Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, Holloway AJ, Johnstone RW. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keen JC, Yan L, Mack KM, Pettit C, Smith D, Sharma D, Davidson NE. A novel histone deacetylase inhibitor, scriptaid, enhances expression of functional estrogen receptor alpha (ER) in ER negative human breast cancer cells in combination with 5-aza 2’-deoxycytidine. Breast Cancer Res. Treat. 2003;81:177–186. doi: 10.1023/A:1026146524737. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Q, Atadja P, Davidson NE. Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor alpha (ER) gene expression without loss of DNA hypermethylation. Cancer Biol. Ther. 2007;6:64–69. doi: 10.4161/cbt.6.1.3549. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Ferguson AT, Nass SJ, Phillips DL, Butash KA, Wang SM, Herman JG, Davidson NE. Transcriptional activation of estrogen receptor alpha in human breast cancer cells by histone deacetylase inhibition. Cancer Res. 2000;60:6890–6894. [PubMed] [Google Scholar]
- 21.Sharma D, Saxena NK, Davidson NE, Vertino PM. Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer Res. 2006;66:6370–6378. doi: 10.1158/0008-5472.CAN-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung JW, Lee S, Seo MS, Park SB, Kurtz A, Kang SK, Kang KS. Histone deacetylase controls adult stem cell aging by balancing the expression of polycomb genes and jumonji domain containing 3. Cell Mol. Life Sci. 2010;67:1165–1176. doi: 10.1007/s00018-009-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q, Chaerkady R, Shaw PG, Kensler TW, Pandey A, Davidson NE. Screening for therapeutic targets of vorinostat by SILAC-based proteomic analysis in human breast cancer cells. Proteomics. 2010;10:1029–1039. doi: 10.1002/pmic.200900602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 26.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 27.Gnad F, Ren S, Choudhary C, Cox J, Mann M. Predicting Posttranslational Lysine Acetylation Using Support Vector Machines. Bioinformatics. 2010;26:1666–1668. doi: 10.1093/bioinformatics/btq260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beck HC. Mass spectrometry in epigenetic research. Methods Mol Biol. 593:263–282. doi: 10.1007/978-1-60327-194-3_13. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 30.Cousens LS, Gallwitz D, Alberts BM. Different accessibilities in chromatin to histone acetylase. J. Biol. Chem. 1979;254:1716–1723. [PubMed] [Google Scholar]
- 31.Jackson V, Shires A, Chalkley R, Granner DK. Studies on highly metabolically active acetylation and phosphorylation of histones. J. Biol. Chem. 1975;250:4856–4863. [PubMed] [Google Scholar]
- 32.Rodriguez-Collazo P, Leuba SH, Zlatanova J. Robust methods for purification of histones from cultured mammalian cells with the preservation of their native modifications. Nucleic Acids Res. 2009;37:e81. doi: 10.1093/nar/gkp273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong Y, Falk J. Genome-wide analysis for protein-DNA interaction: ChIP-chip. Methods Mol Biol. 2009;590:235–251. doi: 10.1007/978-1-60327-378-7_15. [DOI] [PubMed] [Google Scholar]
- 35.Reimer JJ, Turck F. Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip). Part A: ChIP-chip molecular methods. Methods Mol Biol. 2010;631:139–160. doi: 10.1007/978-1-60761-646-7_12. [DOI] [PubMed] [Google Scholar]
- 36.Gobel U, Reimer J, Turck F. Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip). Part B: ChIP-chip data analysis. Methods Mol. Biol. 2010;631:161–184. doi: 10.1007/978-1-60761-646-7_13. [DOI] [PubMed] [Google Scholar]
- 37.Keen JC, Garrett-Mayer E, Pettit C, Mack KM, Manning J, Herman JG, Davidson NE. Epigenetic regulation of protein phosphatase 2A (PP2A), lymphotactin (XCL1) and estrogen receptor alpha (ER) expression in human breast cancer cells. Cancer Biol. Ther. 2004;3:1304–1312. doi: 10.4161/cbt.3.12.1458. [DOI] [PubMed] [Google Scholar]
- 38.Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 39.Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]