Abstract
A number of studies are now collecting diffusion tensor imaging (DTI) data across sites. While the reliability of anatomical images has been established by a number of groups, the reliability of DTI data has not been studied as extensively. In this study, five healthy controls were recruited and imaged at eight imaging centers. Repeated measures were obtained across two imaging protocols allowing intra-subject and inter-site variability to be assessed. Regional measures within white matter were obtained for standard rotationally invariant measures: fractional anisotropy, mean diffusivity, radial diffusivity, and axial diffusivity. Intra-subject coefficient of variation (CV) was typically <1% for all scalars and regions. Inter-site CV increased to ∼1%–3%. Inter-vendor variation was similar to inter-site variability. This variability includes differences in the actual implementation of the sequence.
Key words: diffusion tensor, fractional anisotropy, magnetic resonance, mean diffusivity, reliability, white matter
Introduction
Diffusion weighted imaging is a magnetic resonance (MR) imaging technique that is sensitive to the random thermal motion of water. The data resulting from diffusion weighted sequences can be used to define rotationally invariant scalar measures, insensitive to the relative orientation of the sample within the magnetic field, or to define fiber tracts using the directional information available in the eigen decomposition. Common rotationally invariant scalar metrics include fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD), and radial diffusivity (RD). Diffusion tensor imaging (DTI) allows one to study the microstructure of white matter, providing the ability to quantitatively study white matter integrity to investigate changes in fiber tract organization, density, diameter, or myelination. It is likely that changes in white matter microstructure can be measured using diffusion weighted imaging before gross white matter changes will become apparent using volumetric analysis.
Diffusion weighted imaging has been used to study white matter changes associated with most neurological and psychiatric disorders. Our interest is in the study of Huntington disease (HD) and the development of sensitive biomarkers to examine the nature and pattern of neurobiological and neurobehavioral changes that occur in the period leading up to a diagnosis of HD. Because of the rare occurrence of HD, the Neurobiological Predictors of Huntington's Disease (PREDICT-HD) is a consortium of 32 sites throughout the world that have joined forces to study this disease using a variety of the tools. Imaging in the PREDICT-HD project is being conducted at 30 of these sites.
Progressive volumetric reduction in the caudate and putamen have been the classic sign of HD progression using MR imaging. Based on previous literature, approximately a 2%–4% reduction per year in the size of these striatal structures has been found (Aylward et al., 2000) in HD. In work by the PREDICT group, these structures had the largest effect size throughout the progression of the disease when compared to a control population. When comparing subjects near disease onset to normal controls, the effect size for the volume of the putamen and caudate was −2.46 and −2.29, respectively (Paulsen et al., 2010). Since the brain is a highly interconnected neural network, it is not surprising that the volume of white matter also shows significant differences between subjects with the huntingtin gene expansion as compared to control subjects (effect size −1.45). Several investigators have also reported changes in the white matter associated with HD progression (Aylward et al., 1998; Beglinger et al., 2005; Rosas et al., 2006).
Recently, a number of investigators have employed diffusion imaging to study white matter changes in HD. In a previous study by Magnotta and colleagues (2009) using DTI, we found significant correlations of FA and MD with the 5-year probability of disease onset. Rosas and colleagues (2006) found significantly reduced FA within the corpus callosum in both subjects with HD (genu, body, and splenium) as well as prodromal HD subjects (body). Increases in FA were found in the internal capsule (anterior limb, genu) and decreases found in the posterior limb. Significant increases in FA were seen in the putamen and globus pallidus. A longitudinal study in subjects with presymptomatic and early stage HD by Weaver and colleagues (2009) found significant changes over the course of 1 year in FA located within subcortical grey matter structures, callosal and frontostriatal tracts, while longitudinal changes in controls was restricted to a few very small, nonlocalized regions in the brain. A significant laterality effect was found with greater changes in the right hemisphere. Animal models of HD have also shown white matter changes in similar regions in the internal and external capsules as well as caudate (Van Camp et al., 2010). As a result of these findings in white matter, DTI is included as part of the second generation PREDICT-HD imaging protocol. Unlike many of the MR imaging sequences that are employed for anatomical imaging, diffusion weighted images tend to have many differences in their implementation that are specific to the vendor. Most notable are the types of diffusion encoding sequence used (Stejskal-Tanner vs. double refocused), gradient directions (number and sampling scheme), number of b=0 images, echo-time, and gradient strength. A number of studies have evaluated the number of gradient directions and how they affect the resulting rotationally invariant scalar measurements. Work by Jones found that 20 directions were required to get a robust estimate of anisotropy, while 30 directions are required for robust estimation of the tensor orientation and MD (Jones, 2004).
There have been a relatively small number of studies that have reported reliability data for multivendor and multisite studies in diffusion weighted imaging. The recent work by Pagani and colleagues (2010) has been the most comprehensive study to date, evaluating the reliability of rotationally invariant scalar measures across multiple sites using both 1.5 and 3T field strengths on both Siemens and Philips scanners. In this study, they found the intra-site coefficient of variation (CV) to be between 5.1% and 5.7% for the FA within the corpus callosum and 6.2%–7.9% for MD. They found a significant influence of magnetic field strength and scanner manufacturer on the MD and AD measures. The work by Pfefferbaum and colleagues (2003) evaluated the reproducibility of FA and trace measurements across two different 1.5T GE scanners with different gradient systems using the same pulse sequence and gradient directions. Relatively thick slices (4 mm) were used in this work as compared to imaging sequences now employed at 3T (2 mm). Pfefferbaum found a scanner effect that was statistically significant, but this resulted in only a 2% difference in the FA values and a 1% difference in the trace values. Cercignani and colleagues (2003) evaluated reliability across two scanner vendors and found coefficients of variability between 5.4%–7.3% for FA and 1.7%–5.6% for MD. A recent study by Vollmar and colleagues (2010) assessed reliability across two 3T scanners from the same vendor and found a CV of 1.1% across the whole brain and 1.2% with the corpus callosum for FA measures. They also generated reproducibility maps that showed the variation within the main white matter structures was low (less than 5%), but was larger in gray matter regions (10%–15%) (Vollmar et al., 2010). Danielian and colleagues (2009) found intra-class correlations of 0.8 for MD, FA, and RD up to 1 year apart in several fiber tracts, including the corticospinal tract, uncinate fasciculus, and corpus callosum using deterministic fiber tracking. Cheng and colleagues (2006) evaluated the reliability of FA measurements within fiber tracts between the cerebellum and thalamus in scans collected on the same scanner within a 24-h interval. They found correlations of 0.82 between the two time points. Wang and colleagues (2012) performed a comprehensive intra-site reliability study for fiber tracking. They used fiber tracking to define regions of interest, where CV was used to assess the reliability of FA and MD within the fiber tract as well as the tract size (volume, length, and number of fibers). Forty-three of the 60 regions showed CVs less than 10%.
The purpose of this study was to evaluate the site variation of DTI, across eight sites, in a set of human volunteers. The vendors were selected based on a survey submitted to participating sites. At the time the multicenter study was undertaken, 3T scanners from two vendors (Siemens and Philips) were available to PREDICT-HD investigators; since this reproducibility study was undertaken, sites with 3T GE scanners have become available, but data from GE scanners were not collected for this reproducibility study. This study is unique in that it compares several 3T scanners across vendors and evaluates the commonly used rotationally invariant scalar measures (FA, MD, AD, and RD).
Methods
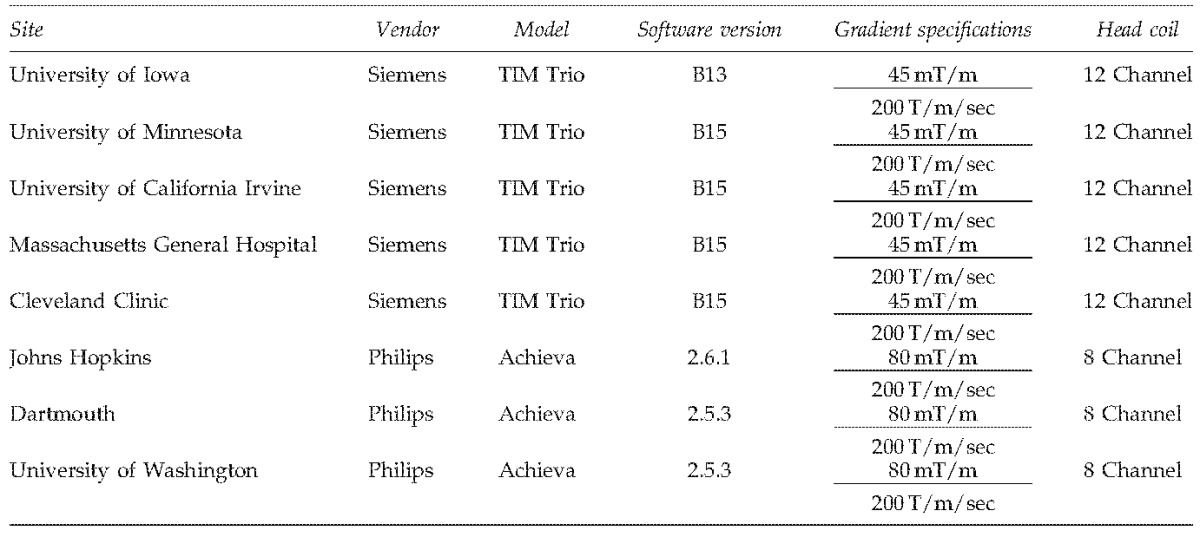
Eight sites participated in this multicenter imaging study to evaluate DTI across multiple centers and vendors (Table 1). The sites involved in this study had either a Siemens 3T TIM Trio scanner (gradient strength=45 mT/m, slew rate=200 T/m/sec) or Philips 3T Achieva scanner (gradient strength=80 mT/m, slew rate=200 T/m/sec). Five healthy control subjects were recruited into this multicenter imaging study after informed consent was obtained in accordance with the Institutional Review Board at each of the imaging sites. All five subjects were imaged at the eight sites within a 30-day period. All DTI scans used the vendor standard DTI sequence for acquisition: for the Philips scanners, a Stejskal-Tanner sequence was used, while the Siemens scanners used a double refocused spin-echo sequence. Two diffusion weighted imaging protocols were evaluated. The first imaging protocol used a vendor-provided diffusion weighted imaging gradient directions table. This sequence consisted of 30 gradient directions for the Siemens scanners and 32 directions for the Philips scanners. A single b=0 image was collected with this sequence and a b-value of 1000 sec/mm2 was used for the diffusion gradient encoding. The second sequence collected the same 71 directions across both the Siemens and Philips scanners using a custom gradient encoding scheme designed using electrostatic repulsion. In this sequence, eight b=0 images were acquired on all scanners. For the diffusion sequence, the same field of view (256×256 mm), matrix size (128×128), echo-time (TE=92 msec), bandwidth (1565 Hz/pixel), and slice thickness/gap (2.0/0.0 mm) were used across all of the sites and protocols. The 30/32 direction sequence collected 70 slices, while the 71 direction sequence collected only 50 slices. The only difference across sites other than the type of sequence was the repetition time (TR) that was employed. Siemens used TR times of 10,000 and 12,000 msec for the 71 and 30/32 direction sequences, respectively. For Philips scanners, TR times of 9750 and 7000 msec were used. Four repetitions of the 30/32 direction sequence and two repetitions of the 71 direction sequence were acquired. All of the diffusion weighted imaging data were collected without the use of cardiac gating. The scan time for each run of the 30/32 direction sequence was ∼6.5 min, while each run of the 71 direction sequence took ∼15 min. In addition to the diffusion weighted sequences, anatomical images were acquired using three-dimensional (3D) T1 weighted (MP-RAGE) and T2 (SPACE) sequences at each center. The anatomical images collected at The University of Iowa were the only anatomical data utilized for this study.
Table 1.
Sites and Scanners Used in the Diffusion Tensor Imaging Reliability Study
 |
Processing of the anatomical images was performed using the Brain Research: Analysis of Images, Networks, and Systems (BRAINS) software (Andreasen et al., 1992, 1993; Magnotta et al., 2002). The anatomical images were processed using a fully automated pipeline (Pierson et al., 2010) that includes anterior commissure–posterior commissure alignment, defining of Talairach parameters to warp the Talairach grid onto the subject of interest, tissue classification (Harris et al., 1999), and skull stripping using an artificial neural network (Magnotta et al., 1999). The following regions of interest based on the Talairach atlas were used in this study (Andreasen et al., 1996): cerebrum, frontal lobe, temporal lobe, parietal lobe, occipital lobe, and subcortical regions.
For the 30/32 direction protocol, data from each single run were analyzed individually as well as concatenating the data from two, three, and four runs. For the 71 direction protocol, a single run was analyzed as well as the combination of both runs. The analysis of the different number of concatenations was carried out separately. The diffusion weighted images were processed with and without the use of DTIPrep (Liu et al., 2010), an automated quality control tool, to remove artifacts typically seen in diffusion weighted images. For analysis without DTIPrep, the diffusion weighted images were corrected for motion and eddy current artifacts before generating the tensor as described below.
DTIPrep is a quality assurance tool that eliminates volumes of the acquisition that have artifacts or may be of poor quality as a result of subject motion, gradient hardware performance, or RF noise. The number of volumes eliminated as a result of this quality assurance check was evaluated in this study. The DTIPrep pipeline begins by first verifying that the protocol used to collect the diffusion weighted data was consistent across all subjects and sites. Image information was checked for mismatches in image size, origin, and voxel spacing. The pipeline terminated if mismatches in image size and voxel spacing were found. Diffusion information was checked to detect scans with incorrect numbers of diffusion gradients, diffusion gradient directions, and the applied b-value.
After the imaging protocol parameter checks, DTIPrep assessed intensity-related artifacts across all diffusion weighted images using a slice-wise checking algorithm. In slice-wise checking, normalized correlation values were calculated between successive slices for each gradient. It is assumed that these correlation values form a normal distribution across the diffusion gradient directions. The user can define the number of standard deviations (SDs) used to define an outlier for the correlation values. In this study, 3.1 and 3.6 SDs were used for the b=0 and diffusion weighted images, respectively. This slice-wise intensity check was used to remove gradient directions that exhibit large changes in signal intensity that was not related to the diffusion encoding gradients, such as table vibrations and spike noise. Next, venetian blind artifacts resulting from subject motion between the interleaves of a multipass acquisition were assessed in two steps. The first step involved computing normalized correlation values similar to those in the slice-wise checking step between the interleaving parts of each gradient. In this study, 2.5 and 3.0 SDs were used for b=0 and diffusion weighted images, respectively. For the second step, motion parameters were obtained by performing a rigid registration between the even and odd slices of the dataset for each diffusion encoding gradient. The resulting estimates for translation and rotation were compared to user defined thresholds (2.0 mm translation and 0.5° rotation in this study). Any gradient volumes that exceeded the specified threshold values at each step are removed from the analysis.
Motion between baseline scans was then estimated by DTIPrep and removed by aligning all of the b=0 images together via a mutual information metric (Mattes et al., 2001) with a stop condition of less than 0.02. The resulting average baseline image was then used as a reference for subsequent motion and eddy-current correction for the diffusion weighted images. The gradient directions were updated based on the rotation component of the affine transformation. The gradient-wise check was the final check of DTIPrep and is meant to remove residual motion artifacts after the eddy-current and head motion correction. DTIPrep allowed the user to remove a diffusion volume when the estimated translation or rotation exceeds a user-defined threshold (2.0 mm translation and 0.5° rotation in this study) relative to the (averaged) b=0 image. The final step of DTIPrep performed a postregistration step that retrospectively computed a rigid rotation to bring all gradients into anatomical space. The transformation into anatomical space accounted for the scan's individual measurement frame and transformations that occurred during the DTIPrep pipeline. The DTIPrep pipeline was automatically terminated at any step if less than six diffusion gradients remained or all of the baseline images were removed.
After the data were preprocessed with or without DTIPrep, the diffusion weighted images were analyzed using the Guided Tensor Restored Anatomical Connectivity Tractography (GTRACT) software (Cheng et al., 2006). The diffusion tensor was estimated from the diffusion weighted images with and without applying a 3×3×3 voxel median filter to the b=0 and diffusion weighted images. Diffusion tensor rotationally invariant scalar images of FA, MD, RD, and AD were generated. Regional measures of the scalar images were obtained within the cerebral white matter defined as the intersection between the white matter defined via tissue classified images and thresholding the FA image at 0.1. This allowed white matter regions to be defined in an automated fashion while eliminating regions that were lost due to susceptibility artifacts. Regional white matter anisotropy measures were obtained for the entire cerebrum, subcortical area, frontal, temporal, parietal, and occipital lobes based on the Talairach atlas (Andreasen et al., 1996). Separate measurements were obtained for the right and left hemisphere as well as the combined hemispheres. Regional measures of FA and MD were compared across all of the sites and vendors. In addition, we evaluated the number of gradient directions that were eliminated for each step of DTIPrep.
Statistical analysis
In this study, both within-subject reliability and between-site reliability of DTI scalar values were evaluated. Within-subject reliability reflects whether DTI scalar values can be reliably reproduced in repeated scans, while between-site reliability indicates whether DTI scalar values can be reliably measured across sites. Reliability was quantified using the CV. A lower CV value indicates better reliability. CVs for within-subject reliabilities were calculated using repeated scans from each subject using the same scanner and scanning protocol within the same site. They were compared across scalar types, brain regions, scanner types, protocols, number of gradient directions, and sites. CVs for between-site reliabilities were calculated using scans obtained from each subject across all eight sites. They were compared for each type of scalar value and in each brain region. The DTI scalar values considered were AD, MD, RD, and FA. Covariates that were thought to affect the CV's included scanner type (Siemens and Philips), sites (eight sites), whether DTIPrep was applied (yes, no), protocol type (high, low number of gradients per scan), median spatial filtering (yes, no), and number of concatenations (1, 2, 3, 4). Mean and SD of CVs were calculated to quantify the central tendency and variability of the study sample. Linear mixed-effects models were employed to evaluate the covariate effects. For the plots of CVs, the same range on the y-axis was used for all plots to facilitate comparison between plots. Statistical analyses were conducted using SAS and plots were generated using R (http://cran.r-project.org/).
Results
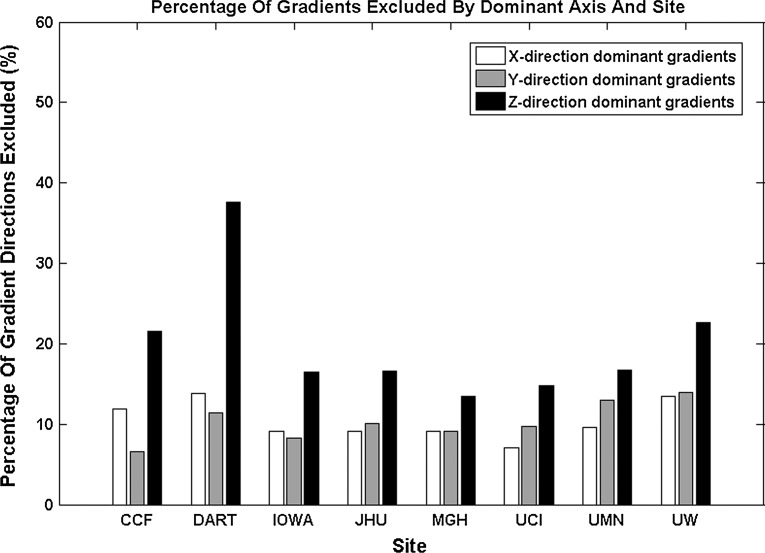
Five scans were eliminated for the low direction protocol from a single vendor (two from Dartmouth and three from Johns Hopkins) due to an insufficient number of gradients per scan after applying DTIPrep. These scans were removed for all of the statistical analyses. These scans suffered from significant subject motion that eliminated too many gradient directions to include that particular run. DTIPrep on average eliminated 12.76% of the 3D volumes (each b=0 and diffusion weighted image was considered a volume) per site after excluding the five runs that were completely eliminated from the analysis (Table 2). The percentage range of 3D volumes removed was 9.57%–20.47% per site. Of the gradient directions eliminated, nearly all were eliminated based on slice-wise checking. The venetian blind artifact check removed a small number of gradient directions, and the motion checking only eliminated three gradient directions across all sites (Fig. 1). From this data, it is clear that most motion-related artifacts are eliminated by the slice-wise and venetian blind artifact detection and only a small number of gradient directions are retained that exhibit a large amount of motion. An evaluation of diffusion encoding directions removed revealed that gradients with the predominant axes along the z-direction were removed more frequently than those for the x- and y-direction (Fig. 2).
Table 2.
Summary of Number of Gradient Directions Eliminated for Each Site
| Site | Slice-wise check | Venetian blind check | Motion check | Total |
|---|---|---|---|---|
| University of Iowa | 136 (10.15%) | 1 (0.075%) | 0 (0%) | 137 (10.22%) |
| University of Minnesota | 167 (11.84%) | 3 (0.21%) | 0 (0%) | 170 (12.06%) |
| University of California Irvine | 133 (9.43%) | 2 (0.14%) | 0 (0%) | 135 (9.57%) |
| Massachusetts General Hospital | 136 (9.65%) | 1 (0.071%) | 0 (0%) | 137 (9.72%) |
| Cleveland Clinic | 171 (12.13%) | 1 (0.071%) | 0 (0%) | 172 (12.20%) |
| Johns Hopkins | 143 (11.16%) | 2 (0.16%) | 3 (0.23%) | 148 (11.55%) |
| Dartmouth | 269 (20.47%) | 0 (0%) | 0 (0%) | 269 (20.47%) |
| University of Washington | 222 (16.08%) | 3 (0.22%) | 0 (0%) | 225 (16.30%) |
FIG. 1.

Examples of gradients containing artifacts that were detected and removed by DTIPrep. (A) An example of a volume removed due to a slice-wise intensity artifact. (B) An example of a volume removed due to a venetian blind artifact. The slice-wise checking step often removed gradients containing venetian blind artifacts before the venetian blind artifact check was executed. DTI, diffusion tensor imaging.
FIG. 2.
Percentages of gradient excluded by DTIPrep for diffusion gradients along the dominant axis.
The regions of interest were analyzed separately for the right and left hemisphere and similar results were obtained. Therefore, we are presenting only the combined results. The mean FA values measured across the brain regions ranged from 0.218 to 0.372 across the various regions of interest (Table 3). The occipital lobe had the smallest FA values, while the subcortical region had the largest values. Without the use of the median filter, the FA value was higher and ranged from 0.279 to 0.437. The ordering of the FA values across brain regions, however, remained the same. The mean FA values measured across sites ranged from 0.295 to 0.309 (0.347–0.377 without median filtering) and the SDs for FA were ∼0.04 for all sites (Table 4). The mean FA value for Siemens scanners was slightly less than the FA value for Philips scanners (Table 5). The mean FA calculated based on the 71 gradient directions was slightly lower to those based with the 30/32 gradient directions. Although the above differences were small, due to the smaller standard errors (SEs), a mixed-effects model analysis indicated that they were all statistically significant. MD measures ranged from 0.766×10−3 to 0.774×10−3 mm2/sec and were not affected by the median filter. However, application of the median filter increased the RD and decreased the AD. Philips scanners had a smaller MD as compared to the Siemens scanners. Again, because of the relatively small SEs, mixed-effects model analysis indicated that these differences were all statistically significant. MD values were also similar between scanning protocols and their SDs were also similar.
Table 3.
Mean and Standard Deviation of Rotationally Invariant Scalar Measures by Region Across All Sites
| Rotationally invariant scalar | Region | Mean with median filter | Standard deviation with median filter | Mean without median filter | Standard deviation without mean filter |
|---|---|---|---|---|---|
| MD | Cerebrum | 0.767×10−3 | 1.87×10−5 | 0.773×10−3 | 1.86×10−5 |
| Frontal | 0.766×10−3 | 1.87×10−5 | 0.773×10−3 | 1.82×10−5 | |
| Occipital | 0.767×10−3 | 2.98×10−5 | 0.767×10−3 | 2.58×10−5 | |
| Parietal | 0.774×10−3 | 2.63×10−5 | 0.776×10−3 | 2.77×10−5 | |
| Subcortical | 0.773×10−3 | 2.11×10−5 | 0.773×10−3 | 2.19×10−5 | |
| Temporal | 0.771×10−3 | 2.06×10−5 | 0.785×10−3 | 1.59×10−5 | |
| RD | Cerebrum | 0.653×10−3 | 1.81×10−5 | 0.608×10−3 | 2.22×10−5 |
| Frontal | 0.642×10−3 | 1.74×10−5 | 0.603×10−3 | 1.99×10−5 | |
| Occipital | 0.694×10−3 | 2.75×10−5 | 0.636×10−3 | 3.13×10−5 | |
| Parietal | 0.653×10−3 | 2.60×10−5 | 0.608×10−3 | 3.25×10−5 | |
| Subcortical | 0.612×10−3 | 2.05×10−5 | 0.566×10−3 | 2.53×10−5 | |
| Temporal | 0.665×10−3 | 1.48×10−5 | 0.624×10−3 | 1.65×10−5 | |
| AD | Cerebrum | 1.00×10−3 | 2.27×10−5 | 1.07×10−3 | 2.27×10−5 |
| Frontal | 1.02×10−3 | 2.43×10−5 | 1.08×10−3 | 2.56×10−5 | |
| Occipital | 0.925×10−3 | 2.66×10−5 | 0.984×10−3 | 2.50×10−5 | |
| Parietal | 1.02×10−3 | 3.01×10−5 | 1.08×10−3 | 3.16×10−5 | |
| Subcortical | 1.10×10−3 | 2.97×10−5 | 1.16×10−3 | 2.99×10−5 | |
| Temporal | 1.01×10−3 | 2.42×10−5 | 1.08×10−3 | 2.56×10−5 | |
| FA | Cerebrum | 0.295 | 0.0078 | 0.349 | 0.0155 |
| Frontal | 0.309 | 0.0081 | 0.364 | 0.0139 | |
| Occipital | 0.218 | 0.0116 | 0.279 | 0.0200 | |
| Parietal | 0.305 | 0.0116 | 0.355 | 0.0196 | |
| Subcortical | 0.372 | 0.0149 | 0.437 | 0.0232 | |
| Temporal | 0.289 | 0.0086 | 0.345 | 0.0160 |
MD, mean diffusivity; RD, radial diffusivity; AD, axial diffusivity; FA, fractional anisotropy.
Table 4.
Mean and Standard Deviation of Rotationally Invariant Scalar Measures Across All Anatomical Regions by Site
| Type | Site | Mean with median filter | Standard deviation with median filter | Mean without median filter | Standard deviation without mean filter |
|---|---|---|---|---|---|
| MD | Cleveland Clinic | 0.791×10−3 | 2.15×10−5 | 0.795×10−3 | 2.19×10−5 |
| Dartmouth | 0.753×10−3 | 1.68×10−5 | 0.755×10−3 | 1.97×10−5 | |
| University of Iowa | 0.779×10−3 | 1.76×10−5 | 0.782×10−3 | 1.71×10−5 | |
| Johns Hopkins | 0.754×10−3 | 1.45×10−5 | 0.764×10−3 | 1.87×10−5 | |
| Massachusetts General Hospital | 0.780×10−3 | 2.56×10−5 | 0.783×10−3 | 2.01×10−5 | |
| University of California Irvine | 0.780×10−3 | 1.70×10−5 | 0.784×10−3 | 1.55×10−5 | |
| University of Minnesota | 0.770×10−3 | 1.54×10−5 | 0.774×10−3 | 1.24×10−5 | |
| University of Washington | 0.759×10−3 | 1.68×10−5 | 0.762×10−3 | 1.92×10−5 | |
| RD | Cleveland Clinic | 0.672×10−3 | 2.76×10−5 | 0.630×10−3 | 2.56×10−5 |
| Dartmouth | 0.634×10−3 | 2.72×10−5 | 0.580×10−3 | 2.81×10−5 | |
| University of Iowa | 0.659×10−3 | 2.95×10−5 | 0.617×10−3 | 2.71×10−5 | |
| Johns Hopkins | 0.635×10−3 | 2.54×10−5 | 0.586×10−3 | 2.78×10−5 | |
| Massachusetts General Hospital | 0.661×10−3 | 2.72×10−5 | 0.619×10−3 | 2.36×10−5 | |
| University of California Irvine | 0.660×10−3 | 3.01×10−5 | 0.619×10−3 | 2.81×10−5 | |
| University of Minnesota | 0.655×10−3 | 2.47×10−5 | 0.614×10−3 | 2.24×10−5 | |
| University of Washington | 0.641×10−3 | 2.60×10−5 | 0.587×10−3 | 2.58×10−5 | |
| AD | Cleveland Clinic | 1.03×10−3 | 5.52×10−5 | 1.10×10−3 | 5.57×10−5 |
| Dartmouth | 0.993×10−3 | 4.57×10−5 | 1.07×10−3 | 5.29×10−5 | |
| University of Iowa | 1.02×10−3 | 5.03×10−5 | 1.11×10−3 | 5.06×10−5 | |
| Johns Hopkins | 1.00×10−3 | 4.48×10−5 | 1.10×10−3 | 4.98×10−5 | |
| Massachusetts General Hospital | 1.03×10−3 | 5.17×10−5 | 1.08×10−3 | 5.20×10−5 | |
| University of California Irvine | 1.03×10−3 | 4.78×10−5 | 1.09×10−3 | 4.66×10−5 | |
| University of Minnesota | 1.01×10−3 | 5.01×10−5 | 1.07×10−3 | 4.94×10−5 | |
| University of Washington | 1.01×10−3 | 4.70×10−5 | 1.07×10−3 | 5.08×10−5 | |
| FA | Cleveland Clinic | 0.295 | 0.0403 | 0.348 | 0.0421 |
| Dartmouth | 0.304 | 0.0381 | 0.374 | 0.0439 | |
| University of Iowa | 0.300 | 0.0437 | 0.353 | 0.0452 | |
| Johns Hopkins | 0.309 | 0.0368 | 0.377 | 0.0403 | |
| Massachusetts General Hospital | 0.297 | 0.0400 | 0.352 | 0.0403 | |
| University of California Irvine | 0.301 | 0.0439 | 0.353 | 0.0449 | |
| University of Minnesota | 0.294 | 0.0423 | 0.347 | 0.0435 | |
| University of Washington | 0.302 | 0.0373 | 0.370 | 0.0426 |
Table 5.
Mean and Standard Deviation of Rotationally Invariant Scalar Measures Across All Anatomical Regions by Vendor
| Type | Manufacturer | Mean with median filter | Standard deviation with median filter | Mean without median filter | Standard deviation without mean filter |
|---|---|---|---|---|---|
| MD | Philips | 0.755×10−3 | 1.63×10−5 | 0.760×10−3 | 1.96×10−5 |
| Siemens | 0.780×10−3 | 2.09×10−5 | 0.783×10−3 | 1.90×10−5 | |
| RD | Philips | 0.638×10−3 | 2.59×10−5 | 0.586×10−3 | 2.67×10−5 |
| Siemens | 0.657×10−3 | 2.99×10−5 | 0.613×10−3 | 3.00×10−5 | |
| AD | Philips | 1.00×10−3 | 4.59×10−5 | 1.08×10−3 | 5.09×10−5 |
| Siemens | 1.02×10−3 | 5.19×10−5 | 1.08×10−3 | 5.24×10−5 | |
| FA | Philips | 0.305 | 0.0375 | 0.374 | 0.0424 |
| Siemens | 0.297 | 0.0421 | 0.351 | 0.0433 |
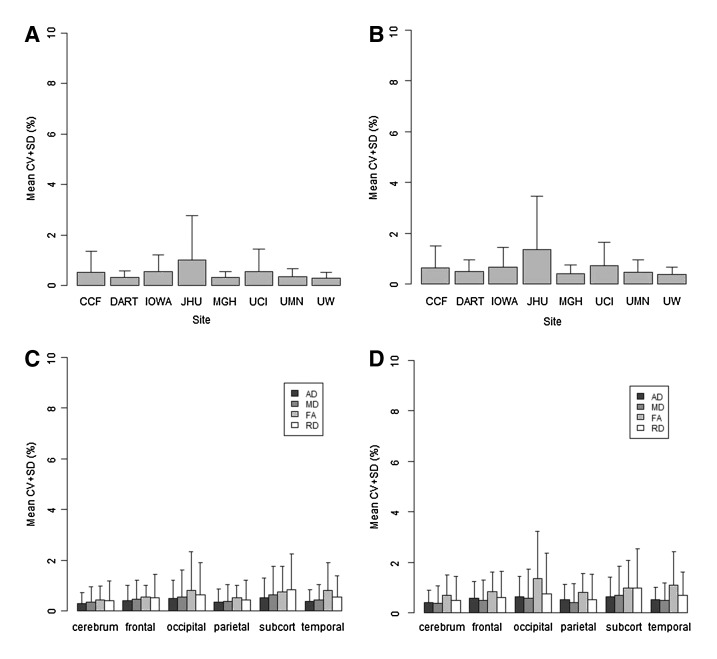
From the within-subject reliability analysis (Fig. 3A), we found that overall the CVs were less than 4.25% across all of the scalar measures and sites when median filtering was used for all brain regions (Fig. 3C). There was an increase in the CV without the application of median filtering (3D). The mean CVs were less than or equal to 1% across all sites and scalar measures except at Johns Hopkins, which had CV values for FA and RD above 1% (Table 6). Without the application of median filtering, all scalar measures at Johns Hopkins were above 1% and FA values at Iowa and University of California, Irvine were also above 1%. The mean CV for protocol with 71 gradient directions (0.46% with median filtering and 0.60% without) was slightly smaller than that for protocol with 30/32 gradient directions (0.56 with median filtering and 0.71% without). Mean CVs were comparable between data analyzed with and without DTIPrep. A mixed-effects model analysis indicated that DTIPrep status did not have a significant effect on the within-subject CV, but the protocol type (71 vs. 30/32 gradient directions), median filtering, and vendor did.
FIG. 3.
Intra-site reliability analysis showing the mean CV. The top row show CV by site for FA: (A) with median filtering and (B) without median filtering. The bottom row shows the CV by region and scalar measure: (C) with median filtering and (D) without median filtering. CV, coefficient of variation; FA, fractional anisotropy.
Table 6.
Mean and Standard Deviation of Within-Subject Coefficient of Variation by Site
| Site | Coefficient of variation median filtering | Standard deviation with median filtering | Coefficient of variation without median filtering | Standard deviation without median filtering |
|---|---|---|---|---|
| Cleveland Clinic | 0.528 | 0.826 | 0.632 | 0.869 |
| Dartmouth | 0.328 | 0.249 | 0.494 | 0.446 |
| University of Iowa | 0.554 | 0.681 | 0.673 | 0.779 |
| Johns Hopkins | 1.000 | 1.760 | 1.359 | 2.104 |
| Massachusetts General Hospital | 0.305 | 0.237 | 0.411 | 0.333 |
| University of California Irvine | 0.556 | 0.873 | 0.708 | 0.945 |
| University of Minnesota | 0.348 | 0.301 | 0.473 | 0.472 |
| University of Washington | 0.297 | 0.210 | 0.372 | 0.281 |
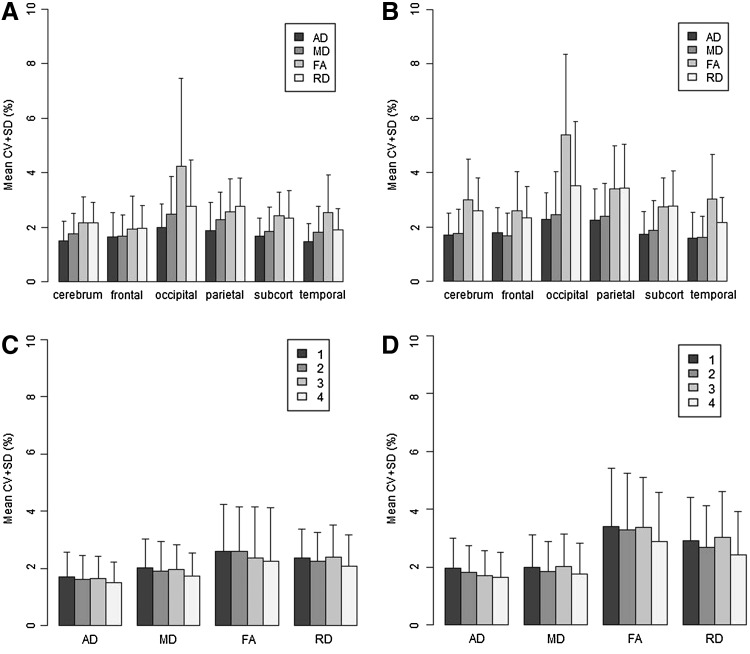
From the between-site reliability analysis (Fig. 4), we found increased CV as compared to the within-subject analysis. The CVs were ∼2% across all of the scalar measures (Fig. 4C, D). The CVs were typically less than 3% across all brain regions (Fig. 4A), except FA within the occipital lobe where the CV was 3.2%. The mean CV in the occipital lobe was higher across all scalar measures as compared to the other brain regions. The mean CV for the 71 direction protocol (2.15%) was higher than that for the 30/32 direction protocol (1.78%).
FIG. 4.
Inter-site reliability analysis showing the mean CV. The top row shows the CV by brain region and scalar type (A) with median filtering and (B) without median filtering. The bottom row shows the CV based on the number of concatenations and scalar measure for the low-diffusion gradient protocol (C) with median filtering and (D) without median filtering.
Mixed-effect model analysis indicated that the protocol type, vendor, median filtering, and concatenation had significant effects on between-site CVs, but DTIPrep did not. We found an interaction effect between the scanner and protocol: for the 71-direction protocol, Siemens had lower CV than Philips, but for the 30/32 direction protocol, the relationship was reverse.
Discussion
Using DTIPrep to perform automated quality assurance for this study eliminated, on average, 13.4% of the gradient directions. This resulted in ∼26 directions being used for tensor estimation for the 30/32 directions sequence and 62 directions for the 71 directions sequence. Based on the work by Jones (2004), reliable estimates are possible and this is close to the 30 directions required for reliable estimation of the tensor orientation and MD. We did not evaluate tensor estimation, but we found similar CV (<1%) for both the FA and MD measures suggesting that MD can be reliably estimated with 26 directions. While the use of DTIPrep did not significantly improve the results, it did identify a number of gradient directions that we identified upon visual inspection as exhibiting image artifacts. While making the measurements for regional FA values, we required that the tissue classification identify voxels as white matter as well as having a FA value larger than 0.1. This may have made our analysis somewhat immune to certain artifacts. Based on our review of the resulting FA maps, it was clear that we could obtain elevated FA values in data that had not undergone quality assurance using DTIPrep. These artifacts tended to be visible primarily in gray matter regions (Fig. 5) that were eliminated from the analysis due to the requirement that the voxels had to be classified as white matter based on the anatomical imaging data. In addition, it excluded five runs of diffusion tensor data that did not have a sufficient number of diffusion directions after removal of gradient directions with artifacts. These data sets were removed for both the analysis with and without DTIPrep.
FIG. 5.
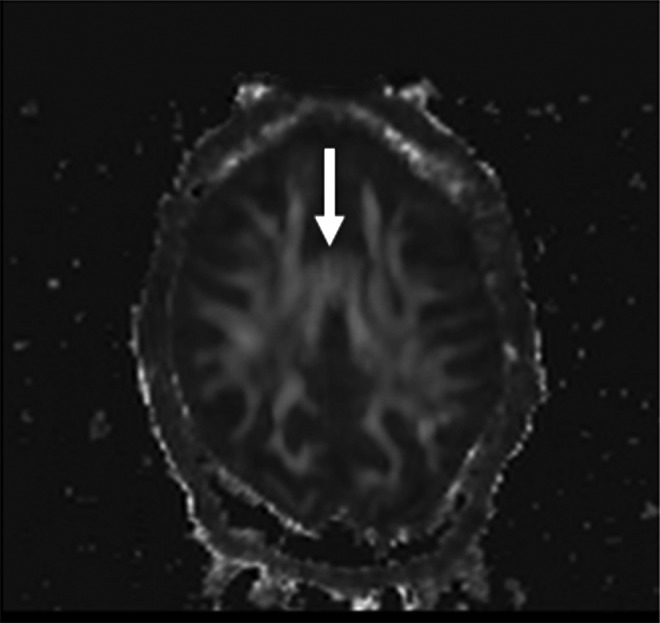
FA artifact resulting from table vibrations that appears in the images not corrected by DTIPrep. The increased FA appears in the anterior cingulate gray matter as denoted by the arrow.
In this study, we found that the application of a median filter significantly improved the reliability of the DTI rotationally invariant scalar measures. The CV was ∼20% smaller using the median filter as compared to no filtering. The median filter decreased the FA values by ∼15%. The MD remained constant, while the AD was decreased by ∼3% and the RD was increased by ∼8%.
The data acquired in this study showed similar reliability when compared to the DTI reliability study performed by Pagani and colleagues (2010). In this previous study, the average CV across centers was ∼5% for all scalar measures across the various regions studied. In our study, we found that the mean CV within subject was ∼0.5% and that between sites was ∼2%. The major difference between the current study and that conducted by Pagani and colleagues (2010) was that we restricted our analysis to 3T scanners, while Pagani and colleagues (2010) studied reliability across different field strengths (1.5T and 3T).
Based on the difference between the mean FA and MD measures obtained across scanners, it is likely that the vendors employ slightly different formulas for determining the gradient magnitude to be applied based on the b-value specified at the scanner console. These different imaging sequences may account for some of the variability. The Siemens scanner utilizes a dual-echo approach, while the Philips scanners used a Stejskal-Tanner sequence. In addition, subjective reports by the participants reported more vibration from the Siemens scanners as compared to the Philips scanners. We did not directly measure this through a formal debriefing session. Instead, subjects were interviewed informally to identify what the site differences were. A number of subtle differences were identified, including the comfort of the pad used for the head and how the head was restrained inside of the head coil.
This reliability study found that within subject variability was small (CV ∼0.5%) across all of the scanners and regions evaluated. This is relatively small as compared to the variation that we have seen in subjects with prodromal HD. In a prior study, we found that the FA value change was ∼0.1 across the 5-year probability of onset (Magnotta et al., 2009). The CV is relatively small compared to this. The reliability study contains a number of sources that may be contributing to variability in the diffusion tensor scalar estimates that include diffusion encoding, susceptibility artifacts, and image registration. We tried to minimize the effects of noise by applying a median filter before tensor estimation across all of the images. This increases the signal-to-noise in the images while preserving the white matter tracks. However, the estimated FA was reduced by 20% with reductions in the AD and increases in the RD values. This study looked at fairly large regions of interest to estimate the reliability and studies focused on small regions of interest or a specific fiber track may want to assess reliability within these specific regions. To facilitate such an analysis, we have made this data available to the general public (https://predict-hd.net/xnat/). As mentioned above, we have focused our analysis on white matter regions.
In summary, this study shows that the estimation of diffusion tensor rotationally invariant scalar measures can be robustly estimated within a site with very little variation (CV ∼0.5%) using a standard diffusion encoding scheme (30/32 gradient directions) provided by the scanner vendors. This suggests that subtle changes in the white matter architecture can be studied longitudinally if the subjects are scanned on the same scanner. We found that four averages of the diffusion encoding scheme were needed to significantly improve reliability. A fourfold increase in variability was observed as multiple sites and multiple vendors were included in the analysis. We also found approximately a 3% difference in the rotationally invariant scalar measures between vendors. This is likely due to variability in how the b-value entered by the user is converted into the gradient amplitude and the different diffusion encoding schemes used by the two vendors in this study. Therefore, to minimize the number of subjects required for a longitudinal study, the same scanner should be utilized throughout the study. Finally, in any longitudinal study of sufficient duration, it is likely that the scanner will undergo upgrades. Software variations across scanners were included in this analysis, but hardware upgrades, such as gradient coils, were not evaluated in this study. Therefore, we expect the reliability resulting from software upgrades will be smaller than the measured inter-site variability measured within vendor, but larger than the intra-site reliability estimated in this study. Further study is needed to assess how major hardware changes will impact the reliability measures. However, one would expect the inter-site reliability estimated in this study would serve as an upper bound for the CV.
Acknowledgments
This work was supported, in part, by awards from CHDI Foundation, Inc.; NIH R01NS050568; NINDS NS40068 Neurobiological Predictors of Huntington's Disease; and NINDS R01 NS054893 Cognitive and Functional Brain Changes in Preclinical HD. Dr. Turner was supported by NCRR 1 U24 RR021992 from the Functional Imaging Biomedical Informatics Network (S.G. Potkin, PI) for her work on this project while at the University of California, Irvine. This publication was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1RR024979. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author Disclosure Statement
No competing financial interests exist.
References
- Andreasen NC. Cizadlo T. Harris G. Swayze V. O'Leary DS. Cohen G, et al. Voxel processing techniques for the antemortem study of neuroanatomy and neuropathology using magnetic resonance imaging. J Neuropsychiatry Clin Neurosci. 1993;5:121–130. doi: 10.1176/jnp.5.2.121. [DOI] [PubMed] [Google Scholar]
- Andreasen NC. Cohen G. Harris G. Cizadlo T. Parkkinen J. Rezai K, et al. Image processing for the study of brain structure and function: problems and programs. J Neuropsychiatry Clin Neurosci. 1992;4:125–133. doi: 10.1176/jnp.4.2.125. [DOI] [PubMed] [Google Scholar]
- Andreasen NC. Rajarethinam R. Cizadlo T. Arndt S. Swayze VW. Flashman LA, et al. Automatic atlas-based volume estimation of human brain regions from MR images. J Comput Assist Tomogr. 1996;20:98–106. doi: 10.1097/00004728-199601000-00018. [DOI] [PubMed] [Google Scholar]
- Aylward EH. Anderson NB. Bylsma FW. Wagster MV. Barta PE. Sherr M, et al. Frontal lobe volume in patients with Huntington's disease. Neurology. 1998;50:252–258. doi: 10.1212/wnl.50.1.252. [DOI] [PubMed] [Google Scholar]
- Aylward EH. Codori AM. Rosenblatt A. Sherr M. Brandt J. Stine OC, et al. Rate of caudate atrophy in presymptomatic and symptomatic stages of Huntington's disease. Mov Disord. 2000;15:552–560. doi: 10.1002/1531-8257(200005)15:3<552::AID-MDS1020>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Beglinger LJ. Nopoulos PC. Jorge RE. Langbehn DR. Mikos AE. Moser DJ, et al. White matter volume and cognitive dysfunction in early Huntington's disease. Cogn Behav Neurol. 2005;18:102–107. doi: 10.1097/01.wnn.0000152205.79033.73. [DOI] [PubMed] [Google Scholar]
- Cercignani M. Bammer R. Sormani M. Fazekas F. Filippi M. Inter-sequence and inter-imaging unit variability of diffusion tensor MR imaging histogram-derived metrics of the brain in healthy volunteers. AJNR Am J Neuroradiol. 2003;24:638–643. [PMC free article] [PubMed] [Google Scholar]
- Cheng P. Magnotta VA. Wu D. Nopoulos P. Moser DJ. Paulsen J, et al. Evaluation of the GTRACT diffusion tensor tractography algorithm: a validation and reliability study. Neuroimage. 2006;31:1075–1085. doi: 10.1016/j.neuroimage.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Danielian L. Iwata N. Thomasson D. Floeter M. Reliability of fiber tracking measurements in diffusion tensor imaging for longitudinal study. Neuroimage. 2009;49:1572–1580. doi: 10.1016/j.neuroimage.2009.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris G. Andreasen NC. Cizadlo T. Bailey JM. Bockholt HJ. Magnotta VA, et al. Improving tissue classification in MRI: a three-dimensional multispectral discriminant analysis method with automated training class selection. J Comput Assist Tomogr. 1999;23:144–154. doi: 10.1097/00004728-199901000-00030. [DOI] [PubMed] [Google Scholar]
- Jones DK. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: a Monte Carlo study. Magn Reson Med. 2004;51:807–815. doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- Liu Z. Wang Y. Gerig G. Gouttard S. Tao R. Fletcher T, et al. Quality control of diffusion weighted images. Paper presented at the SPIE Medical Imaging; San Diego, CA: 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnotta VA. Harris G. Andreasen NC. O'Leary DS. Yuh WT. Heckel D. Structural MR image processing using the BRAINS2 toolbox. Comput Med Imaging Graph. 2002;26:251–264. doi: 10.1016/s0895-6111(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Magnotta VA. Heckel D. Andreasen NC. Cizadlo T. Corson PW. Ehrhardt JC, et al. Measurement of brain structures with artificial neural networks: two- and three-dimensional applications. Radiology. 1999;211:781–790. doi: 10.1148/radiology.211.3.r99ma07781. [DOI] [PubMed] [Google Scholar]
- Magnotta VA. Kim J. Koscik T. Beglinger LJ. Espinso D. Langbehn D, et al. Diffusion tensor imaging in Preclinical Huntington's disease. Brain Imaging Behav. 2009;3:77–84. doi: 10.1007/s11682-008-9051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes D. Haynor D. Vesselle H. Lewellyn T. Eubank W. Nonrigid multimodality image registration. Paper presented at the Proceedings of SPIE; San Diego, CA: 2001. [Google Scholar]
- Pagani E. Hirsch JG. Pouwels PJ. Horsfield MA. Perego E. Gass A, et al. Intercenter differences in diffusion tensor MRI acquisition. J Magn Reson Imaging. 2010;31:1458–1468. doi: 10.1002/jmri.22186. [DOI] [PubMed] [Google Scholar]
- Paulsen JS. Nopoulos PC. Aylward E. Ross CA. Johnson H. Magnotta VA, et al. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res Bull. 2010;82:201–207. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A. Adalsteinsson E. Sullivan E. Replicability of diffusion tensor imaging measurements of fractional anisotropy and trace in brain. J Magn Reson Imaging. 2003;18:427–433. doi: 10.1002/jmri.10377. [DOI] [PubMed] [Google Scholar]
- Pierson R. Johnson H. Harris G. Keefe H. Paulsen JS. Andreasen NC, et al. Fully automated analysis using BRAINS: autoworkup. Neuroimage. 2010;54:328–336. doi: 10.1016/j.neuroimage.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD. Tuch DS. Hevelone ND. Zaleta AK. Vangel M. Hersch SM, et al. Diffusion tensor imaging in presymptomatic and early Huntington's disease: selective white matter pathology and its relationship to clinical measures. Mov Disord. 2006;21:1317–1325. doi: 10.1002/mds.20979. [DOI] [PubMed] [Google Scholar]
- Van Camp N. Blockx I. Camon L. de Vera N. Verhoye M. Veraart J, et al. A complementary diffusion tensor imaging (DTI)-histological study in a model of Huntington's disease. Neurobiol Aging. 2010;33:945–959. doi: 10.1016/j.neurobiolaging.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Vollmar C. O'Muircheartaigh J. Barker GJ. Symms MR. Thompson P. Kumari V, et al. Identical, but not the same: intra-site and inter-site reproducibility of fractional anisotropy measures on two 3.0T scanners. Neuroimage. 2010;51:1384–1394. doi: 10.1016/j.neuroimage.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY. Abdi H. Bakhadirov K. Diaz-Arrastia R. Devous MD., Sr. A comprehensive reliability assessment of quantitative diffusion tensor tractography. Neuroimage. 2012;60:1127–1138. doi: 10.1016/j.neuroimage.2011.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver KE. Richards TL. Liang O. Laurino MY. Samii A. Aylward EH. Longitudinal diffusion tensor imaging in Huntington's Disease. Exp Neurol. 2009;216:525–529. doi: 10.1016/j.expneurol.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]