The Ndc80 outer kinetochore complex plays a critical role in kinetochore–microtubule attachment. A ubiquitous internal loop of Ndc80 acts as a structural platform by which to recruit the Alp7/TACC-Alp14/TOG microtubule-binding complex to the outer kinetochore. This interaction ensures proper chromosome attachment and segregation.
Abstract
The Ndc80 outer kinetochore complex plays a critical role in kinetochore–microtubule attachment, yet our understanding of the mechanism by which this complex interacts with spindle microtubules for timely and accurate chromosome segregation remains limited. Here we address this issue using an ndc80 mutant (ndc80-NH12) from fission yeast that contains a point mutation within a ubiquitous internal loop. This mutant is normal for assembly of the Ndc80 complex and bipolar spindle formation yet defective in proper end-on attachment to the spindle microtubule, with chromosome alignment defects and missegregation happening later during mitosis. We find that ndc80-NH12 exhibits impaired localization of the microtubule-associated protein complex Alp7/transforming acidic coiled coil (TACC)-Alp14/tumor-overexpressed gene (TOG) to the mitotic kinetochore. Consistently, wild-type Ndc80 binds these two proteins, whereas the Ndc80-NH12 mutant protein displays a substantial reduction of interaction. Crucially, forced targeting of Alp7–Alp14 to the outer kinetochore rescues ndc80-NH12-mutant phenotypes. The loop was previously shown to bind Dis1/TOG, by which it ensures initial chromosome capture during early mitosis. Strikingly, ndc80-NH12 is normal in Dis1 localization. Genetic results indicate that the loop recruits Dis1/TOG and Alp7/TACC-Alp14/TOG independently. Our work therefore establishes that the Ndc80 loop plays sequential roles in spindle–kinetochore attachment by connecting the Ndc80 complex to Dis1/TOG and Alp7/TACC-Alp14/TOG.
INTRODUCTION
The ultimate goal of mitotic spindle formation is to capture individual chromosomes and pull each pair of sister chromatids to the opposite poles of the dividing cell. Chromosome attachment is achieved on the kinetochore, a large, proteinaceous structure assembling around centromeres (Rieder and Salmon, 1998; Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). Any errors in this process lead to miscarriage, birth defects, and production of aneuploid progeny, which is the hallmark of human cancer. Recent advances in our knowledge of kinetochore components and their organization provided molecular insights into the structure and function of this megaprotein complex (Westermann et al., 2007; Takeuchi and Fukagawa, 2012). Among the kinetochore components identified, the Ndc80 complex is of particular significance in chromosome attachment (Ndc80 is also known as Hec1 in humans; Chen et al., 1997; Ciferri et al., 2007; Vorozhko et al., 2008; Tooley and Stukenberg, 2011; Foley and Kapoor, 2012). This complex comprises a part of the KMN network (KNL-1/Mis12 complex/Ndc80 complex; Cheeseman et al., 2006) and locates to the kinetochore–microtubule interface on the outer kinetochore (Joglekar et al., 2009; Wan et al., 2009). KNL-1 and the Ndc80 complex individually possess microtubule-binding activities in vitro (Cheeseman et al., 2006; Wei et al., 2007; Ciferri et al., 2008; Guimaraes et al., 2008; Miller et al., 2008; Wilson-Kubalek et al., 2008; Powers et al., 2009; Alushin et al., 2010, 2012; Umbreit et al., 2012), in which the Ndc80 complex exhibits a much stronger affinity to microtubules than KNL-1 (Cheeseman et al., 2006).
The Ndc80 complex consists of four conserved subunits—Ndc80, Nuf2, Spc24 and Spc25—in a 1:1:1:1 stoichiometry, forming a ∼57-nm, elongated, rod-like structure containing internal coiled-coil domains and globular regions at both ends (Ciferri et al., 2005; Wei et al., 2005). It is the Ndc80–Nuf2 subcomplex that binds microtubules, whereas Spc24–Spc25 interacts with other kinetochore components, including the Mis12 complex and CENP-T (Cheeseman et al., 2006; Wei et al., 2006; Ciferri et al., 2008; Wang et al., 2008; Maskell et al., 2010; Petrovic et al., 2010; Hornung et al., 2011; Bock et al., 2012; Schleiffer et al., 2012). In fact, the N-terminal region of Ndc80–Nuf2 contains a calponin-homology domain (Korenbaum and Rivero, 2002) and an unstructured N-tail that are responsible for direct binding to the microtubule (Cheeseman et al., 2006; Deluca et al., 2006; Guimaraes et al., 2008; Miller et al., 2008; Alushin et al., 2012).
In addition, Ndc80, but not Nuf2, contains a unique short loop consisting of 50–100 amino acid residues that is sandwiched by its adjacent coiled coils (Ciferri et al., 2008; Wang et al., 2008). Although this loop may play a structural role, such as in the introduction of the coiled-coil kink to the rod region of the Ndc80 complex (Wang et al., 2008; Wan et al., 2009; Varma et al., 2012; Zhang et al., 2012), most recent results from yeasts and humans have shown that this domain also acts as a protein–protein interaction motif (Hsu and Toda, 2011; Tang and Toda, 2013; Maure et al., 2011; Nilsson, 2012; Varma et al., 2012; Varma and Salmon, 2012; Zhang et al., 2012). We previously showed that the Ndc80 loop in fission yeast recruits the Dis1 microtubule-associated protein (Ohkura et al., 1988; Nabeshima et al., 1995) to the mitotic outer kinetochore via direct interaction (Hsu and Toda, 2011). The Dis1 protein belongs to the conserved Dis1/XMAP215/tumor-overexpressed gene (TOG) protein family (colonic hepatic TOG/ch-TOG in humans; Ohkura et al., 2001; Kinoshita et al., 2002). These proteins track on the plus end of microtubules (plus end–tracking proteins [+TIPs]; Carvalho et al., 2003; Akhmanova and Steinmetz, 2008) and, more importantly, possess microtubule polymerase activities (Brouhard et al., 2008; Widlund et al., 2011; Al-Bassam et al., 2012). We showed that the temperature-sensitive mutant ndc80-21, which contains a point mutation within the loop (L405P; Supplemental Figure S1), fails to establish spindle–kinetochore attachment ascribable to extremely unstable kinetochore microtubules. These phenotypes are mainly, if not entirely, derived from the inability of Dis1 to be recruited to the kinetochore, as the Ndc80-21 mutant protein is impaired in binding to Dis1 and, critically, artificial tethering of Dis1 to the outer kinetochore rescued ndc80-21 (Hsu and Toda, 2011).
In the present study, we isolated an additional loop mutant called ndc80-NH12 that contains a point mutation within the loop but in a position different from that of the previously isolated ndc80-21. Unexpectedly, ndc80-NH12 displays novel defects in spindle–kinetochore attachment that are not observed in ndc80-21. We present evidence that ndc80-NH12 is normal in the recruitment of Dis1 but instead is defective in recruiting another member of the Dis1/XMAP215/TOG family, Alp14/Mtc1 (Garcia et al., 2001; Nakaseko et al., 2001). Alp14/TOG forms a stable complex with Alp7/Mia1 (Radcliffe et al., 1998; Oliferenko and Balasubramanian, 2002; Sato et al., 2003, 2004), which is a member of the transforming acidic coiled-coil (TACC) protein family (Gergely, 2002; Raff, 2002; Peset and Vernos, 2008), and we show that the Alp7–Alp14 complex directly binds the loop, independent of Dis1. We discuss the functional significance of these findings for the establishment of spindle–kinetochore attachment and accurate chromosome segregation.
RESULTS
Isolation of a new Ndc80-loop mutant
To further explore the roles of Ndc80 in chromosome attachment, we conducted screening for temperature-sensitive (ts) ndc80 mutants using the error-prone PCR mutagenesis method. This approach yielded 18 additional mutants (see Materials and Methods for more details). Nucleotide sequencing of these alleles showed that one mutant, ndc80-NH12, contained a single point mutation within the loop (F420S; Figure 1A and Supplemental Figure S1). The ndc80-NH12 mutant was hypersensitive to the microtubule-depolymerizing drug thiabendazole (TBZ) at the permissive temperature (Figure 1B), indicative of mitotic defects. Introduction of multicopy plasmids containing wild-type ndc80+ rescued temperature and TBZ sensitivity of ndc80-NH12 (Figure 1B), indicating that the defective phenotypes of this mutant is indeed due to the F420S point mutation within Ndc80. The structural integrity of the Ndc80 complex remained intact, as Nuf2 and Spc25 colocalized to the kinetochore normally at the restrictive temperature (Figure 1C). This suggested that the defective phenotypes of ndc80-NH12 would not be ascribable to gross malfunctioning such as the disassembly of the Ndc80 complex and delocalization from kinetochores.
FIGURE 1:
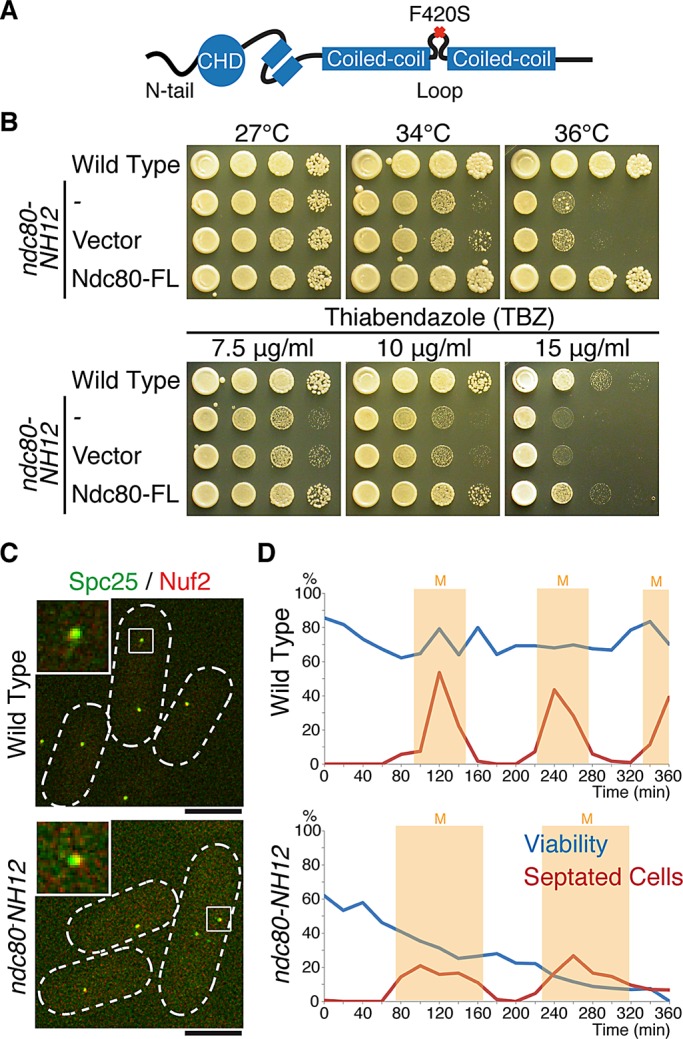
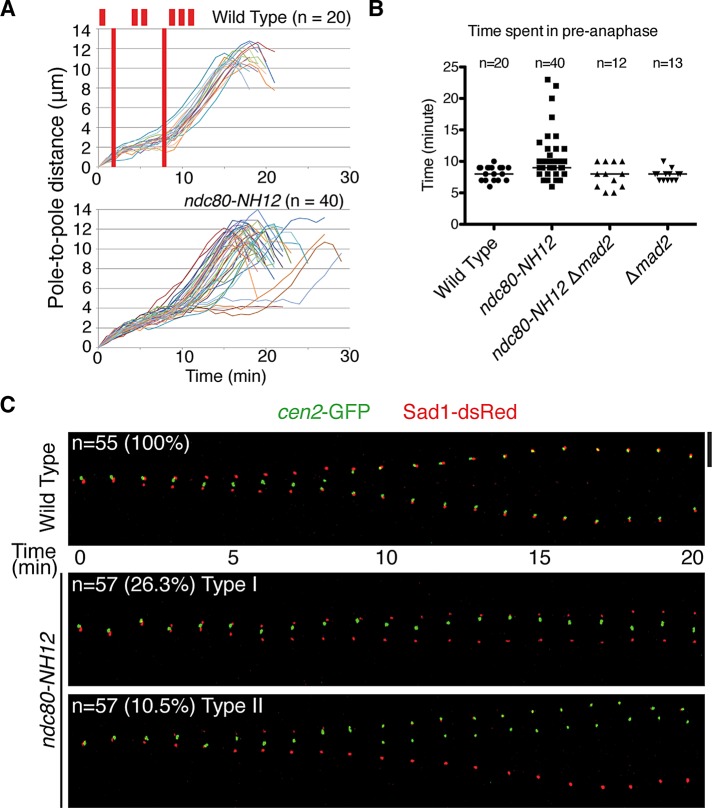
Isolation of the temperature-sensitive ndc80-NH12 mutant, which contains a single point mutation (F420S) within the loop. (A) Schematic presentation of the mutation found in ndc80-NH12. The oval shows the calponin-homology domain (CHD; Wei et al., 2007; Ciferri et al., 2008) preceded by the N-terminal extension; two boxes represent internal coiled-coil domains, which are interrupted by the internal loop. The F420S mutation is located within the loop. See Supplemental Figure S1 for more information on the mutation site. (B) Temperature and thiabendazole sensitivity. Tenfold serial dilutions of wild-type or ndc80-NH12 cells carrying an empty vector (pREP1) or a plasmid carrying the full-length ndc80+ gene (pREP1-Ndc80-FL) were spotted onto rich agar media (5 × 104 cells in the first spot) and incubated at the indicated temperatures for 3 d. The same set of strains were spotted on rich media containing the anti-microtubule drug TBZ (7.5, 10, or 15 μg/ml) and incubated at 27°C for 3 d. (C) Normal kinetochore localization of the Ndc80 complex in the ndc80-NH12 mutant. Wild-type or ndc80-NH12 mutant cells containing Spc25-YFP and Nuf2-mCherry were incubated at 36°C for 4 h. Merged images of fluorescence signals (green for Spc25 and red for Nuf2) are shown. Top left, enlarged images of kinetochore signals (squares). Scale bar, 5 μm. (D) Viability of cells (wild type, top; ndc80-NH12, bottom) in synchronous culture analyses. Small G2 cells grown at 27°C were collected by centrifugal elutriation and shifted to 36°C at time 0. At each time point, aliquots of cells were taken and cell number counted. Cell viability (blue lines) was measured by spreading 100–300 cells on rich agar media and incubated at 27°C. The percentage of septated cells (red lines, n > 100) was also counted by staining cells with Calcofluor (a dye staining septa). Light yellow columns mark periods in mitosis.
Synchronous culture analysis using centrifugal elutriation showed that ndc80-NH12 cells lost viability by 35% during the first mitosis (viability at the permissive temperature also decreased by 20% compared with wild-type cells), followed by a further 20% reduction in the second mitosis (Figure 1D). These results indicate that ndc80-NH12 is a novel loop mutant with mitotic defects.
Preanaphase mitotic delay is observed in some but not all populations of ndc80-NH12 cells
We sought to delineate the defective mitotic phenotypes of ndc80-NH12. To this end, we monitored mitotic progression at the restrictive temperature using a strain containing fluorescently tagged markers for the spindle pole body (SPB; the yeast equivalent of the animal centrosome) Sad1-dsRed (Hagan and Yanagida, 1995) and sister centromere cen2–green fluorescent protein (GFP; Nabeshima et al., 1998; Yamamoto and Hiraoka, 2003; Hauf et al., 2007). Mitotic phases of fission yeast wild-type cells are composed of the three distinct stages, depending upon the spindle length: phase I, in which spindles elongate as the two SPBs start to separate, corresponding to the duration from prophase to prometaphase; phase II, in which the spindle length is constant, corresponding to the duration from prometaphase to anaphase A; and phase III, in which the spindle length again starts to elongate, corresponding to anaphase B (Nabeshima et al., 1998).
Whereas wild-type cells at 36°C spent 8.2 ± 1.0 min in phases I and II (n = 20; Figure 2, A–C, and Supplemental Movie S1), 25% of ndc80-NH12 cells showed varied degrees of mitotic delay during this period (15.5 ± 4.4 min, n = 40; type I). Time-lapse imaging of individual type I cells indicated that cen2-GFP did not segregate toward the two poles but instead underwent prolonged oscillation between the two SPBs as a single dot (Figure 2C, middle; 26.3%, n = 57; and Supplemental Movie S2). This indicated that kinetochores attached to the spindle microtubules, but amphitelic attachment was not established, and hence sister chromatid cohesion was retained in these cells.
FIGURE 2:
The ndc80-NH12 mutant displays two types of mitotic abnormalities. (A) Profiles of mitotic progression. Wild-type and ndc80-NH12 cells that contained Sad1-dsRed (an SPB marker) and cen2-GFP (a GFP-tagged centromere on chromosome II) were grown at 27°C and shifted to 36°C. Mitotic cells were recorded under a fluorescence microscope, by which the distance of the two SPBs was measured and plotted against time (n = 20 for wild type and n = 40 for ndc80-NH12). Phase I is the period of initial spindle elongation (I), whereas phase II represents the duration in which the spindle length is constant (II). Phase III corresponds to anaphase B (III), when spindles elongate toward the cell tips. (B) The duration of the preanaphase stage. The duration of phases I and II was measured in four indicated strains and plotted. The data in A were used for wild type and ndc80-NH12. As for the mad2 deletion (Δmad2) and ndc80-NH12 Δmad2 strains, similar analysis as shown in A was performed (n = 13 and 12, respectively). Note that mitotic delay observed in ndc80-NH12 was abolished by the mad2 deletion. (C) Time-lapse live image analysis of sister centromere behavior. Mitotic wild-type cells (top) or ndc80-NH12 cells (bottom two rows) containing cen2-GFP (green) and Sad1-dsRed (red) cultured at 36°C were recorded (1-min intervals) and converted to kymograph. Top left, sample number (n = 55 for wild type and n = 57 for ndc80-NH12) and percentage of cells showing each phenotype (type I, 26.3%; type II, 10.5%). Type I (middle) displayed mitotic arrest during phase II, whereas type II (bottom) exhibited sister chromatid segregation errors during anaphase without mitotic delay. Scale bar, 5 μm.
Further analysis showed that the delay was ascribable to the activation of the spindle assembly checkpoint (SAC), a mitotic surveillance mechanism by which to monitor bipolar attachment at the kinetochore (Musacchio and Salmon, 2007), as deletion of its core component Mad2 abolished the mitotic delay (Figure 2B). Time-lapse imaging of ndc80-NH12mad2 double-mutant cells indicated the increase in the frequency of chromosome missegregation; 4 of the 12 cells observed displayed cen2-GFP segregation errors. These results suggested that one-fourth of mitotic ndc80-NH12 cells failed to establish bivalent attachment of the kinetochore to the spindle microtubule, resulting in SAC-mediated mitotic delay.
Other populations of ndc80-NH12 cells exhibit chromosome segregation defects during anaphase
As shown in Figure 2, A and B, three-fourths of ndc80-NH12 cells did not display mitotic delay, and yet overall cell viability dropped to ∼30% after the first mitosis (Figure 1D), suggesting additional defects in this mutant. Visual inspection of numerous live images led us to the identification of the second phenotype (type II). In this class (10.5%, n = 57), cells proceeded to anaphase similar to wild type, as the distance of the two SPBs started to increase (Figure 2C, bottom, 10 min, and Supplemental Movie S3). On anaphase onset, however, unlike wild-type cells, in which each sister chromatid reached and remained associated with the individual poles (top), in the mutant, cen2-GFP signals moved toward a single pole, resulting in chromosome missegregation. Furthermore, these cen2-GFP dots could not reach or be associated stably with the pole, thereby exhibiting lagging chromosome–like phenotypes during anaphase. Considering that cen2-GFP visualizes only one pair of sister chromatids of the three fission yeast chromosomes, this value (10.5%) would be an underestimate with regard to the total frequency of chromosome missegregation. Thus ndc80-NH12 cells showed novel anaphase defects that have not been seen in either the previously identified Ndc80-loop mutant ndc80-21 (Hsu and Toda, 2011) or mutants of the fission yeast KMN network (Goshima et al., 1999; Nabetani et al., 2001; Kerres et al., 2004).
Overall mitotic spindle morphology is normal in ndc80-NH12 cells
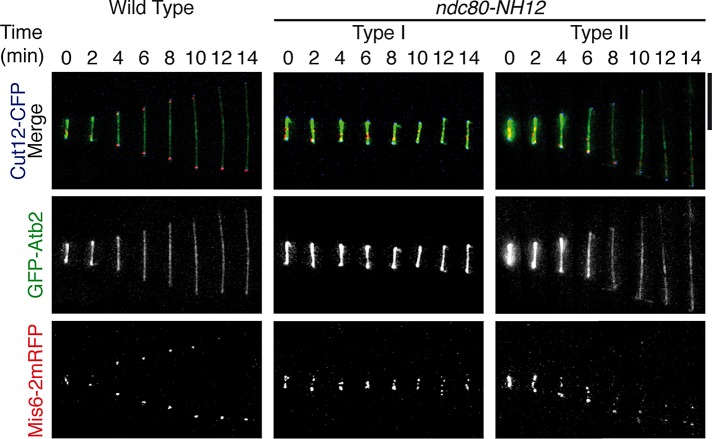
We previously showed that one of the most dramatic phenotypes in ndc80-21 was the appearance of unstable spindle microtubules (Hsu and Toda, 2011); this loop mutant could not form intact bipolar spindles, resulting in persistent mitotic arrest during the premetaphase stage. Given the apparent phenotypic differences between ndc80-21 and ndc80-NH12, we examined the spindle morphology in ndc80-NH12 that contained GFP-Atb2 (α2-tubulin; Toda et al., 1984), Cut12–cyan fluorescent protein (CFP; an SPB marker; Bridge et al., 1998), and Mis6-2-monomeric red fluorescent protein (mRFP; a kinetochore marker; Saitoh et al., 1997). As shown in Figure 3, bipolar spindles looked normal in both type I (middle) and type II cells (right), and we did not see any obvious abnormalities compared with wild-type cells (left). Nonetheless, chromosome missegregation was induced, in which kinetochores moved toward the one pole. Thus ndc80-NH12 is defective in the establishment (defective in type I) and maintenance (defective in type II) of proper attachment of the kinetochore to the spindle microtubule upon assembly of bipolar spindles.
FIGURE 3:
Mitotic spindles are morphologically normal in ndc80-NH12, yet chromosome missegregation is induced. (A) Time-lapse fluorescence montages of the spindle microtubule (GFP-Atb2, green, middle), kinetochore (Mis6-2mRFP, two copies of monomeric RFP, red, bottom), and the SPB (Cut12-CFP, shown in the merged images on the top) are shown in wild-type cells (left) and type I (middle) and type II (right) ndc80-NH12 cells incubated at 36°C for 1 h. Representative images of each strain are shown. Unlike wild-type cells, chromosomes (kinetochores) segregate unequally in ndc80-NH12 cells. Overall spindle microtubule structures (0–6 min), including their intensities and morphologies, are indistinguishable between wild-type and ndc80-NH12 cells. Note that GFP-Atb2 signals in wild-type (left) and type II (right) cells became dim after 6 min, ascribable to fluorescence bleaching. Scale bars, 5 μm.
Alp7/TACC and Alp14/TOG, but not Dis1/TOG, delocalize from the kinetochore in ndc80-NH12 cells
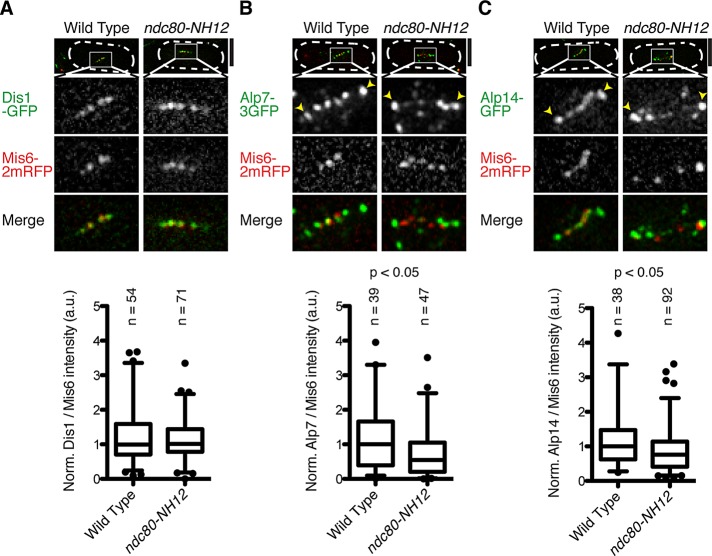
Our previous study showed that the major role of the loop lies in the recruitment of the microtubule-associated protein Dis1, a founding member of the TOG/XMAP215 microtubule-associated protein family (Ohkura et al., 2001; Kinoshita et al., 2002; Al-Bassam and Chang, 2011), and in the ndc80-21 mutant (L405P), Dis1 delocalized from the mitotic kinetochore (Hsu and Toda, 2011). Accordingly, we examined Dis1 localization in the ndc80-NH12 mutant and quantified its intensity. Intriguingly, the signal intensities of Dis1 at the mitotic kinetochore were indistinguishable from those of wild-type cells (Figure 4A).
FIGURE 4:
Alp7 and Alp14, but not Dis1, delocalize from the mitotic kinetochore in ndc80-NH12. (A–C) Wild-type or ndc80-NH12 mutant cells that contain Mis6-2mRFP (kinetochore, red) and Dis1-GFP (A, green), Alp7-3GFP (B, green), or Alp14-GFP (C, green) were incubated at 36°C for 1 h and fixed with methanol. Top, images of individual mitotic cells (outlined with discontinuous lines); bottom three rows, enlarged images that cover the kinetochore regions (squares shown on top). Bottom graphs, quantification of GFP signals at kinetochores vs. those of Mis6-2mRFP. The box-and-whisker plot indicates 5th–95th percentiles, 25th and 75th percentiles, and median. Filled circles indicate outliers. The number of samples (n) is indicated for each strain. Statistical significance was determined by the Student's t test. Note that in wild-type mitotic cells, Alp7 and Alp14 foci localize to both kinetochores (internal dots) and SPBs (external dots marked by arrowheads) as previously reported (Sato et al., 2004). In sharp contrast, in ndc80-NH12 mutants, these two proteins do not colocalize with kinetochores, whereas their localization to SPBs (arrowheads) is retained. Scale bars, 5 μm.
Fission yeast contains another member of the TOG/XMAP215 family, Alp14, which acts as a stable complex with Alp7/TACC (Sato et al., 2004; Sato and Toda, 2007). Previous work showed that Alp7/TACC and Alp14/TOG also localize to the mitotic kinetochore, as well as to the SPB and spindle microtubules (Garcia et al., 2001; Sato et al., 2004). We examined colocalization of these two microtubule-associated proteins with mitotic kinetochores in the mutant. As shown in Figure 4, B and C, signal intensities of both proteins at the mitotic kinetochores were reduced by 30–40% (p < 0.05). The Alp7 and Alp14 dots that did not colocalize with either the kinetochores or the SPBs most likely represented signals localizing to the spindle microtubules (Garcia et al., 2002; Sato et al., 2004). Decreased kinetochore localization of Alp7 and Alp14 raised the interesting possibility that the loop recruits not only Dis1/TOG but also the Alp7/TACC-Alp14/TOG complex to the mitotic kinetochore.
The Ndc80 loop directly binds Alp7/TACC and Alp14/TOG, and F420 in the loop is essential for Alp7’s interaction
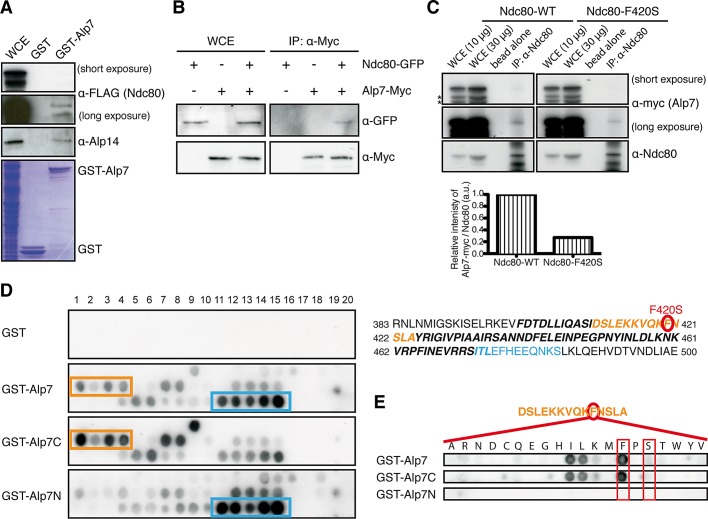
To explore the possibility that the loop interacts with Alp7/TACC and Alp14/TOG, we performed a series of binding experiments. First, bacterially produced and purified GST-Alp7 proteins pulled down Ndc80-FLAG from fission yeast mitotic extracts (Figure 5A; note that Alp14 was also pulled down). Second, immunoprecipitation experiments were performed using a mitotically arrested strain in which Ndc80 (tagged with GFP) and Alp7 (tagged with the Myc epitope) were produced from their native loci under endogenous promoters. We found that Ndc80-GFP coprecipitated with Alp7-Myc (Figure 5B). We then repeated the immunoprecipitations using strains in which either wild-type Ndc80 or mutant Ndc80-F420S proteins were overproduced. As shown in Figure 5C (left), Ndc80 again coimmunoprecipitated with Alp7-Myc. By contrast, in the case of Ndc80-F420S mutant proteins coimmunoprecipitation was still observed, but the intensities of precipitated Alp7-Myc was substantially reduced by ∼75% (Figure 5C, top right and bottom). Having seen physical interaction between Alp7–Alp14 and Ndc80, we asked whether ectopically overproduced Ndc80 is capable of localizing to the mitotic SPB in addition to the kinetochore. However, GFP-tagged Ndc80 still localized only to the kinetochore (Supplemental Figure S2).
FIGURE 5:
Alp7 binds Ndc80, and its C-terminal region interacts specifically with F420-containing peptides within the internal loop. (A) Pull down of Ndc80 by GST-Alp7. Bacterially purified GST-Alp7 on GSH beads was passed through fission yeast protein extracts prepared from mitotic cells (cut9-665 cells) containing Ndc80-3FLAG. Eluted fractions were immunoblotted with anti-FLAG (α-FLAG; top) or anti-Alp14 antibody (middle). The same fraction was stained with Coomassie blue (bottom). GST was used as a control. Immunoblots obtained from two different exposures (short and long) against anti-FLAG antibody are shown. WCE, whole-cell extract (25 μg). (B) Coimmunoprecipitation between Alp7 and Ndc80. Cell extracts were prepared from metaphase-arrested cut9-665 mutants containing Ndc80-GFP only, Alp7-13Myc only, or both Ndc80-GFP and Alp7-13Myc, and immunoprecipitation was performed with anti-Myc antibody, followed by immunoblotting with anti-GFP and anti-Myc antibodies (WCE, 20 μg). (C) Coimmunoprecipitation between Alp7 and Ndc80 wild-type or Ndc80-F420 mutant proteins. Cell extracts were prepared from mitotic cells containing Alp7-13Myc that carry pREP1-Ndc80 (left) or pREP1-Ndc80-F420S (right), and immunoprecipitation was performed with anti-Ndc80 antibody (Hsu and Toda, 2011), followed by immunoblotting with anti-Ndc80 and anti-myc antibodies. Immunoblots obtained from two different exposures (short and long) against anti-myc antibody are shown. Asterisks show degradation products of Alp7-Myc. Bottom, band intensities of Alp7-Myc quantified using those corresponding to Ndc80 as a control. (D) Peptide array assay. GST-fusion proteins containing full-length Alp7 (GST-Alp7), the N-terminal half (GST-Alp7N), or the C-terminal half (GST-Alp7C) were produced in bacteria and purified. GST is used as a negative control (top). Note that the membrane was spotted with 20-residue peptides covering the loop sequence with a 2-residue start increment per spot. Peptides that showed positive interactions (at least four consecutive spots) are boxed in orange and blue. Amino acid sequence of the loop region (bold and italics) is shown on the right with the two regions corresponding to positive peptides (411–424 in orange and 473–484 in blue). The position of F420 is marked with a red circle that is mutated in ndc80-NH12 (F420S). (E) F420 is essential for Alp7 binding. The 420th residue within a peptide (411–424) was mutated to all the possible amino acid residues, and peptide array assay was performed using GST-Alp7, Alp7C, and Alp7N. Note that the replacement with serine (boxed) substantially reduced binding capabilities of Alp7 and Alp7C. Phenylalanine is also boxed for comparison. Only isoleucine and leucine retain a degree of binding abilities similar to that of phenylalanine.
Finally, a peptide array assay, which was previously successfully applied to demonstrate a direct interaction between the loop and Dis1/TOG (Hsu and Toda, 2011), was performed using bacterially produced glutathione S-transferase (GST)–Alp7 proteins and peptide arrays that encompass the whole loop region of Ndc80 (20 amino acid residues per peptide spot, with two residues shifted per spot). As shown in Figure 5D, GST-Alp7 recognized the two regions within the loop (shown in orange and blue boxes and letters), one of them containing F420 (circled in red). Alp7/TACC consists of two structural domains. The N-terminal half contains the nuclear localization signal (NLS) with no homology to the other TACC members (Ling et al., 2009; Sato et al., 2009; Sato and Toda, 2010). On the other hand, the remaining C-terminal half consists of the conserved coiled-coil TACC domain (Sato et al., 2004). To address which region of Alp7 is responsible for binding the loop, we individually expressed each domain in bacteria with GST (GST-Alp7N, containing the N-terminal 1–218 amino acid resides, and GST-Alp7C, encompassing the C-terminal part including 219–474 residues) and repeated the peptide array assay against the loop region as before using purified fusion proteins. This showed that the C-terminal TACC domain is responsible for binding largely to the peptide containing F420, whereas the N-terminal half is capable of interacting with the other region (473–484; Figure 5D). We also performed a peptide array assay using GST-Alp14 or combinations of GST-Alp7 and GST-Alp14 and found that Alp14 did not recognize F420 but instead interacted with the other peptide region (473–484; Supplemental Figure S3).
To substantiate the notion that F420 is critical for binding of Alp7 to the loop, we replaced this residue with all the other amino acids and performed a peptide array assay against these peptides. This experiment unequivocally showed that F420 is essential for interaction, and the replacement with any amino acid, including serine, that mimics the situation of ndc80-NH12 substantially reduced the binding capability (Figure 5E). The only exceptions were conservative hydrophobic amino acids, including isoleucine and leucine. Taken together, these binding assays firmly established that Alp7 is a factor that directly binds the loop, in which F420 is essential.
Forced tethering of Alp7 to the Ndc80 complex rescues the ndc80-NH12 mutant
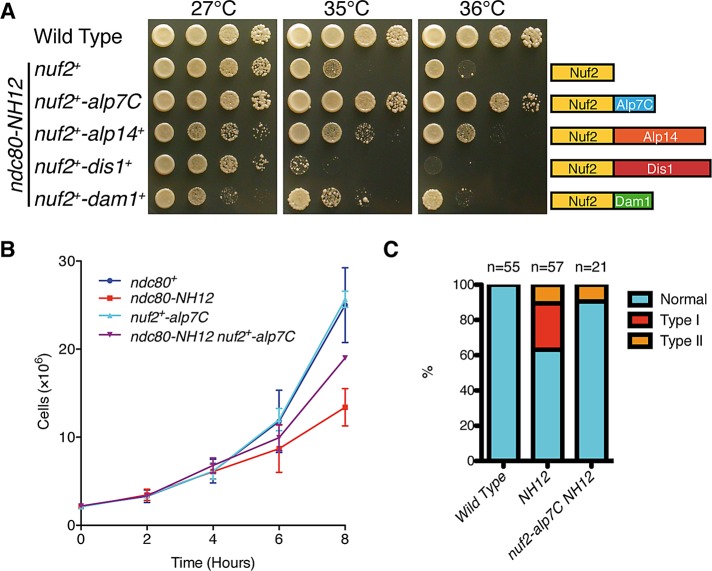
If the delocalization of Alp7 (and Alp14) from the kinetochore were the major reason for the mitotic defects observed in ndc80-NH12 cells, artificial targeting of Alp7 (and Alp14) to the vicinity of Ndc80 would rescue temperature sensitivity of this mutant. To this end, we created strains in which Alp7 was fused to the C-terminus of Nuf2 in its endogenous locus, the strategy we previously implemented for ndc80-21 (in which we constructed Nuf2-Dis1; Hsu and Toda, 2011). Despite repeated trials, we could not obtain a strain containing Nuf2-Alp7 in the ndc80-NH12 background, implying that this fusion protein compromised essential Nuf2 function. Consequently, we instead created a fusion construct in which the C-terminal Alp7 (219–474) was fused to Nuf2 (designated Nuf2-Alp7C). As shown earlier (Figure 5D), Alp7C is sufficient to recognize the Ndc80-loop peptide containing F420, and, moreover, previous studies showed that Alp7C binds Alp14 (Ling et al., 2009; Sato et al., 2009). This fusion construct was viable in ndc80-NH12, and, remarkably, rescued temperature sensitivity on solid plates incubated at the restrictive temperatures (Figure 6A). Similarly, liquid culture analysis showed that the chromosome missegregation phenotype, in particular the appearance of type I cells (see Figure 2C), was effectively suppressed (Figure 6, B and C). Consistent with the binding of Alp7C to Alp14, we found that Alp14 was properly recruited to the mitotic kinetochore in ndc80-NH12 containing Alp7C (Supplemental Figure S4).
FIGURE 6:
Tethering of the C-terminal half of Alp7 to the Ndc80 complex rescues ndc80-NH12. (A) Rescue of ndc80-NH12 by targeting the C-terminal half of Alp7 (Alp7C) to the Ndc80 complex at the kinetochore. Alp7C, Alp14, Dis1 or Dam1 was fused to the C-terminus of Nuf2 on the native locus and produced in the ndc80-NH12 mutant under the native promoter of the nuf2+ gene. Both Nuf2-Alp7C and Nuf2-Alp14 rescued temperature sensitivity of ndc80-NH12, although suppression by Nuf2-Alp14 was more modest. Spot tests were performed as in Figure 1A. (B, C) Rescue in liquid culture. Four strains indicated were grown at 27°C and shifted to 36°C (B). Cell number was counted every 2 h. Experiments were repeated twice, and the mean value and SDs are shown. The percentage of cells displaying mitotic defects was observed as in Figure 2 and quantified (C). Data for wild type and ndc80-NH12 are from Figure 2C.
A similar targeting of Alp14 (Nuf2-Alp14) also rescued ndc80-NH12, albeit less effectively (Figure 6A). In sharp contrast, Nuf2-Dis1, which rescues ndc80-21 (Hsu and Toda, 2011), did not suppress ndc80-NH12. Nor did Nuf2-Dam1 show suppression (Dam1 is a nonessential kinetochore component of the Dam1/DASH complex; Sanchez-Perez et al., 2005), indicating the specificity of Alp7 and Alp14 for the suppression of ndc80-NH12. These results strongly substantiate the notion that the loop, in particular F420, is responsible for Alp7/TACC and Alp14/TOG proteins localizing to the mitotic kinetochore and that the major reason for the defect of ndc80-NH12 is ascribable to their delocalization from the kinetochore.
Genetic data support the specific defect of ndc80-NH12 in Alp7–Alp14 malfunction
It is known that Dis1 and Alp7–Alp14 share essential roles for cell viability (Nakaseko et al., 2001; Garcia et al., 2002; Sato et al., 2004). We expected that if ndc80-NH12 was defective specifically in kinetochore recruitment of Alp7 and Alp14, ndc80-NH12 would be synthetically lethal with the dis1 deletion but not with the alp7 or alp14 deletion. Genetic crosses verified that this was exactly the case; as shown in Figure 7A, we could not obtain any viable colonies of double mutants between ndc80-NH12 and the dis1 deletion, whereas ndc80-NH12 was viable in the absence of Alp7 or Alp14 (summarized in Figure 7B, left). Markedly, in sharp contrast, ndc80-21 displayed synthetic lethality not only with dis1, but also with either alp7 or alp14 (right). This result firmly established the quantitative differences between these two ndc80 mutant alleles and further supported the notion that ndc80-NH12 is specifically defective in the roles of the Alp7/TACC-Alp14/TOG complex at the kinetochore.
FIGURE 7:
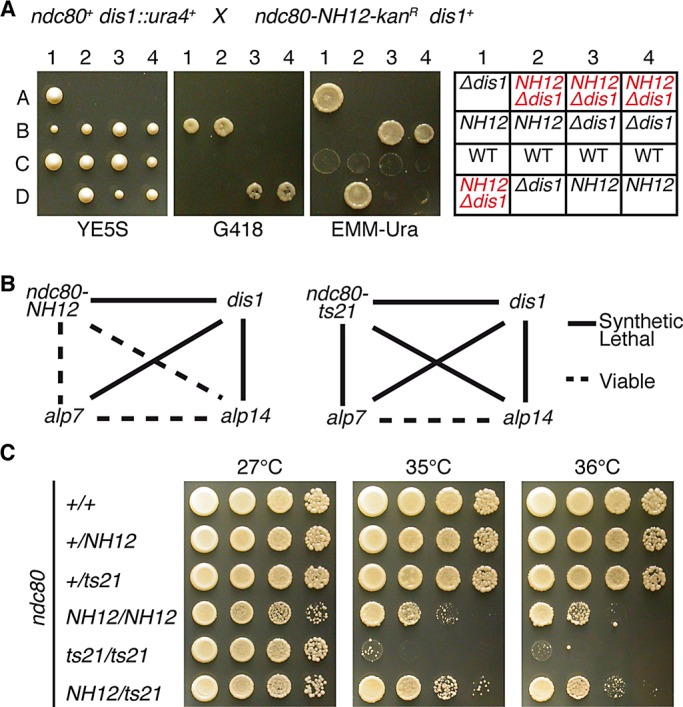
ndc80-NH12 genetically behaves like the alp7 or alp14 mutant, and the loop is a composite platform for Dis1 and Alp7–Alp14. (A) Tetrad analysis. Tetrad dissection was performed between a dis1-deletion strain (dis1::ura4+) and ndc80-NH12 (marked with kan). Four representative tetrads (tetratype) are shown. Far right, assigned genotypes of each segregant (Δdis1 = dis1::ura4+, NH12 = ndc80-NH12, and wild type [WT]). In all four tetrads, double mutants (red, predicted based on Mendelian segregation of tetrads) were inviable. (B) Summary of genetic interactions. Solid lines, synthetically lethal; dotted lines, viable. (C) Intragenic complementation between the two loop mutants. Indicated diploid strains were constructed, and spot tests were performed at various temperatures. Note that a heterozygous diploid (ndc80-NH12/ndc80-21) ameliorated growth at 35 and 36°C compared with each homozygous diploid.
Two loop mutants partially complement each defect
Having obtained data showing the distinct molecular defects between ndc80-NH12 and ndc80-21, we next addressed a potential complementation of these two mutant alleles in a diploid cell. If mutations occurred in the same gene, usually no complementation would be observed. However, there are a number of cases, known as intragenic complementation, in which two different mutant alleles in heterozygous diploids complement each other (Carlson et al., 1981; Moreno et al., 1991). In particular, if proteins play multiple roles with structurally distinct domains and/or act as dimers or multimers, intragenic complementation could arise.
We constructed a series of diploid strains between ndc80-NH12 and ndc80-21 and examined temperature sensitivity of these diploids. We found that heterozygosity in a diploid strain (ndc80-NH12/ndc80-21) ameliorated growth at the restrictive temperatures compared with that of each homozygous diploid (35 and 36°C; Figure 7C). This result implied that the loop consists of at least two independent functional regions; one region, including F420, is required for Alp7 (and Alp14) binding, and the other region, including L405, is essential to interact with Dis1. Our study therefore establishes that the Ndc80 loop provides a composite platform on the outer kinetochore for Dis1/TOG and Alp7/TACC-Alp14/TOG and thereby plays an essential role in proper spindle–kinetochore attachment.
DISCUSSION
In this work, we showed that the internal loop of fission yeast Ndc80 found in all homologues across species acts as a structural platform for the recruitment of the Alp7/TACC and Alp14/TOG. Of importance, Alp14 belongs to the conserved TOG/XMAP215 family members that are +TIPs and the major regulators of microtubule dynamics inside the cell (Gard and Kirschner, 1987; Gard et al., 2004; Ohkura et al., 2001; Kinoshita et al., 2002; Al-Bassam and Chang, 2011). We propose that the Ndc80 loop regulates spindle microtubule dynamics at the kinetochore–microtubule interface, by which it ensures proper attachment of the outer kinetochore to the spindle microtubule and coupling of the kinetochore with destabilizing microtubules during sister chromatid segregation.
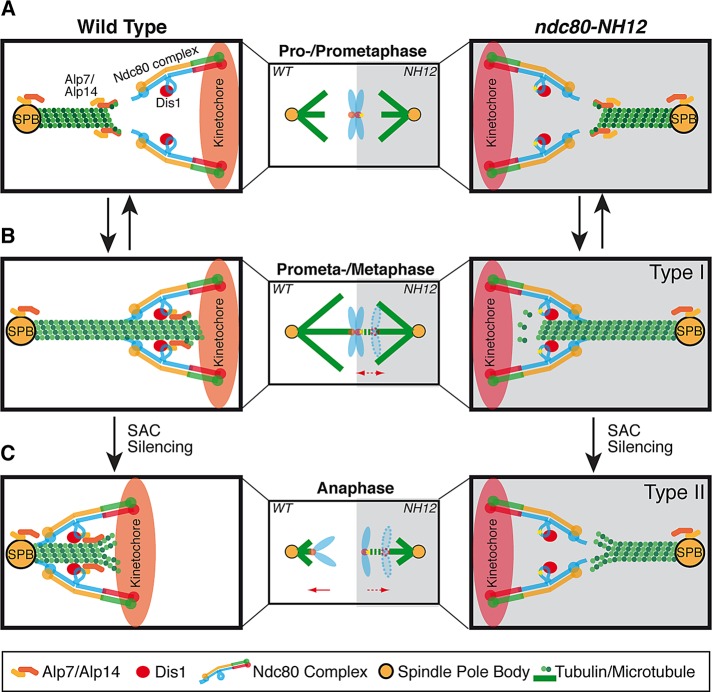
Our previous study identified the Ndc80 loop as a Dis1/TOG-binding domain (Hsu and Toda, 2011). The present work shows that the loop in fact recruits additional proteins, Alp7/TACC and Alp14/TOG, to the outer kinetochore. In the absence of Dis1 at the kinetochore, bipolar spindle formation is defective, by which chromosomes are retained in the vicinity of the SPBs often in an unattached state (Hsu and Toda, 2011). This situation is analogous to mitotic phase I (early prometaphase; Funabiki et al., 1993; Nabeshima et al., 1998; Figure 8, top). By contrast, in ndc80-NH12, which fails to recruit Alp7-Alp14, bipolar spindles are formed, but defects are observed at the later stage. Type I cells (Figures 2C and 3) show that sister centromeres undergo interpolar oscillation. This is analogous to early phase II (Figure 8, middle, prometaphase to metaphase), during which robust end-on attachment is established simultaneously with SAC silencing.
FIGURE 8:
A model of the roles of the Ndc80 loop and TOG–TACC proteins in spindle–kinetochore attachment. On onset of mitosis, Dis1/TOG is recruited to the outer kinetochore through interaction with the Ndc80 loop in a microtubule-independent manner (Nakaseko et al., 2001), whereas the Alp7/TACC-Alp14/TOG complex localizes to the SPB and promotes spindle assembly (top, prophase to prometaphase; Sato et al., 2004; Sato and Toda, 2007). ndc80-NH12 is not defective at this stage (right, gray background). When the spindle microtubule reaches the outer kinetochore, it is stabilized through interaction with Dis1 (Hsu and Toda, 2011), and simultaneously Alp7–Alp14 is loaded to the outer kinetochore in a microtubule-dependent manner (Garcia et al., 2001; Sato et al., 2004) via interaction with the Ndc80 loop (middle, prometaphase to metaphase). This ensures end-on attachment of the kinetochore to the spindle microtubule, leading to silencing of the SAC. Type I cells of ndc80-NH12 (right, the mutation within the loop is depicted by an asterisk) are defective in this step, by which chromosomes remain oscillating between the two poles under SAC activation. On SAC silencing and subsequent anaphase onset, Alp7–Alp14 continues to be required for maintenance of stable attachment, which secures equal partition of sister chromatids to the opposite poles (bottom, anaphase). In type II cells (right), end-on attachment is not maintained during this stage, resulting in chromosome missegregation. Note that it remains to be established whether Dis1 stays at the kinetochore during this stage (Nabeshima et al., 1995; Aoki et al., 2006). Alp7 and Alp14 are shown as light- and dark-orange rods, respectively, and Dis1 is depicted by red ovals.
In type II cells, on the other hand, the SAC is satisfied, and mitotic cells proceed from metaphase to anaphase without apparent delay, yet equal chromosome segregation is impeded during anaphase A (late phase II; Figure 8, bottom). An intriguing possibility is that Alp7–Alp14 may directly promote microtubule depolymerization or recruit the third molecule that is responsible for microtubule depolymerization. This would sustain chromosome attachment during anaphase A; its absence from the loop would result in failure to uphold end-on attachment during chromosome segregation. Although Alp14 is a microtubule polymerase (Al-Bassam et al., 2012), frog XMAP215 also exhibits a converse microtubule-depolymerizing activity under certain experimental conditions (Brouhard et al., 2008). Hence our data indicate that the Alp7/TACC-Alp14/TOG complex at the kinetochore is required for both establishment (defective in type I) and maintenance (defective in type II) of end-on attachment of the kinetochore to the spindle microtubule.
Roles of Alp7 and Alp14 during midmitosis described in the foregoing have not been defined before, for the following reasons. The Alp7–Alp14 complex is first delivered to the mitotic SPB, where this complex promotes spindle assembly and moves toward kinetochores as the spindle microtubule elongates (Figure 8A, top; Sato et al., 2004, 2009; Sato and Toda, 2007). Accordingly, deletion mutants of Alp7 or Alp14 display short, unstable spindles (Garcia et al., 2002; Sato et al., 2004). This defect during early mitosis hinders the involvement of Alp7–Alp14 at the kinetochore, and therefore appears quite different from those seen in ndc80-NH12, in which bipolar spindle assembly is normal but Alp7–Alp14 delocalizes selectively from the kinetochore but not the SPB or spindle microtubules (Figure 8, middle and bottom). Thus the specific roles of Alp7-Alp14 during midmitosis at the kinetochore could be uncovered only through analysis of ndc80-NH12. Consistent with delocalization from the mitotic kinetochore, either Alp7 (the C-terminal half) or Alp14 suppressed temperature sensitivity of ndc80-NH12 when artificially fused to Nuf2. However, Nuf2-Alp7C rescued this mutant more efficiently than Nuf2-Alp14 (Figure 6A). Given the notion that Alp14/TOG is a plus end–tracking microtubule polymerase (Al-Bassam et al., 2012) whereas Alp7/TACC is a recruitment factor for Alp14 (Sato et al., 2004, 2009; Sato and Toda, 2007), this result appears puzzling. We envision the following two possibilities. One is that Alp7/TACC plays additional roles (Sato et al., 2004; Zheng et al., 2006) in spindle–kinetochore attachment independent of Alp14 recruitment. The other is that the Nuf2-Alp14 fusion protein (a sole source of Nuf2 in this construct) may interfere with Nuf2’s own function in the ndc80-NH12 background.
We do not know whether Dis1 and Alp7–Alp14 interact with the loop within the same Ndc80 molecule or whether the binding is in fact competitive, in which case these proteins would interact with different Ndc80 molecules. We consider that either scenario could be envisioned. What we know from the peptide array assay is that these molecules recognize distinct yet overlapping regions. We previously showed that Dis1 interacts with the Ndc80 loop at the three regions within the loop (395–408, 431–436, and 473–484), in which L405 is critical for binding (Hsu and Toda, 2011). Here we show that Alp7 interacts with two regions (411–424 and 473–484) and F420 is crucial (Figure 5D). Furthermore, Alp14 also recognizes one region (473–484). It therefore appears that the Ndc80 loop in fission yeast consists of multiple functional modules; two regions (395–408 and 431–436) are specific for Dis1 binding, the third region (411–424) is specific for Alp7 binding, and the fourth region (473–484), which includes the adjacent coiled-coil domain, is recognized by Alp7, Alp14, and Dis1. These results, however, do not distinguish the validity of either possibility. Nor does intragenic complementation between ndc80-NH12 and ndc80-21 (Figure 7C) favor one possibility over the other. To answer this issue explicitly, we need to establish an in vitro binding assay system consisting of Ndc80 (or the Ndc80 complex), Alp7–Alp14, and Dis1, which is underway. Whichever mode of protein–protein interaction happens within the Ndc80 loop, our results show that the fission yeast TACC- and TOG-family microtubule–associated proteins regulate spindle–kinetochore attachment throughout early to late mitotic stages by interacting with the Ndc80 loop.
It has become clear that the Ndc80 loop plays a common role among eukaryotes as a protein–protein interaction site in recruiting other proteins to the mitotic outer kinetochore (Nilsson, 2012; Varma and Salmon, 2012; Tang and Toda, 2013). In human cells, a DNA replication licensing factor Cdt1 (Machida et al., 2005; Sclafani and Holzen, 2007) and the Ska1 complex (also called the Ska complex; Guimaraes and Deluca, 2009) are recruited to the kinetochore directly or indirectly via the Ndc80 loop, and this step is critical for establishing proper spindle–kinetochore attachment (Chan et al., 2012; Jeyaprakash et al., 2012; Schmidt et al., 2012; Varma et al., 2012; Zhang et al., 2012). By contrast, in budding yeast the Dam1 complex (Miranda et al., 2005; Asbury et al., 2006; Westermann et al., 2006; Welburn et al., 2009) is recruited to the mitotic kinetochore in an Ndc80 loop–dependent manner (Maure et al., 2011), although precise structural domains with which the Dam1 complex interacts within Ndc80 are under scrutiny (Lampert et al., 2013). Collectively, it appears that the role of the Ndc80 loop as a landing pad for multiple proteins required for spindle–kinetochore attachment has been conserved. Yet, interacting molecules seemingly have undergone significant evolutionary diversification among different species as the length and amino acid sequence of the loop have also become varied and less conserved, respectively.
MATERIALS AND METHODS
Yeast genetics, strains, and general methodology
The strains used in this study are listed in Supplemental Table S1. Standard methods for yeast genetics and molecular biology were used (Moreno et al., 1991; Bähler et al., 1998; Sato et al., 2005). The plasmids used in this study are listed in Supplemental Table S2.
Isolation of ts ndc80 mutants
The kanamycin-resistance marker gene cassette (kanr) was inserted in the 3′ flanking region of the ndc80+ gene (ndc80+-kanr; Hsu and Toda, 2011). The ndc80+-kanr fragment purified from this strain was amplified with error-prone PCR using Vent DNA polymerase (New England BioLabs, Ipswich, MA) supplemented with 10× dGTP. Pools of mutagenized PCR fragments were ethanol precipitated and transformed into a wild-type strain (513; Supplemental Table S1). Approximately 5000 transformants were screened for temperature sensitivity at 36°C, and 18 ts mutants were isolated. These mutants were crossed with a wild-type strain, and random spore analysis was performed. In all the segregants (>1000 colonies), the ts phenotype cosegregated with kanamycin resistance.
Determination of the mutation site in ndc80-NH12
Nucleotide sequencing of genomic DNA isolated from all 18 ts mutants was performed. In the ndc80-NH12 allele, a single point mutation was found within the ndc80 open reading frame (ORF), T1351C (the first adenine for the initiator methionine is denoted as 1), leading to a missense mutation F420S. To construct plasmids for expressing GST-fusion proteins, ORFs encoding full-length Alp7 or Alp14, the N-terminal part of Alp7 (1–218), or the C-terminal part of Alp7 (219–474) were amplified with PCR and cloned into the pGEX-KG vector (GE Healthcare Life Sciences, Piscataway, NJ). Nucleotide sequencing was performed to verify the correct insertion of each PCR-amplified fragment.
Preparation of synchronous mitotic cultures
To accumulate mitotic cells, we used hydroxyurea (HU)-induced S-phase arrest and release (Figures 2, A–C, 3, 4, A–C, and 6C). Cultures of ndc80-NH12 were treated with 12.5 mM HU at 25°C for 4 h, filtered, and incubated in HU-free media at 36°C for 1–2 h. For immunoprecipitation experiments (Figure 5, B and C), cut9-665 mutant strains (defective in Cdc16/anaphase-promoting complex component; Yamashita et al., 1996) were used that contain Alp7-Myc and Ndc80-GFP (Figure 5B) or Alp7-Myc carrying either pREP1-Ndc80 or pREP1-Ndc80-F420S (Figure 5C). A cut9 mutant containing Alp7-Myc and Ndc80-GFP was grown in rich YE5S media at 25°C and then shifted to 36°C for 3 h. Plasmid-borne strains were grown at 25°C in liquid minimal media in the absence of thiamine for 14 h to induce the nmt1 promoter (ndc80+ and ndc80-F420S are connected; Supplemental Table S2), followed by incubation at 36°C for an additional 3 h.
Microscopy
Cells were imaged on lectin-coated, glass-bottom microwell dishes (MatTek, Ashland, MA) supplemented with rich medium. Images were taken using an Olympus IX70 wide-field inverted epifluorescence microscope with an Olympus PlanApo 100×, NA 1.4, oil immersion objective (Olympus, Tokyo, Japan). DeltaVision image acquisition software (softWoRx 3.3.0; Applied Precision, Issaquah, WA) equipped with a CoolSNAP HQ (Roper Scientific, Tucson, AZ) was used. The sections of images at each time point were compressed into a two-dimensional (2D) projection using the DeltaVision maximum-intensity algorithm. Deconvolution was applied before the 2D projection. Kymograph pictures derived from 2D-projected time-lapse images were constructed using softWoRx 3.3.0. Images were taken as 14 sections along the z-axis at 0.3-μm intervals; they were then deconvolved and merged into a single projection. Captured images were processed with Photoshop CS4 (version 11.0.1; Adobe, San Jose, CA).
Fluorescence signal intensity quantification
Images were merged into a single projection using the maximum-intensity algorithm in DeltaVision-softWoRx (Olympus and Applied Precision). Fluorescence signals were then quantified using maximum intensity after subtracting background signals in the vicinity of the fluorescent spot. In Figure 4, cells were fixed with methanol before data acquisition for quantification.
GST pull-down assay
GST-Alp7 fusion protein (and other GST-fused proteins; see later description) was produced in Escherichia coli and purified with glutathione–Sepharose beads as recommended by the manufacturer (Invitrogen, Carlsbad, CA). Gel filtration was further applied to increase the protein purity. Yeast cell extracts were prepared from a cut9-665 strain containing Ndc80-3FLAG arrested at 36°C for 3 h. Glutathione–Sepharose beads bound with GST alone or GST-Alp7 were incubated with the cell extracts (5 mg) at 4°C for 1 h. After washing four times with IP buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, pH 8.0, 500 mM NaCl, 0.05% NP-40, 0.1% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 15 mM p-nitrophenyl phosphate, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Sigma-Aldrich, Gillingham, United Kingdom]), the bound proteins were eluted with reduced glutathione, separated by SDS–PAGE, and immunoblotted with anti-FLAG (M2; Sigma-Aldrich) or anti-Alp14 antibody (Garcia et al., 2001).
Immunoprecipitation
Immunoprecipitation was performed using standard procedures. Briefly, proteins were extracted in IP buffer by breaking the cells at 4°C with glass beads (setting 5.5, 25 s, 2×) in a FastPrep FP120 apparatus (Savant, Albertville, MN). The protein extracts were collected after 15 min of centrifugation at 13,000 × g at 4°C. The coimmunoprecipitations were performed by adding 1.5 mg of protein extract to protein A Dynabeads (Invitrogen) bound with anti-Ndc80 antibody (Hsu and Toda, 2011). For immunoblotting, antibodies against Ndc80, Myc peptides (MMS-150R; Covance, Berkeley, CA), and GFP (1814 460; Roche Diagnostics, Indianapolis, IN) were used at 1:1000. Films were scanned by using an Epson V700 Photo scanner (Epson, Long Beach, CA), and band intensities were quantified by ImageJ 1.42q software (National Institutes of Health, Bethesda, MD).
Peptide array assay
We basically followed previously described procedures (Maskell et al., 2010; Hsu and Toda, 2011). Peptides were synthesized and spotted onto a cellulose membrane. The membrane was activated by treatment with 50% ethanol + 10% glacial acetic acid for 1 h and then washed with IP buffer. Then 1 μg/ml GST-Alp7, GST-Alp14, GST-Alp7, and GST-Alp14 or GST alone was added to the solution. After overnight incubation at 4°C, the membrane was blocked with 3% skim milk in the same buffer, followed by incubation with anti-GST antibody (27457701V; GE Healthcare Life Science) for 2 h at room temperature. After further washes, bound spots were detected using bovine anti–goat immunoglobulin G/horseradish peroxidase–conjugated secondary antibody (sc-2378; Santa Cruz Biotechnology, Santa Cruz, CA) and visualized by chemiluminescence.
For the assessment of positive or negative binding, we performed the following quantification. First, we quantified spot intensities with the ImageJ 1.42q software as described earlier. Next we judged regions displaying >33,000 values (positive, 33,822; negative, 31,897) derived from four consecutive spots as interacting peptides.
Construction of strains containing fusion genes between nuf2+ and alp7C or alp14+
Construction of nuf2+-dis1+ and nuf2+-dam1+ was previously described (Hsu and Toda, 2011). DNA fragments encoding Nuf2-Alp7C-kan (amino acids 219–474) or Nuf2-Alp14-hph were amplified by using two-step PCR. These fragments were transformed into a wild-type strain (513; Supplemental Table S1), and integrants were selected on kanamycin or hygromycin B–containing plates respectively. Correct integrations were verified by colony PCR and/or nucleotide sequencing.
Supplementary Material
Acknowledgments
We thank Yasushi Hiraoka, Jonathan Millar, Osami Niwa, Ayumu Yamamoto, and Mitsuhiro Yanagida for the gift of reagents used in this study. We are grateful to the Peptide Synthesis Unit of Cancer Research UK for preparation of the peptide arrays. We thank Martin Singleton for critical reading of the manuscript and useful comments. H.T. was supported by the Uehara Memorial Foundation. This work was supported by Cancer Research UK (T.T.).
Abbreviations used:
- SAC
spindle assembly checkpoint
- TACC
transforming acidic coiled coil
- TBZ
thiabendazole
- +TIP
plus end–tracking protein
- TOG
tumor-overexpressed gene
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0817) on February 20, 2013.
Present addresses: *Department of Pharmacy, Osaka Rosai Hospital, 1179-3 Nagasone-Cho, Kita-Ku, Sakai, Osaka 591-8025, Japan
†Dyson Vision Research Institute, Department of Ophthalmology, Weill Cornell Medical College, New York, NY 10065.
REFERENCES
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Flor-Parra I, Lal N, Velji H, Chang F. Fission yeast Alp14 is a dose dependent plus end tracking microtubule polymerase. Mol Biol Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol. 2012;19:1161–1167. doi: 10.1038/nsmb.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Nakaseko Y, Kinoshita K, Goshima G, Yanagida M. Cdc2 phosphorylation of the fission yeast Dis1 ensures accurate chromosome segregation. Curr Biol. 2006;16:1627–1635. doi: 10.1016/j.cub.2006.06.065. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bock LJ, et al. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat Cell Biol. 2012;14:614–624. doi: 10.1038/ncb2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge AJ, Morphew M, Bartlett R, Hagan IM. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Osmond BC, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Tirnauer JS, Pellman D. Surfing on microtubule ends. Trends Cell Biol. 2003;13:229–237. doi: 10.1016/s0962-8924(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol. 2012;27:563–571. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A. Architecture of the human Ndc80-Hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Musacchio A, Petrovic A. The Ndc80 complex: hub of kinetochore activity. FEBS Lett. 2007;581:2862–2869. doi: 10.1016/j.febslet.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2012;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan IM, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002;21:6015–6024. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Vardy L, Koonrugsa N, Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Becker BE, Romney SJ. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int Rev Cytol. 2004;239:179–272. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F. Centrosomal TACCtics. Bioessays. 2002;24:915–925. doi: 10.1002/bies.10162. [DOI] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Deluca JG. Connecting with Ska, a key complex at the kinetochore-microtubule interface. EMBO J. 2009;28:1375–1377. doi: 10.1038/emboj.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Biswas A, Langegger M, Kawashima SA, Tsukahara T, Watanabe Y. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 2007;26:4475–4486. doi: 10.1038/sj.emboj.7601880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung P, Maier M, Alushin GM, Lander GC, Nogales E, Westermann S. Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J Mol Biol. 2011;405:548–559. doi: 10.1016/j.jmb.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell. 2012;46:274–286. doi: 10.1016/j.molcel.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres A, Vietmeier-Decker C, Ortiz J, Karig I, Beuter C, Hegemann J, Lechner J, Fleig U. The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association. Mol Biol Cell. 2004;15:5255–5267. doi: 10.1091/mbc.E04-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Habermann B, Hyman AA. XMAP215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 2002;6:267–273. doi: 10.1016/s0962-8924(02)02295-x. [DOI] [PubMed] [Google Scholar]
- Korenbaum E, Rivero F. Calponin homology domains at a glance. J Cell Sci. 2002;115:3543–3545. doi: 10.1242/jcs.00003. [DOI] [PubMed] [Google Scholar]
- Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol. 2013;200:21–30. doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling YC, Vjestica A, Oliferenko S. Nucleocytoplasmic shuttling of the TACC protein Mia1p/Alp7p is required for remodeling of microtubule arrays during the cell cycle. PLoS One. 2009;4:e6255. doi: 10.1371/journal.pone.0006255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Maskell DP, Hu XW, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol. 2010;190:823–834. doi: 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maure JF, Komoto S, Oku Y, Mino A, Pasqualato S, Natsume K, Clayton L, Musacchio A, Tanaka TU. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80Hec1. Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JL, De Wulf P, Sorger P, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analyses of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:773–782. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Kurooka H, Takeuchi M, Kinoshita K, Nakaseko Y, Yanagida M. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–1585. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M. M phase-specific kinetochore proteins in fission yeast microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol. 2001;11:537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Nilsson J. Looping in on Ndc80—how does a protein loop at the kinetochore control chromosome segregation? Bioessays. 2012;34:1070–1077. doi: 10.1002/bies.201200096. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Adachi Y, Kinoshita N, Niwa O, Toda T, Yanagida M. Cold-sensitive and caffeine supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 1988;7:1465–1473. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Garcia MA, Toda T. Dis1/TOG universal microtubule adaptors—one MAP for all. J Cell Sci. 2001;114:3805–3812. doi: 10.1242/jcs.114.21.3805. [DOI] [PubMed] [Google Scholar]
- Oliferenko S, Balasubramanian MK. Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat Cell Biol. 2002;4:816–820. doi: 10.1038/ncb861. [DOI] [PubMed] [Google Scholar]
- Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18:379–388. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Petrovic A, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe P, Hirata D, Childs D, Vardy L, Toda T. Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol Biol Cell. 1998;9:1757–1771. doi: 10.1091/mbc.9.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff JW. Centrosomes and cancer: lessons from a TACC. Trends Cell Biol. 2002;12:222–225. doi: 10.1016/s0962-8924(02)02268-7. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Sato M, Okada N, Kakui Y, Yamamoto M, Yoshida M, Toda T. Nucleocytoplasmic transport of Alp7/TACC organizes spatiotemporal microtubule formation in fission yeast. EMBO Rep. 2009;10:1161–1167. doi: 10.1038/embor.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–337. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- Sato M, Toda T. Space shuttling in the cell: nucleocytoplasmic transport and microtubule organization during the cell cycle. Nucleus. 2010;1:231–236. doi: 10.4161/nucl.1.3.11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Vardy L, Garcia MA, Koonrugsa N, Toda T. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol Biol Cell. 2004;15:1609–1622. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Vardy L, Koonrugsa N, Tournier S, Millar JBA, Toda T. Deletion of Mia1/Alp7 activates Mad2-dependent spindle assembly checkpoint in fission yeast. Nat Cell Biol. 2003;5:764–766. doi: 10.1038/ncb0903-764. [DOI] [PubMed] [Google Scholar]
- Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol. 2012;14:604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- Schmidt JC, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–980. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Fukagawa T. Molecular architecture of vertebrate kinetochores. Exp Cell Res. 2012;318:1367–1374. doi: 10.1016/j.yexcr.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Tang NH, Toda T. Ndc80 Loop as a protein-protein interaction motif. Cell Div. 2013;8:2. doi: 10.1186/1747-1028-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Adachi Y, Hiraoka Y, Yanagida M. Identification of the pleiotropic cell division cycle gene NDA2 as one of two different α-tubulin genes in Schizosaccharomyces pombe. Cell. 1984;37:233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Tooley J, Stukenberg PT. The Ndc80 complex: integrating the kinetochore's many movements. Chromosome Res. 2011;19:377–391. doi: 10.1007/s10577-010-9180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Gestaut DR, Tien JF, Vollmar BS, Gonen T, Asbury CL, Davis TN. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci USA. 2012;109:16113–16118. doi: 10.1073/pnas.1209615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Chandrasekaran S, Sundin LJ, Reidy KT, Wan X, Chasse DA, Nevis KR, Deluca JG, Salmon ED, Cook JG. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol. 2012;14:593–603. doi: 10.1038/ncb2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Salmon ED. The KMN protein network—chief conductors of the kinetochore orchestra. J Cell Sci. 2012;125:5927–5936. doi: 10.1242/jcs.093724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorozhko VV, Emanuele MJ, Kallio MJ, Stukenberg PT, Gorbsky GJ. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma. 2008;117:169–179. doi: 10.1007/s00412-007-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, Howard J. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci USA. 2011;108:2741–2746. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature. 1996;384:276–279. doi: 10.1038/384276a0. [DOI] [PubMed] [Google Scholar]
- Zhang G, Kelstrup CD, Hu XW, Hansen MJ, Singleton MR, Olsen JV, Nilsson J. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125:3243–3253. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
- Zheng L, Schwartz C, Wee L, Oliferenko S. The fission yeast TACC-related protein, Mia1p/Alp7p, is required for formation and maintenance of persistent microtubule-organizing centers at the nuclear envelope. Mol Biol Cell. 2006;17:2212–2222. doi: 10.1091/mbc.E05-08-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.