There are conflicting models regarding the role of the Ypt11 GTPase in mitochondrial inheritance during yeast budding. This study demonstrates that Ypt11 function requires mitochondrial membrane targeting and GTPase domain–dependent effector interactions. In addition, the abundance of active Ypt11 forms is controlled by phosphorylation and degradation.
Abstract
The Rab GTPase Ypt11 is a Myo2-binding protein implicated in mother-to-bud transport of the cortical endoplasmic reticulum (ER), late Golgi, and mitochondria during yeast division. However, its reported subcellular localization does not reflect all of these functions. Here we show that Ypt11 is normally a low-abundance protein whose ER localization is only detected when the protein is highly overexpressed. Although it has been suggested that ER-localized Ypt11 and ER–mitochondrial contact sites might mediate passive transport of mitochondria into the bud, we found that mitochondrial, but not ER, association is essential for Ypt11 function in mitochondrial inheritance. Our studies also reveal that Ypt11 function is regulated at multiple levels. In addition to membrane targeting and GTPase domain–dependent effector interactions, the abundance of active Ypt11 forms is controlled by phosphorylation status and degradation. We present a model that synthesizes these new features of Ypt11 function and regulation in mitochondrial inheritance.
INTRODUCTION
Mitochondrial transport from mother to daughter (bud) during asymmetric cell division in Saccharomyces cerevisiae ensures that the healthiest organelles are inherited by the new generation (McFaline-Figueroa et al., 2011; Rafelski et al., 2012). This inheritance is critical, as buds lacking mitochondria do not separate from the mother and thus cannot survive independently (McConnell et al., 1990; Garcia-Rodriguez et al., 2009). This process is mediated by the type V myosin motor Myo2, which transports mitochondrial membranes along actin cables into the bud (Altmann et al., 2008; Förtsch et al., 2011). Two additional proteins, Ypt11 and Mmr1, interact with the cargo-binding domain on the Myo2 tail and participate in mitochondrial inheritance (Itoh et al., 2002, 2004; Eves et al., 2012). Although Mmr1 colocalizes with mitochondria (Itoh et al., 2004; Swayne et al., 2011), Ypt11 does not, raising questions about how Ypt11 promotes mitochondrial partitioning.
Ypt11 is a Rab GTPase implicated in the bud-directed movement of multiple organelles. Although initially linked to mitochondrial inheritance, subsequent studies showed it also functions in the transport of cortical endoplasmic reticulum (cER) and late Golgi membranes into yeast buds (Buvelot Frei et al., 2006; Arai et al., 2008; Frederick et al., 2008). Curiously, Ypt11 localization studies are not entirely consistent with these functions. Different studies have reported Ypt11 on the perinuclear and cortical ER (Buvelot Frei et al., 2006), as well as at the bud tip and mother–bud neck (Itoh et al., 2002). Several scenarios could explain how Ypt11 acts on mitochondria. A small (undetected) fraction of cellular Ypt11 could associate directly with the organelle and recruit the Myo2 motor to the membrane. Alternatively, the effect of Ypt11 on mitochondrial distribution could be indirect. A protein complex called ER mitochondria encounter structure (ERMES) was recently shown to mediate direct physical contact between the yeast ER and mitochondria (Kornmann et al., 2009). ERMES complexes were observed moving from the mother cell into the bud during division (Nguyen et al., 2012). Thus it is theoretically possible that mitochondria “hitch a ride” with ER membranes that are actively transported during cell division. It has also been proposed that, instead of recruiting Myo2 to mitochondria, Ypt11 mediates bud-directed transport of a factor or factors associated with the cER and/or Golgi, which anchor mitochondria in the daughter cell (Pon, 2008; Swayne et al., 2011).
Although initially characterized as regulators of membrane trafficking and fusion, members of the Rab GTPase family also act directly in membrane transport by tethering vesicles and organelles to motor proteins (Seabra and Coudrier, 2004; Hutagalung and Novick, 2011). Consistent with its organelle inheritance function, Ypt11 is the only yeast Rab that displays periodic transcription, with expression peaking during the G1 phase of the cell cycle, immediately before bud emergence (Cho et al., 1998; Spellman et al., 1998; Pramila et al., 2006). Ypt11 function may also be posttranscriptionally regulated, since proteomic studies indicate that Ypt11 is phosphorylated (Albuquerque et al., 2008; Bodenmiller et al., 2008, 2010; Holt et al., 2009). In addition, Ypt11 contains unique sequence features not found in other Rabs. Whether and how these sequence features and protein modifications contribute to Ypt11 function in vivo, and specifically in mitochondrial inheritance, are not known.
In this study, we explore the relationship between Ypt11 expression level and cellular localization. We present new evidence that Ypt11 acts directly on mitochondria, rather than indirectly through ER–mitochondrial connections, to promote mitochondrial inheritance. We describe a novel interaction between Ypt11 and Mmr1, indicating that the activities of these two Myo2-binding partners may be physically coordinated in vivo. Our studies also reveal an uncommon mode of Rab regulation in which phosphorylation status and degradation contribute to the selective turnover of active forms of the Ypt11 GTPase.
RESULTS
The N-terminus of Ypt11 contributes to mitochondrial inheritance
The YPT11 sequence initially deposited in genomic databases was missing 185 nucleotides at the 5′ end of the gene (Saccharomyces Genome Database [www.yeastgenome.org/], YNL304W Locus History). As a result, initial studies on Ypt11 either used the shorter sequence (Itoh et al., 2002) or potentially both short and long versions of the sequence (Buvelot Frei et al., 2006). Subsequent studies used the updated sequence with the 5′ extension (Arai et al., 2008; Eves et al., 2012). This 5′ extension creates two potential initiation codons for the Ypt11 protein (Met1 or Met63, Figure 1A). Members of the Rab superfamily typically contain a short (up to 20 amino acids [aa]) variable region N-terminal to the conserved GTPase domain. In the case of Ypt11, translation initiation at Met-63 would generate an N-terminus of comparable length (28-aa variable region). By contrast, initiation at Met-1 would add 62 aa to this variable region (90-aa variable region). As described later, we used two complementary approaches to determine whether the longer N-terminus was important for Ypt11 function. This analysis was performed in a ypt11Δ mmr1Δ strain. Although Ypt11 and Mmr1 are not essential, mitochondrial transport into buds is delayed when either of the proteins is absent, and cells lacking both proteins have severe mitochondrial inheritance and growth defects. As shown previously (Itoh et al., 2004; Frederick et al., 2008), ∼70% of medium and large buds in a ypt11Δ mmr1Δ strain fail to receive mitochondria from the mother cell during division. This defect can be rescued by overexpression of Ypt11 alone, providing a sensitive assay for function of this protein.
FIGURE 1:
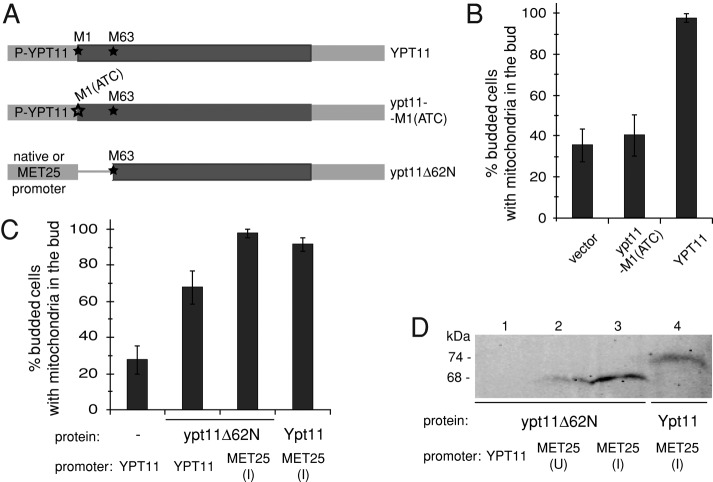
The N-terminus of Ypt11 contributes to mitochondrial inheritance. (A) Constructs used to test codons M1 and M63 (marked with asterisks) as potential translation start sites. The YPT11 ORF is dark gray; the thin line in ypt11Δ62N marks the sequence missing in this construct; noncoding regions are light gray. P-YPT11 is the native YPT11 promoter (see Supplemental Methods). (B, C) Quantification of mitochondrial inheritance in medium- and large-budded cells of the mmr1Δ ypt11Δ strain containing the indicated plasmids. Bars, mean and SD of three independent experiments (n = 100). (D) Steady-state abundance of GFP-tagged ypt11Δ62N or Ypt11 in the mmr1Δypt11Δ strain assayed by Western blotting using an anti-GFP antibody. I, induced; U, uninduced. Predicted molecular weights for fusion proteins are indicated.
We began by mutating the initial ATG codon (Met-1) to ATC, creating ypt11-M1(ATC) (Figure 1A). This construct allowed us to test whether the downstream Met63 codon could serve as a translational start site in vivo and whether the resulting protein was functional. Mitochondrial inheritance is only observed in 30–35% of ypt11Δ mmr1Δ cells containing vector alone (Figure 1, B and C). This defect was fully rescued by the full-length YPT11 gene flanked by native upstream and downstream sequence. This rescue depended on the presence of the Met-1 initiation codon, since rescue was not observed with ypt11-M1(ATC). Thus either Met-63 cannot be used as an initiation codon or the short form of Ypt11 is not functional for mitochondrial inheritance.
To test the latter possibility, we cloned the short form of YPT11 (encoding ypt11Δ62N) downstream of the native YPT11 or MET25 promoter (Figure 1A, bottom). ypt11Δ62N expressed from the YPT11 promoter partially rescued mitochondrial inheritance (68% of buds contained mitochondria; Figure 1C), even though a green fluorescent protein (GFP)–tagged version of the protein could not be detected by immunoblotting (Figure 1D, lane 1; note that full-length Ypt11 protein expressed from the native promoter is also not detected by immunoblotting; see Figure 2C). When induced from the MET25 promoter, ypt11Δ62N abundance increased (Figure 1D, compare lanes 2 and 3), and the protein fully restored mitochondrial inheritance in ypt11Δ mmr1Δ (Figure 1C). Although these results demonstrate that ypt11Δ62N can promote mitochondrial inheritance, overexpression is required for full rescue, indicating that this shorter form is only partially functional. Because these findings indicate that the N-terminal 62 amino acids of Ypt11 are important for its function in mitochondrial inheritance, we used the longer form for all experiments in this study.
FIGURE 2:
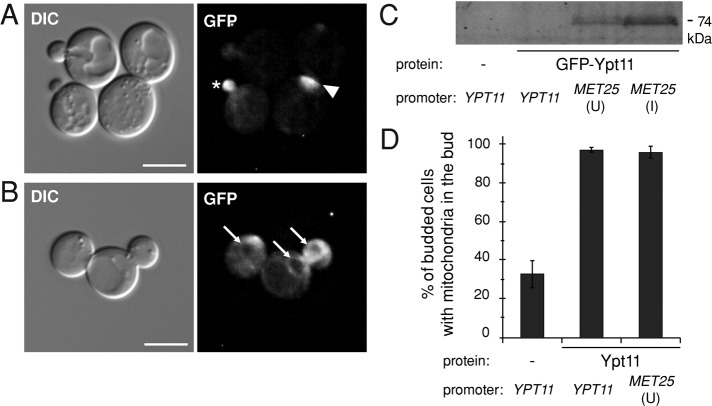
Subcellular localization of Ypt11 is affected by expression level. (A, B) Representative DIC and fluorescence images of ypt11Δ cells expressing GFP-Ypt11 from the MET25 promoter, grown on synthetic media with (A, uninduced) or without (B, induced) methionine. An asterisk marks the tip of a small bud; an arrowhead points to the neck between the mother cell and a large bud. Perinuclear staining is marked with arrows in B. Bar, 5 μm. (C) Expression of GFP-Ypt11 under the indicated conditions was analyzed by Western blotting using anti-GFP antibody. I, induced; U, uninduced. (D) Quantification of mitochondrial inheritance in medium- and large-budded cells of the mmr1Δ ypt11Δ strain expressing Ypt11 from the indicated promoters. Bars, mean and SD of three independent experiments (n = 100).
ER-localized Ypt11 does not participate in mitochondrial inheritance
To determine whether the different subcellular localizations reported for Ypt11 could be due, in part, to the presence or absence of the N-terminal extension, we investigated the localization of the longer, fully functional form of the protein. Of interest, the subcellular localization observed for this protein depended on its level of expression. GFP-Ypt11 expressed from the native YPT11 promoter was at the limit of detection by fluorescence microscopy and could not be detected by immunoblotting (unpublished data; Figure 2C). When overexpressed from the MET25 promoter under noninducing conditions (Figure 2C, MET25/U), GFP-Ypt11 localized to bud tips and necks, mirroring Myo2 localization (Figure 2A). This localization is consistent with that previously reported for the overexpressed short form of Ypt11 (Itoh et al., 2002). Induction of the MET25 promoter (Figure 2C, MET25/I) shifted the localization to the cER and perinuclear ER. Similar ER localization was reported previously for Ypt11 (Buvelot Frei et al., 2006). Despite the fact that the only known adaptor for Ypt11, Ret2, is associated with Golgi compartments (Arai et al., 2008), we did not see a pattern resembling Golgi localization. We also did not detect Ypt11 on mitochondrial networks (compare Figure 2, A and B, with Supplemental Figure S1C), although Ypt11 expressed from either the native or the MET25 promoter was functional for mitochondrial inheritance (Figure 2D). Thus Ypt11 is present on mitochondrial membranes at a very low level, or acts indirectly, via ER–mitochondrial contacts, for example, to promote mitochondrial inheritance.
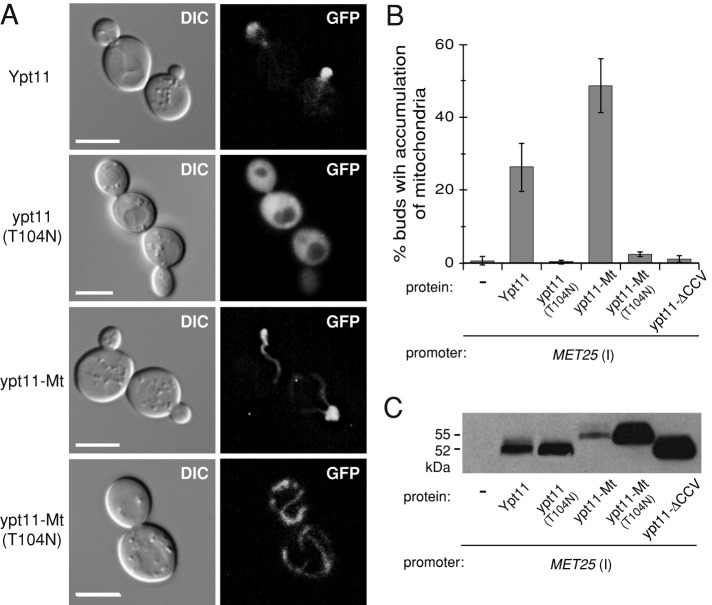
To determine whether Ypt11 can influence mitochondrial inheritance from the ER, the mitochondrion, or both locations, we created localization-restricted variants of the protein. First, we removed the last three amino acids (CCV) from Ypt11, which form a prenylation site necessary for membrane targeting. The GFP-tagged form of this ypt11ΔCCV variant localized to the cytoplasm (Figure 3A). Second, we replaced the Ypt11 CCV motif with the transmembrane domain of Fis1 (a tail-anchored protein of the outer mitochondrial membrane), creating ypt11-Mt. GFP-ypt11-Mt colocalized with red fluorescent protein (RFP)–labeled mitochondrial networks in yeast cells (Figure 3B). Third, we replaced the Ypt11 CCV motif with the 35–amino acid C-terminal transmembrane domain of the ER protein Frt1 (Beilharz et al., 2003), creating ypt11-ER. GFP-ypt11-ER colocalized with RFP-labeled perinuclear ER and cER in yeast (Figure 3C).
FIGURE 3:
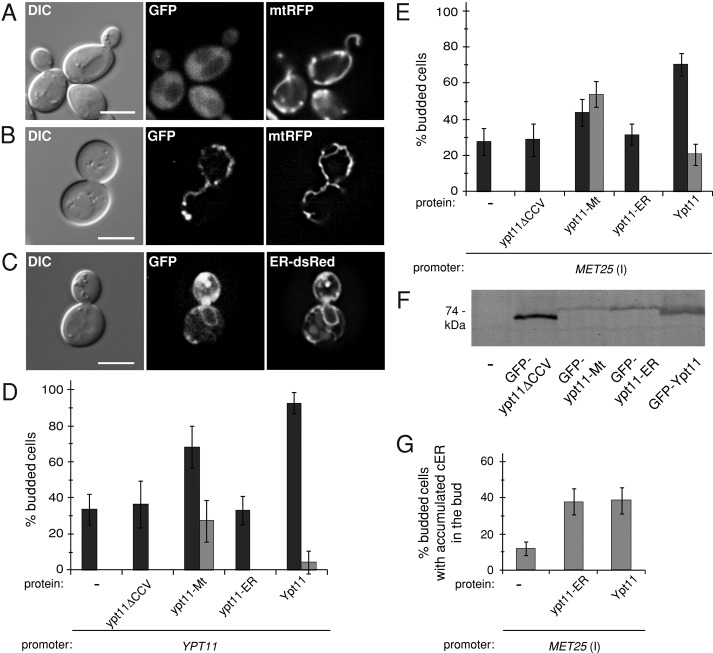
Mitochondrial association is essential for Ypt11 function in mitochondrial inheritance. (A–C) Representative DIC and fluorescence images of ypt11Δ cells coexpressing GFP-tagged Ypt11 variants (uninduced MET25 promoter) and the indicated red fluorescent protein markers. Bar, 5 μm. (D) Quantification of mitochondrial inheritance in medium- and large-budded cells of the mmr1Δ ypt11Δ strain expressing mtGFP and either WT untagged Ypt11 or its variants from the native YPT11 promoter. Both normal distribution (black bars) and mitochondria accumulated in the bud (gray bars) were scored (black plus gray is total inheritance). (E) Same as D, except that Ypt11 variants were expressed from the MET25 promoter under inducing conditions. (F) Expression of GFP-Ypt11 variants expressed from the MET25 promoter was analyzed by Western blotting using anti-GFP antibody. (G) Accumulation of cER in small yeast buds was quantified in ypt11Δ cells coexpressing ER-dsRed and either Ypt11 or ypt11-ER (MET25 promoter, inducing conditions). The graph shows percentage of cells displaying excessive amount of cER in the bud as compared with normal distribution during cell division. Bars, mean and SD of three independent experiments (n = 100).
When untagged versions of these variants were expressed from the YPT11 promoter, neither the cytoplasmic nor the ER-targeted proteins complemented the mitochondrial inheritance defect of the ypt11Δ mmr1Δ strain compared with vector alone (Figure 3D). By contrast, ypt11-Mt not only rescued the defect but also was much more efficient in driving mitochondria into the bud than the wild-type (WT) protein. This effect was observed as an increase in the accumulation of excess mitochondria in yeast buds (Figure 3D, gray bars, and Supplemental Figure S1, A and B). Although the phenomenon was also observed to some extent in cells expressing WT Ypt11 (4%), the effect was more pronounced in cells expressing ypt11-Mt (28%).
Additional control studies were performed to verify that the ypt11-ER protein was expressed and that the Frt1-derived transmembrane domain did not interfere with protein's activity. Although MET25-overexpressed GFP-ypt11-ER could be detected by immunoblotting (Figure 3F), the protein was not functional for mitochondrial inheritance (only 31% of buds contained mitochondria; Figure 3E). Conversely, overexpression of ypt11-Mt or Ypt11 from the MET25 promoter rescued the inheritance defect fully and induced the mitochondrial accumulation phenotype in which excess organelles accumulate in the bud (in 56 and 21% of budded cells, respectively; Figure 3E, gray bars). The fact that ypt11-ER is less abundant than WT Ypt11 (threefold lower; Figure 3F) does not explain its inability to rescue mitochondrial inheritance. Even though ypt11-Mt steady-state levels were 10-fold lower than the WT protein (Figure 3F), its function in mitochondrial inheritance was 2.5-fold higher.
WT Ypt11 has also been shown to mediate cER inheritance during yeast budding (Buvelot Frei et al., 2006; Frederick et al., 2008). We verified that this activity had not been compromised by irreversibly tethering Ypt11 to the ER. When overexpressed from the MET25 promoter, ypt11-ER caused the accumulation of cER in yeast buds, similar to WT Ypt11 (Figure 3G). Thus addition of an ER transmembrane anchor to Ypt11 did not disrupt the protein's ER inheritance function. Our combined results demonstrate that Ypt11 localized to the ER cannot drive mitochondrial inheritance and suggest that Ypt11 needs to be localized to mitochondria to perform that function.
Rab characteristics of Ypt11 are essential for mitochondrial inheritance
Comparison of the Ypt11 sequence with other Rab proteins singles it out as a unique family member. Several stretches of the Ypt11 sequence either are not found or are much longer than those in other Rabs. In addition to the unusually long N-terminal extension (Figure 4A, region I), Ypt11 contains an 83-aa insert separating the P-loop from the switch I region of the GTPase (Figure 4A, region II). In addition, the C-terminal unstructured region is significantly longer in Ypt11 than in other Rabs (Figure 4A, region III). As a result of these additions, Ypt11 is twice as long (417 aa) as a typical Rab (200+ aa). However, all GTPase and Rab-specific motifs (Figure 4A, G1-G3, PM1-PM3, and RabF1-5) can be identified in Ypt11 with significant conservation of these regions of the protein.
FIGURE 4:
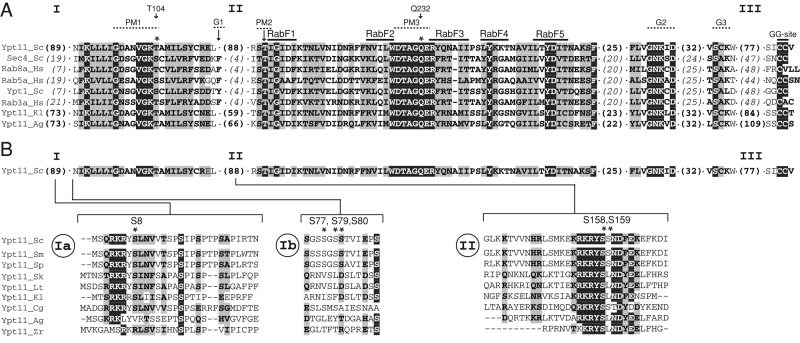
Multiple sequence alignment of Ypt11 with fungal and mammalian Rabs. (A) The alignment highlights similarities and differences between S. cerevisiae Ypt11 and other members of the Rab superfamily. Sequences unique to Ypt11 and excluded from the alignment are marked I (for the N-terminal extension) and II (for an insert within the switch I region of the GTPase). The highly variable C-terminal region (III) is significantly longer in Ypt11 orthologues than in other Rabs. Numbers in parentheses in the alignment denote the number of amino acids hidden from view. The positions of the conserved GTPase motifs for nucleotide binding (PM1-3 required for phosphate and/or Mg2+ binding; G1-3 required for guanine binding), Rab-family signature motifs (RabF1-5; Pereira-Leal and Seabra, 2000), and the geranyl-geranylation site (GG site) are indicated above the alignment. Asterisks mark residues changed in the GTPase-domain mutants ypt11(T104N) and ypt11(Q232L). (B) Alignment of fungal Ypt11 sequences that are conserved in regions marked I and II in A. Serine residues (S8, S77, S79, S80, S158, S159) mutated to alanine in this study are marked with asterisks. Residues shaded black are identical, and those shaded gray are similar. Ag, Ashbya gossypii; Cg, Candida glabrata; Hs, Homo sapiens; Kl, Kluyveromyces lactis; Lt, Lachancea thermotolerans; Sc, S. cerevisiae; Sk, Saccharomyces kluyveri; Sm, Saccharomyces mikatae; Sp, Saccharomyces paradoxus; Zr, Zygosaccharomyces rouxii.
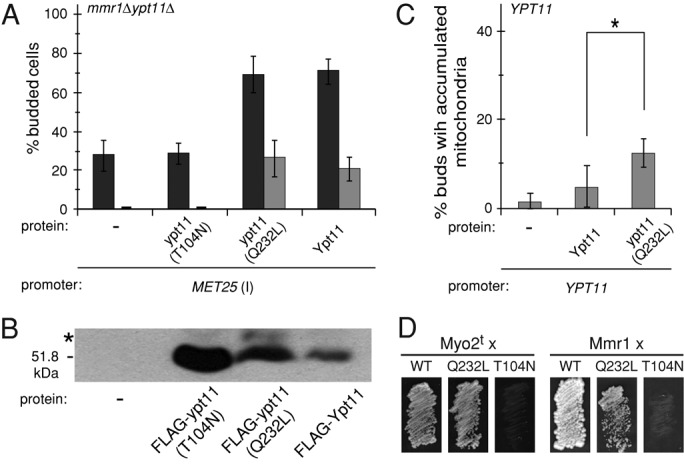
Several mutations are known to affect the GTPase activities of Ras and Rabs. Although predicted inactivating mutations in the Ypt11 GTPase domain have been analyzed (Itoh et al., 2002), activating mutations have not been studied. To address this issue, we replaced the glutamine in the switch II region, which coordinates a water molecule during GTP hydrolysis, with a leucine residue (Figure 4A, motif PM3, ypt11(Q232L)). As a negative control, we created a ypt11(T104N) mutant. The substitution of asparagine for serine/threonine in the P-loop (Figure 4A, motif PM1) is predicted to cause preferential GDP binding, inactivating the downstream signaling by Ras and Rab proteins (Tisdale et al., 1992; Stenmark et al., 1994). As expected, the ypt11(T104N) mutant protein was unable to rescue the mitochondrial inheritance defect (Figure 5A, black bars), even though the protein was expressed (Figure 5B). Of interest, the steady-state abundance of the inactive protein was much higher than that of the WT protein. This was the first indication that the cellular abundance of Ypt11 might be linked to (and constrained by) its activity, an idea we test in greater detail later in this study.
FIGURE 5:
Rab properties of Ypt11 are important for mitochondrial inheritance. (A) Mitochondrial inheritance was quantified in mmr1Δ ypt11Δ cells expressing either WT Ypt11 or the GTPase-mutant variants from MET25 promoter under inducing conditions. Both normal distribution (black bars) and mitochondria accumulated in the bud (gray bars) were scored (black plus gray is total inheritance). (B) Expression of FLAG-tagged Ypt11 variants (51.8 kDa) from the MET25 promoter analyzed by Western blotting using anti-FLAG antibody. The asterisk marks a slower-migrating molecular weight species predicted to be the phosphorylated form of the protein. (C) Quantification of the mitochondrial accumulation phenotype in WT cells expressing either Ypt11 or ypt11(Q232L) mutant from the native YPT11 promoter. *p < 0.005 in Student's t test. (D) Interaction of WT and mutant Ypt11 variants with the Myo2 tail (Myo2t, left) or Mmr1 (right) was tested using a two-hybrid assay on SD‑Leu-Trp-His medium containing 10 mM (Myo2t) or 2 mM (Mmr1) 3-AT. Bars, mean and SD of three independent experiments (n = 100; A, C).
The ypt11(Q232L) allele rescued the mitochondrial inheritance defect fully, indicating that the protein is active (Figure 5A). Although overexpression of ypt11(Q232L) also promoted accumulation of mitochondria in buds (Figure 5A, gray bars), the extent of this effect was similar to that caused by overexpression of WT Ypt11. This result suggested that ypt11(Q232L) was not a constitutively active protein, as a more pronounced accumulation phenotype would be expected in this case. To test whether differences between ypt11(Q232L) and Ypt11 were masked by overexpression, we analyzed the accumulation phenotype in ypt11Δ cells expressing either protein from the native YPT11 promoter. This assay showed a small but statistically significant difference between ypt11(Q232L) and WT Ypt11 activity (Figure 5C). Although neither WT Ypt11 (Figure 2C) nor ypt11(Q232L) (unpublished results) could be detected by Western blotting when expressed from the YPT11 promoter, MET25-expressed ypt11(Q232L) had a higher steady-state abundance than WT Ypt11 (Figure 5B). Thus differences in the mitochondrial accumulation phenotypes noted in Figure 5C are likely caused by differences in protein abundance rather than intrinsic differences in activities of the proteins.
Consistent with the observation that ypt11(Q232L) is active in mitochondrial inheritance, the mutant protein interacted with the Myo2 tail in a yeast two-hybrid assay (Figure 5D), similar to WT Ypt11 (Itoh et al., 2002; Figure 5D). By contrast, the T104N mutation abolished the interaction with Myo2. Of importance, we identified a novel interaction of Ypt11 with Mmr1 (Figure 5D; note that the Ypt11–Mmr1 interaction was not as robust as that between Ypt11 and Myo2 and was more sensitive to the presence of 3‑aminotriazole in the medium). This interaction also required a functional Ypt11 GTPase domain, since Mmr1 binding occurred with the Q232L but not the T104N allele.
Our combined results establish that a functional Ypt11 GTPase domain is important for interaction with two distinct binding partners needed for mitochondrial inheritance. Moreover, tethering Ypt11 to the membrane enhances its effect on mitochondrial distribution significantly more than a mutation predicted to lock the enzyme in the GTP-bound state (compare Figures 3E and 5A). Our data also suggest that less active forms of Ypt11 are maintained in the cell at a higher steady-state level than active forms.
A conserved phosphorylation site controls Ypt11 abundance in cells
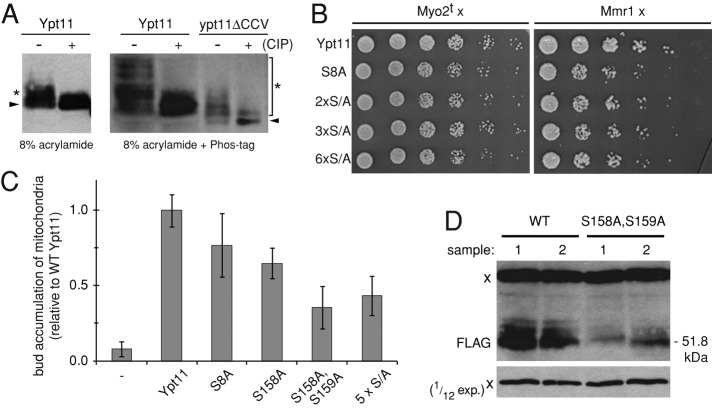
Phosphorylation has been shown to regulate the membrane localization or effector interactions of a handful of yeast and mammalian Rabs (van der Sluijs et al., 1992; Fitzgerald and Reed, 1999; Ding et al., 2003; Heger et al., 2011). Our immunoblotting studies reproducibly showed a weaker upper protein band migrating close to the predominant Ypt11 species, suggesting that this Rab might also be phosphorylated (see Figures 5B and 6A, asterisk). We used a phosphatase assay in combination with a Phos-tag gel system to test whether the high–molecular weight Ypt11 species were phosphoproteins. As shown in Figure 6A, the slower-migrating species were reduced or disappeared completely when extracts from cells expressing WT Ypt11 were treated with phosphatase. In addition, the main protein band shifted to a lower molecular weight upon CIP treatment in all samples (Figure 6A and Supplemental Figure S2), suggesting that Ypt11 is constitutively phosphorylated in vivo. We also noticed that the extent and pattern of phosphorylation was different for the inactive, cytoplasmic variant ypt11ΔCCV, with the major dephosphorylated band migrating faster than the equivalent WT Ypt11 protein. This altered mobility is not a consequence of the C-terminal amino acid deletion in ypt11ΔCCV, since we observed the same pattern for the inactive ypt11(T104N) mutant form of the full-length protein (Supplemental Figure S2). Thus WT Ypt11 is likely subject to additional posttranslational modification(s) in addition to phosphorylation.
FIGURE 6:
A conserved phosphorylation site modulates Ypt11 abundance. (A) Whole-cell extracts from ypt11Δ cells expressing FLAG-Ypt11 or FLAG-ypt11ΔCCV from the MET25 promoter were incubated with or without calf alkaline phosphatase (CIP). Samples were separated in 16-cm 8% acrylamide (left) or 8% acrylamide + Phos-tag minigels (right), and Ypt11 bands were visualized by anti-FLAG immunoblotting. Threefold less material was loaded for ypt11ΔCCV samples. Arrowheads mark the presumed dephosphorylated protein bands; asterisks mark modified species. (B) Interaction of serine-to-alanine Ypt11 variants with Myo2 tail (Myo2t) and Mmr1 was tested using a two hybrid assay. Fivefold serial dilutions were spotted on SD‑Leu-Trp-His and incubated for 2 d at 30°C. (C) Quantification of mitochondrial accumulation in buds of ypt11Δ cells overexpressing WT Ypt11 and the indicated serine-to-alanine variants from the MET25 promoter. Results are normalized to WT. Bars, mean and SD of three independent experiments (n = 100). Student's t test: S8A, p < 0.05; S158A, p < 0.001; S158A,S159A, p < 0.001; 5xS/A, p < 0.001). (D) The expression of FLAG-Ypt11 or FLAG-ypt11(S158A,S159A) in ypt11Δ cells analyzed by anti-FLAG Western blotting. Two replicates are shown for each protein. A nonspecific band reacting with anti-FLAG antibody (marked with x) was used as a loading control. Bottom, a shorter exposure of the nonspecific band. 2xS/A = ypt11(S158A,S159A); 3xS/A = ypt11(S8A,S158A, S159A); 5xS/A = ypt11(S77A,S79A,S80A,S158A, S159A); 6xS/A = ypt11(S8A,S77A,S79A,S80A,S158A, S159A).
The multiple high–molecular weight species observed in the untreated WT Ypt11 extract (Figure 6A) may indicate that multiple sites are phosphorylated in the protein. Alternatively, Ypt11 proteins with differing phosphorylation status at individual sites may coexist in the cell. Phosphorylation prediction algorithms (NetPhos, Blom et al., 1999; DISPHOS, Iakoucheva et al., 2004; pkaPS, Neuberger et al., 2007) detected a number of potential phosphorylation sites in Ypt11. Two of these (S77, S158–S159) were identified by mass spectrometry (Albuquerque et al., 2008; Bodenmiller et al., 2008, 2010; Holt et al., 2009). Residues S158 and S159 lie within a unique Ypt11 insert and are part of an RKRYS motif conserved among fungi (Figure 4B, II). A similar motif (containing S8) is found in the N-terminal extension specific for Ypt11 (Figure 4B, Ia). Although the region surrounding S77 is not conserved, it contains several serine residues with high phosphorylation-prediction scores, which we included in our analysis (S77, S79, S80; Figure 4B, Ib).
We used the two-hybrid assay to evaluate the effect of phosphorylation-site mutation(s) on Ypt11 effector interactions. Even when a serial dilution assay was applied to test for subtle differences in growth rates, none of the mutant proteins, including the one combining all six sites (S8A, S77A, S79A, S80A, S158A, S159A), exhibited interaction defects with the Myo2 tail or Mmr1 (Figure 6B). In spite of this, some of the substitutions decreased Ypt11’s ability to promote mitochondrial accumulation in buds when overexpressed (Figure 6C). The strongest defect was observed for the ypt11(S158A,S159A) double mutant. However, this mutant was significantly less abundant than the WT protein (Figure 6D). Thus the decrease in mitochondrial accumulation phenotypes for both mutant proteins (Figure 6C) is probably due to their decreased abundance. The fact that we observed slower-migrating bands for ypt11(S158A,S159A) (and the 5xS/A ypt11 mutant; unpublished results) in separating gels suggests that Ypt11 may be phosphorylated on some lower-probability sites that were not included in our analysis.
Taken together, our findings demonstrate that Ypt11 is a phosphoprotein. For the sites we queried, Ypt11’s phosphorylation status does not affect its interaction with effectors required for mitochondrial inheritance. However, mutation of predicted phosphorylation sites decreases Ypt11 abundance, suggesting that this modification plays a role in regulating Ypt11 availability in the cell.
Ypt11 activity and abundance are tightly linked
We and others showed previously that overexpression of WT Ypt11 is lethal in yeast, in part because excessive mitochondrial transport depletes mitochondria from the mother cell (Itoh et al., 2002; Frederick et al., 2008). We found that Ypt11-variant abundance and activity are inversely correlated when proteins are overexpressed from the MET25 promoter (high abundance: ypt11ΔCCV and ypt11(T104N) >> ypt11(Q232L) ≥ WT Ypt11 >>> ypt11-Mt: low abundance; Supplemental Figure S3). Thus the pool of active Ypt11 appears to be tightly regulated. A scenario in which active forms of the protein are rapidly turned over to prevent their accumulation could explain these results. To test this idea, we analyzed protein stability under conditions in which translation was blocked by cycloheximide (CHX). As shown in Figure 7, degradation of active variants WT Ypt11 and ypt11-Q232L occurred in the first 30 min of the experiment, although a fraction of the protein lingered throughout the 2-h time course. By contrast, the inactive cytoplasmic variant ypt11ΔCCV was stable throughout the time course. The very low abundance of the ypt11-Mt variant prevented its analysis by this method. These CHX-chase experiments confirmed that active variants of the protein are maintained at a lower level in cells and indicate that this is achieved through degradation.
FIGURE 7:
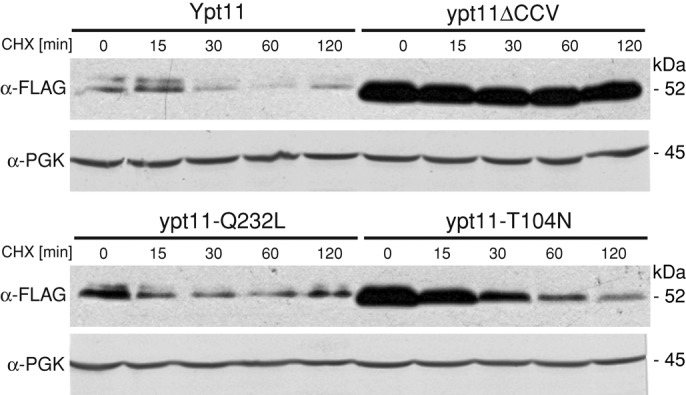
Ypt11 abundance is controlled by degradation. FLAG-tagged Ypt11 and mutant variants were expressed from the MET25 promoter in the ypt11Δ strain. Protein stability was assessed in whole-cell extracts prepared at the indicated time points after addition of cycloheximide. Ypt11 variants and a phosphoglycerate kinase (PGK) loading control were detected by immunoblotting with anti-FLAG or anti-PGK antibodies. See Materials and Methods for details.
Although the behavior of inactive ypt11ΔCCV is entirely consistent with our model (the protein is not degraded), the results for inactive ypt11(T104N) are less straightforward. Although ypt11(T104N) was degraded in the presence of CHX, the abundance of ypt11(T104N) at the start of the chase was similar to that of ypt11ΔCCV. These findings are consistent with the idea that inactive Ypt11 variants are less deleterious to the cell and can be tolerated at higher steady-state abundance.
As shown in Figure 8A, overexpressed ypt11(T104N) is predominantly cytoplasmic. However, this variant contains the C-terminal prenylation site and has the potential to interact with membranes. We introduced the T104N mutation into ypt11-Mt to test whether membrane association (in addition to protein activity) determines protein abundance in cells. In contrast to the nontethered form, ypt11-Mt(T104N) localized exclusively to mitochondria (Figure 8A). Similar to the inactive ypt11(T104N) and ypt11ΔCCV variants, overexpression of ypt11-Mt(T104N) did not induce mitochondrial accumulation in buds (Figure 8B). Despite its membrane localization, the abundance of ypt11-Mt(T104N) remained high, similar to that of the inactive ypt11(T104N) and ypt11ΔCCV proteins (Figure 8C). Thus the localization of ypt11-Mt(T104N) to membranes is not sufficient to decrease its abundance in the cell.
FIGURE 8:
Membrane targeting does not control the abundance of ypt11(T104N). (A) Representative DIC and fluorescence images of ypt11Δ cells expressing GFP-tagged Ypt11 and mutant variants from the MET25 promoter. Bar, 5 μm. (B) Quantification of the mitochondrial accumulation phenotypes in ypt11Δ cells expressing mtGFP and the indicated Ypt11 variants. Bars, mean and SD of three independent experiments (n = 100). (C) Whole-cell extracts from ypt11Δ cells expressing FLAG-tagged Ypt11 variants from the MET25 promoter were analyzed by Western blotting using anti-FLAG antibody. Two exposures of the same membrane are shown.
DISCUSSION
Our studies advance the understanding of key aspects of Ypt11 function. First, our findings provide an explanation for the different subcellular localizations reported for Ypt11 in the literature. Second, we provide evidence that direct association of Ypt11 with mitochondria is essential for its role in mitochondrial inheritance. Third, we demonstrate that ER-localized Ypt11 cannot support mitochondrial inheritance, arguing against the idea that ER–mitochondrial contacts allow passive movement of mitochondria during polarized ER transport. Fourth, we show that Ypt11 function is regulated at multiple levels. The C-terminal prenylation motif in Ypt11, which controls its membrane association, is required for its mitochondrial inheritance function. The Ypt11 GTPase cycle controls interaction with Myo2 and a novel interacting partner, Mmr1. In addition, active forms of Ypt11 are selectively degraded in vivo. Finally, Ypt11 is a phosphoprotein. We show that a conserved phosphorylation motif within a unique Ypt11 insert regulates its cellular abundance.
The function of Ypt11 in organelle inheritance has been assigned, in part, by in vivo localization studies (Itoh et al., 2002; Buvelot Frei et al., 2006). Ypt11 is expressed at very low levels from its native promoter and cannot be definitively localized. When highly overexpressed, Ypt11 clearly localizes to the perinuclear and cortical ER (Buvelot Frei et al., 2006; this study). The physiological relevance of this localization is supported by studies showing that Ypt11 promotes cER inheritance (Buvelot Frei et al., 2006; Frederick et al., 2008; Swayne et al., 2011) and can drive excess cER into buds (Frederick et al., 2008). Collins and colleagues have shown that Ypt11 interacts with a host of ER- and Golgi-associated membrane proteins, which bind a broad range of yeast Rabs (Calero and Collins, 2002; Calero et al., 2002). The availability of less specific binding partners at the ER may explain why this localization predominates when Ypt11 is highly overexpressed in cells. The identification of Ret2 as an adaptor for Ypt11 validated the Golgi as a physiological target for this Rab (Arai et al., 2008). Nevertheless, clear Golgi localization of Ypt11 is not observed at endogenous expression levels or after overexpression (Arai et al., 2008; this study). As shown by us and others (Itoh et al., 2002; Frederick et al., 2008), there is compelling evidence that Ypt11 acts on mitochondrial membranes to promote inheritance, even though fluorescence imaging studies have not revealed Ypt11 on these compartments. This inability to detect Ypt11 at cellular membranes where it clearly functions may reflect a transient interaction of Ypt11 at these sites or a requirement for very few molecules of Ypt11 to exert its function. Alternatively, Ypt11 could act indirectly on mitochondria from another cellular locale, as will be discussed.
If Ypt11 directly transports ER membranes, mitochondria could be moved indirectly via stable ER–mitochondrial contact sites. We previously showed that one such linkage (formed by the ERMES complex) was not essential for mitochondrial inheritance, since overexpression of Ypt11 partially rescued mitochondrial transmission to buds in ERMES mutants (Nguyen et al., 2012). These findings established that ERMES contact sites are not required for Ypt11 action on mitochondria but did not rule out the possibility that Ypt11 might mediate tandem organelle transport through other types of ER–mitochondrial linkages. We tested this idea by expressing an ER-tethered form of Ypt11. Although Ypt11-ER was active and promoted cER transport, it was unable to rescue mitochondrial inheritance defects in ypt11Δ mmr1Δ. Thus it is unlikely that Ypt11 acts from the ER membrane to promote mitochondrial movement into buds.
Recent studies outlined how a yeast Rab cascade regulates Myo2 binding and transport of cargo through the secretory pathway (Jin et al., 2011; Santiago-Tirado et al., 2011; Donovan and Bretscher, 2012). Based on its involvement in cER inheritance (Buvelot Frei et al., 2006; Frederick et al., 2008) and late Golgi transport (Arai et al., 2008), Ypt11 could act early in this cascade to help deliver cargo to the growing bud. It has been proposed that, instead of recruiting Myo2 to mitochondria, Ypt11 mediates bud-directed transport of ER- and Golgi-associated factor(s) that retain mitochondria in the daughter cell once they arrive (Boldogh et al., 2004; Pon, 2008). If this was the case, the ypt11-ER construct should enhance delivery of this factor(s) to buds and indirectly improve mitochondrial inheritance. However, this did not occur. Although ER-anchored Ypt11 was active and caused accumulation of cER in buds, mitochondrial bud accumulation was not observed. We favor a model in which Ypt11-dependent, bud-directed transport relies on localization of a fraction of this Rab GTPase to mitochondria. This model is supported by our finding that Ypt11 tethered to mitochondria promotes mitochondrial inheritance and the observation that Myo2 localizes to the surface of mitochondria (Förtsch et al., 2011). Although it is clear that mitochondria transported from the mother cell are retained at the bud tip (Yang et al., 1999) and that this is critical for successful mitochondrial inheritance, the components required for this anchoring step are only beginning to be identified (Swayne et al., 2011).
Rab GTP binding and hydrolysis control the effector interactions through which Rabs carry out their cellular functions. Our study is the first to analyze a putative activating allele of Ypt11. Of interest, the ypt11(Q232L) allele maintains mitochondrial inheritance activity but does not behave as a constitutively activated protein, which would be expected to significantly enhance organelle accumulation in buds. This finding is not necessarily unexpected, since the effect of the Q-to-L mutation is less predictable in Rabs than for the prototypical Ras GTPase. A similar mutation in the secretory Rab Ypt1 reduced GTP hydrolysis of the protein but did not cause the predicted dominant phenotype (Richardson et al., 1998). A corresponding mutation in Sec4 blocks its GTP hydrolysis ability but has a negative rather than positive effect on its function (Walworth et al., 1992). Only active Ypt11 was able to interact with the Myo2 tail, consistent with the idea that Ypt11-GTP binds the motor and GTP hydrolysis releases the motor (this study and Itoh et al., 2002). Of importance, we found that Mmr1, the only other known Myo2-binding partner involved in mitochondrial inheritance, can also interact with Ypt11. The interaction with Mmr1 was also restricted to active forms of Ypt11 (WT and Q232L). The binding sites for Mmr1 and Ypt11 on Myo2 do not overlap and lie on opposite sides of the Myo2 cargo-binding domain (Eves et al., 2012). Thus the Ypt11–Mmr1 interaction could be direct or could reflect simultaneous binding of both proteins to Myo2. In either case, Ypt11 and Mmr1 might act together or sequentially to modulate mitochondrial binding to Myo2 or the activity of the Myo2 motor. Sequential functions for Ypt11 and Mmr1 seem more likely since Ypt11 is expressed early in G1 of the cell cycle, whereas Mmr1 transcription peaks at the G2/M transition (Cho et al., 1998; Spellman et al., 1998; Pramila et al., 2006).
The finding that active forms of Ypt11 are subject to degradation was unexpected. The lethal effects of WT Ypt11 overexpression are an indication that cells need to tightly control the abundance of this Rab. Although Ypt11 expression is regulated at the transcriptional level (Cho et al., 1998; Spellman et al., 1998; Pramila et al., 2006), degradation provides an additional mechanism to posttranslationally fine tune protein abundance. Ypt11 degradation may occur constitutively or selectively in the bud after the Rab has completed its function in organelle inheritance. The latter scenario would be similar to the degradation reported for the Myo2 vacuole adaptor Vac17 after bud delivery (Tang et al., 2003). To our knowledge, this is the first report of selective degradation of active forms of a Rab.
We identified an 84–amino acid insert in Ypt11 orthologues that is not found in other Rabs. The localization of the insert, directly upstream of the critical switch I region in Rabs, suggested it might affect binding partner interactions (Merithew et al., 2001; Pfeffer, 2005; Cherfils and Zeghouf, 2011; Wittinghofer and Vetter, 2011). Highly conserved residues within this insert form a predicted phosphorylation motif common among substrates of Ca2+-calmodulin and PKA kinases. However, mutation of these predicted phosphorylation sites had no effect on Myo2 or Mmr1 binding. Instead, these mutations significantly decreased Ypt11 protein abundance. It is possible that phosphorylation at these sites stabilizes Ypt11 during key stages of the cell cycle. In addition, this phosphorylation could regulate Ypt11 interactions with binding partners other than Myo2 and Mmr1.
On the basis of our new findings, we propose a working model for Ypt11-mediated mitochondrial inheritance (Figure 9). Borrowing from what is known about the regulation of other Rabs, the model indicates that Ypt11-GDP is recruited from the cytoplasm to the mitochondrial surface. Subsequent interaction with an unknown exchange factor converts the Rab to its GTP-bound form, promoting binding to the Myo2 motor tail and bud-directed mitochondrial transport. Phosphorylation of Ypt11 after membrane recruitment might stabilize Ypt11 and prevent degradation during mitochondrial inheritance. After mitochondrial delivery to the tip of the bud, Ypt11 dephosphorylation and degradation would remove Ypt11 (and Myo2) from the membrane and release mitochondria from the anchoring site at the bud tip. This would allow the organelle to interact with downstream factors that spread the network at the cell cortex as bud growth shifts from apical to isotropic expansion mode. In the future, this model will provide an important framework for testing and ordering critical Ypt11 functions required for polarized mitochondrial movement.
FIGURE 9:
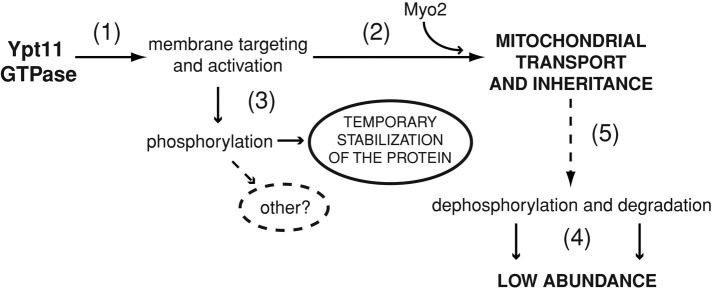
Multiple inputs combine to regulate availability of the active Ypt11 GTPase for mitochondrial inheritance. A working model. The ability of Ypt11 to associate with mitochondrial membranes (1) and interact with Myo2 in a GTP-dependent manner (2) is a prerequisite for its function in mitochondrial inheritance. Phosphorylation of serines 158 and 159 is not essential for mitochondrial inheritance. Instead, phosphorylation of the active protein on these (and perhaps other) sites occurs after membrane targeting and stabilizes the membrane-bound complex (3). As shown in this study, membrane association is not sufficient for degradation of the inactive GTPase. Thus both membrane association and a functional GTPase domain are required for Ypt11 degradation and a return to low steady-state abundance (4). Because active, membrane-associated Ypt11 is required to complete mitochondrial bud delivery, it seems likely that the protein is targeted for degradation after its inheritance function is completed (5). Consistent with this interpretation, we observed that Ypt11 variants that cannot mediate mitochondrial inheritance (either due to a defective GTPase domain or the inability to bind membranes) are not deleterious to the cell and are not targeted for selective degradation.
MATERIALS AND METHODS
Yeast strains and plasmid construction
The yeast strains used in this study were created in the W303 genetic background (ade2-1 leu2-3 his3-11,15 trp1-1 ura3-1 can1-100) and are listed in Supplemental Table S1. All mutations, disruptions, and constructs were confirmed by PCR and DNA sequencing. Construct generation is described in Supplemental Methods, and the plasmids used in this study are listed in Supplemental Table S2. Standard methods were used for transformation and growth of S. cerevisiae (Sherman et al., 1986; Guthrie and Fink, 1991) and Escherichia coli (Maniatis et al., 1982).
Microscopy and imaging
Cells were visualized by an Axioplan 2 microscope equipped with Zeiss Plan-Apochromat 100×/numerical aperture 1.4 objective (Carl Zeiss, Jena, Germany) and differential interference contrast (DIC) optics. Images were captured using a monochrome camera (AxioCam Mm; Carl Zeiss) and AxioVision 3.1 software and assembled using Photoshop (Adobe, San Jose, CA) with linear adjustments of brightness and contrast applied. Organelle markers used in this study were Su9(1-69)-GFP or Su9(1-69)-RFPff (referred to as mtGFP or mtRFP, respectively) for mitochondria and DsRed-HDEL (ER-DsRed) for the endoplasmic reticulum.
Mitochondrial inheritance assays
The inheritance of mitochondria labeled with mtGFP was quantified in medium- and large-budded cells in cultures grown to logarithmic phase in synthetic dextrose (SD) media as described previously (Frederick et al., 2008). For Ypt11 overexpression experiments strains were grown overnight at 30°C in SD media lacking appropriate amino acids for selection and containing 0.085 mg/ml methionine. Ypt11 expression was induced for 30 min or 2 h by transferring cells into SD medium lacking methionine. Mitochondrial accumulation phenotype is defined as the presence of a disproportionately large amount of mitochondria in the bud (Frederick et al., 2008).
Protein expression analysis
Whole-cell extracts were prepared by incubating cells in 0.1 M NaOH (100 μl per 1 OD600 unit of cells) on ice for 10 min. Cells were pelleted, resuspended in SDS–PAGE loading buffer, and boiled (Kushnirov, 2000). Unless otherwise indicated, samples (extract volume equivalent to 0.2–0.5 OD600 unit of cells per lane for WT Ypt11, amounts adjusted as needed for mutants) were run in 10% acrylamide minigels under standard conditions. Membranes were decorated with mouse monoclonal antibodies generated against FLAG (Agilent Technologies/Stratagene, Santa Clara, CA), GFP (Covance, Berkeley, CA), and 3-phosphoglycerate kinase (Molecular Probes, Eugene, OR), followed by a goat anti-mouse immunoglobulin G (IgG)–peroxidase (Sigma-Aldrich, St. Louis, MO) or IRDye 800CW donkey anti-mouse IgG (LI-COR Biosciences, Lincoln, NE) secondary antibody. Signal was detected using ECL Plus (Thermo Scientific/Pierce, Rockford, IL) or a digital imaging system (Odyssey; LI-COR Biosciences). Quantitative analysis was performed using ImageJ software (National Institutes of Health, Bethesda, MD).
Protein stability assay (cycloheximide block)
We collected 20 OD units of cells grown in SD medium to mid–logarithmic phase, resuspended them in SD lacking methionine, and incubated them with shaking for 30 min to induce YPT11 overexpression. To stop induction, cells were washed once with water and resuspended in SD medium (0.5 mg/ml methionine). Cycloheximide was added to 200 μg/ml, and aliquots equal to 1 OD600 unit were collected at indicated time points and processed as described.
Phosphatase treatment
Protein extraction and dephosphorylation reactions were performed as described previously (Peng and Weisman, 2008; Fagarasanu et al., 2009) with minor modifications. Yeast were grown to mid–logarithmic phase and switched to medium lacking methionine for 30 min, and 20 OD600 units of cells were collected. Cells were resuspended in 1 ml of ice-cold 0.2 M NaOH and 0.5% 2-mercaptoethanol (2-ME) and incubated on ice for 10 min. Trichloroacetic acid (TCA) was added to 5% final concentration, followed by 15-min incubation on ice. Protein pellets were collected by centrifugation (10 min, 14,000 × g, 4°C) and resuspended in 140 μl of buffer S (0.3 M sorbitol, 10 mM Tris, pH 7.5, 0.1 M NaCl) plus 60 μl of 1 M Tris base, 133.4 μl 10% SDS, and 7 μl or 2-ME. For alkaline phosphatase treatment, 100 μl of precleared lysates and 30 U of calf intestine phosphatase (CIP; New England Biolabs, Ipswich, MA) were added to tubes containing 900 μl of NEB3 buffer (1× in final volume; New England Biolabs). Reactions were incubated for 1.5 h at 37°C and terminated by adding 50% TCA (15% final concentration), followed by 20 min of incubation on ice. Protein pellets were dissolved in 100 μl of 2× SDS loading buffer (150 mM Tris, pH 6.8, 15% glycerol, 2% SDS, 4% 2-ME, and bromophenol blue) and 20 μl of 1 M Tris base. Aliquots equivalent to 1 OD600 unit of starting material were loaded on 8% polyacrylamide gel (30:1 acrylamide:bisacrylamide ratio) containing 50 μg/ml Phos-tag acrylamide (Nard Institute, Amagasaki, Japan) and 10 mM MnCl2, followed by immunoblotting with mouse anti-FLAG antibody (Agilent Technologies/Stratagene).
Two-hybrid assay
MMR1, YPT11, or its variants were PCR amplified and cloned into pGAD-c1 and pGBD-c1 (James et al., 1996). The two-hybrid tests were performed in the yeast strain PJ69-4A, which contains HIS3, ADE2, and lacZ as chromosomally integrated reporter genes. Interactions shown were tested by plating on a solid dropout medium agar without histidine containing 3-amino-1,2,4-triazole (2 and 10 mM for Mmr1 and Myo2t interaction, respectively). BD-Mmr1 fusion was not functional (lack of interaction with AD-Myo2), and thus only interactions with Ypt11 fused to the binding domain are shown.
Supplementary Material
Acknowledgments
We thank current Shaw lab members for stimulating discussions and former members Jason Singer and Rebecca Frederick for providing several plasmids used in this study. We also thank Steven Gygi for discussions regarding Ypt11 phosphorylation. Support for this work was provided by National Institutes of Health Grants GM084970 and GM53466 to J.M.S. and American Heart Association Postdoctoral Fellowship 0825067F to A.L.
Abbreviations used:
- aa
amino acid
- cER
cortical endoplasmic reticulum
- CHX
cycloheximide
- DIC
differential interference contrast
- SD
synthetic dextrose (medium)
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-12-0848) on February 20, 2013.
REFERENCES
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann K, Frank M, Neumann D, Jakobs S, Westermann B. The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J Cell Biol. 2008;181:119–130. doi: 10.1083/jcb.200709099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, Noda Y, Kainuma S, Wada I, Yoda K. Ypt11 functions in bud-directed transport of the Golgi by linking Myo2 to the coatomer subunit Ret2. Curr Biol. 2008;18:987–991. doi: 10.1016/j.cub.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Ramcharan SL, Yang H-C, Pon LA. A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol Biol Cell. 2004;15:3994–4002. doi: 10.1091/mbc.E04-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot Frei S, et al. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10p and Ypt11p. Mol Cell Biol. 2006;26:7299–7317. doi: 10.1128/MCB.02405-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero M, Collins RN. Saccharomyces cerevisiae Pra1p/Yip3p interacts with Yip1p and Rab proteins. Biochem Biophys Res Commun. 2002;290:676–681. doi: 10.1006/bbrc.2001.6242. [DOI] [PubMed] [Google Scholar]
- Calero M, Winand NJ, Collins RN. Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett. 2002;515:89–98. doi: 10.1016/s0014-5793(02)02442-0. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Chronicles of the GTPase switch. Nat Chem Biol. 2011;7:493–495. doi: 10.1038/nchembio.608. [DOI] [PubMed] [Google Scholar]
- Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Ding J, Soule G, Overmeyer JH, Maltese WA. Tyrosine phosphorylation of the Rab24 GTPase in cultured mammalian cells. Biochem Biophys Res Commun. 2003;312:670–675. doi: 10.1016/j.bbrc.2003.10.171. [DOI] [PubMed] [Google Scholar]
- Donovan KW, Bretscher A. Myosin-V is activated by binding secretory cargo and released in coordination with rab/exocyst function. Dev Cell. 2012;23:769–781. doi: 10.1016/j.devcel.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves PT, Jin Y, Brunner M, Weisman LS. Overlap of cargo binding sites on myosin V coordinates the inheritance of diverse cargoes. J Cell Biol. 2012;198:69–85. doi: 10.1083/jcb.201201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, et al. Myosin-driven peroxisome partitioning in S. cerevisiae. J Cell Biol. 2009;186:541–554. doi: 10.1083/jcb.200904050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald ML, Reed GL. Rab6 is phosphorylated in thrombin-activated platelets by a protein kinase C-dependent mechanism: effects on GTP/GDP binding and cellular distribution. Biochem J. 1999;342:353–360. [PMC free article] [PubMed] [Google Scholar]
- Förtsch J, Hummel E, Krist M, Westermann B. The myosin-related motor protein Myo2 is an essential mediator of bud-directed mitochondrial movement in yeast. J Cell Biol. 2011;194:473–488. doi: 10.1083/jcb.201012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RL, Okamoto K, Shaw JM. Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics. 2008;178:825–837. doi: 10.1534/genetics.107.083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ, Crider DG, Gay AC, Salanueva IJ, Boldogh IR, Pon LA. Mitochondrial inheritance is required for MEN-regulated cytokinesis in budding yeast. Curr Biol. 2009;19:1730–1735. doi: 10.1016/j.cub.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Heger CD, Wrann CD, Collins RN. Phosphorylation provides a negative mode of regulation for the yeast Rab GTPase Sec4p. PLoS One. 2011;6:e24332. doi: 10.1371/journal.pone.0024332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Toh-e A, Matsui Y. Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J. 2004;23:2520–2530. doi: 10.1038/sj.emboj.7600271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Watabe A, Toh-e A, Matsui Y. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:7744–7757. doi: 10.1128/MCB.22.22.7744-7757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Sultana A, Gandhi P, Franklin E, Hamamoto S, Khan AR, Munson M, Schekman R, Weisman LS. Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev Cell. 2011;21:1156–1170. doi: 10.1016/j.devcel.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10:885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merithew E, Hatherly S, Dumas JJ, Lawe DC, Heller-Harrison R, Lambright DG. Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition. J Biol Chem. 2001;276:13982–13988. doi: 10.1074/jbc.M009771200. [DOI] [PubMed] [Google Scholar]
- Neuberger G, Schneider G, Eisenhaber F. pkaPS: prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biol Direct. 2007;2:1. doi: 10.1186/1745-6150-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Lewandowska A, Choi JY, Markgraf DF, Junker M, Bilgin M, Ejsing CS, Voelker DR, Rapoport TA, Shaw JM. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 2012;13:880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Weisman LS. The cyclin-dependent kinase Cdk1 directly regulates vacuole inheritance. Dev Cell. 2008;15:478–485. doi: 10.1016/j.devcel.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Structural clues to Rab GTPase functional diversity. J Biol Chem. 2005;280:15485–15488. doi: 10.1074/jbc.R500003200. [DOI] [PubMed] [Google Scholar]
- Pon LA. Golgi inheritance: rab rides the coat-tails. Curr Biol. 2008;18:R743–R745. doi: 10.1016/j.cub.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafelski SM, Viana MP, Zhang Y, Chan Y-HM, Thorn KS, Yam P, Fung JC, Li H, Costa Lda F, Marshall WF. Mitochondrial network size scaling in budding yeast. Science. 2012;338:822–824. doi: 10.1126/science.1225720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CJ, Jones S, Litt RJ, Segev N. GTP hydrolysis is not important for Ypt1 GTPase function in vesicular transport. Mol Cell Biol. 1998;18:827–838. doi: 10.1128/mcb.18.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Legesse-Miller A, Schott D, Bretscher A. PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Dev Cell. 2011;20:47–59. doi: 10.1016/j.devcel.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra MC, Coudrier E. Rab GTPases and myosin motors in organelle motility. Traffic. 2004;5:393–399. doi: 10.1111/j.1398-9219.2004.00190.x. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lütcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne TC, et al. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr Biol. 2011;21:1994–1999. doi: 10.1016/j.cub.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kauffman EJ, Novak JL, Nau JJ, Catlett NL, Weisman LS. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature. 2003;422:87–92. doi: 10.1038/nature01453. [DOI] [PubMed] [Google Scholar]
- Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs P, Hull M, Huber LA, Mâle P, Goud B, Mellman I. Reversible phosphorylation-dephosphorylation determines the localization of rab4 during the cell cycle. EMBO J. 1992;11:4379–4389. doi: 10.1002/j.1460-2075.1992.tb05538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Brennwald P, Kabcenell AK, Garrett M, Novick P. Hydrolysis of GTP by Sec4 protein plays an important role in vesicular transport and is stimulated by a GTPase-activating protein in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2017–2028. doi: 10.1128/mcb.12.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- Yang HC, Palazzo A, Swayne TC, Pon LA. A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr Biol. 1999;9:1111–1114. doi: 10.1016/s0960-9822(99)80480-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.