The LIM-domain protein ZYX-1 is the unique zyxin-like protein in Caenorhabditis elegans. In striated body-wall muscles, ZYX-1 localizes at dense bodies/Z-discs and M-lines, as well as in the nucleus, and its localization is highly dynamic. ZYX-1 plays an unexpected role in dystrophin-dependent muscle degeneration.
Abstract
In vertebrates, zyxin is a LIM-domain protein belonging to a family composed of seven members. We show that the nematode Caenorhabditis elegans has a unique zyxin-like protein, ZYX-1, which is the orthologue of the vertebrate zyxin subfamily composed of zyxin, migfilin, TRIP6, and LPP. The ZYX-1 protein is expressed in the striated body-wall muscles and localizes at dense bodies/Z-discs and M-lines, as well as in the nucleus. In yeast two-hybrid assays ZYX-1 interacts with several known dense body and M-line proteins, including DEB-1 (vinculin) and ATN-1 (α-actinin). ZYX-1 is mainly localized in the middle region of the dense body/Z-disk, overlapping the apical and basal regions containing, respectively, ATN-1 and DEB-1. The localization and dynamics of ZYX-1 at dense bodies depend on the presence of ATN-1. Fluorescence recovery after photobleaching experiments revealed a high mobility of the ZYX-1 protein within muscle cells, in particular at dense bodies and M-lines, indicating a peripheral and dynamic association of ZYX-1 at these muscle adhesion structures. A portion of the ZYX-1 protein shuttles from the cytoplasm into the nucleus, suggesting a role for ZYX-1 in signal transduction. We provide evidence that the zyx-1 gene encodes two different isoforms, ZYX-1a and ZYX-1b, which exhibit different roles in dystrophin-dependent muscle degeneration occurring in a C. elegans model of Duchenne muscular dystrophy.
INTRODUCTION
Zyxin is a LIM domain–containing protein and belongs to the zyxin family, which in vertebrates is composed of seven members: ajuba, LIM domain–containing protein 1 (LIMD1), lipoma preferred partner (LPP), migfilin, thyroid receptor–interacting protein 6 (TRIP 6), Wilms tumor interacting protein (WTIP), and zyxin (Renfranz et al., 2003; Kadrmas and Beckerle, 2004; Zheng and Zhao, 2007). These proteins share the same overall protein organization, characterized by N-terminal proline-rich regions (PRRs), three C-terminal LIM domains (Sadler et al., 1992), and a nuclear export signal (NES; Beckerle, 1997; Nix and Beckerle, 1997). LIM (Lin-11, Isl-1, and Mec-3) domains are cysteine- and histidine-rich motifs that form tandem zinc finger structures implicated in protein interaction. LIM domain–containing proteins are found in all eukaryotic organisms from yeast to human and are implicated in different biological processes (Kadrmas and Beckerle, 2004; Zheng and Zhao, 2007).
In mice, the genetic inactivation of zyxin has no obvious consequences for viability, embryonic development, fertility, morphology, or behavior. A functional redundancy between zyxin family proteins has been hypothesized, suggesting that their simultaneous inactivation might be necessary to produce a mutant phenotype (Hoffman et al., 2003).
Many studies have been conducted in different vertebrate cell lines to decipher the function of the zyxin protein. Zyxin is a cytoskeletal protein shown to be associated with cell adhesion complexes and to shuttle between these sites and the nucleus (Nix and Beckerle, 1997; Nix et al., 2001; Wang and Gilmore, 2003; Hervy et al., 2006). Zyxin regulates actin assembly and remodeling, cell mobility, and cell adhesion, as well as gene expression (Yamada et al., 2003; Hoffman et al., 2006, 2012; Janssen and Marynen, 2006; Martynova et al., 2008; Fraley et al., 2012). Zyxin was further shown to promote apoptosis (Hervy et al., 2010; Crone et al., 2011) and to be implicated in mechanotransduction and stretch-induced gene expression (Cattaruzza et al., 2004; Wójtowicz et al., 2010). In humans, skeletal muscle expresses zyxin, which interacts with several sarcomeric proteins (Morgan et al., 1995; Linnemann et al., 2010; Sun et al., 2010). However, the physiological role of zyxin in skeletal muscle has not been reported.
The fruit fly Drosophila melanogaster has two zyxin-like proteins, ZYX102 and CG11063, containing the characteristic motifs of vertebrate zyxins. The zyx102 gene is expressed during oogenesis and embryogenesis, which involve major cytoskeletal reorganizations (Renfranz et al., 2003). In follicular cells of egg chambers subjected to disruption of actin bundles by laser nanosurgery, the ZYX102 protein rapidly relocalizes at anchor points, suggesting a mechanosensitive role, as in vertebrates (Colombelli et al., 2009). Both Drosophila zyxin-like proteins seem to be required during development, as the knockdown of the corresponding genes results in lethality during the pharate adult stage (Das Thakur et al., 2010; Renfranz et al., 2010) and are also reported to be components of the Hippo signaling pathway implicated in growth control (Das Thakur et al., 2010; Rauskolb et al., 2011).
The nematode Caenorhabditis elegans has a homologue of the zyxin protein named ZYX-1. This protein was first identified in a yeast two-hybrid screen for proteins that bind to germline RNA helicase (GLH) proteins (Smith et al., 2002). A functional interaction between GLHs and ZYX-1 seems, however, unlikely, since the zyx-1 gene is not expressed in the germline. We previously showed that ZYX-1 interacts with the DYC-1 protein known to be functionally related to DYS-1, the C. elegans orthologue of dystrophin (Bessou et al., 1998; Gieseler et al., 2000; Lecroisey et al., 2008). In humans, mutations of the dystrophin gene cause Duchenne muscular dystrophy (DMD), the major X-linked muscular disorder (Koenig et al., 1987; Ahn and Kunkel, 1993). In C. elegans, loss-of-function mutations of the dys-1 gene do not lead to obvious muscle degeneration unless placed in the sensitized genetic background of a mild mutation affecting the hlh-1 gene (Gieseler et al., 2000). HLH-1 is the homologue of MyoD, a muscle-specific transcription factor (Chen et al., 1994). The dys-1(cx-18); hlh-1(cc561) double mutant exhibits a time-dependent muscle degeneration phenotype and is a powerful C. elegans model for dystrophin-dependent muscle degeneration, mimicking DMD (Gieseler et al., 2000).
The overall sarcomeric composition and organization, as well as the function of the muscle cells, was well conserved between nematodes and vertebrates during evolution. The actin-containing thin filaments are anchored at Z-discs, myosin-containing thick filaments are anchored at M-lines, and both M-lines and Z-discs are connected to the extracellular matrix. In vertebrates, this connection is provided by costameres, which are muscle-specific integrin adhesion complexes (Ervasti, 2003; Samarel, 2005). In C. elegans, the Z-disc analogues, named dense bodies, and the M-lines are directly anchored into the sarcolemma via integrin-containing complexes (Moerman and Williams, 2006; Lecroisey et al., 2007). Genetic inactivation of zyx-1 either by mutation or RNA interference (RNAi) is not lethal and does not lead to any obvious phenotype except for a locomotion defect detected in a body-bending assay (Smith et al., 2002; Lecroisey et al., 2008; Nahabedian et al., 2012).
By sequence and phylogenetic analysis, we found that, in contrast to vertebrates and other animal species, ZYX-1 is the unique representative of zyxin protein family in C. elegans and appears to be the orthologue of the vertebrate zyxin subfamily composed of zyxin, migfilin, TRIP6, and LPP. This prompted us to decipher the function of this protein, and we investigated in detail the localization, dynamics, and protein interactions of ZYX-1 within the striated body-wall muscles of C. elegans. Using a genetic analysis, we established that ZYX-1 plays a role in dystrophin-dependent muscle degeneration.
RESULTS
The C. elegans ZYX-1 is a zyxin-like protein
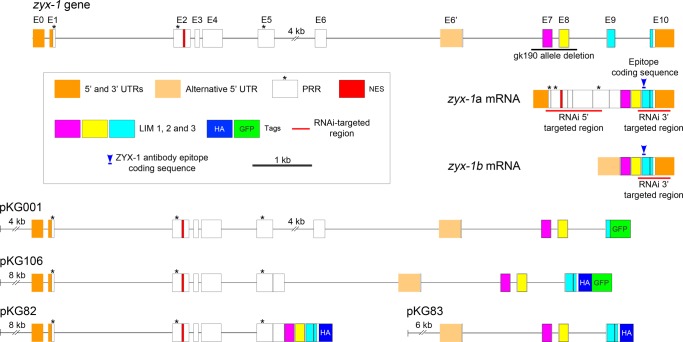
The zyx-1 gene (also known as F42G4.3) is located on chromosome II and is predicted to produce different transcripts encoding putative proteins of 200–647 amino acids (WormBase, www.wormbase.org). All predicted ZYX-1 isoforms contain three C-terminal LIM domains. The first isoform that was predicted on WormBase, F42G4.3a (ZYX-1a), is composed of 603 amino acids (Figure 1), and the LIM domains extend, respectively, over amino acids 411–463, 471–522, and 531–592 (Figure 2A). The shortest predicted isoform, F42G4.3b (ZYX-1b), composed of 200 amino acid residues, is almost identical to the C-terminus of the ZYX-1a isoform and contains the three LIM domains. The five most N-terminal amino acids are encoded by an alternative exon (Figure 1).
FIGURE 1:
Organization of the zyx-1 gene, predicted mRNAs, and transgene constructs. The organization of the zyx-1 (F42G4.3) gene was retrieved from WormBase (www.wormbase.org). Exons are indicated as rectangles numbered from E0 to E10. The gk190 deletion is indicated by a black line, and the sequence used for RNAi constructs targeting either the 5′ or the 3′ region of the zyx-1 transcripts is shown by a red line. The predicted zyx-1a and zyx-1b mRNAs are shown. The predicted protein encoded by the zyx-1b mRNA is almost identical to the C-terminus of the ZYX-1a isoform containing the three LIM domains, except for the five N-terminal amino acids encoded by exon E6′. A blue line with a blue arrowhead indicates the coding sequence of the peptide used to produce the monoclonal ZYX-1 antibody. The four constructs used in this work (pKG001, pKG106, pKG82, and pKG83) are presented, including the upstream regulatory and promoter sequences. Predicted 5′ and 3′ UTRs are in orange, the alternative 5′ UTR of zyx-1b (exon E6′) is in light orange, the PRR-encoding sequences are indicated by asterisks, the NES is in red, and LIM domains 1–3, respectively, are in purple, yellow, and cyan. GFP and HA tags are indicated as green and blue squares, respectively, without respecting the scale.
FIGURE 2:
Analysis of the ZYX-1 protein sequence. (A) Predicted amino acid sequence of the C. elegans ZYX-1 protein encoded by the F42G4.3/zyx-1 gene (UniProt accession number Q9U3F4). This sequence corresponds to the putative 603–amino acid isoform (F42G4.3a). The characteristic motifs of zyxin family proteins are indicated as follows: PRRs in boldface, the NES in red, the three successive LIM domains in purple (LIM domain 1), yellow (LIM domain 2), and cyan (LIM domain 3), and the cysteine or histidine residues within the LIM domains predicted to form zinc fingers in white letters on black background. (B) Comparison of characteristic motifs of zyxin family protein with known consensus sequences. The three LIM domains of C. elegans ZYX-1 perfectly match the consensus sequence proposed by Kadrmas and Beckerle (2004), where X is any amino acid. The class 1 NES consensus sequence proposed by Kosugi et al. (2008) is ΨX1–2[^P]Ψ [^P]2–3Ψ [^P]Ψ, where Ψ is an aliphatic residue (L, I, V, M, F, C, W, A or T), [^P] is any amino acid except proline, and [^P]2–3 are any two or three amino acids except proline. It correctly matches the C. elegans ZYX-1 sequence, except at the carboxy-terminal end, where four amino acid residues are present instead of one between the two last aliphatic residues.
The LIM-domain consensus sequence is defined as CX2CX16–23HX2CX2CX2CX16–21CX2(C/H/D), where X is any amino acid (reviewed in Kadrmas and Beckerle, 2004). This consensus perfectly matches all three LIM domains of the C. elegans ZYX-1 proteins (Figure 2B). The high sequence conservation of these regions in ZYX-1 suggests that they form LIM domains, each consisting of two adjacent zinc (or other metal) fingers, a structure that mediates protein–protein interactions.
The ZYX-1a isoform contains two other characteristic motifs of zyxin-like proteins in its N-terminal region: a potential NES and three PRRs, which are putative sites for protein–protein interactions (reviewed in Kay et al., 2000).
The putative NES sequence extends from amino acid 69 to 81 and is in agreement with the generally accepted NES consensus sequence (Nix and Beckerle, 1997). In particular, the ZYX-1 NES sequence matches the class1 NES consensus sequence, for which nuclear export activity has been demonstrated (Kosugi et al., 2008; Figure 2B). No nuclear localization signal (NLS) has been reported in zyxins, and none was found in the ZYX-1 sequence.
The PRRs in ZYX-1a consist of two sequences of, respectively, nine and six contiguous proline residues, located at amino acids 3–11 and 283–288, and a third region expanding from amino acids 36–46 with the sequence PAPAPPKPSRP (Figure 2A). It is noteworthy that none of these three PRRs contains the consensus sequence F/LPPPP characterized as ActA repeats that were identified in vertebrate zyxins to link EVH1 domains of the Ena/VASP family (Renfranz and Beckerle, 2002).
ZYX-1 is the unique zyxin-like protein in C. elegans
To investigate whether other members of the zyxin-like protein family exist in C. elegans, we used the seven human members of the zyxin family to search for similarities in the nematode proteome (as available from WormBase) with PsiBLAST. The best hit was ZYX-1 and then, decreasing by rank, PXL-1, LIM-9, ALP-1, and UNC-97 (Figure 3). Each of these five C. elegans proteins was used as a query for a BLASTp search in the human protein database. For ZYX-1, the seven best hits were the seven members of the zyxin family, with the following decreasing rank: LPP, TRIP-6, zyxin, WITP, LIMD1, ajuba, and migfilin. The best hits for PXL-1, LIM-9, ALP-1, and UNC-97 were, respectively, paxillin, FHL-2, ALP, and PINCH (Figure 3). The C. elegans PXL-1, LIM-9, ALP-1, and UNC-97 are all LIM-domain proteins, expressed in the nematode body wall muscles, and for some in other tissues, and were previously described as being orthologues of, respectively, paxillin, FHL, ALP/enigma, and PINCH (McKeown et al., 2006; Qadota and Benian, 2010; Warner et al., 2011) and belonging to other LIM-domain protein families. Thus ZYX-1 is the unique representative of the zyxin protein family in C. elegans.
FIGURE 3:
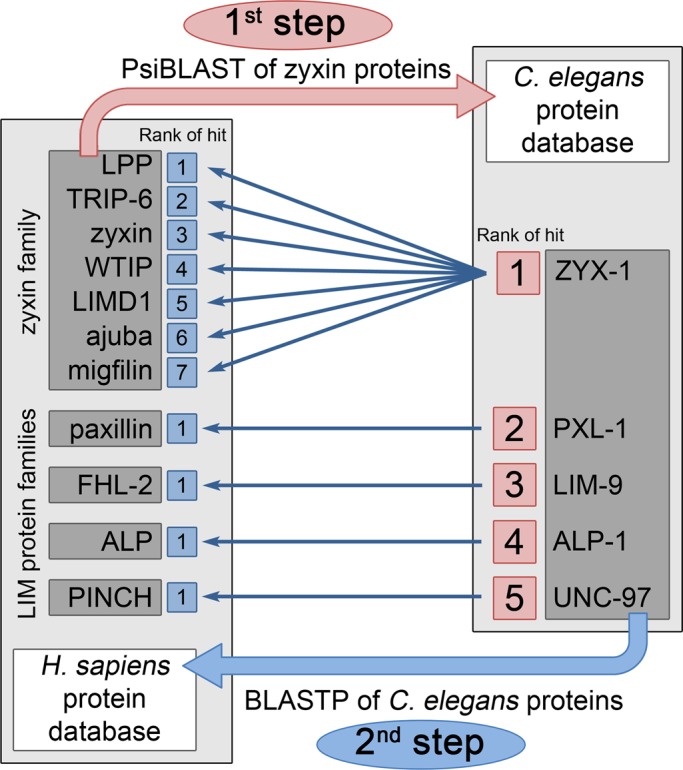
BLAST analyses of the zyxin protein family and other LIM-domain protein families between human and C. elegans protein databases. PsiBLAST analysis (first step, in pink) was performed using the sequences of the seven human members of the zyxin family to search for homologous proteins in the C. elegans protein database. The best hit corresponds to the highest similarity score. The five best hits that were selected are indicated by their decreasing rank. Each of these five closest C. elegans homologues was used as query for a BLASTp search (second step, in blue) against the human protein database. The best hits for ZYX-1 were the seven members of the zyxin family, which are indicated with their respective rank. The best hits of the C. elegans PXL-1, LIM-9, ALP-1, and UNC-97 were, respectively, human paxillin, FHL-2, ALP, and PINCH.
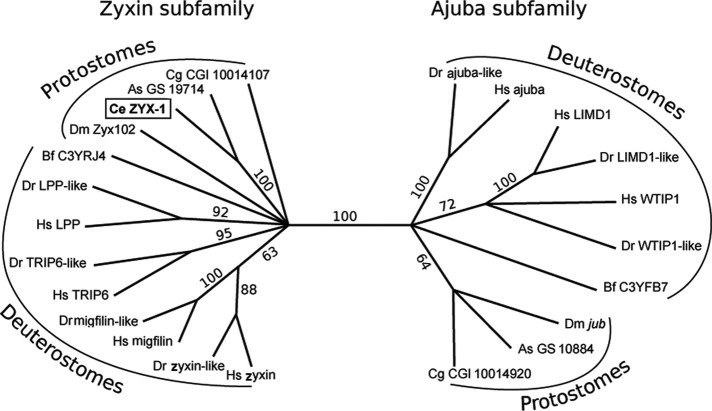
We then investigated the relationship between ZYX-1 and the seven human zyxin proteins by means of molecular phylogeny. For this purpose, zyxin-like proteins were searched in other deuterostomes: the zebrafish Danio rerio and the amphioxus Branchiostoma floridae, and in protostomes: another nematode, Ascaris suum; an arthropod, the fruit fly Drosophila melanogaster; and a lophotrocozoan, the oyster Crassostrea gigas. Twenty-three sequences of zyxin-like proteins were retrieved: one from C. elegans, respectively two from A. suum, D. melanogaster, C. gigas, and B. floridae, and respectively seven from H. sapiens and D. rerio. All of these sequences were aligned using Multiple Sequence Comparison by Log Expectation (MUSCLE) software. The N-terminal part of the sequences was too divergent to be reliably aligned; therefore the analysis was based on the conserved C-terminal region containing the three LIM domains. A maximum-likelihood tree was built using RAxML software (Figure 4).
FIGURE 4:
Maximum-likelihood phylogenetic tree of zyxin-like proteins. Bootstrap values (in percentage) are displayed on branches indicating their statistical support. Branches with <60% support were collapsed. The ajuba and zyxin subfamilies and the taxonomic groups (protostomes and deuterostomes) are indicated. Ce, C. elegans; As, A. suum; Dm, D. melanogaster; Cg, C. gigas; Hs, H. sapiens; Dr, D. rerio; Bf, B. floridae.
First, our analysis clearly demonstrates that the zyxin family is divided into two subfamilies with a 100% bootstrap support, which were named the ajuba and zyxin subfamilies. In both deuterostomes and protostomes, proteins of each subfamily are present, suggesting that their common ancestor already had two zyxin-like proteins. The D. melanogaster Zyx102 and jub (or CG11063), respectively, belong to the zyxin and the ajuba subfamilies, which is in agreement with a previous report (Renfranz et al., 2003). Second, the seven zyxin-like proteins of the two vertebrate species appeared to be arranged in pairs, all of which had high bootstrap support, except for the WTIP proteins. This suggests a one-to-one orthology relationship and the presence of a family composed of seven zyxin-like proteins in the vertebrate ancestor. Third, the unique C. elegans ZYX-1 fell into the zyxin subfamily and appeared to be the orthologue of the human zyxin, migfilin, TRIP6, and LPP proteins.
Expression pattern and subcellular localization of the ZYX-1 protein
To analyze the expression pattern of the ZYX-1 protein in C. elegans, we generated the pKG106 transgene, which contains the entire ZYX-1–encoding sequence fused to the green fluorescent protein (GFP)–encoding sequence (Figure 1). The expression of this ZYX-1-GFP protein was observed in spermathecae, body-wall muscles, and neurons (Figure 5, A–D). Using confocal analysis of transgenic worms expressing the pKG106 transgene, we detected the ZYX-1–GFP protein at dense bodies and M-lines, as well as in the nucleus of body-wall muscle cells (Figure 5C) and in both cell bodies and axons of neurons (Figure 5D). These observations confirm previously published results using transgenic worms carrying the pKG001 construct, which encodes a ZYX-1–GFP protein lacking the 39 C-terminal amino acids of the third LIM domain (Figure 1; Lecroisey et al., 2008). Thus the absence of the third LIM domain (in pKG001) does not significantly alter the subcellular localization of the ZYX-1–GFP fusion protein.
FIGURE 5:
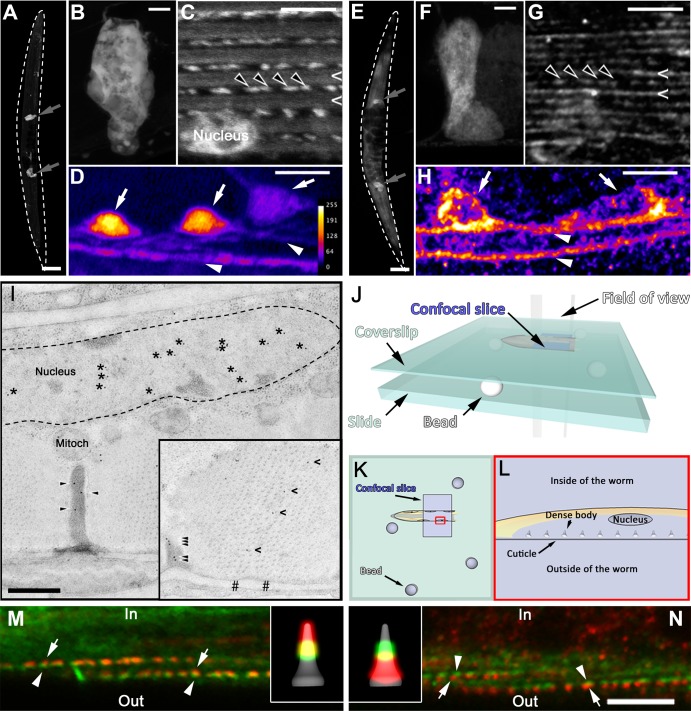
Localization of ZYX-1–GFP and ZYX-1 proteins. (A–H) Confocal analysis using antibodies directed against GFP in transgenic worms carrying the pKG106 transgene (A–D) or against ZYX-1 in wild-type worms (E–H). ZYX-1–GFP and ZYX-1 are present in spermathecae (gray arrows in A and E; the worm is surrounded by a dotted line; scale bar, 100 μm; B and F, scale bar, 10 μm), in body-wall muscle cells (C and G, scale bar, 5 μm) with a subcellular localization in the nucleus, at dense bodies (empty arrowheads) and M-lines (chevrons), and in nerve cells (D and H, scale bar, 5 μm) in both cell bodies (filled arrows) and axons (filled arrowheads). In D and H the “fire” artificial colors were used to enhance the differences of signal intensity between axons and cell bodies. The corresponding “fire” scale shown on the right in D allows for the observation of the relative intensity of the artificial colors in D (GFP antibody) and H (ZYX-1 antibody). Note that the ZYX-1 antibody leads to an important background signal (E and H). (I) Immuno–electron microscopy analysis of body-wall muscle cells in a transgenic line carrying the pKG001 transgene using an antibody directed against GFP. In transversal sections of muscle cells, ZYX-1–GFP localizes at dense bodies (arrowheads) and nucleus (asterisks inside the dotted line highlighting the nuclear membrane). Mitoch, mitochondria. Inset, localization of ZYX-1–GFP along the M-line (chevrons). The anchoring site of the M-line is located between the two hashes, and the basal part of a dense body is indicated by arrowheads. Scale bar, 0.5 μm for both images. (J–N) Confocal analysis of dense bodies in body-wall muscle cells in the same transgenic line as in A–D. (J, K) Schematic view of the method used to mount worms; lateral view in J, top view in K. Microbeads were used to prevent flattening of the worms. We focused on muscle cells oriented as schematically presented in L so as to obtain a lateral view of dense bodies. (M, N) Localization of ZYX-1–GFP within dense bodies. The ZYX-1–GFP fusion protein was detected with an antibody directed against GFP (green), and antibodies directed against ATN-1 (M) and DEB-1 (N) were used to detect these two major components of the dense bodies (red). As schematically represented in L, the orientation of the worm allows us to observe the dense bodies from their side and to locate ZYX-1–GFP (white arrowheads) with respect to ATN-1 and DEB-1 (white arrows). Scale bar, 5 μm in M and N. Insets, schematic views of the green (ZYX-1–GFP) and red (ATN-1 or DEB-1) fluorescence on a dense body backbone in gray.
We further examined the expression pattern of the ZYX-1 protein with a monoclonal antibody directed against 20 amino acids located at the beginning of the third LIM domain. In wild-type N2 worms, this antibody detects the ZYX-1 protein in spermathecae, body-wall muscle cells (at dense bodies and M-lines), and cord neurons (in cell bodies and axons; Figure 5, E–H). The signal observed with the monoclonal antibody is much weaker, however, than the GFP signal in transgenic worms. In particular, we were unable to detect a signal in muscle cell nuclei. This could be explained by a lower efficiency of antibodies or most likely by the lower amounts of the endogenous proteins compared with overexpressed transgenes. Other studies reported a discrepancy between the localization of the GFP-tagged protein and antibody localization. For example, UNC-95-GFP is found at M-lines, dense bodies, and nuclei (Broday et al., 2004), but anti–UNC-95 stained only M-lines and dense bodies (Qadota et al., 2007). In addition, it should be pointed out that in vertebrate tissues, zyxin is also expressed at low amounts (Crawford and Beckerle, 1991). Thus we performed most of the following experiments on transgenic worms expressing a ZYX-1–GFP fusion protein.
We particularly focused on the precise localization of ZYX-1–GFP in body-wall muscles. To this end, we performed immuno–electron microscopy experiments on transgenic zyx-1::gfp–expressing worms. We observed a localization of the ZYX-1–GFP protein at dense bodies, over the whole length of M-lines, and in the nucleus (Figure 5I).
Using confocal analysis, we determined the localization of the ZYX-1–GFP protein relative to two major dense body proteins, ATN-1 and DEB-1, orthologues of α-actinin and vinculin, respectively (Barstead and Waterston, 1989, 1991). We stained zyx-1::gfp transgenic worms for DEB-1 or ATN-1 using monoclonal antibodies MH24 and MH35, respectively (Francis and Waterston, 1991), and mounted them with microbeads, which prevent flattening of the worms. This approach allowed us to acquire a lateral view of the dense bodies in order to take advantage of the three-times-higher x/y resolution compared with the z resolution (Figure 5, J–L). ATN-1 and DEB-1, respectively, occupied the apical and basal regions of the dense body, as previously described (Francis and Waterston, 1991). The ZYX-1–GFP protein mainly localizes in the middle region and partially overlaps with the ATN-1– and the DEB-1–containing regions (Figure 5, M and N).
Dynamics of ZYX-1 localization in body-wall muscle cells
In vertebrates, zyxin moves from focal adhesions to the nucleus and is proposed to have a signaling function (Nix and Beckerle, 1997; Nix et al., 2001). However, the dynamics of zyxin had not been analyzed in skeletal muscles. We investigated the dynamics of the ZYX-1 protein within the C. elegans striated body-wall muscle cells, using fluorescence recovery after photobleaching (FRAP) experiments on living zyx-1::gfp transgenic worms (Figure 6).
FIGURE 6:
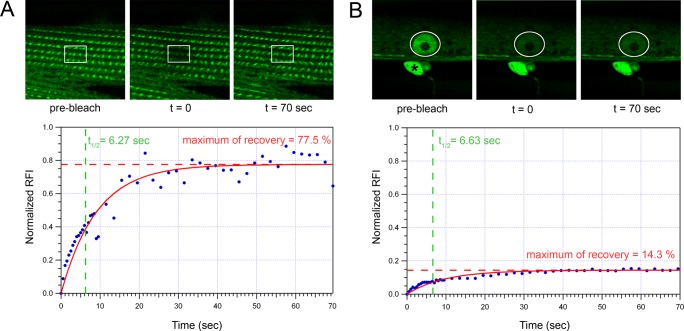
FRAP experiments of ZYX-1–GFP in body-wall muscle cells. FRAP experiments were performed on zyx-1::gfp transgenic worms carrying the pKG001 plasmid. Two FRAP experiments were performed in a cytoplasmic zone containing contractile filaments (A) and in a nucleus (B) of body-wall muscle cells. Top, fluorescence images of photobleaching and recovery of the signal. The asterisk in B indicates a cord neuron expressing the zyx-1-gfp transgene. The square in A and the circle in B indicate the photobleached regions. Images were taken before bleaching (prebleach), just after bleaching (t = 0), and after 70 s of recovery (t = 70 s). Bottom, graphs showing normalized RFI vs. time in seconds and calculated parameters of the respective experiences. After 20 and 8 bleaching laser iterations on, respectively, sarcomeric and nuclear regions, a reduction by 60 and 70% of the initial prebleach fluorescence was observed. On the y-axis, 1.0 represents the RFI of the bleached zone before bleaching steps. Here t = 0 corresponds to the first acquisition after bleaching steps, and RFI was normalized to be zero at t = 0. For each time point, the fluorescence intensity was measured (blue dots) and the corresponding fitted curve calculated (in red). The maximum recovery of fluorescence (red dotted line) and half-time for maximum recovery (green dotted line) were calculated. The percentage of maximum fluorescence recovery corresponds to the mobile fraction of the ZYX-1–GFP protein.
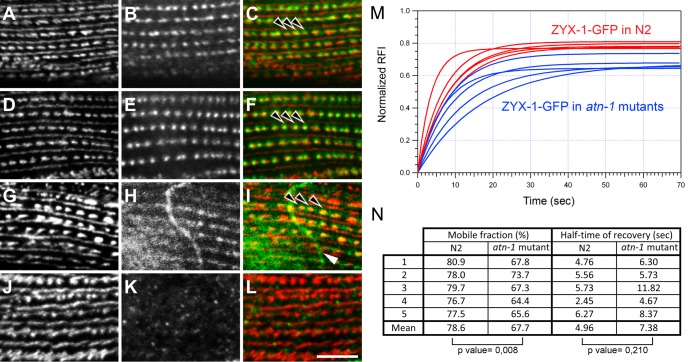
FRAP experiments were performed in cytoplasmic regions containing contractile filaments of body-wall muscle cells (Figure 6A). Twenty successive fluorescence-bleaching laser iterations led to a reduction of the initial GFP fluorescence signal by ∼60%. In five independent repeats of the experiment the half-time of fluorescence recovery ranged from 2.45 to 6.27 s, with an average half-time recovery of 4.96 s (Figure 7N). The maximal recovery of the initial fluorescence was reached after 70 s. The mobile fraction of the ZYX-1–GFP protein, which corresponds to the percentage of maximal recovery, was 76.7–80.9%, with an average of 78.6%. In comparison, a previous FRAP study in C. elegans reported a half-time of fluorescence recovery of ∼90 s for the most dynamic analyzed proteins associated with dense bodies and/or M-lines like T03G6.3 and UNC-95. These proteins exhibited mobile fractions of, respectively, 90 and 50%, and the maximal recovery of the initial fluorescence was observed after 900 s, whereas for the dense body protein UNC-112 a complete lack of recovery was reported (Ghosh and Hope, 2010). Thus the extremely rapid and nearly complete recovery of the ZYX-1–GFP signal in the sarcomeric zone reflects a high rate of exchange of ZYX-1–GFP in the cytoplasm of muscle cells. It also indicates a high mobility of this protein within the muscle cell, as well as a dynamic association of the ZYX-1 protein with muscle adhesion structures.
FIGURE 7:
Localization of ATN-1 and ZYX-1 in body-wall muscle cells of wild-type and mutants and FRAP experiments of ZYX-1–GFP in wild-type and the atn-1(ok84) mutant. (A–F) Localization of ATN-1 in wild type (A–C) and in the zyx-1(gk190) mutant (D–F). UNC-95 antibody was used as a positive control for dense bodies and M-line organization (A, D). ATN-1 staining (B, E) and the merged images (C, F) show that the localization of ATN-1 at dense bodies (arrowheads) is not disturbed in the zyx-1(gk190) mutant. (G–L) Localization of ZYX-1 in wild type (G–I) and in the atn-1(ok84) mutant (J, K). UNC-95 staining broadened at dense bodies in the atn-1(ok84) mutant (J) compared with wild-type (G). ZYX-1 antibody (H, K) and merged images (I, L) show that the localization of ZYX-1 is lost at dense bodies (arrowheads in I) in the atn-1(ok84) mutant. The white arrow in I points to an axon of a nerve cell stained with the ZYX-1 antibody. Scale bar, 10 μm for all images. (M, N) Comparison of FRAP experiments of sarcomeric ZYX-1–GFP performed on body-wall muscle cells on wild-type and atn-1(ok84) worms carrying pKG001 plasmid. (M) Fitted curves of five representative FRAP experiment data sets of ZYX-1–GFP in wild-type (red) and atn-1(ok84) worms (blue). (N) Comparison of the mobile fraction (i.e., maximum of recovery) and the half-time recovery for each sarcomeric bleached zone in N2 (wild type) or in atn-1(ok84) worms. The mobile fraction of ZYX-1–GFP is slightly but significantly lower in the atn-1 mutant than in wild type (Mann–Whitney test). No significant difference was observed for the half-time recovery. Note that data set #5 in wild type corresponds to the experiment presented in Figure 6A.
We further performed FRAP experiments on the muscle cell nucleus (Figure 6B). Eight successive bleaching laser iterations were sufficient to reduce the initial fluorescence by 70%. In four independent replicates of this experiment a mobile fraction of 14.3–26.3% was observed, with an average of 19.9%. The half-time of fluorescence recovery of ZYX-1–GFP in the nucleus ranged from 4.5 to 11.2 s, with an average half-time recovery of 7.57 s (data not shown). This result indicates that the recovery of ZYX-1–GFP in the nucleus is slightly slower than at the sarcomere, which is in agreement with an active transport mechanism. Indeed, LIM domains and the N-terminal region of zyxin can independently target the protein into the nucleus (Nix et al., 2001). Because zyxin-family proteins lack a NLS, it has been suggested that their import into the nucleus may involve protein interactions with proteins carrying a NLS (reviewed in Wang and Gilmore, 2003). Our results also suggest that the larger portion of nuclear ZYX-1–GFP (∼80%) may be permanently localized in the nucleus.
Taken together, our FRAP experiments indicate that ZYX-1 is a highly dynamic protein within the muscle cell and may shuttle between muscle adhesion complexes and the nucleus, thus suggesting a signaling function for the protein.
Interaction of ZYX-1 with muscle dense body and M-line proteins
To identify the molecular partners of the ZYX-1 protein at dense bodies and M-lines of C. elegans body-wall muscle cells, we investigated whether ZYX-1 interacts with proteins of these two structures, using the yeast two-hybrid system.
The ZYX-1a protein was used as a bait to test its interaction with a collection of known dense body and M-line proteins (Qadota et al., 2007, 2008; Xiong et al., 2009). Among the tested components of the dense body (Table 1), ZYX-1 interacted with ATN-1 (α-actinin) and DEB-1 (vinculin), which is in agreement with our confocal analysis reported earlier. ZYX-1 also interacted with ALP-1 (Enigma; McKeown et al., 2006), as well as with UIG-1, an UNC-112–interacting guanine nucleotide exchange factor specific for Cdc42 (Hikita et al., 2005), and DYC-1, as previously described (Lecroisey et al., 2008). Among the tested M-line components, ZYX-1 interacted with the LIM-domain proteins LIM-8 and LIM-9 (FHL; Qadota et al., 2007) and the CTD-type protein phosphatase domain–containing SCPL-1 (SCP) protein (Qadota et al., 2008).
TABLE 1:
Dense body and M-line proteins tested for interaction with ZYX-1.
| Dense body | ||||||
|---|---|---|---|---|---|---|
| C. elegans protein | Vertebrate orthologue or related family | Top | Mid | Base | M-line | Interaction with ZYX-1 |
| ATN-1 | α-Actinin | X | X | Yes | ||
| ALP-1 | ALP/Enigma | X | X | Yes | ||
| DEB-1 | Vinculin | X | Yes | |||
| DYC-1 | CAPON | X | Yes | |||
| UIG-1 | Cdc42 GEF | X | Yes | |||
| DYS-1 | Dystrophin | X | No | |||
| DIM-1 | Immunoglobulin-domain family | * | No | |||
| TLN-1 | Talin | X | X | No | ||
| UNC-97 | Pinch | X | X | No | ||
| PAT-4 | ILK | X | X | No | ||
| PAT-6 | Actopaxin | X | X | No | ||
| UNC-112 | Kindilin | X | X | No | ||
| PXL-1 | Paxilin | X | X | No | ||
| UNC-95 | LIM-domain family | X | X | No | ||
| LIM-8 | LIM-domain family | X | Yes | |||
| LIM-9 | FHL | X | Yes | |||
| SCPL-1 | SCP | X | Yes | |||
| HUM-6 | MYO7A | Unknown | No | |||
| KIN-32 | FAK | Unknown | No | |||
| MLP-1 | MLP | Unknown | No | |||
| F42H10.3 | LASP | Unknown | No | |||
| ZYX-1 | Yes | |||||
| ZYX-1 LIM domains | Yes | |||||
| ZYX-1 ΔLIM domains | No | |||||
| pGAD-C1 (empty vector) | No | |||||
All results obtained by yeast two-hybrid assays using the ZYX-1 bait protein (isoform ZYX-1a) against a prey protein collection of known dense body and M-line proteins. The first column gives the tested C. elegans proteins, and the second column their vertebrate orthologues or related families. The third and fourth columns summarize the localization of the concerned proteins at dense bodies and M-lines, respectively. For dense body localization three subcolumns indicate the position of the respective protein within the dense body along the basoapical axis. Last four lines: the ZYX-1a bait protein was also tested against itself (ZYX-1), ZYX-1 LIM domains, and ZYX-1 without LIM domains (ZYX-1 ΔLIM domains). The empty vector (pGAD-C1) was used as the negative control. The last column recapitulates the interactions: Yes, interaction; No, no interaction. *DIM-1 localizes near the muscle cell membrane around and between the dense bodies.
ZYX-1 did not interact with the C-terminal domain of DYS-1 (dystrophin), a scaffold protein of the subsarcolemmal cytoskeleton (Bessou et al., 1998; and our unpublished results); DIM-1 (disorganized muscle), necessary for proper myofilament anchoring (Rogalski et al., 2003); TLN-1 (talin); and members of a four-protein complex associated with the cytoplasmic tail of integrin: UNC-97 (PINCH), PAT-4 (ILK), PAT-6 (actopaxin or parvin), and UNC-112 (kindlin; Moulder et al., 1996; Rogalski et al., 2003; Norman et al., 2007; Qadota et al., 2012). ZYX-1 also did not interact with the LIM-domain proteins PXL-1 (paxillin) and UNC-95, which are both present at the bases of dense bodies and M-lines (Broday et al., 2004; reviewed in Qadota and Benian, 2010; Warner et al., 2011), HUM-6 (myosin VIIa; Baker and Titus, 1997), KIN-32 (focal adhesion kinase; Cram et al., 2008), MLP-1 (muscle LIM-domain protein), and F42H10.3 (LASP; Shaye and Greenwald, 2011).
We further observed that the ZYX-1a bait protein was able to interact with the ZYX-1a prey protein, indicating a possible homodimerization of ZYX-1 molecules. This interaction requires the LIM domains because a ZYX-1 prey protein deleted for the three LIM domains was no longer able to interact with the ZYX-1 bait protein. Accordingly, a prey protein composed of only the LIM domains interacted with the full-length ZYX-1a bait protein (Table 1).
Localization and dynamics of ZYX-1 at dense bodies is ATN-1 (α-actinin) dependent
To analyze the functional relevance of the protein interactions detected in yeast two-hybrid assays, we further asked whether the localization of ZYX-1 at dense bodies or M-lines depends on the presence of its two-hybrid partners and vice versa. For this purpose, using appropriate antibodies, we analyzed the localization of ATN-1, ALP-1, DEB-1, DYC-1, UIG-1, LIM-8, LIM-9, and SCPL-1 proteins in the body-wall muscles of zyx-1(gk190) mutants. The zyx-1(gk190) mutation deletes the coding sequences of the first two LIM domains (Figure 1), leading to a frameshift, followed by a premature stop codon, thus preventing the translation of all the three LIM domains (www.celeganskoconsortium.omrf.org). No significant modification of the localization of any of the analyzed proteins was detected in zyx-1(gk190) mutants compared with wild type (Figure 7 and Supplemental Figure S1). Thus it appears that ZYX-1 is not required for the proper assembly and morphology of muscle cell adhesion complexes.
Conversely, the ZYX-1 antibody was used to detect the protein in the muscle cells of atn-1(ok84), alp-1(tm1137), dyc-1(cx32), uig-1(ok884), lim-8(ok941), lim-9(gk106), and scpl-1(gk283) mutants. Because deb-1 mutants are embryonic lethal, we were unable to establish whether ZYX-1 requires DEB-1 for its localization. The localization of the ZYX-1 protein was diffuse or even absent in the body-wall muscles of the atn-1(ok84) mutant (Figure 7, K and L) but remained unchanged in all the other tested mutants when compared with wild-type muscles (data not shown). These observations suggest that ALP-1, UIG-1, DYC-1, LIM-8, and LIM-9, and SCLP-1 are not essential for the localization of ZYX-1 at dense bodies or M-lines, whereas ATN-1 is required for proper ZYX-1 localization at dense bodies.
FRAP experiments performed on atn-1(ok84) mutants expressing ZYX-1–GFP further revealed a significant modification of the ZYX-1–GFP dynamic when compared with wild-type (Figure 7, M and N). We observed that after the bleaching of a sarcomeric zone, ZYX-1–GFP fluorescence recovery had the same half-time of fluorescence recovery in wild-type and the atn-1(ok84) mutant; however, the mobile fraction of ZYX-1–GFP was significantly decreased in the atn-1(ok84) mutant compared with wild type. This indicates that the interaction of ZYX-1 with ATN-1 creates a mobile pool of ZYX-1 and thus that ATN-1 is required for the turnover of ZYX-1 at dense bodies. Moreover, we observed that in the absence of ATN-1, the DEB-1 and ZYX-1–GFP proteins were still present at dense bodies, but their localizations were enlarged compared with wild type (Supplemental Figure S2). A similar broadening of the localization of ALP-1 was reported in atn-1(ok84) mutants (Han and Beckerle, 2009). The broader localization of ZYX-1-GFP and DEB-1 is consistent with shorter and broader dense bodies observed by electron microscopy in atn-1(ok84) mutants (Moulder et al., 2010).
ZYX-1 participates in dystrophin-dependent muscle degeneration
We previously showed that transgenic overexpression of the ZYX-1 binding partner DYC-1 in the dystrophic dys-1(cx-18); hlh-1(cc561) double mutant significantly reduced muscle degeneration compared with nontransgenic dys-1(cx-18); hlh-1(cc561) worms (Gieseler et al., 2000). We thus investigated whether ZYX-1 plays a similar role in dystrophin-dependent muscle degeneration. To this end, we quantified abnormal body-wall muscle cells stained with phalloidin–rhodamine in different genetic contexts and normalized to 100% the muscle degeneration observed in the dystrophic dys-1(cx-18); hlh-1(cc561) mutant (Figure 8).
FIGURE 8:
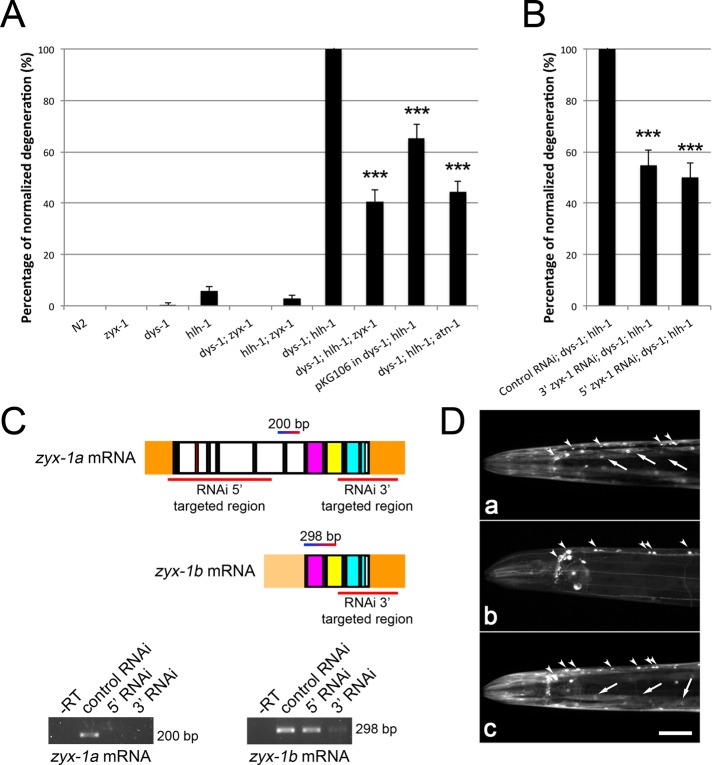
Quantification of muscle degeneration in different genetic contexts. Body-wall muscle cells were observed on adults (L4 stage plus 3 d), fixed, and stained with phalloidin–rhodamine. Abnormal muscle cells were quantified in two of the four muscle quadrants in each worm. The genetic contexts are indicated on the abscissa, and ordinates show percentages of degeneration with respect to the degeneration observed in the dys-1(cx18); hlh-1(cc561) double mutant (A) and the dys-1(cx18); hlh-1(cc561) double mutant fed with HT115 bacteria carrying the empty L4440 plasmid (control RNAi) (B), which were both set at 100%. For A and B, error bars correspond to SEM (standard error of the mean). Significant reduction of muscle degeneration compared with the corresponding dys-1; hlh-1 control strain is indicated with asterisks (***p < 0.001). Statistical tests were performed with Mann–Whitney test. (C) zyx-1a and zyx-1b mRNAs are presented with the RNAi-targeted regions in red, as well as with the sequence amplified by RT-PCR (200 and 298 base pairs, respectively). Agarose gels of the resulting PCR products are shown below. The left gel corresponds to the zyx-1a mRNA–specific RT-PCRs, the right gel to the zyx-1b mRNA–specific RT-PCRs. –RT, negative controls with PCR carried out on RT-untreated total RNA. Other lanes: RT-PCRs carried out on total RNA extracts from wild-type worms treated by the indicated RNAi (control or 5′ or 3′ RNAi). Control RNAi corresponds to wild-type worms fed with HT115 bacteria carrying the empty L4440 plasmid. Note the absence of amplification of the zyx-1a mRNA after both treatments with RNAi and a faint amplification of the zyx-1b mRNA after treatment with 3′ RNAi. (D) RNAi experiments targeting zyx-1 mRNA sequences in zyx-1::gfp transgenic worms. The images show the anterior region of living zyx-1::gfp–expressing worms carrying the pKG106 transgene. (a) Transgenic worm on control RNAi. The ZYX-1–GFP protein is detected in body-wall muscle cells (arrows) and in the ventral nerve cord (arrowheads). (b) Transgenic worm subjected to RNAi-mediated down-regulation of both zyx-1 isoforms (3′ RNAi). Note the complete extinction of the ZYX-1–GFP signal in body-wall muscles. The transgene is still detected in the ventral nerve cord (arrowheads). (c) Worm subjected to specific RNAi-mediated down-regulation of the zyx-1a isoform (5′ RNAi). No obvious difference of the ZYX-1–GFP signal is observed compared with control worms. Scale bar, 50 μm for all images.
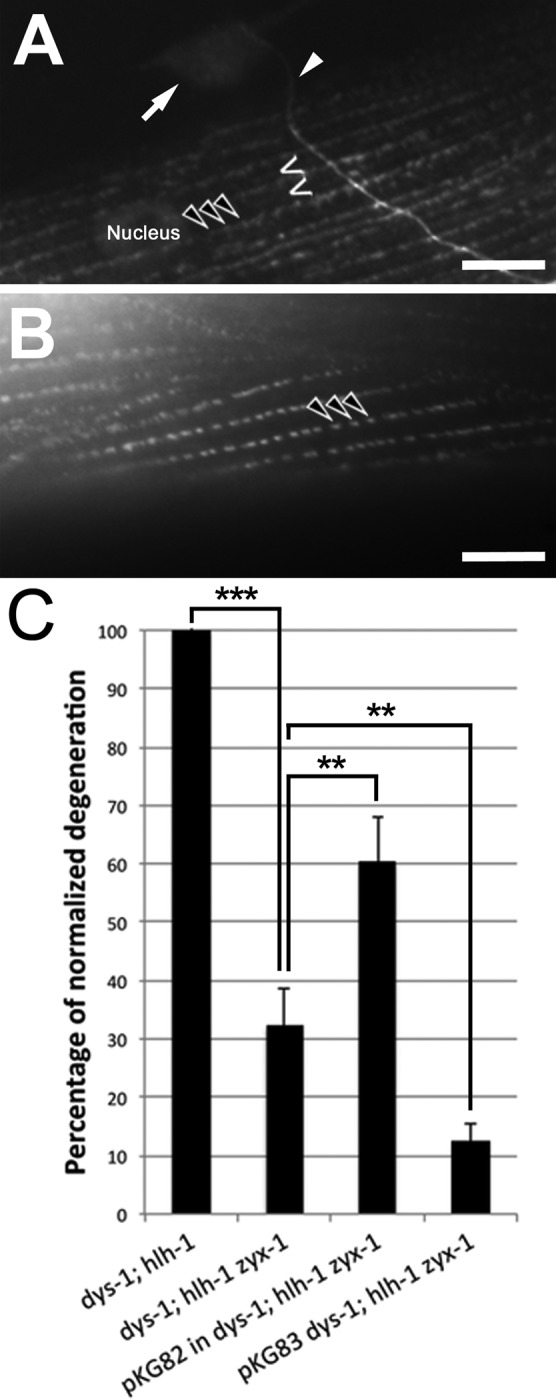
The pKG106 transgene (Figure 1) was introduced in the dystrophic dys-1(cx18); hlh-1(cc561) mutant background in order to overexpress the ZYX-1 protein. These transgenic worms exhibited ∼35% less muscle degeneration compared with nontransgenic dystrophic dys-1(cx-18); hlh-1(cc561) worms. This observation suggests that the overexpression of zyx-1 is beneficial to dystrophin-dependent muscle degeneration. We then genetically introduced the zyx-1(gk190) loss-of-function allele into the hlh-1(cc561) and the dys-1(cx18) mutant background. The zyx-1(gk190) hlh-1(cc561) or the zyx-1(gk190); dys-1(cx18) double mutant did not present obvious muscle degeneration (Figure 8A). However, the introduction of the zyx-1(gk190) allele in the dys-1(cx-18); hlh-1(cc561) double mutant background led to a strong reduction of muscle degeneration. The dys-1(cx-18); hlh-1(cc561) zyx-1(gk190) triple mutant exhibited ∼60% fewer degenerated muscle cells compared with the dys-1(cx-18); hlh-1(cc561) double mutant (Figure 8A; representative pictures of phalloidin-rhodamine–stained muscles of the dys-1(cx-18); hlh-1(cc561) double mutant and the dys-1(cx-18); hlh-1(cc561) zyx-1(gk190) triple mutant are shown in Supplemental Figure S3). Thus the overexpression of zyx-1 and the loss of zyx-1 function are both beneficial to dystrophin-dependent muscle degeneration in the dystrophic dys-1(cx-18); hlh-1(cc561) genetic background; the beneficial effect, however, is more important when zyx-1 function is lost (Figure 8A). We confirmed this result using systemic RNAi-mediated inactivation of the zyx-1 gene by targeting the 3′ sequence of the mRNA in the dys-1(cx18); hlh-1(cc561) double mutant. The muscle degeneration was significantly reduced by ∼45% compared with untreated dys-1(cx-18); hlh-1(cc561) control animals (Figure 8B), indicating that ZYX-1 actively contributes to muscle degeneration in dystrophic dys-1(cx-18); hlh-1(cc561) worms.
The role of ZYX-1 in muscle degeneration depends on its muscular localization
Because ZYX-1 is present in both muscle cells and motor neurons, we further asked whether the observed effect on muscle degeneration was due to the loss of zyx-1 function in muscles and/or motor neurons. Worms carrying the zyx-1::gfp pKG106 transgene were exposed to RNAi-mediated inactivation of the zyx-1 gene. In transgenic worms grown on control RNAi, the ZYX-1–GFP signal was detected as expected in body-wall muscle cells and in neurons (Figure 8D, a). In transgenic worms treated with systemic zyx-1 RNAi targeting the 3′ sequence of the mRNA, the ZYX-1–GFP signal could no longer be detected in body-wall muscle cells, whereas the neuronal signal was still present (Figure 8D, b). This observation, taken together with the previously reported resistance of neurons to systemic RNAi (Maeda et al., 2001; Timmons et al., 2001; Kamath et al., 2003), indicates that the loss of ZYX-1 in muscles, but not in neurons, is responsible for the reduction of muscle degeneration observed in dystrophic dys-1(cx-18); hlh-1(cc561) worms exposed to zyx-1 RNAi.
In addition, we showed earlier that ZYX-1 is mislocalized in the muscles of the atn-1(ok84) mutant. We therefore wondered whether the loss of atn-1 function would be beneficial to dystrophin-dependent muscle degeneration. Thus we introduced the atn-1(ok84) allele in the dys-1(cx-18); hlh-1(cc561) double-mutant background. The dys-1(cx-18); hlh-1(cc561); atn-1(ok84) triple mutant exhibited nearly 60% less muscle degeneration compared with the dys-1(cx-18); hlh-1(cc561) double mutant (Figure 8A). This observation indicates a functional link between ATN-1 and ZYX-1 and suggests that proper ZYX-1 localization at the dense body may be required for muscle degeneration to occur.
The ZYX-1a and ZYX-1b isoforms play distinct roles in dystrophin-dependent muscle degeneration
To decipher the dual effect of zyx-1 overexpression and loss of function in the dystrophic dys-1(cx-18); hlh-1(cc561) mutant background, we investigated whether and how both isoforms are involved in the muscle degenerative process.
Western blot analysis using an antibody directed against GFP performed on protein extracts of transgenic worms carrying either the pKG001 or the pKG106 constructs revealed the production of two different isoforms: one weakly expressed, long isoform and one strongly expressed, short isoform (Supplemental Figure S4A). The size of these proteins suggests that they correspond, respectively, to the predicted ZYX-1a and ZYX-1b isoforms fused to GFP (WormBase). Western blots performed with the monoclonal antibody directed against ZYX-1 revealed the expression of a small protein (∼23 kDa) in wild-type worms, which was absent in zyx-1(gk190) mutants. This protein is likely to correspond to the ZYX-1b isoform (200 amino acids; Supplemental Figure S4B). With this monoclonal ZYX-1 antibody, however, we were unable to detect the long ZYX-1a isoform. Reverse transcription (RT)-PCR and real-time quantitative PCR (RT-qPCR) performed on RNA extracts of wild-type worms confirmed the expression of zyx-1a and zyx-1b mRNAs (Figure 8C and Supplemental S4). This suggests that wild-type worms produce two isoforms, ZYX-1a and ZYX-1b; the ZYX-1a isoform, however, is much less abundant than the ZYX-1b isoform. Because the amounts of zyx-1a and zyx-1b mRNAs are not significantly different from each other (Supplemental Figure S4D), we hypothesize that the ZYX-1a protein may be translated at a lower level or be less stable than the ZYX-1b isoform.
The mRNAs of the ZYX-1a and ZYX-1b isoforms share in common the LIM domains encoding the 3′ region (Figure 1). We thus could not specifically inactivate the short isoform without affecting the expression of the long isoform. Semiquantitative RT-PCRs performed on worms subjected to systemic RNAi targeting the 3′ sequence of the mRNA revealed strongly reduced amounts of both zyx-1a and the zyx-1b mRNAs. Specific targeting of the 5′ coding sequence of the long zyx-1a transcripts specifically down-regulated the amount of the zyx-1a mRNA without affecting the amount of the zyx-1b mRNA (Figure 8C).
The specific RNAi-mediated down-regulation of the zyx-1a mRNA in the dystrophic dys-1(cx-18); hlh-1(cc561) mutant led to a reduction of muscle degeneration by ∼50% compared with untreated dys-1(cx-18); hlh-1(cc561) control animals and is almost identical to that observed after RNAi-mediated down-regulation of both zyx-1a and zyx-1b mRNAs (Figure 8B). This result suggests that only the long ZYX-1a isoform contributes to muscle degeneration in the dys-1(cx-18); hlh-1(cc561) mutant background.
To further test this hypothesis, we constructed two transgenes coding either the long ZYX-1a (pKG82) or the short ZYX-1b (pKG83) isoform (Figure 1). In transgenic worms carrying the pKG83 transgene, the ZYX-1b–hemagglutinin (HA) protein presented the same localization in muscle and neuronal tissues as the ZYX-1–GFP fusion protein in transgenic worms carrying the pKG001 and pKG106 transgenes (Figure 9A). In contrast, in transgenic worms carrying the pKG82 transgene the ZYX-1a–HA protein could be detected only in body-wall muscle cells with a localization that seemed restricted to dense bodies (Figure 9B). Thus the long and short isoforms exhibit different expression patterns. Both are coexpressed in body-wall muscle, where they colocalize at dense bodies. Accordingly, we observed that the specific RNAi-mediated down-regulation of the ZYX-1a isoform in transgenic worms expressing both isoforms (pKG106 transgene) did not abolish the muscular ZYX-1–GFP signal, suggesting that this remaining signal corresponds to the short ZYX-1b–GFP isoform (Figure 8D, c).
FIGURE 9:
Expression of isoform-specific zyx-1::ha transgenes in wild-type worms. (A) Transgenic worms expressing the pKG83 zyx-1b::ha transgene encoding the short ZYX-1b isoform. The ZYX-1b–HA fusion protein is detected by the use of an antibody directed against the HA tag in muscle cells with a subcellular localization in the nucleus, at dense bodies (empty arrowheads) and M-lines (chevrons), and in cell bodies (filled arrows) and axons of nerve cells (filled arrowhead). (B) Transgenic worms expressing the pKG82 zyx-1a::ha transgene encoding the ZYX-1a isoform. The ZYX-1a–HA fusion is detected in muscle cells with a subcellular localization restricted to dense bodies (open arrowheads). Scale bar, 5 μm for both images. (C) Quantification of muscle degeneration in the dys-1(cx18); hlh-1(cc561ts) zyx-1(gk190) triple mutant and in the triple mutant overexpressing isoform-specific zyx-1 transgenes (pKG82 and pKG83). Muscle degeneration is normalized, as in Figure 8, to that of the dys-1(cx18); hlh-1(cc561ts) double mutant set at 100%. Levels of muscle degeneration were compared between strains linked by brackets, and significant differences are indicated by asterisks (**p < 0.01; ***p < 0.001). Statistical tests were performed with Mann–Whitney test.
To investigate the role of the two ZYX-1 isoforms and their contribution to muscle degeneration, we introduced each transgene into the dys-1(cx18); hlh-1(cc561) zyx-1(gk190) triple mutant. As expected, the overexpression of the long isoform (pKG82) increased the muscle degeneration phenotype compared with the triple mutant, reaching 60% of muscle degeneration with respect to the dystrophic dys-1(cx18); hlh-1(cc561) mutant (Figure 9C). The overexpression of the short isoform (pKG83), in contrast, reduced muscle degeneration to 10% compared with the dystrophic mutant. In the genetic context of the dys-1(cx18); hlh-1(cc561) zyx-1(gk190) triple mutant, the overexpression of the long ZYX-1a isoform thus seemed to abolish the beneficial effect of the zyx-1(gk190) mutation on muscle degeneration, whereas the short isoform instead enhanced the beneficial effect.
DISCUSSION
Phylogeny of the C. elegans ZYX-1 protein
We demonstrated that ZYX-1 is the unique representative of the zyxin-like protein family in C. elegans. Our phylogenetic analysis indicated that zyxin family proteins are distributed into two distinct subfamilies: the ajuba and the zyxin subfamilies. Because these two subfamilies exist in both deuterostomes (amphioxus, zebrafish, and human) and protostomes (fruit fly, oyster, and the nematode Ascaris), we propose that two zyxin-like proteins were present in their last common ancestor and probably in the bilaterian ancestor. During the evolution of vertebrates, the genes encoding the two ancestor zyxin-like proteins were duplicated several times in a pattern that coincides with the two successive whole-genome duplications (Dehal and Boore, 2005). It is noteworthy that seven of the eight paralogues were maintained in the vertebrate lineage, suggesting neofunctionalization or subfunctionalization of these proteins (Conant and Wolfe, 2008; Kassahn et al., 2009) or dosage compensation requirements (Makino and McLysaght, 2010).
In protostomes and amphioxus (in which no genome duplications occurred), the two zyxin-like proteins were maintained, one in the ajuba subfamily and the other in the zyxin subfamily. Each of these zyxin-like proteins can be considered as the functional orthologues of, respectively, vertebrate ajuba-LIMD1-WTIP1 and zyxin-migfilin-TRIP6-LPP. The nematode C. elegans represents an exception since, in contrast to another nematode, A. suum, it has a unique zyxin protein belonging to the zyxin subfamily and lost the zyxin-like protein from the ajuba subfamily. This suggests that ZYX-1 fulfills the zyxin-subfamily functions in C. elegans and might compensate for the loss of ajuba-subfamily functions.
Function of ZYX-1 in striated muscle cells
We focused our analysis on ZYX-1 in body-wall muscle cells, where it is localized at dense bodies, M-lines, and nuclei. In C. elegans, muscle adhesion structures (i.e., dense bodies and M-lines) have been suggested not only to play a structural role, but also to serve as signaling platforms within the muscle cell (reviewed in Lecroisey et al., 2007; Qadota and Benian, 2010). Our results suggest that ZYX-1 is involved in these two functions.
ZYX-1 interacts with the dense body proteins ATN-1, ALP-1, and DEB-1, respectively orthologous to vertebrate α-actinin, ALP/enigma, and vinculin (Barstead and Waterston, 1989, 1991; McKeown et al., 2006). This suggests that ZYX-1 participates—although indirectly—in the anchoring of actin filaments to dense bodies via its interaction with these proteins known to cross-link actin filaments. ZYX-1 probably has a functional link with ATN-1, since the loss of ATN-1 leads to ZYX-1 mislocalization and to the modification of the dynamics of ZYX-1 at dense bodies. In addition, loss of either zyx-1 or atn-1 function in the dystrophic dys-1; hlh-1 mutant leads to a reduction of muscle degeneration, and atn-1 and zyx-1 loss-of-function mutants exhibit the same body-bending phenotype (Moulder et al., 2010; Nahabedian et al., 2012).
ZYX-1 was found not to directly interact with the C-terminal domain of DYS-1. However, our results indicate that the function of ZYX-1 may be linked to the function of DYS-1. It is therefore interesting to note that, like ZYX-1, the C-terminal domain of DYS-1 interacts with DEB-1 (our unpublished results). The DYS-1 protein is abundantly expressed in striated body-wall muscles and localizes in broad bands overlapping actin containing thin filaments, which are anchored at dense bodies (our unpublished results and Supplemental Figure S2). In addition, the localization of both DYS-1 and ZYX-1 partially overlaps with the dense body protein DEB-1 (Supplemental Figure S2). Thus ZYX-1 and DYS-1 may be physically linked at dense bodies through their common binding partner DEB-1.
No interaction was detected between ZYX-1 and UNC-112, PAT-4, PAT-6, or UNC-97. These four conserved proteins form a complex that is directly involved in the anchorage of dense bodies and M-lines to the extracellular matrix (Qadota and Benian, 2010). Consequently, ZYX-1 may not directly participate in the adhesive function to the extracellular matrix of these two sarcomeric structures.
A signaling function for the ZYX-1 protein within the muscle cell is suggested by its interaction with 1) UIG-1 (Hikita et al., 2005), a Cdc42-specific guanine nucleotide exchange factor (GEF), suggesting participation of ZYX-1 in Cdc42 signaling during sarcomere maintenance (in fact, participation of ZYX-1 in sarcomeric integrin attachment complex maintenance was recently proposed; Etheridge et al., 2012); 2) LIM-8 and LIM-9 (FHL), two other LIM proteins of M-lines both binding UNC-97 (PINCH; Qadota and Benian, 2010), providing a basis for a potential mechanotransducing function in muscle cells as described for zyxin in smooth muscles and nonmuscle cells (Cattaruzza et al., 2004; Hirata et al., 2008; Wolfenson et al., 2011); and 3) SCPL-1, a CTD-type phosphatase, which itself interacts with the giant protein UNC-89 (obscurin; Qadota et al., 2008; Xiong et al., 2009).
Our FRAP experiments also agree with a signaling function for ZYX-1. Indeed, in C. elegans muscle, ZYX-1 seems to shuttle between adhesion complexes (i.e., dense bodies and M-lines) and the nucleus, in accordance with observations of vertebrate zyxin shuttling (Nix and Beckerle, 1997; Nix et al., 2001; Wang and Gilmore, 2003; Hervy et al., 2006). In addition, gold beads observed by immuno–electron microscopy in the cytoplasm between the dense body and the nucleus may correspond to staining of shuttling ZYX-1–GFP proteins (Figure 5I).
Previous studies using FRAP experiments on C. elegans body-wall muscle cells suggested that the exchange rates of proteins may reflect the position of the corresponding protein within muscle adhesion structures, peripheral proteins like T03G6.3 or UNC-95 being more dynamic than central proteins, providing a more solid point of anchorage, like UNC-112 (Ghosh and Hope, 2010). The high rate of replacement observed in our experiments strongly suggests localization of ZYX-1–GFP at the periphery of sarcomeric adhesion structures. High dynamics and mobility were also reported for vertebrate zyxin, suggesting that this protein is continuously and rapidly (within seconds) recruited from the cytosol into focal adhesions and able to “hop” between adjacent focal adhesions (Welman et al., 2010).
Differential involvement of the ZYX-1a and ZYX-1b isoforms in dystrophin-dependent muscle degeneration
We showed that the loss of function, as well as overexpression, of the zyx-1 gene reduced dystrophin-dependent muscle degeneration. This reveals a dual and paradoxical function of ZYX-1 in the dystrophic dys-1(cx18); hlh-1(cc561) genetic context. Previous examples were reported in which the same C. elegans muscle phenotypes can be obtained by loss or gain of function: unc-96 (Mercer et al., 2006; Qadota et al., 2007), unc-45 (Barral et al., 1998; Landsverk et al., 2007), and mel-26 and mei-1 (Wilson et al., 2012).
The zyx-1 gene produces at least two isoforms, a long, ZYX-1a, and a short, ZYX-1b, isoform, which are differently involved in dystrophin-dependent muscle degeneration. We propose that their functional domains dictate their different roles in muscle degeneration. LIM domains in vertebrate zyxins were shown to be essential for targeting the protein to focal adhesions and stress fibers. PRRs, α-actinin and Ena/Vasp–binding sites located in the N-terminal region, can independently direct the protein to the nucleus, whereas the NES is required to export zyxin from the nucleus; however, at steady state, no zyxin is detected in cell nuclei (Nix et al., 2001; Hoffman et al., 2012).
We hypothesized that in C. elegans muscle cells, the LIM domains would have a mechanical function to stabilize dense bodies and M-lines via interactions with several components of these structures. The N-terminal region, present only in the long, ZYX-1a isoform, would have a signaling function, sensing cytoplasmic stress such as disorganized sarcomeres, and, once within the nucleus, would activate transcription of genes involved in repair of the damage and if necessary, induce a death signal. This hypothesis would explain why loss and gain of function of the zyx-1 gene in dystrophic worms are both beneficial for muscles.
In the case of zyx-1 loss of function, the absence of the ZYX-1a isoform would prevent the induction of a death signal. Of interest, dystrophic dys-1(cx18); hlh-1(cc561) double mutants overexpressing either the full-length ZYX-1a protein or the N-terminal domain of the ZYX-1a isoform lacking the LIM-domains are not viable (our unpublished results), arguing for the presence of a sequence in the N-terminal region able to induce a death response. The overexpression of the N-terminal region of vertebrate zyxin was also shown to be lethal in cultured cells (Nix et al., 2001). A death signal or survival function of zyxin was reported in cells subjected to various stresses (Cerisano et al., 2004; Kato et al., 2005; Hervy et al., 2010). Although there is no evidence for a direct binding of LIM proteins to DNA (Li et al., 2012), vertebrate zyxin has been shown to interact with transcription factors such as ZNF384 and ZHX1 (Yamada et al., 2003; Janssen and Marynen, 2006). In C. elegans, the ZYX-1a protein might modify gene expression through an interaction with transcription factors involved in the stress-induced gene expression.
In the case of the overexpression of both ZYX-1 isoforms in the muscle of dystrophic worms, the large amounts of LIM domains would stabilize dense bodies and M-lines, thereby reducing the ZYX-1a death signal. LIM domains were described to act as negative regulators of zyxin–VASP complexes by intramolecular interactions between head and tail of the protein, preventing subsequent binding to other partners (Moody et al., 2009). Thus LIM domains of the very abundant short, ZYX-1b isoform could also interact with those of the long isoform, forming a heterodimer, and could exhibit a dominant-negative effect by inhibiting the signaling function of the ZYX-1a isoform. Accordingly, a dominant-negative role of the LIM domains has been suggested for zyxin function in cell–cell adhesion (Hansen and Beckerle, 2006).
Zyxin, cell survival, and muscle pathologies
Members of the zyxin family were suggested to act as sensors at cell adhesion sites and to serve as putative transcriptional coregulators in different cell survival–signaling processes. In humans, several mutations affecting genes encoding LIM proteins, most notably the FHL-1 and ALP-1 proteins, cause muscular dystrophies (Gueneau et al., 2009; Ohsawa et al., 2011; reviewed in Schessl et al., 2011). However, none of the members of the zyxin family has been associated with a human myopathy. In C. elegans, the genetic inactivation of the zyx-1 gene does not cause any obvious structural muscle phenotype, but the loss of the ZYX-1 protein is beneficial to dystrophin-dependent muscle degeneration, thus suggesting for the first time that this protein actively contributes to the muscle degenerative process. With regard to our results, it would be of great interest to analyze the potential contribution of zyxin-family proteins to dystrophin-dependent muscle degeneration in mammalian models for DMD to establish whether targeting of zyxin proteins would be effective strategies for treatment of DMD.
MATERIALS AND METHODS
C. elegans strains
C. elegans strains were cultured as described (Brenner, 1974) and grown at 15°C on OP50 bacteria. The N2 Bristol strain was used as the wild-type control. The PD4613 hlh-1(cc561ts) strain (Harfe et al., 1998) was obtained from B. D. Harfe (University of Florida, Gainesville, FL) and A. Fire (Stanford University School of Medicine, Stanford, CA). The LS292 dys-1(cx18) strain was described in Bessou et al. (1998), the LS587 dys-1(cx18); hlh-1(cc561) and the LS396 dyc-1(cx32) strains were described in Gieseler et al. (2000), the VC299 zyx-1(gk190), RB1812 atn-1(ok84), RB978 uig-1(ok884), VC654 lim-8(ok941), VC209 lim-9(gk106), and VC612 scpl-1(gk283) strains were obtained from the Caenorhabditis Genetic Center (University of Minnesota, St. Paul, MN), and the alp-1(tm1137) allele was obtained from the Mitani laboratory at the Tokyo Women's Medical University School of Medicine (Tokyo, Japan). Classic genetics methods were used to construct double and triple mutants: LS898 hlh-1(cc561) zyx-1(gk190), LS901 dys-1(cx18); hlh-1(cc561); atn-1(ok84), LS925 dys-1(cx18; zyx-1(gk190), and LS936 dys-1(cx18); hlh-1(cc561) zyx-1(gk190). The reference transgenic strains presented in this study are LS770 pKG001(zyx-1::gfp); pRF4, LS918 pKG001(zyx-1::gfp); pRF4; atn-1(ok84), LS1028 cxIs19(pKG001 zyx-1::gfp; pKP13), LS1128 pKG82(zyx-1a::HA), pCFJ(Punc-122::gfp), LS1130 pKG83(zyx-1b::ha), pCFJ(Punc-122::gfp), LS1056 cxIs19(pKG001 zyx-1::gfp; pKP13); dys-1(cx18); hlh-1(cc561ts), LS1150 pKG106 (zyx-1::ha::gfp), pCFJ(Punc-122::gfp), LS1176 pKG106(zyx-1::ha::gfp), pCFJ(Punc-122::gfp); dys-1(cx18); hlh-1(cc561), KAG124 pKG83(zyx-1b::ha), pCFJ(Punc-122::gfp); dys-1(cx18); hlh-1(cc561) zyx-1(gk190), and KAG126 pKG82(zyx-1a::ha), pCFJ(Punc-122::gfp); dys-1(cx18); hlh-1(cc561) zyx-1(gk190).
Sequence comparison and phylogenetic analyses
Human zyxin-family member sequences were retrieved from UniProt (www.uniprot.org). They were aligned using MUSCLE, version 3.7 (Edgar, 2004), and used as seed for a PsiBLAST (Altschul et al., 1997) search against the WormBase C. elegans protein database (www.wormbase.org, version WB234). The five proteins with the highest scores were retained and used as seeds for BLASTp (Altschul et al., 1997) searches against RefSeq (www.ncbi.nlm.nih.gov/refseq/ [accessed November 2012]; Pruitt et al., 2009) H. sapiens proteins.
This procedure was repeated to find zyxin-family proteins in the proteomes of D. rerio (RefSeq), B. floridae (UniProt), A. suum (WormBase, version 233), D. melanogaster (RefSeq), and C. gigas (http://gigadb.org/pacific_oyster/). A. suum protein GS_14729 was discarded, as it is likely a pseudogene of GS 19714, as suggested by its increased substitution rate and by the absence of hits when using tBLASTn in the National Center for Biotechnology Information Expressed Sequence Tag database.
The multiple alignment of all zyxin sequences was computed using MUSCLE and refined using BMGE (Criscuolo and Gribaldo, 2010). Maximum-likelihood tree estimation and parametric bootstrap were performed using RAXML, version 7.2 (Stamatakis, 2006), under model PROTCATLG, with 100 bootstrap replicates.
zyx-1 constructs and transgenesis
The different zyx-1 constructs are presented in Figure 1. The plasmid pKG001 was described in Lecroisey et al. (2008). Briefly, this plasmid contains a 16,928–base pair PstI–NcoI fragment of cosmid F42G3 encompassing the genomic sequence of the zyx-1 gene cloned into the PstI–MscI sites of pPD95.75 plasmid (kindly provided by A. Fire) in frame with the gfp-encoding sequence. This fragment starts 4171 base pairs upstream of the predicted initiation codon of the zyx-1a isoform–containing promoter sequences and ends at the NcoI site within exon 9, thus lacking the coding sequences of the 50 most C-terminal amino acids of the ZYX-1 protein (including the 39 C-terminal amino acids of the third LIM domain).
The pKG82 construct was obtained in different steps. 1) The pPD118.20 plasmid (kindly provided by A. Fire) was digested with AgeI and EcoRI to removed the GFP-coding sequence. The resulting plasmid was named pKG74. 2) The zyx-1a cDNA was amplified by PCR on the yk247e1 zyx-1a cDNA clone (kindly provided by Y. Kohara, National Institute of Genetics, Mishima, Japan), using forward primer tta agg tac cct tgg gtc gtg ctc gct cct tc, which contains a KpnI site, and reverse primer tta agg tac ctt aag cgt aat ctg gaa cat cgt atg ggt acg tgg agc tga cca cgc ggag, containing an HA-coding sequence followed by a stop codon and a KpnI site. This fragment was inserted into the KpnI site of pKG74, resulting in plasmid pKG79. 3) A 11,471–base pair SgrAI–XhoI fragment from cosmid F42G4 containing 7618 base pairs of upstream regulatory and promoter sequences and 3853 base pairs encompassing the genomic zyx-1a sequence from 1 to exon 5 was inserted into the XmaI and XhoI sites of pBluescript, resulting in plasmid pKG80. 4) A 1523–base pair XhoI–ApaI fragment from plasmid pKG79 was inserted into the XhoI and ApaI sites of pKG80, resulting in plasmid pKG82 (Figure 1).
The pKG83 plasmid was obtained in two steps: 1) a 12,182–base pair NheI–FspI genomic fragment from cosmid F42G4 starting in intron 4 and ending 1762 base pairs after the zyx-1 stop codon was inserted into the XbaI–EcoRV sites of pBluescript, resulting in plasmid pKG81; 2) a 659–base pair NcoI–ApaI zyx-1 cDNA (fused to the HA tag–coding sequence) fragment from plasmid pKG79 was inserted into the NcoI and ApaI sites of pKG80, resulting in plasmid pKG83 (Figure 1).
The pKG106 plasmid was constructed as follows. 1) A 1076–base pair PCR fragment was amplified on pKG83 using the forward primer ctc act gga gaa gtg tacc and the reverse primer tta agc tag cag cgt aat ctg gaa cat cgt atg ggta containing HA tag–encoding sequence and an NheI site. An NcoI and NheI fragment of this PCR product replaced the NcoI–NheI fragment of pKG83. The aim of this step was to suppress the stop codon present in pKG83 after the HA tag. The resulting plasmid was named pKG98. 2) A 869–base pair GFP-coding fragment was amplified on plasmid pPD118.20 using forward primer tta agc tag cat gag taa agg aga aga ac and reverse primer tta agc tag cgc cat gtg taa tcc cagc (both primers containing an NheI site) and inserted into the NheI site of pKG98, resulting in plasmid pKG103. 3) The NruI–ApaI fragment of pKG82 was replaced by the NruI–ApaI fragment from pKG103 (corresponding to the 3′ genomic sequence including introns 6–8, as well as exons 6–10 with the HA tag and GFP fused in frame at the end of exon 10. The resulting plasmid was named pKG106 (Figure 1).
The constructs of interest were injected into gonads of young adult wild-type or zyx-1(gk190) mutant animals using standard procedures (Mello and Fire, 1995) at a concentration of 10 ng/μl, along with the marker pCFJ68 (Punc-122::gfp) plasmid (Addgene, Cambridge, MA; expressed in coelomocytes) at a concentration of 25 ng/μl; pKG001 was coinjected with the marker pKP13 (100 ng/μl) or pRF4 (150 ng/μl; Ségalat et al., 1995; Mello and Fire, 1995). At least three independent stable transgenic lines were generated for each transgene. The pKG001 transgene was integrated into the genome by ultraviolet irradiation of L4 larvae at 0015 J/cm2. Fluorescent animals of the F1 and F2 generation were individually grown, and animals homozygous for the integrated transgene were selected in the F3 generation. Classic genetic procedures were used to introduce the pKG001and pKG106 transgenes into the dys-1(cx18); hlh-1(cc561) or atn-1(ok84) mutant background or the pKG82, pKG83 transgenes into the dys-1(cx18); hlh-1(cc561) zyx-1 mutant background.
Immunostaining and fluorescence microscopy
Worms were fixed by the constant-spring method unless stated otherwise (Benian et al., 1996). Transgenic zyx-1::gfp worms were observed either directly after fixation on a fluorescence Zeiss LSM510 microscope (Carl Zeiss, Jena, Germany) or stained with a GFP rabbit antibody (1/200; Invitrogen, Carlsbad, CA). Anti-rabbit Alexa 488 (Invitrogen) secondary antibody was used at 1/1000 dilution. Transgenic zyx-1::ha worms were stained with a HA primary mouse antibody (1/100; Santa Cruz Biotechnology, Santa Cruz, CA) and then secondary anti-mouse Alexa 548 (1/1000; Invitrogen).
A ZYX-1 monoclonal antibody was produced by BIOTEM (Apprieu, France) against the peptide CALCSKPIVPQDGEKESVRV of the first part of the third LIM domain (Figure 1), amino acids 531–550 in exon 9 of the ZYX-1-a isoform (F42G4.3a), or amino acids 127–146 in exon 4 of the ZYX-1-b isoform (F42G4.3b). This antibody was used on wild-type worms at a dilution of 1/50. Secondary anti-mouse antibody, conjugated to streptavidin (1/250), and tertiary fluorescein-conjugated biotin (1/400; Vector Laboratories, Burlingame, CA) were used to reveal the ZYX-1 antibody staining.
For costaining of ZYX-1–GFP and ATN-1 or DEB-1, a GFP rabbit antibody (1/200; Invitrogen) was used, along with an ATN-1 antibody (MH35 1/200; kindly provided by Pamela Hoppe, Western Michigan University, Kalamazoo, MI) and a DEB-1 antibody (MH24, 1/200; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA; Francis and Waterston, 1985). A secondary anti-rabbit Alexa 488 (Invitrogen) was used to reveal the GFP rabbit antibody (1/1000). Secondary anti-mouse Alexa 548 (Invitrogen) was used to reveal the MH35 or MH24 (1/1000) staining. Worms were mounted on a glass slide with a coverslip together with 40-μm microbeads (Thermo Scientific, Fremont, CA) and antifading Prolong mounting medium (Invitrogen).
Acquisition of images was done on a Zeiss LSM510 confocal microscope using a 63× oil immersion objective with the LSM software. Image postprocessing was performed in ImageJ (National Institutes of Health, Bethesda, MD), with the Grays LUT in Figure 5, A–C and E–G, and Fire LUT in Figure 5, D and H.
For immunolocalization of ZYX-1 interactors, wild-type and zyx-1(gk190) worms were fixed by the Nonet method (Nonet et al., 1993), and antibodies to the following proteins were used: ATN-1 (MH35, 1/200 dilution), ALP-1 (B74 at 1/100; Han and Beckerle, 2009), DEB-1 (MH24, dilution 1/200), DYC-1 (dilution 1/20; Lecroisey et al., 2008), UIG-1 (dilution 1/100; kindly provided by Kozo Kaibuchi, Nagoya University, Nagoya, Japan), LIM-8 (dilution 1/100; Qadota et al., 2007), LIM-9 (dilution 1/100; Qadota et al., 2007), and SCPL-1 antibodies (Benian-17 at 1/100 dilution; Qadota et al., 2008). As markers of M-lines and dense bodies, we used a UNC-95 antibody (Benian-13 at 1/100; Qadota et al., 2007). Wild-type, zyx-1(gk190), atn-1(ok84), alp-1(tm1137), dyc-1(cx32), uig-1(ok884), lim-8(ok941), lim-9(gk106), and scpl-1(ok1080) worms were fixed by the constant-spring method (Benian et al., 1996) to determine the effect of these mutations on ZYX-1 localization, as detected by a ZYX-1 antibody (at 1/100; this study). Secondary antibodies, purchased from Invitrogen, were anti-rabbit Alexa 488, anti-mouse Alexa 594, and anti-rat Alexa 594, each used at 1/200 dilution. Stained samples were mounted on a glass slide with a coverslip containing mounting solution (20 mM Tris, pH 8.0), 0.2 M 1,4-diazabicyclo[2.2.2]octane, and 90% glycerol). Images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope and an Apochromat 63×/1.4 oil objective in 2.5× zoom mode. The color balances of the images were adjusted by using Photoshop (Adobe, San Jose, CA)
Electron microscopy
Electron microscopy analysis was performed on zyx-1::gfp worms containing the pKG001 transgene fixed by high-pressure freezing as previously described (Liegeois et al., 2007). The next steps were processed as previously described (Lecroisey et al., 2008).
Fluorescence recovery after photobleaching experiments
FRAP was performed on transgenic wild-type or atn-1(ok84) worms carrying the zyx-1::gfp (pKG001) transgene immobilized by capillarity between slice and large coverslip with 20 μl of M9 buffer under a Leica spectral confocal microscope (TCS SP5 AOBSDM6000; Leica Microsystems, Wetzlar, Germany). Five images were taken before bleaching, and then the region of interest was bleached with the 488-nm laser at full power; subsequent scans were taken at 5% of full laser power. Sarcomeric zones were bleached 20 times and nuclei 8 times to reduce significantly the amount of fluorescence. For each bleaching experiment, 20 frames were acquired every 0.5 s, followed by 30 frames acquired every 2 s. Ten bleaching experiments were performed on sarcomeric zones of zyx-1::gfp–expressing wild-type or atn-1(ok84) worms and four on muscle nuclei of wild-type zyx-1::gfp–expressing worms. One relevant experience performed on zyx-1::gfp–expressing wild-type worms was presented for sarcomeric zones and nucleus bleaching experiments in Figure 6. The five most representative data sets of sarcomeric zone bleaching experiments on zyx-1::gfp–expressing wild-type worms and atn-1(ok84) mutants are presented in Figure 7. Fluorescence intensity was measured using ImageJ software and analyzed using Igor (WaveMetrics, Portland, OR) software. Relative fluorescence intensity (RFI) was calculated as described in Ghosh and Hope (2010). On the y-axis, 1.0 represents the RFI of the bleached zone before bleaching steps. Time origin corresponds to the first acquisition after bleaching steps, and RFI was normalized to be zero at t = 0. Half-time recoveries and mobile fractions were calculated from the fitted curves as described (Ghosh and Hope, 2010). Statistical analyses were based on a Mann–Whitney test carried out using Excel Stat software (Microsoft, Redmond, WA). The p values for the tested data sets are presented below Figure 7N in brackets.
Yeast two-hybrid assay and screening
The yk247e1 zyx-1a cDNA was amplified using T3 and T7 primers, blunted by using the T4 DNA polymerase (New England BioLabs, Ipswich, MA), digested by SacI, and cloned into the SacI/SmaI sites of pBluescript (SK+), resulting in the plasmid named pBSC-zyx-1cDNA. The ZYX-1a bait plasmid (encoding amino acids 68–603 of ZYX-1) was obtained by cloning an AgeI–SalI zyx-1 cDNA fragment from pBSC-zyx-1cDNA into the NcoI–SalI sites of the pAS2-1 bait plasmid (Clontech, Palo Alto, CA), resulting in plasmid pAS2-1-zyx-1. The construction of prey plasmids encoding the C-terminal domain of DYS-1, the DYC-1 protein (pSEB), and ZYX-1a (pSNXX, amino acids 68–603) was described in Lecroisey et al. (2008). The ZYX-1a prey plasmid deleted for the three LIM domains (encoding amino acids 68–265 of ZYX-1a) was obtained by cloning an NdeI–XhoI zyx-1 cDNA fragment from the pAS2-1zyx-1 plasmid into the SmaI–XhoI sites of the pACT2 prey plasmid (Clontech), resulting in plasmid pACT2-zyx-1SNX. The ZYX-1 prey plasmid encoding only the three LIM domains (amino acids 265–603 of ZYX-1a) was obtained by cloning an XhoI–XhoI zyx-1 cDNA fragment from the pAS2-1-zyx-1 plasmid into the XhoI site of the pAct2 prey plasmid (Clontech), resulting in plasmid pACT2-zyx-1XX. Transformations of yeasts were performed as described (Lecroisey et al., 2008). The collection of M-line and dense body prey proteins was described in Qadota et al. (2007, 2008) and Xiong et al. (2009). All two-hybrid assays were conducted using methods described in Mackinnon et al. (2002).
RNA interference
The pBSC-zyx-1cDNA plasmid was digested with EcoRI. The 5′ cDNA EcoRI fragment containing 42 base pairs of the 5′ UTR and ZYX-1a coding sequence (amino acids and 1–303) and the 3′ cDNA EcoRI fragment containing ZYX-1 coding sequences (amino acids and 509–603 of ZYX-1a) and 257 base pairs of the 3′ UTR were cloned into the EcoRI site of the RNAi feeding vector L4440. The 5′ and 3′ sequence–targeting constructs, named pKG116 and pKG117, respectively, were transformed into HT115 Escherichia coli. RNAi was performed by feeding wild-type worms or dys-1(cx18); hlh-1(cc561ts) mutants with double-strand RNA–producing E. coli (Timmons et al., 2001). Negative and positive control experiments were performed, respectively, with the empty L4440 vector and a pos-1 transcript–targeting RNAi clone (Ahringer library; Kamath et al., 2003).
Quantification of muscle degeneration
Animals were fixed and stained 3 d after they reached the L4 stage. Fixation and phalloidin–rhodamine staining (FluoProbes; Interchim, San Pedro, CA) were performed as described (Waterston et al., 1984). Stained body-wall muscles were observed using a Zeiss Axioskop microscope. Only the two most visible quadrants of body-wall muscles in each animal were quantified (47 or 48 cells per animal), and 20 animals were scored for each genotype. The dys-1(cx18); hlh-1(cc561ts) dystrophic mutant present six to nine degenerated muscle cells, corresponding to 12–18% of total muscle cells (Gieseler et al., 2000). We normalized the number of degenerated muscle cells in this double mutant to 100% to compare to other genetic backgrounds. Statistical analyses were carried out between the strain of interest compared with the relative dys-1; hlh-1 strain (unless stated otherwise using brackets) and based on a Mann–Whitney test carried out using Excel Stat software. Significant differences between tested strains are indicated by asterisks (*p < 0.05; **p < 0.01; ***p < 0.001).
Reverse transcription-PCR and RT-qPCR analysis
Total RNA was extracted from worms fed with OP50 or HT115 E. coli. Four-day-old worms were harvested in M9 and washed twice in M9 and once in RNase-free distilled water. Two volumes of TRIzol (Invitrogen) were added to one volume of pelleted worms and frozen at −80°C overnight. While unfreezing, samples were vortexed for 10 min, 0.2 volume of chloroform was added, and they were again vortexed for 1 min. After 3 min, samples were centrifuged. Total RNA was extracted with the RNeasy Mini Binding Kit (Qiagen, Valencia, CA). RNA from each extraction was DNase treated (Invitrogen) and reverse transcripted using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) following provider's instructions. PCR were carried out on 1 μl of cDNA with the following primers: gta atc aga cag gtg ctc tgag and gta att tgc tcg cga cga gtcc for amplification of zyx-1a cDNA (Figure 8C); ctt ggg tcg tgc tcg ctc cttc and tct tac gca gac gtg ctgc for amplification of zyx-1a cDNA (Figure 8C); and aac tat ggc gga tca agaag and ttt gca gga gaa gca cac gaag for amplification of zyx-1b cDNA. PCR was stopped after 25 cycles (a 5-μl sample was removed and frozen) and run again up to 35 cycles. All 25th- and 35th-cycle PCR products were loaded on a 1.5% agarose gel. Acquisitions were processed with Image Lab Bio-Rad Software.
For RT-qPCRs total RNA from wild-type worms were extracted using the TRIzol/chloroform (Merck, Darmstadt, Germany) method. The RNeasy Mini Binding Kit was used to clean up the extracts, and 3 μg were treated with DNase (DNase I, Amp Grade; Invitrogen). A 10-ng amount of total RNA from three independent extractions was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad) and amplified in a CFX96 Realtime System C1000 Thermal Cycler (Bio-Rad) using the iQ SYBR Green SuperMix (Bio-Rad) and 1) the zyx-1a–specific forward primer act ccc atc cgg aga gat act and reverse primer cgt gct gta gtg ccg aag or 2) the zyx-1b–specific forward primer gag aga aac cac gcc ttc tta and reverse primer atc ttc ttg atc cgc cat agtt. The act-1 gene was used as housekeeping gene, using forward primer cca gga att gct gat cgt atg cag aa and reverse primer tgg aga ggg aag cga gga taga.
Western blot analysis
To detect ZYX-1–GFP fusion proteins from transgenic worms, extracts were obtained from cultured worms harvested in M9, pelleted, frozen at −80°C, and boiled in Laemmli buffer. Electrophoresis migration was carried out in a Bio-Rad MiniProtean system using 4–10% gradient gels. After transferral of the proteins on a Protan 0.45-μm polyacrylamide membrane (Whatman, Piscataway, NJ) in an electrophoretic wet chamber (Bio-Rad Mini Trans-Blot), Ponceau's red staining (Sigma-Aldrich, St. Louis, MO) was carried out, and the membrane was scanned. Then the membrane was treated as follows: TBS (Tris 20 mM, NaCl 0.14 mM, pH 7.6) with 5% fat reduced milk powder (Régilait, Saint Martin Belle Roche, France) for 1 h, three washes in TBS 1× and GFP mouse antibody (Santa Cruz Biotechnology) diluted 1/1000 in TBS with 1% fat reduced milk powder, three washes in TBST 1× and horseradish peroxidase (HRP)–conjugated goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted 1/2000 in TBS with 1% fat reduced milk powder, and one wash in TBST 1×. ECL Plus Kit (Pierce, Rockford, IL) was used to reveal the signal. Acquisitions were processed with Image Lab Bio-Rad Software under a Bio-Rad Chemidoc.
To detect endogenous ZYX-1 proteins, we prepared extracts (Hannak et al., 2002) from wild-type and zyx-1(gk190) mutant worms. We loaded approximately equal amounts of protein extract, estimated by finding volumes of extracts that would give equal intensity of banding after Coomassie staining. After separation on a 10% SDS–PAGE and transfer to nitrocellulose, the blot was reacted with the monoclonal ZYX-1 antibody at 1/100 dilution and HRP-conjugated anti-mouse antibody (GE Healthcare, Piscataway, NJ), and the signal was detected by ECL (Pierce).
Supplementary Material
Acknowledgments
We thank Yuji Kohara for the gift of the zyx-1 cDNA clone, B. D. Harfe and A. Fire for the hlh-1(cc561ts) strain, A. Fire for GFP-encoding plasmids, Pamela Hoppe for the gift of the MH35 antibody, Manolo Gouy for helpful discussions about phylogenetic analyses, and Samuel Leroy for help with FRAP data analysis. The MH24 antibody developed by R. H. Waterston was obtained from the Developmental Studies Hybridoma Bank (developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, IA). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. The alp-1(tm1137) mutant strain was provided by the Mitani laboratory at the Tokyo Women's Medical University School of Medicine (Tokyo, Japan). Electron microscopy was performed at the Centre d'Imagerie of the Institut Génétique Biologie Moléculaire Cellulaire (Illkirch, France) with the gracious help of Yannick Schwab. FRAP experiments were performed at the PLATIM IFR128 microscope platform at the Ecole Normale Supérieure Lyon (Lyon, France) with the gracious help of Christophe Chamot. Confocal microscopy was performed at the Center Technologique des Microstructures, Université Claude Bernard Lyon 1 (Villeurbanne, France).
This project was supported by Agence National de la Recherche Grant DUCHENNE ELEGANS 07 MRAR 005 01 and the European Muscle Development Network (MYORES). C.L. and N.B. were funded by Ph.D. fellowships from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche, and C.L. by the Fondation pour la Recherche Médicale en France. H.Q. and G.M.B. were supported at first by National Institutes of Health Grant R01AR052133 and later by American Heart Association Grant 11GRNT7820000.
Abbreviations used:
- DMD
Duchenne muscular dystrophy
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- HA
hemagglutinin
- LIM
Lin-11, Isl-1, and Mec-3
- LIMD1
LIM domain–containing protein 1
- LPP
lipoma preferred partner
- NES
nuclear export signal
- NLS
nuclear localization signal
- PRR
proline-rich region
- RNAi
RNA interference
- SEM
standard error of the mean
- TRIP 6
thyroid receptor–interacting protein 6
- WTIP
Wilms tumor interacting protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-09-0679) on February 20, 2013.
*These authors contributed equally.
REFERENCES
- Ahn AH, Kunkel LM. The structural and functional diversity of dystrophin. Nat Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JP, Titus MA. A family of unconventional myosins from the nematode Caenorhabditis elegans. J Mol Biol. 1997;272:523–535. doi: 10.1006/jmbi.1997.1232. [DOI] [PubMed] [Google Scholar]
- Barral JM, Bauer CC, Ortiz I, Epstein HF. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol. 1998;143:1215. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. Vinculin is essential for muscle function in the nematode. J Cell Biol. 1991;114:715. doi: 10.1083/jcb.114.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerle MC. Zyxin: zinc fingers at sites of cell adhesion. Bioessays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132:835. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessou C, Giugia J-B, Franks CJ, Holden-Dye L, Ségalat L. Mutations in the Caenorhabditis elegans dystrophin-like gene dys-1 lead to hyperactivity and suggest a link with cholinergic transmission. Neurogenetics. 1998;2:61–72. doi: 10.1007/s100480050053. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L, Kolotuev I, Didier C, Bhoumik A, Podbilewicz B, Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaruzza M, Lattrich C, Hecker M. Focal adhesion protein zyxin is a mechanosensitive modulator of gene expression in vascular smooth muscle cells. Hypertension. 2004;43:726–730. doi: 10.1161/01.HYP.0000119189.82659.52. [DOI] [PubMed] [Google Scholar]
- Cerisano V, et al. Molecular mechanisms of CD99-induced caspase-independent cell death and cell–cell adhesion in Ewing's sarcoma cells: actin and zyxin as key intracellular mediators. Oncogene. 2004;23:5664–5674. doi: 10.1038/sj.onc.1207741. [DOI] [PubMed] [Google Scholar]
- Chen L, Krause M, Sepanski M, Fire A. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development. 1994;120:1631–1641. doi: 10.1242/dev.120.6.1631. [DOI] [PubMed] [Google Scholar]
- Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EHK. Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J Cell Sci. 2009;122:1665–1679. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Cram EJ, Fontanez KM, Schwarzbauer JE. Functional characterization of KIN-32, the Caenorhabditis elegans homolog of focal adhesion kinase. Dev Dyn. 2008;237:837–846. doi: 10.1002/dvdy.21457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AW, Beckerle MC. Purification and characterization of zyxin, an 82,000-dalton component of adherens junctions. J Biol Chem. 1991;266:5847–5853. [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone J, Glas C, Schultheiss K, Moehlenbrink J, Krieghoff-Henning E, Hofmann TG. Zyxin is a critical regulator of the apoptotic HIPK2-p53 signaling axis. Cancer Res. 2011;71:2350–2359. doi: 10.1158/0008-5472.CAN-10-3486. [DOI] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM. Costameres: the Achilles’ heel of Herculean muscle. J Biol Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- Etheridge T, Oczypok EA, Lehmann S, Fields BD, Shephard F, Jacobson LA, Szewczyk NJ. Calpains mediate integrin attachment complex maintenance of adult muscle in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002471. doi: 10.1371/journal.pgen.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley SI, Feng Y, Giri A, Longmore GD, Wirtz D. Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nat Commun. 2012;3:719. doi: 10.1038/ncomms1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SR, Hope IA. Determination of the mobility of novel and established Caenorhabditis elegans sarcomeric proteins in vivo. Eur J Cell Biol. 2010;89:437–448. doi: 10.1016/j.ejcb.2009.11.027. [DOI] [PubMed] [Google Scholar]
- Gieseler K, Grisoni K, Ségalat L. Genetic suppression of phenotypes arising from mutations in dystrophin-related genes in Caenorhabditis elegans. Curr Biol. 2000;10:1092–1097. doi: 10.1016/s0960-9822(00)00691-6. [DOI] [PubMed] [Google Scholar]
- Gueneau L, et al. Mutations of the FHL1 gene cause Emery-Dreifuss muscular dystrophy. Am J Hum Genet. 2009;85:338–353. doi: 10.1016/j.ajhg.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H-F, Beckerle MC. The ALP-enigma protein ALP-1 functions in actin filament organization to promote muscle structural integrity in Caenorhabditis elegans. Mol Biol Cell. 2009;20:2361–2370. doi: 10.1091/mbc.E08-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Oegema K, Kirkham M, Gönczy P, Habermann B, Hyman AA. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MDH, Beckerle MC. Opposing roles of zyxin/LPP ACTA repeats and the LIM domain region in cell-cell adhesion. J Biol Chem. 2006;281:16178–16188. doi: 10.1074/jbc.M512771200. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Branda CS, Krause M, Stern MJ, Fire A. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development. 1998;125:2479–2488. doi: 10.1242/dev.125.13.2479. [DOI] [PubMed] [Google Scholar]
- Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr Opin Cell Biol. 2006;18:524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Hervy M, Hoffman LM, Jensen CC, Smith M, Beckerle MC. The LIM protein zyxin binds CARP-1 and promotes apoptosis. Genes Cancer. 2010;1:506–515. doi: 10.1177/1947601910376192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita T, Qadota H, Tsuboi D, Taya S, Moerman DG, Kaibuchi K. Identification of a novel Cdc42 GEF that is localized to the PAT-3-mediated adhesive structure. Biochem Biophys Res Commun. 2005;335:139–145. doi: 10.1016/j.bbrc.2005.07.068. [DOI] [PubMed] [Google Scholar]
- Hirata H, Tatsumi H, Sokabe M. Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures. Commun Integr Biol. 2008;1:192–195. doi: 10.4161/cib.1.2.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Chaturvedi A, Yoshigi M, Beckerle MC. Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol Biol Cell. 2012;23:1846–1859. doi: 10.1091/mbc.E11-12-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Kloeker S, Wang A, Yoshigi M, Beckerle MC. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol. 2006;172:771–782. doi: 10.1083/jcb.200512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, et al. Targeted disruption of the murine zyxin gene. Mol Cell Biol. 2003;23:70–79. doi: 10.1128/MCB.23.1.70-79.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen H, Marynen P. Interaction partners for human ZNF384/CIZ/NMP4—zyxin as a mediator for p130CAS signaling. Exp Cell Res. 2006;312:1194–1204. doi: 10.1016/j.yexcr.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA. Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome Res. 2009;19:1404–1418. doi: 10.1101/gr.086827.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Muraski J, Chen Y, Tsujita Y, Wall J, Glembotski CC, Schaefer E, Beckerle M, Sussman MA. Atrial natriuretic peptide promotes cardiomyocyte survival by cGMP-dependent nuclear accumulation of zyxin and Akt. J Clin Invest. 2005;115:2716–2730. doi: 10.1172/JCI24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic. 2008;9:2053–2062. doi: 10.1111/j.1600-0854.2008.00825.x. [DOI] [PubMed] [Google Scholar]
- Landsverk ML, Li S, Hutagalung AH, Najafov A, Hoppe T, Barral JM, Epstein HF. The UNC-45 chaperone mediates sarcomere assembly through myosin degradation in Caenorhabditis elegans. J Cell Biol. 2007;177:205–210. doi: 10.1083/jcb.200607084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecroisey C, Martin E, Mariol M-C, Granger L, Schwab Y, Labouesse M, Ségalat L, Gieseler K. DYC-1, a protein functionally linked to dystrophin in Caenorhabditis elegans is associated with the dense body, where it interacts with the muscle LIM domain protein ZYX-1. Mol Biol Cell. 2008;19:785–796. doi: 10.1091/mbc.E07-05-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecroisey C, Ségalat L, Gieseler K. The C. elegans dense body: anchoring and signaling structure of the muscle. J Muscle Res Cell Motil. 2007;28:79–87. doi: 10.1007/s10974-007-9104-y. [DOI] [PubMed] [Google Scholar]
- Li A, Dos Ponten F, Remedios CG. The interactome of LIM domain proteins: the contributions of LIM domain proteins to heart failure and heart development. Proteomics. 2012;12:203–225. doi: 10.1002/pmic.201100492. [DOI] [PubMed] [Google Scholar]
- Liegeois S, Benedetto A, Michaux G, Belliard G, Labouesse M. Genes required for osmoregulation and apical secretion in Caenorhabditis elegans. Genetics. 2007;175:709–724. doi: 10.1534/genetics.106.066035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann A, Van der Ven PFM, Vakeel P, Albinus B, Simonis D, Bendas G, Schenk JA, Micheel B, Kley RA, Fürst DO. The sarcomeric Z-disc component myopodin is a multiadapter protein that interacts with filamin and α-actinin. Eur J Cell Biol. 2010;89:681–692. doi: 10.1016/j.ejcb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci USA. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynova NY, Eroshkin FM, Ermolina LV, Ermakova GV, Korotaeva AL, Smurova KM, Gyoeva FK, Zaraisky AG. The LIM-domain protein zyxin binds the homeodomain factor Xanf1/Hesx1 and modulates its activity in the anterior neural plate of Xenopus laevis embryo. Dev Dyn. 2008;237:736–749. doi: 10.1002/dvdy.21471. [DOI] [PubMed] [Google Scholar]
- McKeown CR, Han H-F, Beckerle MC. Molecular characterization of the Caenorhabditis elegans ALP/Enigma gene alp-1. Dev Dyn. 2006;235:530–538. doi: 10.1002/dvdy.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol Biol Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman D, Williams BD. Sarcomere assembly in C. elegans muscle. WormBook: The Online Review of C. elegans Biology. 2006 doi: 10.1895/wormbook.1.81.1. Available at: www.ncbi.nlm.nih.gov/books/NBK19751/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody JD, Grange J, Ascione MPA, Boothe D, Bushnell E, Hansen MDH. A zyxin head–tail interaction regulates zyxin–VASP complex formation. Biochem Biophys Res Commun. 2009;378:625–628. doi: 10.1016/j.bbrc.2008.11.100. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Madgwick AJ, Charleston B, Pell JM, Loughna PT. The developmental regulation of a novel muscle Lim-protein. Biochem Biophys Res Commun. 1995;212:840–846. doi: 10.1006/bbrc.1995.2045. [DOI] [PubMed] [Google Scholar]
- Moulder GL, Cremona GH, Duerr J, Stirman JN, Fields SD, Martin W, Qadota H, Benian GM, Lu H, Barstead RJ. α-Actinin is required for the proper assembly of Z-disk/focal-adhesion-like structures and for efficient locomotion in Caenorhabditis elegans. J Mol Biol. 2010;403:516–528. doi: 10.1016/j.jmb.2010.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder GL, Huang MM, Waterston RH, Barstead RJ. Talin requires beta-integrin, but not vinculin, for its assembly into focal adhesion-like structures in the nematode Caenorhabditis elegans. Mol Biol Cell. 1996;7:1181. doi: 10.1091/mbc.7.8.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahabedian JF, Qadota H, Stirman JN, Lu H, Benian GM. Bending amplitude—a new quantitative assay of C. elegans locomotion: Identification of phenotypes for mutants in genes encoding muscle focal adhesion components. Methods. 2012;56:95–102. doi: 10.1016/j.ymeth.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear–cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, Beckerle MC. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Norman KR, Cordes S, Qadota H, Rahmani P, Moerman DG. UNC-97/PINCH is involved in the assembly of integrin cell adhesion complexes in Caenorhabditis elegans body wall muscle. Dev Biol. 2007;309:45–55. doi: 10.1016/j.ydbio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Ohsawa N, Koebis M, Suo S, Nishino I, Ishiura S. Alternative splicing of PDLIM3/ALP, for α-actinin-associated LIM protein 3, is aberrant in persons with myotonic dystrophy. Biochem Biophys Res Commun. 2011;409:64–69. doi: 10.1016/j.bbrc.2011.04.106. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Benian GM. Molecular structure of sarcomere-to-membrane attachment at M-lines in C. elegans muscle. J Biomed Biotechnol. 2010;2010:864749. doi: 10.1155/2010/864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, McGaha LA, Mercer KB, Stark TJ, Ferrara TM, Benian GM. A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (obscurin) in Caenorhabditis elegans. Mol Biol Cell. 2008;19:2424–2432. doi: 10.1091/mbc.E08-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Moerman DG, Benian GM. A molecular mechanism for the requirement of PAT-4 (ILK) for the localization of UNC-112 (kindlin) to integrin adhesion sites. J Biol Chem. 2012;287:28537–28551. doi: 10.1074/jbc.M112.354852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Pan G, Reddy BVVG, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS Biol. 2011;9:e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfranz PJ, Beckerle MC. Doing (F/L)pppps: EVH1 domains and their proline-rich partners in cell polarity and migration. Curr Opin Cell Biol. 2002;14:88–103. doi: 10.1016/s0955-0674(01)00299-x. [DOI] [PubMed] [Google Scholar]
- Renfranz PJ, Blankman E, Beckerle MC. The cytoskeletal regulator zyxin is required for viability in Drosophila melanogaster. Anat Rec (Hoboken) 2010;293:1455–1469. doi: 10.1002/ar.21193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfranz PJ, Siegrist SE, Stronach BE, Macalma T, Beckerle MC. Molecular and phylogenetic characterization of Zyx102, a Drosophila orthologue of the zyxin family that interacts with Drosophila Enabled. Gene. 2003;305:13–26. doi: 10.1016/s0378-1119(02)01173-3. [DOI] [PubMed] [Google Scholar]
- Rogalski TM, Gilbert MM, Devenport D, Norman KR, Moerman DG. DIM-1, a novel immunoglobulin superfamily protein in Caenorhabditis elegans, is necessary for maintaining bodywall muscle integrity. Genetics. 2003;163:905–915. doi: 10.1093/genetics/163.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler I, Crawford AW, Michelsen JW, Beckerle MC. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol. 1992;119:1573. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarel AM. Costameres, focal adhesions, cardiomyocyte mechanotransduction. Am J Physiol Heart Circ Physiol. 2005;289:H2291–H2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- Schessl J, Feldkirchner S, Kubny C, Schoser B. Reducing body myopathy and other FHL1-related muscular disorders. Semin Pediatr Neurol. 2011;18:257–263. doi: 10.1016/j.spen.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Ségalat L, Elkes DA, Kaplan JM. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Science. 1995;267:1648–1651. doi: 10.1126/science.7886454. [DOI] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Leung-Chiu W-M, Montgomery R, Orsborn A, Kuznicki K, Gressman-Coberly E, Mutapcic L, Bennett K. The GLH proteins, Caenorhabditis elegans P granule components, associate with CSN-5 and KGB-1, proteins necessary for fertility, and with ZYX-1, a predicted cytoskeletal protein. Dev Biol. 2002;251:333–347. doi: 10.1006/dbio.2002.0832. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sun N, Huiatt TW, Paulin D, Li Z, Robson RM. Synemin interacts with the LIM domain protein zyxin and is essential for cell adhesion and migration. Exp Cell Res. 2010;316:491–505. doi: 10.1016/j.yexcr.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gilmore TD. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim Biophys Acta. 2003;1593:115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- Warner A, Qadota H, Benian GM, Vogl AW, Moerman DG. The Caenorhabditis elegans paxillin orthologue, PXL-1, is required for pharyngeal muscle contraction and for viability. Mol Biol Cell. 2011;22:2551–2563. doi: 10.1091/mbc.E10-12-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Hirsh D, Lane TR. Dominant mutations affecting muscle structure in Caenorhabditis elegans that map near the actin gene cluster. J Mol Biol. 1984;180:473–496. doi: 10.1016/0022-2836(84)90023-8. [DOI] [PubMed] [Google Scholar]
- Welman A, Serrels A, Brunton VG, Ditzel M, Frame MC. Two-color photoactivatable probe for selective tracking of proteins and cells. J Biol Chem. 2010;285:11607–11616. doi: 10.1074/jbc.M110.102392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KJ, Qadota H, Mains PE, Benian GM. UNC-89 (obscurin) binds to MEL-26, a BTB-domain protein, and affects the function of MEI-1 (katanin) in striated muscle of Caenorhabditis elegans. Mol Biol Cell. 2012;23:2623–2634. doi: 10.1091/mbc.E12-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójtowicz A, Babu SS, Li L, Gretz N, Hecker M, Cattaruzza M. Zyxin mediation of stretch-induced gene expression in human endothelial cells. Circ Res. 2010;107:898–902. doi: 10.1161/CIRCRESAHA.110.227850. [DOI] [PubMed] [Google Scholar]
- Wolfenson H, Bershadsky A, Henis YI, Geiger B. Actomyosin-generated tension controls the molecular kinetics of focal adhesions. J Cell Sci. 2011;124:1425–1432. doi: 10.1242/jcs.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, Qadota H, Mercer KB, McGaha LA, Oberhauser AF, Benian GM. A LIM-9 (FHL)/SCPL-1 (SCP) complex interacts with the C-terminal protein kinase regions of UNC-89 (obscurin) in Caenorhabditis elegans muscle. J Mol Biol. 2009;386:976–988. doi: 10.1016/j.jmb.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Kawata H, Shou Z, Hirano S, Mizutani T, Yazawa T, Sekiguchi T, Yoshino M, Kajitani T, Miyamoto K. Analysis of zinc-fingers and homeoboxes (ZHX)-1-interacting proteins: molecular cloning and characterization of a member of the ZHX family, ZHX3. Biochem J. 2003;373:167. doi: 10.1042/BJ20021866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Zhao Y. The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein–protein interaction. Biol Cell. 2007;99:489–502. doi: 10.1042/BC20060126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.