Abstract
Anaemia is a chief determinant of globalill health, contributing to cognitive impairment, growth retardation and impaired physical capacity. To understand further the genetic factors influencing red blood cells, we carried out a genome-wide association study of haemoglobin concentration and related parameters in up to 135,367 individuals. Here we identify 75 independent genetic loci associated with one or more red blood cell phenotypes at P <10−8, which together explain 4–9% of the phenotypic variance per trait. Using expression quantitative trait loci and bioinformatic strategies, we identify 121 candidate genes enriched in functions relevant to red blood cell biology. The candidate genes are expressed preferentially in red blood cell precursors, and 43 have haematopoietic phenotypes in Mus musculus or Drosophila melanogaster. Through open-chromatin and coding-variant analyses we identify potential causal genetic variants at 41 loci. Our findings provide extensive new insights into genetic mechanisms and biological pathways controlling red blood cell formation and function.
Haemoglobin, an iron-containing metalloprotein found in the red blood cells of all vertebrates, provides the primary mechanism for oxygen transport in the circulation. Haemoglobin levels and related red blood cell phenotypes are tightly regulated, including an important genetic component1–5. To refine our understanding of the genetic factors influencing red blood cell formation and function, we carried out a meta-analysis of genome-wide association studies (GWAS) and staged follow-up genotyping of six red blood cell phenotypes: haemoglobin, mean cell haemoglobin (MCH), mean cell haemoglobin concentration (MCHC), mean cell volume (MCV), packed cell volume (PCV) and red blood cell count (RBC).
Our study design is summarized in Supplementary Fig. 1. In brief, we combined genome-wide association data from 71,861 individuals of European or South Asian ancestry, with up to 2,644,161 autosomal single-nucleotide polymorphisms (SNPs) and 67,645 X-chromosome SNPs. Characteristics of participants, genotyping arrays and imputation are summarized in Supplementary Tables 1–3. Meta-analysis was carried out among Europeans and South Asians separately, followed by a final combined analysis of results from the two populations. We performed replication testing of 22 loci showing suggestive association (10−8<P <10−7) in a further 63,506 individuals using a combination of in silico data and direct genotyping (Supplementary Tables 1, 2 and Supplementary Note). Genome-wide significance was set at P <1 × 10−8, allowing a Bonferroni correction both for the ~106 independent SNPs tested6, as well as for the six inter-related red blood cell phenotypes (Supplementary Note)7.
Seventy-five independent genetic loci reached genome-wide significance for association with one or more red blood cell phenotypes (Table 1 and Supplementary Fig. 2), 43 of which are novel. For descriptive and downstream purposes, we identified a single ‘sentinel’ SNP for each of the 75 loci, defined as the SNP with the lowest P value against any phenotype at each locus; regional plots for the 75 loci are shown in Supplementary Fig. 3. Full lists of the SNPs associated with phenotype at P <10−6 and of the sentinel SNPs are provided (Supplementary Tables 4 and 5). Of the 38 loci previously reported to be associated with red blood cell traits1–5, we replicate 32 loci (P <10−8) and find three to be nominally associated (P <0.05; Supplementary Table 6). The remaining three loci, initially reported in an East Asian GWAS4, were not associated with red blood cell phenotypes in our sample (Supplementary Fig. 4 and Supplementary Note).
Table 1.
Genomic loci associated with red blood cell phenotypes
| Region | Sentinel SNP | Position (B36) | Alleles (EA/OA) | EAF | Phenotype | Effect (SE) | P | Candidate genes |
|---|---|---|---|---|---|---|---|---|
| 1p36 | rs1175550 | 3,681,388 | G/A | 0.22 | MCHC | 0.008 (0.013) | 8.6 × 10−15 | CCDC27n, LRRC48n |
| 1p34‡ | rs3916164 | 39,842,526 | G/A | 0.71 | MCH | 0.008 (0.004) | 3.1 × 10−10 | HEYLn |
| 1p32 | rs741959 | 47,448,820 | G/A | 0.57 | MCV | 0.157 (0.025) | 6.0 × 10−10 | TAL1n |
| 1q23† | rs857684 | 156,842,353 | C/T | 0.74 | MCHC | −0.006 (0.011) | 3.5 × 10−16 | OR6Y1c, OR10Z1nc, SPTA1ncg |
| 1q32† | rs7529925 | 197,273,831 | C/T | 0.28 | RBC | 0.014 (0.002) | 8.3 × 10−9 | MIR181A1n |
| 1q32 | rs7551442 | 201,921,744 | A/G | 0.09 | MCHC | −0.023 (0.017) | 9.7 × 10−12 | ATP2B4ng |
| 1q32 | rs9660992 | 203,516,073 | G/A | 0.42 | MCH | 0.007 (0.004) | 7.1 × 10−10 | TMCC2n |
| 1q44† | rs3811444 | 246,106,074 | T/C | 0.35 | RBC | 0.018 (0.003) | 4.5 × 10−10 | TRIM58nc |
| 2p21† | rs4953318 | 46,208,555 | A/C | 0.62 | PCV | 0.152 (0.018) | 3.1 × 10−19 | PRKCEn |
| 2p16† | rs243070 | 60,473,790 | T/A | 0.72 | MCV | −0.181 (0.027) | 4.4 × 10−13 | BCL11An |
| 2q13 | rs10207392 | 111,566,130 | G/A | 0.44 | MCV | −0.132 (0.025) | 4.4 × 10−11* | ACOXLn |
| 3p24† | rs9310736 | 24,325,815 | A/G | 0.35 | MCV | −0.210 (0.026) | 6.1 × 10−16 | THRBn |
| 3q22 | rs6776003 | 142,749,183 | A/G | 0.44 | MCV | −0.138 (0.026) | 3.7 × 10−11* | RASA2n |
| 3q23 | rs13061823 | 143,603,476 | T/C | 0.56 | MCV | −0.168 (0.025) | 4.7 × 10−13 | XRN1n |
| 3q29† | rs11717368 | 197,318,754 | C/G | 0.52 | MCH | 0.008 (0.004) | 6.6 × 10−19 | TFRCng |
| 4q11† | rs218238 | 55,089,781 | A/T | 0.78 | RBC | 0.033 (0.003) | 2.8 × 10−39 | KITn |
| 4q27 | rs13152701 | 122,970,511 | A/G | 0.37 | MCV | 0.150 (0.026) | 9.0 × 10−10 | BBS7n, CCNA2ne |
| 6p23 | rs6914805 | 16,389,166 | C/T | 0.75 | MCH | 0.012 (0.004) | 1.2 × 10−19 | GMPRne |
| 6p21† | rs1408272 | 25,950,930 | G/T | 0.07 | MCH | 0.033 (0.009) | 4.8 × 10−67 | HFEcg, SLC17A3n |
| 6p22 | rs13219787 | 27,969,649 | A/G | 0.09 | MCH | 0.023 (0.007) | 5.9 × 10−17 | HIST1H2AMn, HIST1H2BOn, HIST1H3Jn |
| 6p22 | rs2097775 | 30,462,282 | A/T | 0.15 | HB | 0.055 (0.008) | 1.3 × 10−10 | TRIM39-RPP21n |
| 6p21 | rs9272219 | 32,710,247 | G/T | 0.72 | RBC | 0.015 (0.002) | 4.3 × 10−10 | HLA-DQA1nce, HLA-DQA2e |
| 6p21† | rs9349204 | 42,022,356 | G/A | 0.27 | MCV | −0.367 (0.028) | 2.4 × 10−40 | CCND3n |
| 6p12 | rs9369427 | 43,919,408 | A/C | 0.68 | HB | 0.042 (0.006) | 5.6 × 10−12 | VEGFAn |
| 6q21† | rs1008084 | 109,733,658 | G/A | 0.56 | MCH | −0.010 (0.003) | 6.4 × 10−26 | CCDC162Pn |
| 6q23† | rs9389269 | 135,468,852 | T/C | 0.72 | MCV | −0.600 (0.028) | 2.6 × 10−19 | HBS1Ln |
| 6q24† | rs590856 | 139,886,122 | G/A | 0.43 | MCV | 0.313 (0.026) | 5.0 × 10−36 | CITED2n |
| 6q26 | rs736661 | 164,402,826 | A/G | 0.62 | MCH | 0.007 (0.004) | 1.6 × 10−11 | QKIn |
| 7p13† | rs12718598 | 50,395,939 | T/C | 0.51 | MCV | −0.204 (0.030) | 1.6 × 10−13 | IKZF1n |
| 7q22† | rs2075672 | 100,078,232 | A/G | 0.39 | RBC | 0.022 (0.003) | 1.9 × 10−20 | ACTL6Bn, TFR2ng |
| 7q36† | rs10480300 | 151,036,938 | C/T | 0.72 | HB | 0.052 (0.007) | 7.8 × 10−15 | PRKAG2ng |
| 8p11 | rs4737009 | 41,749,562 | G/A | 0.74 | MCHC | −0.014 (0.013) | 4.9 × 10−11 | ANK1ng |
| 8p11 | rs6987853 | 42,576,607 | C/T | 0.62 | MCHC | −0.002 (0.010) | 6.1 × 10−11 | C8orf40ne |
| 9p24† | rs2236496 | 4,834,265 | C/T | 0.22 | MCV | −0.279 (0.031) | 1.4 × 10−19 | RCL1n |
| 9q34† | rs579459 | 135,143,989 | T/C | 0.8 | RBC | 0.021 (0.003) | 9.3 × 10−18 | ABOn |
| 10q11† | rs901683 | 45,286,428 | A/G | 0.08 | MCV | 0.364 (0.050) | 1.5 × 10−16 | MARCH8nce |
| 10q22† | rs10159477 | 70,769,894 | A/G | 0.16 | HB | 0.087 (0.010) | 4.4 × 10−20 | HK1ng |
| 10q24 | rs11190134 | 101,272,190 | G/A | 0.6 | MCH | −0.011 (0.004) | 1.3 × 10−10* | NKX2-3n |
| 11p15 | rs11042125 | 8,894,625 | A/T | 0.6 | HB | 0.032 (0.006) | 1.5 × 10−9 | AKIP1ne, C11orf16ne, NRIP3e, ST5n |
| 11p15 | rs7936461 | 9,997,462 | C/T | 0.75 | PCV | 0.121 (0.021) | 1.0 × 10−9 | SBF2n |
| 11q13 | rs2302264 | 66,964,002 | G/A | 0.58 | MCV | 0.140 (0.025) | 1.3 × 10−10 | CORO1Bne, PTPRCAPne, RPS6KB2nce |
| 11q13 | rs7125949 | 72,686,732 | A/G | 0.11 | HB | 0.053 (0.010) | 2.1 × 10−9 | ARHGEF17ce, P2RY6n |
| 12p13 | rs7312105 | 2,393,616 | G/A | 0.36 | PCV | 0.104 (0.019) | 3.2 × 10−9* | CACNA1Cn |
| 12p13† | rs10849023 | 4,202,739 | C/T | 0.79 | MCH | −0.008 (0.005) | 7.5 × 10−12 | CCND2ng |
| 12q22 | rs11104870 | 87,353,425 | C/T | 0.3 | RBC | 0.013 (0.002) | 6.2 × 10−11 * | KITLGn |
| 12q24† | rs3184504 | 110,368,991 | T/C | 0.48 | HB | 0.051 (0.006) | 4.3 × 10−19 | ATXN2n, SH2B3nc |
| 12q24 | rs3829290 | 119,610,821 | C/T | 0.44 | MCV | −0.153 (0.026) | 2.1 × 10−9 | ACADSc, MLECn |
| 14q23† | rs7155454 | 64,571,992 | A/G | 0.51 | MCH | 0.002 (0.004) | 1.8 × 10−12 | FNTBn, MAXn |
| 14q24 | rs11627546 | 69,435,677 | C/A | 0.84 | MCV | 0.162 (0.032) | 1.1 × 10−9* | SMOC1n |
| 14q32‡ | rs17616316 | 102,892,515 | G/C | 0.07 | MCH | 0.014 (0.009) | 8.2 × 10−11* | EIF5n |
| 15q21‡ | rs1532085 | 56,470,658 | G/A | 0.59 | HB | 0.034 (0.006) | 6.7 × 10−11* | LIPCn |
| 15q22† | rs2572207 | 63,857,747 | C/T | 0.74 | MCV | 0.153 (0.029) | 3.4 × 10−9 | DENND4An, PTPLAD1e |
| 15q24 | rs8028632 | 73,108,315 | T/C | 0.8 | MCV | 0.188 (0.032) | 6.9 × 10−10 | PPCDCn, SCAMP5n |
| 15q24 | rs11072566 | 74,081,026 | A/G | 0.48 | HB | 0.028 (0.006) | 3.0 × 10−10* | NRG4n |
| 15q25 | rs2867932 | 76,378,092 | G/A | 0.61 | MCHC | −0.021 (0.010) | 3.3 × 10−9 | DNAJA4e, WDR61n |
| 16p11† | rs11248850 | 103,598 | G/A | 0.5 | MCH | 0.007 (0.004) | 6.3 × 10−23 | NPRL3n |
| 16q22 | rs2271294 | 66,459,827 | T/A | 0.15 | RBC | 0.017 (0.003) | 1.1 × 10−9 | CTRLc, DUS2Le, EDC4n, NUTF2n, PSMB10c |
| 16q24† | rs10445033 | 87,367,963 | G/A | 0.37 | MCHC | 0.020 (0.012) | 1.5 ×10−22 | PIEZO1n |
| 17p11 | rs888424 | 19,926,019 | A/G | 0.43 | MCH | 0.006 (0.004) | 5.4 × 10−20 | SPECC1n |
| 17q11 | rs2070265 | 24,099,550 | T/C | 0.2 | MCH | 0.013 (0.004) | 5.1 × 10−14 | C17orf63n, ERAL1e, NEK8n, TRAF4ne |
| 17q12 | rs8182252 | 34,981,476 | C/T | 0.18 | RBC | 0.016 (0.003) | 5.9 × 10−9 | CDK12e, NEUROD2n |
| 17q21 | rs2269906 | 39,649,863 | C/A | 0.36 | MCHC | 0.027 (0.010) | 2.0 × 10−11 | SLC4A1g, UBTFn |
| 17q21 | rs12150672 | 41,182,408 | A/G | 0.23 | RBC | 0.017 (0.003) | 4.7 × 10−12 | ARHGAP27e, ARL17Be, C17orf69ce, CRHR1nc, SPPL2Cc, KANSL1c, MAPTc, STHc |
| 17q25 | rs4969184 | 73,905,008 | G/A | 0.53 | HB | 0.031 (0.006) | 7.0 × 10−9 | PGS1ne |
| 18q21 | rs4890633 | 42,087,276 | G/A | 0.27 | MCH | 0.005 (0.004) | 1.9 × 10−23 | C18orf25ne |
| 19p13 | rs2159213 | 2,087,102 | C/T | 0.5 | HB | 0.032 (0.006) | 1.9 × 10−9 | AP3D1n |
| 19p13 | rs732716 | 4,317,219 | A/G | 0.71 | MCV | 0.201 (0.028) | 1.5 × 10−14 | MPNDn, SH3GL1n, UBXN6c |
| 19p13† | rs741702 | 12,885,250 | A/C | 0.35 | MCH | 0.006 (0.004) | 8.2 × 10−20 | CALRe, FARSAne, SYCE2n |
| 19q13 | rs3892630 | 37,873,324 | T/C | 0.18 | MCV | 0.176 (0.034) | 1.0 × 10−10* | NUDT19nc |
| 20q13† | rs737092 | 55,423,811 | C/T | 0.49 | MCV | 0.216 (0.033) | 4.0 × 10−13 | RBM38n |
| 21q22‡ | rs2032314 | 34,276,393 | T/C | 0.08 | PCV | 0.154 (0.034) | 7.5 × 10−10* | ATP5On |
| 22q11† | rs5754217 | 20,269,675 | G/T | 0.83 | MCV | 0.194 (0.031) | 8.6 × 10−10 | UBE2L3ne, YDJCc |
| 22q12† | rs5749446 | 31,210,585 | T/C | 0.62 | MCH | 0.007 (0.004) | 3.3 × 10−13 | FBXO7ncg |
| 22q12† | rs855791 | 35,792,882 | G/A | 0.57 | MCH | 0.012 (0.004) | 1.0 × 10−69 | KCTD17n, TMPRSS6nc |
| 22q13† | rs140522 | 49,318,132 | C/T | 0.67 | MCV | 0.287 (0.030) | 4.5 × 10−23 | TYMPne, NCAPH2n, ODF3Bn, SCO2n |
Candidate gene superscripts indicate the method of identification.
Replication testing performed.
Previously reported.
Discovered from combined analysis of European and South Asian genome-wide association data.
c, coding variant; e, eQTL; EA, effect allele; EAF, effect allele frequency; g, GRAIL; HB, haemoglobin; n, nearby; OA, other allele; SE, standard error.
Among the 75 genomic loci identified, we found that 31 were associated with one red blood cell phenotype, and 44 with two or more phenotypes, at P <10−8. The total number of locus–phenotype associations identified at P <10−8 was 156, of which 92 are novel (Supplementary Fig. 5 and Supplementary Table 7). In addition, at 8 of the 75 loci we found evidence for multiple SNPs independently associated with red blood cell phenotype at P <10−8 in conditional analyses8, suggesting the presence of possible secondary genetic mechanisms at these loci (Supplementary Table 8).
Identification of candidate genes
There are >3,000 protein-coding genes within 1 megabase (Mb) of the sentinel SNPs from the 75 genetic loci associated with red blood cell phenotypes. We prioritized genes as probable candidates underlying the observed genetic associations using the following criteria: (1) gene nearest to the sentinel SNP, and any other gene within 10 kilo-bases (kb) (97 genes; Table 1); (2) gene containing a non-synonymous SNP in high linkage disequilibrium (r2 >0.8) with the sentinel SNP (24 genes; Supplementary Table 9); (3) gene with expression quantitative trait loci (eQTL) associated with sentinel SNP in peripheral blood lymphocytes (27 genes; Supplementary Table 10); and (4) gene relationships among implicated loci (GRAIL) literature analysis9 (9 genes; Supplementary Table 11). This strategy identified 121 candidate genes (Table 1 and Supplementary Fig. 6).
Pathway analysis revealed that the list of candidate genes is strongly enriched for genes known to be involved in haematological development and function (P = 10−63), as well as in cellular proliferation, development and death, and immunological processes (Supplementary Tables 12 and 13). Current knowledge of gene function for all 121 candidates is summarized in Supplementary Table 14. Of note, some of the genes within these regions are known to underlie the Mendelian red blood cell disorders of elliptocytosis, ovalocytosis and spherocytosis (ANK1, SLC4A1, SPTA1)10, haemolytic anaemia (HK1)11 and iron deficiency or overload (TMPRSS6, HFE, TFR2)12. Furthermore, somatic mutations of IKZF1, KIT, SH2B3, SH3GL1 and TAL1 (also known as SCL) underlie several haematologic proliferative disorders (Supplementary Table 14).
Gene expression during haematopoiesis
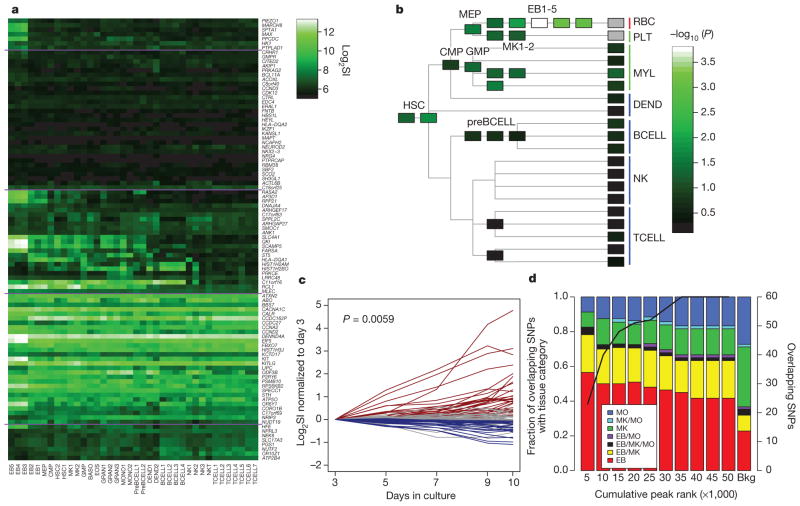
We next explored expression of the 121 candidate genes using an atlas of 38 different haematopoietic cell types (Supplementary Table 15)13. Ninety-seven genes could be reliably assigned a probe on the Affymetrix HG_U133AAofAv2 array (Fig. 1a); these transcripts were, on average, expressed at higher levels in late erythroblasts (or the precursors of red blood cells, EB3–EB5) compared to other transcripts in the same cell type (P <0.01 after Bonferroni correction; Fig. 1b). Furthermore, expression was more likely to be upregulated in EB3–5 relative to other cell types (P = 1.2 × 10−6, rank-sum test).
Figure 1. Gene-expression patterns for 121 putative candidate genes, and tissue distribution of NDRs.
a, Heat-map of candidate genes in the Differentiation Map of Hematology13. Cell acronyms refer to original source (summarized in Supplementary Table 15). Expression above a log2 signal intensity (SI) of 6 is consistently above background. b, −log10 P of the signed-rank test for candidate genes being more highly expressed in each cell type than non-candidate genes. c, Time-course of differentiation of cord-blood haematopoietic stem cells cultured along the erythroid lineage. Putative candidate genes are shown as upregulated (red), downregulated (blue) or with the slope not being significantly different from zero (grey). d, Tissue distribution of NDRs containing a potential causal variant. NDRs were ranked by peak score (proportional to their peak height in FAIRE-seq). The rankings were then used to divide the NDRs into cumulative tranches to explore the effect of calling-thresholds on results (left bar, tranche containing the 5,000 top-ranked NDRs of each cell type; penultimate bar, tranche containing the 50,000 top-ranked NDRs of each cell type). The solid line indicates the number of SNPs overlapping the tranche-specific NDRs that are potential causal variants (defined as a sentinel SNP from the red blood cell GWAS, or a SNP in high linkage disequilibrium (r2 >0.8) and located within 1 Mb of a sentinel SNP; right-hand y axis); the bar summarizes the tissue distribution of these SNPs (as a percentage of tranche-specific total). The right-hand bar represents the expected tissue distribution for the SNPs under the null hypothesis. Results show that the potential causal variants are most commonly found in erythroblast-specific NDRs, and that this is true across the spectrum of peak-calling thresholds.
To further investigate lineage-specific effects, we assessed temporal patterns of gene expression during in vitro differentiation of haematopoietic stem cells to erythroblasts14. On average, candidate genes have increasing expression over time along the erythroid lineage (P =0.006, rank-sum test; Fig. 1c). These data support the view that the gene set identified here is enriched for genes relevant to red blood cell biology, including a number of candidate genes differentially regulated to increase their expression in late erythropoiesis.
Coding and regulatory sequence variants
To better capture common sequence variation at the 75 loci, we searched the 1000 Genomes Project data set (www.1000genomes.org) and identified 39 non-synonymous SNPs that are in high linkage disequilibrium (r2 >0.8) with sentinel SNPs at the red blood cell loci (Supplementary Table 9). This represents a ~sixfold enrichment compared to the expectation under the null hypothesis (P =0.01; Supplementary Note). Although re-sequencing will be needed to obtain a complete assessment of variants at these loci, these non-synonymous sites represent an initial set of candidates for genetic variants underlying the observed associations with red blood cell phenotypes, potentially mediated through changes in protein function.
We next searched for sequence variants at the red blood cell loci that might influence gene regulation. We used formaldehyde-assisted isolation of regulatory elements followed by next-generation sequencing (FAIRE-seq) to identify nucleosome-depleted regions (NDRs) that may represent active regulatory elements15. We studied three haematologic cell types, and found 103,308 unique NDRs, of which 38,014 were present in erythroblasts, 50,372 in megakaryocytes and 34,833 in monocytes. We then searched the 1000 Genomes Project data set and found 60 SNPs located within one of these NDRs that are either: (1) one of the 75 sentinel SNPs from the red blood cell GWAS, or (2) in high linkage disequilibrium (r2>0.8) and located within 1 Mb of a sentinel SNP (Supplementary Table 16). The NDRs overlapping these 60 SNPs were more likely to be erythroblast specific than expected by chance (1.8-fold enrichment compared to background distribution of NDRs; P =0.007, Bonferroni-adjusted binomial test); by contrast, there were fewer megakaryocyte-specific NDRs coinciding with red blood cell SNPs (0.4-fold enrichment; P =0.007; Fig. 1d). This pattern of erythroblast enrichment and megakaryocyte depletion was robust to the stringency of NDR peak-calling (Supplementary Table 17). Our results indicate that regulatory variation within the erythroid lineage may underlie the associations observed at several of the loci identified in our red blood cell GWAS. The 19 genes closest to the 25 erythroblast-specific NDRs were more likely to be upregulated during erythropoiesis compared to all other expressed transcripts (P=6.3×10−6, rank-sum test; Supplementary Table 18), lending further support to the view that the NDRs identified have a role in the regulation of genes involved in erythropoiesis16,17. Interestingly, the SNPs associated with MCH at 16p11 overlap an erythroblast-specific NDR that coincides with the NPRL3 regulatory element in the locus control region of the downstream haemoglobin-α locus18,19.
Together our coding- and regulatory-variant analyses thus identify a set of ~100 SNPs across 41 regions that are candidates for functional genomic elements influencing red blood cell formation and function, and which constitutes a priority set for future experimental evaluation.
Insights from mouse models
A systematic search of the Mouse Genome Informatics database reveals haematologic phenotypes for 29 of the 100 candidate genes that have mouse homologues (Supplementary Fig. 6 and Supplementary Tables 14, 19), including genes involved in cell cycle regulation: CCNA2 (4q27), CCND2 (12p13) and CCND3 (6p21); genes coding for transcription factors and their interacting proteins: BCL11A (2p16), CITED2 (6q24), IKZF1 (7p13) and TAL1 (1p32); and genes involved in growth factor or cytokine signalling: KIT (4q11), KITLG (12q22), SH2B3 (12q24) and PTPRCAP (11q13). Among the gene products encoded at the newly identified loci, KITLG, also known as stem cell factor, is the cognate ligand for the KIT tyrosine kinase receptor20. KIT signalling is involved in the perinatal transition from fetal to adult haemoglobin, in addition to maintenance, proliferation and differentiation of haematopoietic stem cells21. Kitlg−/− and Kit−/− mice have low red blood cell concentrations, anaemia and other haematological abnormalities. CCNA2, CCND2 and CCND3 are cyclin-dependent kinases that contribute to initiation and progression of cell division22. Knock-out models of these genes have a number of haematological abnormalities, including reduced stem cell and red blood cell concentrations, and anaemia22. Of the 29 candidate genes with a blood phenotype in mouse, 25 were identified as the genes nearest to the sentinel SNP, and 15 through the eQTL (n =2), coding-variant (n = 6) or GRAIL (n = 8) analyses (Supplementary Table 19).
RNAi silencing in D. melanogaster
We used haemocyte-specific RNA interference (RNAi) silencing in D. melanogaster to further evaluate the candidate genes for their role in blood cell formation. We first carried out permutation testing in a genome-wide D. melanogaster RNAi silencer screen (Supplementary Note). Results confirmed that the 121 candidates are enriched for genes with a blood cell phenotype in D. melanogaster, supporting the view that our GWAS identifies a set of genes conserved across phyla and involved in blood cell formation or survival.
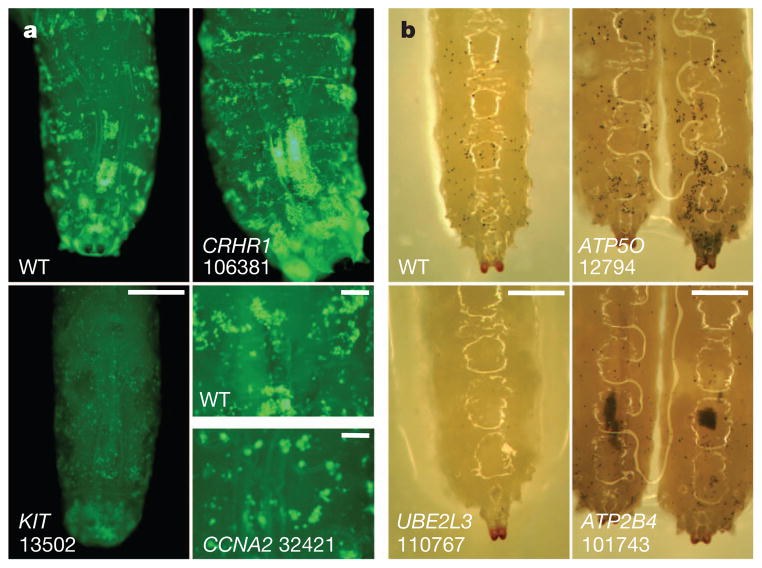
We next created haemocyte-specific RNAi knockdowns for 96 D. melanogaster genes that are orthologues for 74 of the 121 candidate genes, and assessed blood cell formation (crystal cells and plasmatocytes) in early- and late-stage L3 larvae23. We found 19 out of the 74 candidate genes with orthologues in D. melanogaster to have a blood cell phenotype, of which 5 also have a haematological phenotypes in mouse models: KIT, HK1, CCNA2, AP3D1 and PSMB10 (Supplementary Tables 19 and 20). Among the genes highlighted, RNAi silencing of KIT and CCNA2 orthologues was associated with a profound reduction in plasmocyte formation (Fig. 2), consistent with their established role in cytokinesis20,22. AP3D1 is involved in vesicular trafficking and dense granule formation in platelets24, whereas PSMB10 is a component of a widely distributed proteasome linked to inflammation and ubiquitin signalling25. UBE2L3 is also involved in ubiquitin signalling and immune regulation26, and genetic variants in UBE2L3 are strongly associated with several autoimmune diseases known to influence blood cell counts27,28. EIF5 (14q32) is involved in activation of the ribosomal initiation complex29, whereas RPS6KB2 (22q11) is a key component of growth factor and other signalling cascades that regulate ribosomal function, cellular proliferation and survival30. For most of the genes identified, the mechanisms underlying their potential relationship to red cell biology remain to be elucidated; our gene set thus provides a rich resource for future experimental evaluation and discovery.
Figure 2. RNAi silencing in D. melanogaster.
a, Plasmatocytes imaged by green fluorescent protein expression (light green spots on posterior dorsal end of L3 larvae) from wild-type (WT) cells and cells with RNAi silencing of orthologues of the following human genes: CRHR1 (106381, increased cell counts (CC)), KIT (13502, decreased CC) and CCNA2 (32421, increased CC). Numbers represent the unique Flybase IDs corresponding to the D. melanogaster orthologues. Scale bar, 0.5 mm. Bottom right, plasmatocyte size is also increased in CCNA2 compared to wild type. Scale bars, 0.1 mm. b, Crystal cells (black spots visualized by heating larvae to 60 °C) in wild-type larvae, and in RNAi silencing of ATP5O (12794, increased CC), UBE2L3 (110767, decreased CC) or ATP2B4 (101743, aggregated). Scale bars, 0.5 mm.
Contribution to clinical phenotype
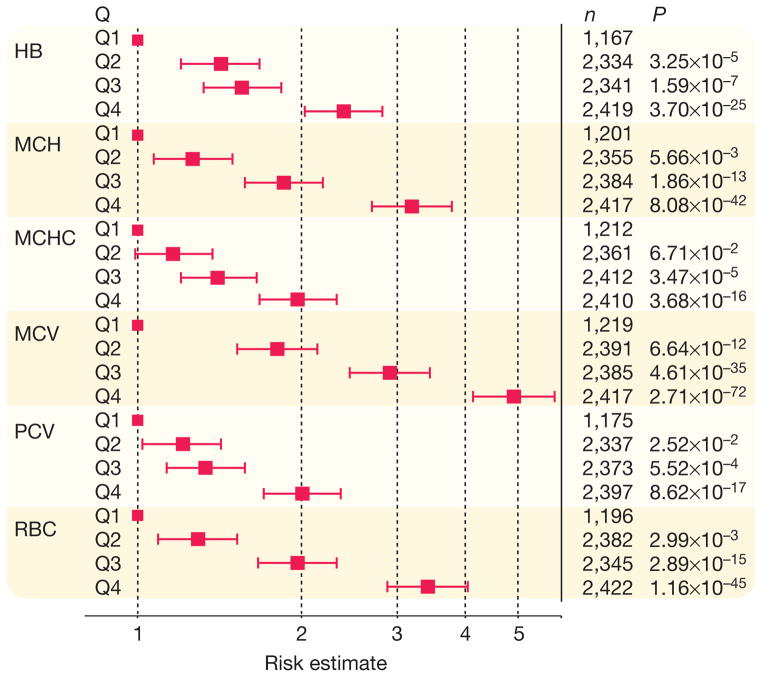
The 75 sentinel SNPs together account for between 3.9% (PCV) and 8.9% (MCV) of population variation in red blood cell phenotypes (Supplementary Table 21). Individuals in the highest quartile of genetic risk score (GRS; on the basis of weighted effect of the 75 sentinel SNPs) are 3–5-fold more likely to be in the highest quartile for population distribution of MCH, MCV and RBC (Fig. 3). GRS is associated with haemoglobin concentrations across the physiological range, including at haemoglobin levels that predict adverse outcomes in pregnancy, cardiovascular and neurologic disease, in addition to mortality in the elderly31–34.
Figure 3. Association of SNP score with red blood cell phenotypes.
Results presented as odds ratio (95% confidence interval) for participants in each SNP score quartile (Q) having phenotype level in the top quartile versus the lowest quartile of the respective population distribution, compared to people in the lowest quartile of SNP score (Q1, reference group). HB, haemoglobin; n, number of participants in the respective comparison of SNP score quartiles.
We next investigated the association of the 75 sentinel SNPs with red blood cell phenotypes in thalassaemia, a group of genetic disorders characterized by defects in haemoglobin synthesis and anaemia. We confirmed association of several of the sentinel SNPs with respective blood cell trait, and found that GRS predicts phenotype similarly, among 460 β-thalassaemia heterozygotes (Supplementary Table 22 and Supplementary Note). In separate experiments, GRS predicts time to first blood transfusion among 495 patients with thalassaemia major (P =6.9 × 10−4); however, this effect was fully accounted for by the MYB-HBS1L locus, which modifies the severity of thalassaemia major through its effect on fetal haemoglobin levels (Supplementary Note)35. Together, our findings demonstrate that the common genetic variants identified contribute to phenotypic variation in the general population, and suggest that they may also act as genetic modifiers in clinically relevant red blood cell abnormalities.
Conclusions
Our genome-wide association and replication study in 135,367 individuals identifies 75 genetic loci influencing red blood cell phenotypes, and 156 locus–phenotype associations; most of these discoveries are novel. Through open-chromatin and coding-variant studies, we identify a first set of SNPs as potential causal variants. In parallel, our bioinformatic strategies identify a core set of genes, differentially regulated in haematologic precursor cells, which are candidates for mediating the effects on red blood cell phenotypes. However, despite our extensive GWAS, bioinformatic and experimental data, the precise identities of the causal variants, regulatory regions and genes remain to be determined; definitive identification will require further detailed experimental evaluation. Our results thus provide new insights into the genes and gene variants that may influence haemoglobin levels and related red blood cell indices, and will underpin a deeper knowledge of the biological mechanisms involved in haematopoiesis and red blood cell function.
METHODS
Genome-wide association
Genome-wide association was carried out in 62,553 people of European ancestry and 9,308 people of South Asian ancestry, using up to 2,644,161 autosomal and 67,645 X-chromosome SNPs. Imputation was done using haplotypes from HapMap Phase 2. Characteristics of participants, genotyping arrays and imputation are summarized in Supplementary Tables 1 and 2. Participants with extreme measurements (> ± 3 s.d. from mean) were excluded on a per-phenotype basis. Each population cohort was approved by a research ethics committee, and all participants gave informed consent.
SNP associations with each phenotype were tested by linear regression using an additive genetic model. Associations were tested separately in men and women in each cohort, with principal components and other study-specific factors as covariates to account of population substructure as described in Supplementary Table 2. Test statistics from each cohort were then corrected for their respective genomic-control inflation factor to adjust for residual population sub-structure; genomic-control inflation factors are summarized in Supplementary Table 3. We then carried out a meta-analysis of results from the individual cohorts using Z-scores weighted by the square root of sample size. The meta-analysis was varied out among Europeans and South Asians separately. There were no South-Asian-specific discoveries, but also little evidence for heterogeneity of effect at known or new genetic loci (Supplementary Table 23); we therefore carried out a final combined analysis of results for the two populations. SNPs with minor allele frequency <1% (weighted average across cohorts) were removed, as were SNPs with weight <50% of phenotype sample size. There was no evidence for inflation of test statistics at SNPs not known to be associated with red blood cell phenotypes (Supplementary Table 3), and genomic control was not applied to the final meta-analysis results. We used the function ‘clump’ implemented in PLINK to cluster the SNPs into genomic loci using a 2-Mb window; clustering was done separately for each phenotype. Inverse variance meta-analysis was used to quantify effect sizes for SNPs of interest.
Genome-wide significance was inferred at P <1 × 10−8. This choice of statistical threshold was grounded on the guidelines derived from studies of the ENCODE (encyclopedia of DNA elements) regions6, combined with results of permutation testing to determine the additional adjustment needed for the six red blood cell phenotypes studied (Supplementary Tables 24, 25 and Supplementary Note). As an alternative strategy, a P-value threshold of P <3.2 × 10−9 would provide correction for the number of SNP–phenotype combinations tested without any adjustment for the correlations between the SNPs or phenotypes tested. We note that 70 of the 75 loci identified would exceed such a highly stringent threshold, including all four of the loci identified through the joint analysis of European and South Asian data.
Replication testing
We carried out replication testing of 22 SNPs selected on the basis of the following criteria: (1) the lead SNP from each of 17 loci showing suggestive evidence for association with one or more red blood cell phenotypes in Europeans (P >10−8 and P <10−7), and (2) the lead SNP from each of the loci identified through combined analysis of genome-wide association data for Europeans and South Asians. Replication testing was done using a combination of in silico results and direct genotyping among 63,506 people from four population cohorts.
In silico data were available for 34,843 people from Iceland participating in the deCODE (diabetes epidemiology: collaborative analysis of diagnostic criteria in Europe) study37 (Supplementary Table 1). SNPs were directly genotyped with the Illumina HumanHap300 or CNV370 chips or imputed from one or more of four sources: the HapMap2 CEU sample (60 triads), the 1000 Genomes Project data (179 individuals) and Icelandic samples genotyped with the Illumina Human1 M-Duo (123 triads) or the HumanOmni1-Quad chips (505 individuals), as previously described in ref. 37. The 22 SNPs were tested for association against their respective discovery phenotypes, under an additive genetic model; results were combined with the genome-wide association data by weighted-Z-score meta-analysis.
We found that for 7 of the 22 SNPs carried forward for replication, their associations with phenotype remained inconclusive after in silico testing (P >10−8 but P <10−7). For these SNPs we carried out additional direct genotyping using Sequenom assays, among up to 20,066 people from three population cohorts (Supplementary Table 1). Associations were tested in each cohort separately, and results combined across the replication cohorts, and then with the genome-wide association data, by weighted-Z-score meta-analysis (Supplementary Table 26).
Conditional analysis
We performed conditional-association analysis using the summary statistics from the meta-analysis to test for the association of each SNP while conditioning on the top SNPs, with correlations between SNPs due to linkage disequilibrium estimated from the imputed genotype data from the atherosclerosis risk in communities (ARIC) cohort8,38. Secondary-association signals were selected with conditional-association P <1 × 10−8.
Identification of candidate genes
We considered the nearest gene, and any other gene located within 10 kb of the sentinel SNP, to be a candidate for mediating the association with red blood cell phenotype. We also used coding variant, eQTL and literature analyses to identify candidate genes. On the basis of analysis of linkage-disequilibrium relations at the 75 genetic loci, we defined genomic region as the 1-Mb interval either side of the sentinel SNP for our functional genomic studies (Supplementary Fig. 7).
Coding variation
We identified all non-synonymous SNPs that were in linkage disequilibrium with one or more of the sentinel SNPs at r2 >0.8 in 1000 Genomes Project data set (released in March 2012). We considered the gene to be a candidate when the non-synonymous and sentinel SNPs were in linkage disequilibrium at r2 >0.8 and with no evidence for heterogeneity of effect on phenotype. This strategy identified 39 non-synonymous SNPs distributed between 24 genes (Supplementary Table 9), representing a ~sixfold enrichment compared to the mean number expected under the null hypothesis generated by permutation testing of SNP sets matched for allele frequency (±0.05) and number of genes in proximity (±10 kb), but selected otherwise at random (P =0.01; Supplementary Note).
Expression analyses
To identify the possible genes influencing red blood cell phenotypes at the 75 loci, we examined the association of the sentinel SNPs with eQTL data from two data sets: (1) peripheral blood lymphocytes from 206 families of European descent (830 parents and offspring)39 and (2) peripheral blood lymphocytes from 1,469 unrelated individuals40.
SNPs were tested for association with expression of nearby (1 Mb) genes (P <0.05 after Bonferroni correction for number of SNP–transcript associations tested). Where eQTLs were identified, we used the whole-genome SNP data available in these data sets (imputed with HapMap Phase 2 genotypes), to identify the SNP at the locus most closely associated with transcript level (the transcript SNP). We then tested whether the sentinel SNP and the transcript SNP were coincident, defined as r2 >0.8 with no evidence for heterogeneity of effect on phenotype or transcript level (P >0.05). This strategy identified eQTLs involving 28 genes from 18 loci (Supplementary Table 10).
GRAIL analyses
We carried out a literature analysis using the GRAIL algorithm9, a statistical tool that uses text mining of PubMed abstracts to annotate candidate genes from loci associated with phenotypic traits. We carried out the analysis using the 2006 data set to avoid confounding by subsequent GWAS discoveries; results identified candidate genes at nine loci (P <0.05; Supplementary Table 11). Results are also shown for a GRAIL analysis using the 2011 PubMed data set, although these were not used for the final analysis.
Gene expression in haematopoietic precursors
Cord-blood-derived CD34+ haematopoietic stem cells were differentiated in vitro along the erythroid lineage in the presence of 6 U ml−1 erythropoietin (R&D Systems), 10 ng ml−1 inter-leukin (IL)-3 (Miltenyi Biotec) and 100 ng ml−1 stem cell factor (R&D Systems). Cells were collected at days 3, 5, 7, 9 and 10 in three biological replicates and gene expression was assayed using Illumina human WGv3.0 microarrays41. For each gene, we determined the relationship of gene expression with time using linear regression, and calculated the t-statistic for the difference in β from zero. We then classified gene-expression patterns as increasing, decreasing or unchanged on the basis of the 2.5% and 97.5% quartiles of the t distribution with 4 degrees of freedom. To test whether a gene set was enriched for differentially regulated genes, a Wilcoxon signed-rank test of the t scores in the gene set relative to all others genes that were expressed in at least one time point was calculated.
FAIRE-seq
We generated maps of chromatin accessibility (‘open chromatin’) in primary human erythroblasts and megakaryocytes, and in peripheral blood monocytes using FAIRE-seq. Cord-blood-derived CD34+ haematopoietic progenitor cells from two unrelated individuals were differentiated in vitro into either erythroblasts (in the presence of erythropoietin, IL-3 and stem cell factor) or megakaryocytes (in the presence of thrombopoietin and IL-1β). Monocytes were purified from leukocyte cones of apheresis collections from another two individuals.
FAIRE experiments were performed as previously described in ref. 42. FAIRE DNA was processed following the Illumina paired-end library-generation protocol. Genomic libraries derived from erythroblast and megakaryocyte cultures were sequenced with 54-bp paired-end reads on Illumina Genome Analyzer II. Libraries derived from monocyte extractions were sequenced with 50-bp paired-end reads on Illumina HiSeq. Raw sequence reads were aligned to the human reference sequence (NCBI build 37) using the read mapper Stampy43. Reads were realigned around known insertions and deletions, followed by base-quality recalibration using the Genome Analysis Toolkit (GATK)44. Duplicates were flagged using the software Picard (http://picard.sourceforge.net/) and excluded from subsequent analyses. For each cell type, we merged all read fragments into one data set. NDRs were identified as regions of sequencing enrichment (peaks) using the software F-Seq36. We applied a feature length of L =600 bp and a s.d. threshold of T =8.0 over the mean across a local background. In order to reduce false-positive peak calls, we removed regions of collapsed repeats as recently described, applying a threshold of 0.1%45. For each associated locus, candidate functional SNPs were selected by identifying all biallelic SNPs with an r2 >0.8 and within 1 Mb of the sentinel SNP in the European samples of the 1000 Genomes Project (data released June 2011).
D. melanogaster gene-silencing models
We used haemocyte-specific RNAi silencing to investigate whether the 121 candidate genes identified in the red blood cell GWAS influenced blood cell formation in D. melanogaster. We identified D. melanogaster genes predicted to be orthologues of human genes using the Ensembl v65 Compara pipeline, an established phylogenetic-tree-based approach for orthology prediction46; this revealed 96 D. melanogaster orthologues for 74 of the 121 human candidate genes (Supplementary Table 27). We evaluated each of the 96 orthologues for a blood cell phenotype in D. melanogaster. We obtained all 225 available D. melanogaster lines carrying inducible siRNA constructs from the Vienna Drosophila RNAi Center (VDRC)23. To achieve haemocyte-specific knockdowns, flies were crossed to the blood-specific Hml-Gal4 line driving Gal4 expression under the control of a hemolectin promoter47. Flies were crossed at 29 °C, and early and late L3 larvae analysed 7 days after mating. Upstream activating sequence–green fluorescent protein enabled microscopic visualization of plasmatocytes and evaluation of cell size and cell number (L3 larvae only). Early- and late-stage larvae were incubated at 60 °C for 15 min, a process that turns the crystal cells black and allows quantification of crystal cells microscopically. For each orthologue, all available RNAi silencer constructs were investigated, and in addition, each construct was assayed in duplicate, blind to initial result. Cell counts were quantified visually (0–3, decreased or increased) and the mean of the duplicate measurements calculated.
We separately carried out permutation testing in a genome-wide screen of 5,658 D. melanogaster genes to simulate expectations under the null hypothesis (Supplementary Fig. 8 and Supplementary Note); results confirmed that the 121 candidate genes were enriched for blood cell phenotype in D. melanogaster orthologues (P <0.05), and showed that this was robust to threshold for calling.
Contribution of the genetic loci identified to population variation in red blood cell phenotypes
This was investigated in participants from the Estonian Genome Center of University of Tartu (EGCUT), LIFELINES, Ludwigshafen Risk and Cardiovascular Health Study (LURIC) and Young Finns cohorts using samples that were not included in the discovery experiment (Supplementary Table 1). The contribution of the SNPs to population variation in red blood cell phenotypes was quantified using two models: model 1, limited to SNPs associated with respective phenotype at P <1 × 10−8; and model 2, comprising all of the 75 sentinel SNPs identified. Estimates of population variance explained were made in each study separately, and average values calculated weighted by sample size (Supplementary Table 21).
We then investigated whether the 75 sentinel SNPs influenced the probability of being in the highest versus the lowest quartile for population distribution of phenotype. Two SNP scores were calculated for each phenotype: score 1, limited to SNPs associated with respective phenotype at P <1 × 10−8, and score 2, containing all 75 sentinel SNPs identified. For both, SNP score was calculated as the sum of number of effect (trait raising) alleles present, weighted according to effect size. We then calculated the odds ratio for being in the highest versus the lowest quartile of phenotype, associated with SNP scores in the second, third and fourth quartiles, compared to first quartile of SNP score. Odds ratios were calculated in each study separately, and then combined by inverse variance meta-analysis (Fig. 3).
Supplementary Material
Acknowledgments
A detailed list of acknowledgements is provided in the Supplementary Material.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions Study organisation: J.C.C., C.G., P.v.d.H., J.S.K., W.H.O. and N.S. Manuscript preparation: H.A., J.S.B., J.C.C., G.V.D., P.D., C.G., P.v.d.H., A.A. Hicks, J.S.K., I.M.-L., W.H.O., A. Radhakrishnan, A. Rendon, S.S., J. Sehmi, N.S., D.S.P., M.U., N.V. and W.Z. All authors reviewed and had the opportunity to comment on the manuscript. Data collection and analysis in the participating genome-wide association, replication and phenotype cohorts: ALSPAC: D.M.E., J.P.K., S.M.R., G.D.S; AMISH: Q.D.G., B.D.M., A. Parsa, A.R.S.; Beta-thalassaemia: F.A., F.D., P. Fortina, R.G, L. Perseu, A. Piga, S.S., M.U.; CBR: A. Attwood, J.D., S.F.G., H.L.-J., C. Moore, W.H.O., J. Sambrook; CoLAUS: F.B., J.S.B., M.H., P.V.; DeCODE: G.I.E., D.F.G., H.H., I.O., P.T.O., K.S., P.S., U.T.; DESIR: B. Balkau, C.D., P. Froguel, R. Sladek; EGCUT: T.E., K.F., A.M., E.M., A.S.; EPIC: K.-T.K., C.L., R.J.F.L., N.J.W., J.-H.Z.; Genebank: H.A., J.H., S.L.H., W.H.W.T.; INGI CARL: P.G., G.G., N.P.; INGI CILENTO: M.C., T.N., D.R., R. Sorice.; INGI FVG: A.P.d.A., A. Robino, S.U.; INGI Val Borbera: G.P., C.S., D.T., M.T.; KORA: A.D., C.G., T.I., C. Meisinger, J.S.R.; LBC: I.J.D., S.E.H., L.M.L., J.M.S.; LIFELINES: R.A.d.B., I.P.K., I.M.-L., G.N., P.v.d.H., L.J.v.P., N.V., B.H.R.W.; LOLIPOP: A. Al-Hussani, J.C.C., D.D., P.E., J.S.K., X.L., K.M., J. Scott, J. Sehmi, S.-T.T., W.Z.; LURIC: B.G., B.O.B., M.E.K., W.M., B.R.W.; MDC: A.F.D., G.E., B.H., C.E.H., O.M., S.P., J.G.S.; MICROS: M.G., A. AHicks, A.S.-P., P.P.P.; NESDA: I.M.N., B.W.P., J.H.S., H. Snieder; NFBC1966: A.-L.H., M.-R.J., P.F.O., A. Pouta, A. Ruokonen.; NTR: A. Abdellaoui, D.I.B., E.J.C.d.G., J.-J.H., M.H.d.M., G. Willemsen; OGP: F.M., D.P., L. Portas, M.P.; PREVEND: R.A.d.B., I.M.-L., G.N., P.v.d.H., W.H.v.G., D.J.v.V., N.V.; QIMR: B. Benyamin, M.A.F., N.G.M., S.E.M., G.W.M., C.S.T., P.M.V., J.B.W.; SardiNIA: F.C., E.P., S.S., M.U.; SHIP: A.G., M. Nauck, C.O.S., A. Teumer, U.V.; SMART: A. Algra, F.W.A., P.I.W.d.B., V.T.; SORBS: V.L., I.P., M.S., A. Tönjes.; TwinsUK: Y.M., S.-Y.S., N.S., T.D.S.; UKBS: J.J., W.H.O., N.S., J. Stephens; Young Finns: M.K., T.L., L.-P.L., O.R. Functional studies: Drosophila, U.E., F.S.D., A.A. Hicks, M. Novatchkova, J.M.P., U.P., C.X.W., G. Wirnsberger; expression profiling, W.O.C., L. Franke, L.L., M.F.M., A. Rendon, E.S., H.-J.W.; FAIRE, C.A.A., P.D., W.H.O., D.S.P., A. Rendon, N.S. Data analysis and bioinformatics: A. Al-Hussani, S.B., J.C.C., M.D., L. Ferrucci, P.v.d.H., S.K., X.L., I.M.-L., K.M., S.M., A. Radhakrishnan, A. Rendon, R.R.-S., H. Schepers, J. Sehmi, N.S., H.H.W.S., S.T., T.T., N.V., K.V., P.V., J.Y., W.Z.
Summary statistics from the genome-wide association study are available from the European Genome–Phenome Archive (EGA, http://www.ebi.ac.uk/ega) under accession number EGAS00000000132.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Full Methods and any associated references are available in the online version of the paper.
References
- 1.Chambers JC, et al. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nature Genet. 2009;41:1170–1172. doi: 10.1038/ng.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganesh SK, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nature Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soranzo N, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nature Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamatani Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nature Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 5.Ding K, et al. Genetic loci implicated in erythroid differentiation and cell cycle regulation are associated with red blood cell traits. Mayo Clin Proc. 2012;87:461–474. doi: 10.1016/j.mayocp.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 7.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An X, Mohandas N. Disorders of red cell membrane. Br J Haematol. 2008;141:367–375. doi: 10.1111/j.1365-2141.2008.07091.x. [DOI] [PubMed] [Google Scholar]
- 11.van Wijk R, Rijksen G, Huizinga EG, Nieuwenhuis HK, van Solinge WW. HK Utrecht: missense mutation in the active site of human hexokinase associated with hexokinase deficiency and severe nonspherocytic hemolytic anemia. Blood. 2003;101:345–347. doi: 10.1182/blood-2002-06-1851. [DOI] [PubMed] [Google Scholar]
- 12.Camaschella C, Poggiali E. Inherited disorders of iron metabolism. Curr Opin Pediatr. 2011;23:14–20. doi: 10.1097/MOP.0b013e3283425591. [DOI] [PubMed] [Google Scholar]
- 13.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gieger C, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul DS, et al. Maps of open chromatin guide the functional follow-up of genome-wide association signals: application to hematological traits. PLoS Genet. 2011;7:e1002139. doi: 10.1371/journal.pgen.1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrester WC, Thompson C, Elder JT, Groudine M. A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuan D, Solomon W, Li Q, London IM. The “beta-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalczyk MS, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Baù D, et al. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nature Struct Mol Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zsebo KM, et al. Stem cell factor is encoded at the Sl locus of the mouse and is the ligand for the c-kit tyrosine kinase receptor. Cell. 1990;63:213–224. doi: 10.1016/0092-8674(90)90302-u. [DOI] [PubMed] [Google Scholar]
- 21.Heissig B, et al. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell. 2002;109:625–637. doi: 10.1016/s0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozar K, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 23.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 24.Clark RH, et al. Adaptor protein 3-dependent microtubule-mediated movement of lytic granules to the immunological synapse. Nature Immunol. 2003;4:1111–1120. doi: 10.1038/ni1000. [DOI] [PubMed] [Google Scholar]
- 25.Berhane S, et al. Adenovirus E1A interacts directly with, and regulates the level of expression of, the immunoproteasome component MECL1. Virology. 2011;421:149–158. doi: 10.1016/j.virol.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 26.Tiwari S, Weissman AM. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s) J Biol Chem. 2001;276:16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- 27.Fransen K, et al. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn’s disease. Hum Mol Genet. 2010;19:3482–3488. doi: 10.1093/hmg/ddq264. [DOI] [PubMed] [Google Scholar]
- 28.Zhernakova A, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S, Ghosh R, Maitra U. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J Biol Chem. 2001;276:6720–6726. doi: 10.1074/jbc.M008863200. [DOI] [PubMed] [Google Scholar]
- 30.Fenton TR, Gout IT. Functions and regulation of the 70 kDa ribosomal S6 kinases. Int J Biochem Cell Biol. 2011;43:47–59. doi: 10.1016/j.biocel.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Scanlon KS, Yip R, Schieve LA, Cogswell ME. High and low hemoglobin levels during pregnancy: differential risks for preterm birth and small for gestational age. Obstet Gynecol. 2000;96:741–748. doi: 10.1016/s0029-7844(00)00982-0. [DOI] [PubMed] [Google Scholar]
- 32.Shah RC, Buchman AS, Wilson RS, Leurgans SE, Bennett DA. Hemoglobin level in older persons and incident Alzheimer disease: prospective cohort analysis. Neurology. 2011;77:219–226. doi: 10.1212/WNL.0b013e318225aaa9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabatine MS, et al. Association of hemoglobin levels with clinical outcomes in acute coronary syndromes. Circulation. 2005;111:2042–2049. doi: 10.1161/01.CIR.0000162477.70955.5F. [DOI] [PubMed] [Google Scholar]
- 34.Zakai NA, et al. A prospective study of anemia status, hemoglobin concentration, and mortality in an elderly cohort: the Cardiovascular Health Study. Arch Intern Med. 2005;165:2214–2220. doi: 10.1001/archinte.165.19.2214. [DOI] [PubMed] [Google Scholar]
- 35.Galanello R, et al. Amelioration of Sardinian β0 thalassemia by genetic modifiers. Blood. 2009;114:3935–3937. doi: 10.1182/blood-2009-04-217901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm H, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nature Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon AL, et al. A genome-wide association study of global gene expression. Nature Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 40.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nature Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson RJ, et al. Reduced dependency on arteriography for penetrating extremity trauma: influence of wound location and noninvasive vascular studies. J Trauma. 1990;30:1059–1063. [PubMed] [Google Scholar]
- 42.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickrell JK, Gaffney DJ, Gilad Y, Pritchard JK. False positive peaks in ChIP-seq and other sequencing-based functional assays caused by unannotated high copy number regions. Bioinformatics. 2011;27:2144–2146. doi: 10.1093/bioinformatics/btr354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vilella AJ, et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goto A, et al. A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem J. 2001;359:99–108. doi: 10.1042/0264-6021:3590099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.