Abstract
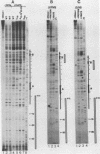
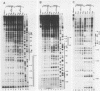
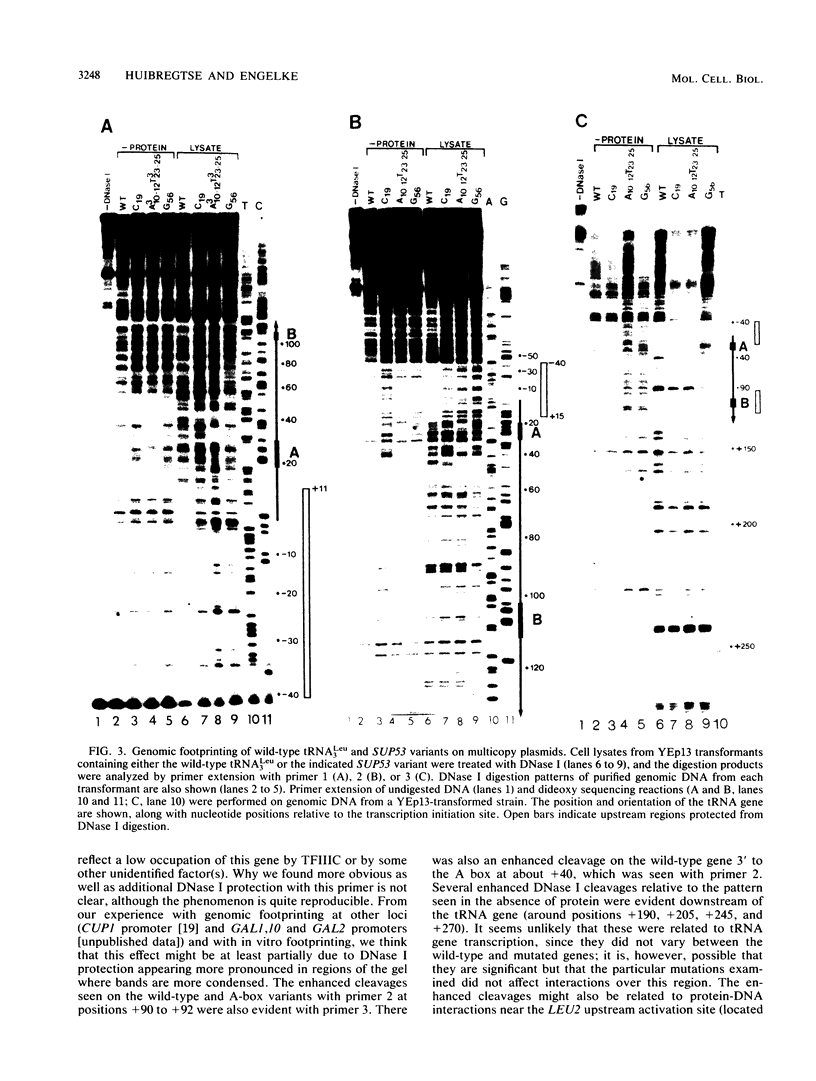
We have shown by genomic footprinting that the 5'-flanking region of the Saccharomyces cerevisiae tRNASUP53 gene is protected from DNase I digestion. The protected region has a 5' boundary at -40 (relative to the transcription initiation site) and extends into the coding region of the gene, with a 3' boundary at approximately +15. Although the DNase I protection over this region was much greater than at the A- and B-box internal promoters, point mutations within the A or B box that reduced transcription in vitro eliminated the upstream DNase I protection. This implies that formation of a stable complex over the 5'-flanking region is dependent on interaction of the gene with transcription factor IIIC but that stability of the complex may not require continued interaction with this factor. The DNase I protection under varied growth conditions further suggested that the upstream complex is composed of two or more components. The region over the transcription initiation site (approximately +15 to -10) was less protected in stationary-phase cultures, whereas the more upstream region (approximately -10 to -40) was protected in both exponential- and stationary-phase cultures.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Camier S., Sentenac A., Hall B. D. Gene size differentially affects the binding of yeast transcription factor tau to two intragenic regions. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8768–8772. doi: 10.1073/pnas.84.24.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E., Paule M. R. Regulation of eukaryotic ribosomal RNA transcription by RNA polymerase modification. Cell. 1986 Nov 7;47(3):445–450. doi: 10.1016/0092-8674(86)90601-x. [DOI] [PubMed] [Google Scholar]
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Murgo S., Ebel J. P., Krol A., Tebb G., Mattaj L. W. A common octamer motif binding protein is involved in the transcription of U6 snRNA by RNA polymerase III and U2 snRNA by RNA polymerase II. Cell. 1987 Oct 9;51(1):71–79. doi: 10.1016/0092-8674(87)90011-0. [DOI] [PubMed] [Google Scholar]
- Carey M. F., Gerrard S. P., Cozzarelli N. R. Analysis of RNA polymerase III transcription complexes by gel filtration. J Biol Chem. 1986 Mar 25;261(9):4309–4317. [PubMed] [Google Scholar]
- Cummins C. M., Donahue T. F., Culbertson M. R. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3565–3569. doi: 10.1073/pnas.79.11.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol Cell Biol. 1987 Aug;7(8):2708–2717. doi: 10.1128/mcb.7.8.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Garcia A. D., O'Connell A. M., Sharp S. J. Formation of an active transcription complex in the Drosophila melanogaster 5S RNA gene is dependent on an upstream region. Mol Cell Biol. 1987 Jun;7(6):2046–2051. doi: 10.1128/mcb.7.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Direct identification of small sequence changes in chromosomal DNA. Gene. 1986;44(1):151–158. doi: 10.1016/0378-1119(86)90056-9. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R., Thiele D. J. Copper-induced binding of cellular factors to yeast metallothionein upstream activation sequences. Proc Natl Acad Sci U S A. 1989 Jan;86(1):65–69. doi: 10.1073/pnas.86.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Evans C. F., Engelke D. R. Comparison of tRNA gene transcription complexes formed in vitro and in nuclei. Mol Cell Biol. 1987 Sep;7(9):3212–3220. doi: 10.1128/mcb.7.9.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. D., Raymond G. J. Three regions of a yeast tRNALeu3 gene promote RNA polymerase III transcription. J Biol Chem. 1984 May 10;259(9):5990–5994. [PubMed] [Google Scholar]
- Johnson P. F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983 Apr 21;302(5910):681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Partial purification and characterization of the Saccharomyces cerevisiae transcription factor TFIIIB. J Biol Chem. 1986 Feb 25;261(6):2819–2827. [PubMed] [Google Scholar]
- Krol A., Carbon P., Ebel J. P., Appel B. Xenopus tropicalis U6 snRNA genes transcribed by Pol III contain the upstream promoter elements used by Pol II dependent U snRNA genes. Nucleic Acids Res. 1987 Mar 25;15(6):2463–2478. doi: 10.1093/nar/15.6.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Lin J. P., Aker M., Sitney K. C., Mortimer R. K. First position wobble in codon-anticodon pairing: amber suppression by a yeast glutamine tRNA. Gene. 1986;49(3):383–388. doi: 10.1016/0378-1119(86)90375-6. [DOI] [PubMed] [Google Scholar]
- Lofquist A., Sharp S. The 5'-flanking sequences of Drosophila melanogaster tRNA5Asn genes differentially arrest RNA polymerase III. J Biol Chem. 1986 Nov 5;261(31):14600–14606. [PubMed] [Google Scholar]
- Majowski K., Mentzel H., Pieler T. A split binding site for TFIIIC on the Xenopus 5S gene. EMBO J. 1987 Oct;6(10):3057–3063. doi: 10.1002/j.1460-2075.1987.tb02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R., Dingermann T. Identification of a protein factor binding to the 5'-flanking region of a tRNA gene and being involved in modulation of tRNA gene transcription in vivo in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14B):6737–6752. doi: 10.1093/nar/16.14.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzouki N., Camier S., Ruet A., Moenne A., Sentenac A. Selective proteolysis defines two DNA binding domains in yeast transcription factor tau. Nature. 1986 Sep 11;323(6084):176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., von Hippel P. H. Rho-dependent termination of transcription. I. Identification and characterization of termination sites for transcription from the bacteriophage lambda PR promoter. J Biol Chem. 1983 Aug 10;258(15):9553–9564. [PubMed] [Google Scholar]
- Morton D. G., Sprague K. U. In vitro transcription of a silkworm 5S RNA gene requires an upstream signal. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5519–5522. doi: 10.1073/pnas.81.17.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S., Di Liegro C., Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987 Oct 9;51(1):81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- Nakajima N., Horikoshi M., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol. 1988 Oct;8(10):4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Page G. S., Sentenac A., Piper P. W., Worthington M., Weiss R. B., Hall B. D. Only one of two closely related yeast suppressor tRNA genes contains an intervening sequence. Nature. 1981 Jun 11;291(5815):464–469. doi: 10.1038/291464a0. [DOI] [PubMed] [Google Scholar]
- Ottonello S., Rivier D. H., Doolittle G. M., Young L. S., Sprague K. U. The properties of a new polymerase III transcription factor reveal that transcription complexes can assemble by more than one pathway. EMBO J. 1987 Jul;6(7):1921–1927. doi: 10.1002/j.1460-2075.1987.tb02452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Schmidt O., Mao J., Ogden R., Beckmann J., Sakano H., Abelson J., Söll D. Dimeric tRNA precursors in yeast. Nature. 1980 Oct 23;287(5784):750–752. doi: 10.1038/287750a0. [DOI] [PubMed] [Google Scholar]
- Schultz L. D. Transcriptional role of yeast deoxyribonucleic acid dependent ribonucleic acid polymerase III. Biochemistry. 1978 Feb 21;17(4):750–758. doi: 10.1021/bi00597a031. [DOI] [PubMed] [Google Scholar]
- Segall J. Assembly of a yeast 5 S RNA gene transcription complex. J Biol Chem. 1986 Sep 5;261(25):11578–11584. [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Selker E. U., Morzycka-Wroblewska E., Stevens J. N., Metzenberg R. L. An upstream signal is required for in vitro transcription of Neurospora 5S RNA genes. Mol Gen Genet. 1986 Oct;205(1):189–192. doi: 10.1007/BF02428052. [DOI] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. E. In vivo DNA-binding properties of a yeast transcription activator protein. Mol Cell Biol. 1987 Sep;7(9):3260–3267. doi: 10.1128/mcb.7.9.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S. J., Garcia A. D. Transcription of the Drosophila melanogaster 5S RNA gene requires an upstream promoter and four intragenic sequence elements. Mol Cell Biol. 1988 Mar;8(3):1266–1274. doi: 10.1128/mcb.8.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Geiduschek E. P. Differential binding of a S. cerevisiae RNA polymerase III transcription factor to two promoter segments of a tRNA gene. EMBO J. 1984 Apr;3(4):847–853. doi: 10.1002/j.1460-2075.1984.tb01895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986 Jul;6(7):2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Huber H. E., Richardson C. C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987 Nov 25;262(33):16212–16223. [PubMed] [Google Scholar]
- Van Arsdell S. W., Stetler G. L., Thorner J. The yeast repeated element sigma contains a hormone-inducible promoter. Mol Cell Biol. 1987 Feb;7(2):749–759. doi: 10.1128/mcb.7.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J., Bull P., Valenzuela P., Venegas A. Nucleotide sequence of a yeast tRNAArg3A gene and its transcription in a homologous in vitro system. FEBS Lett. 1984 Feb 13;167(1):165–169. doi: 10.1016/0014-5793(84)80854-6. [DOI] [PubMed] [Google Scholar]
- Warmington J. R., Anwar R., Newlon C. S., Waring R. B., Davies R. W., Indge K. J., Oliver S. G. A 'hot-spot' for Ty transposition on the left arm of yeast chromosome III. Nucleic Acids Res. 1986 Apr 25;14(8):3475–3485. doi: 10.1093/nar/14.8.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. T., Larson D., Young L. S., Sprague K. U. A large region controls tRNA gene transcription. J Mol Biol. 1985 May 25;183(2):153–163. doi: 10.1016/0022-2836(85)90209-8. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S. K., Boulanger P. A., Berk A. J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. S., Takahashi N., Sprague K. U. Upstream sequences confer distinctive transcriptional properties on genes encoding silkgland-specific tRNAAla. Proc Natl Acad Sci U S A. 1986 Jan;83(2):374–378. doi: 10.1073/pnas.83.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]